Abstract
Porcine epidemic diarrhea, a disease caused by porcine epidemic diarrhea virus (PEDV), results in large economic losses to the global swine industry. To manage this disease effectively, it is essential to detect PEDV early and accurately. We developed a sensitive and accurate droplet digital PCR (ddPCR) assay to detect PEDV. The optimal primer-to-probe concentration and melting temperature were identified as 300:200 nM and 59.2°C, respectively. The specificity of the ddPCR assay was confirmed by negative test results for common swine pathogens. The detection limit for the ddPCR was 0.26 copies/μL, which is a 5.7-fold increase in sensitivity compared to that of real-time PCR (rtPCR). Both ddPCR and rtPCR assays exhibited good linearity, although ddPCR provided higher sensitivity for clinical detection compared to that of rtPCR. Our ddPCR methodology provides a promising tool for evaluating the PEDV viral load when used for clinical testing, particularly for detecting samples with low-copy viral loads.
Keywords: detection, droplet digital PCR, porcine epidemic diarrhea virus, quantification
Porcine epidemic diarrhea virus (PEDV; single-stranded RNA; Coronaviridae) was first observed in Europe in 1971 and then spread throughout Europe during the 1970s and 1980s.7,15 Then, during the 1980s and 1990s, the number of PED outbreaks decreased markedly in Europe, while PED became an endemic disease in Asian countries, such as Korea, China, Japan, the Philippines, and Thailand.7,13 An outbreak of PEDV infection occurred in Ohio in 2013, and spread throughout the United States.5 PEDV infections have been reported to be associated with vomiting, diarrhea, anorexia, dehydration, weight loss, and high mortality, and this disease results in large economic losses to the global swine industry.15,19 The N protein is a phosphoprotein that plays an important role in viral genome transcription and replication; N protein is often used as a target for detecting PEDV infection.14-16
Many PEDV detection methods exist and include clinical observation, histologic observation, neutralization tests, immunofluorescence, and immunohistochemistry. These methods are time-consuming and unsuitable for large-scale clinical testing.14 Molecular methods have become more commonly used for detecting PEDV. A reverse-transcription PCR (RT-PCR) assay has been developed to allow for detection of PEDV, but this method requires in-gel analysis of the PCR products.12 Loop-mediated isothermal amplification is a novel DNA amplification method that has been developed11,14; however, this method produces false-positive results because of self-primer interactions. Real-time PCR (rtPCR) is used widely in clinical testing, and quantification using this method relies on a standard curve, quantification cycles (Cq), and the need to establish a Cq threshold line that can result in data bias. Therefore, a more accurate method needs to be established to detect PEDV infection.
Droplet PCR (dPCR) has come into use for precise quantification, given that this method can provide an absolute measurement of nucleic acid concentration without the use of standard curves.1 Additionally, dPCR is more tolerant to inhibitors, and this allows for improved accuracy and precision of quantification of the target.4 dPCR has been widely used in many areas such as food authentication, identification of genetically modified organisms, and clinical testing.4,6,17 We established a method for detection and quantification of PEDV using droplet digital PCR (ddPCR).
The primers and probes were designed for rtPCR and ddPCR assays (Oligo Primer Analysis software; Molecular Biology Insights), based on the nucleotide sequences of the N gene (GenBank accession NC_003436), and synthesized commercially (Sangon Biotech). The sequences were as follows: PEDV sense (5′-CCGTGGTGAGCGAATTGAA-3′), PEDV antisense (5′-GGTCCTGTTCCGAGGTAGTAGAAA-3′) primers, and hydrolysis probes (FAM-5′-AACCTTCCAATTGGC-3′-MGB). The complete sequence of the N gene (960 bp) was inserted into the PUC57 vector (Sangon Biotech) to generate the PUC57-N plasmid according to the manufacturer’s instructions; the plasmid was then transformed into Escherichia coli DH5α cells. Standard plasmid DNA was serially 10-fold diluted, and was used to generate standard curves for rtPCR, which were also used for ddPCR assays; ddPCR was then performed (QX200 droplet digital PCR system; Bio-Rad). Digital PCR reactions consisted of 10 μL of 2× ddPCR Supermix for Probes (Bio-Rad), 300 nmol/L of PEDV primers, 200 nmol/L of hydrolysis probes, 1 μL of template DNA, and sterile distilled water to provide a final volume of 20 μL. Droplets were generated using the QX200 droplet generator (Bio-Rad) according to the manufacturer’s instructions, and the droplets (~ 40 μL) were transferred to a 96-well plate and heat-sealed for PCR (thermal cycler C100 Touch; Bio-Rad) using the following conditions: 95°C for 10 min, then 40 cycles of 94°C for 30 s, 55°C for 1 min, and 98°C for 10 min. A ramp rate of 2°C/s was used for all steps. After PCR, the droplets were analyzed using a QX200 droplet reader (QuantaSoft software v1.7; Bio-Rad). No-template controls (NTC) used as negative controls allowed for the monitoring of contamination and primer–dimer formation.
For the rtPCR system, the same primers and probe used for ddPCR were used (ABI QuantStudio 6 Flex real-time PCR system; Thermo Fisher Scientific). Real-time PCR reactions contained 2× AceQ qPCR probe master mix (Vazyme Biotech), 300 nmol/L of PEDV primers, 200 nmol/L of hydrolysis probes, 1 μL of template DNA, and sterile distilled water to provide a final volume of 20 μL. The reaction conditions for rtPCR included 95°C for 30 s, 35 cycles of 95°C for 5 s, and 60°C for 35 s.
For comparison, copy numbers were calculated for rtPCR based on the concentration calculated by standard curve using a calculator (http://scienceprimer.com/copy-number-calculator-for-realtime-pcr). Each sample was tested in triplicate to evaluate intra- and inter-assay repeatability. All statistical analyses were performed using the SPSS program (v.13.0; IBM).
To test if the ddPCR reaction system for PEDV could be improved, the primer-to-probe concentration and annealing temperature were optimized.8 To select an optimal annealing temperature, PEDV complementary DNA (cDNA; 3.9 × 107 copies/μL) was annealed at temperatures of 55, 56, 57, 58.2, 59.2, and 60°C. Our results indicated that 59.2°C provided the optimal annealing temperature (Fig. 1). The fluorescence amplitude difference between the positive and the negative droplet clusters was the highest at this temperature. The primer concentration was optimized using PEDV cDNA (3.9 × 106 copies/μL for each reaction mixture). The optimal concentration ratio was 300:200 nM because this ratio of reagents resulted in optimal separation between positive and negative droplets (Fig. 2). When the concentration ratio was 400:400 nM or 900:250 nM, the rain effect (intermediate fluorescence of some droplets) was more pronounced and could affect the accuracy of the results. All results were analyzed using QuantaSoft software.
Figure 1.
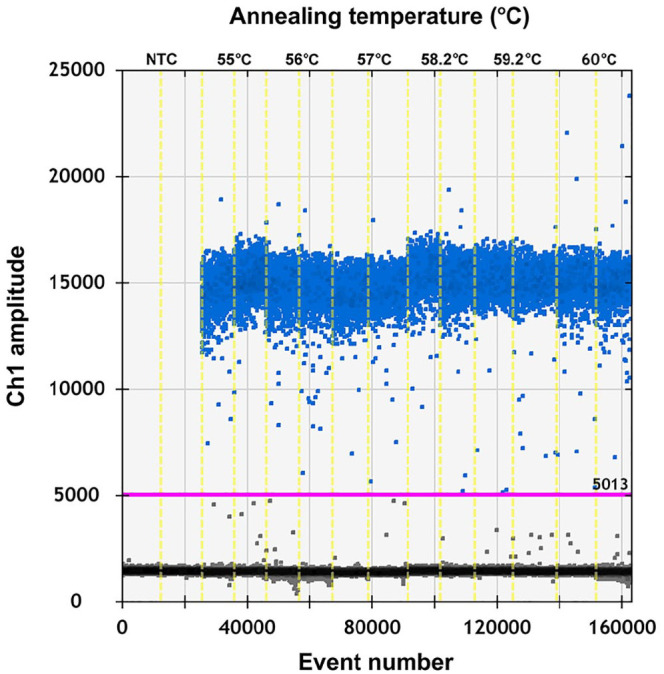
Influence of annealing temperature (55, 56, 57, 58.2, 59.2, and 60°C) on the porcine epidemic diarrhea virus droplet digital PCR system.
NTC = no-template control.
Figure 2.
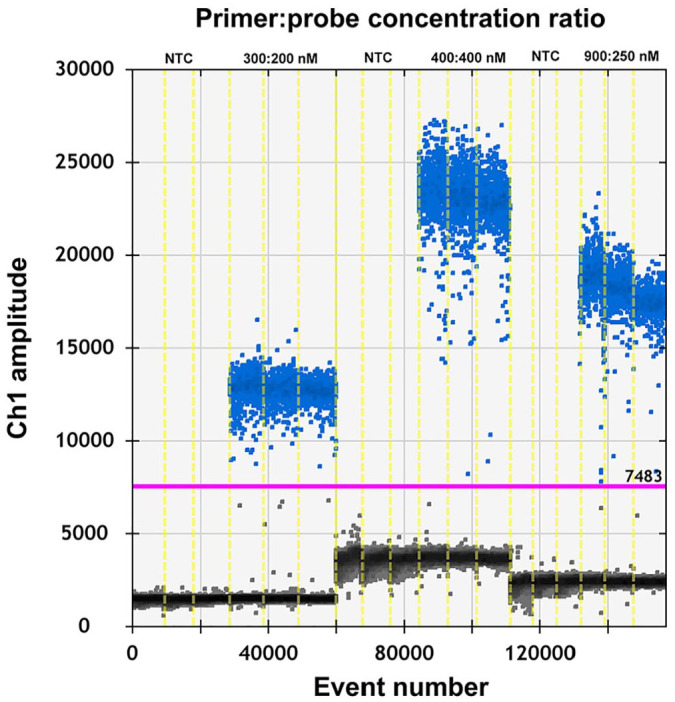
Influence of primer-to-probe concentration ratio (300:200 nM, 400:400 nM, and 900:250 nM) used in the porcine epidemic diarrhea virus droplet digital PCR system.
NTC = no-template control.
In specificity tests of the rtPCR and ddPCR assays, the nucleic acid extracts used as reaction templates included: porcine pseudorabies virus (Suid alphaherpesvirus 1), classical swine fever virus (CSFV; Pestivirus C), porcine circovirus 2, porcine reproductive and respiratory syndrome viruses (PRRSV; Betaarterivirus suid 1 and 2), Actinobacillus pleuropneumoniae, Haemophilus parasuis, Streptococcus suis, porcine deltacoronavirus (PDCoV), transmissible gastroenteritis virus and porcine respiratory coronavirus (TGEV, PRCV; Alphacoronavirus 1), swine acute diarrhea syndrome coronavirus (SADS-CoV), Escherichia coli, Salmonella typhimurium, Clostridium (Clostridioides) difficile, Clostridium perfringens, and PEDV; RNase-free H2O was used as a negative control. All tests correctly identified the target strains without producing false-positive or false-negative results, thereby confirming the specificity of these 2 assays.
A total of 147 clinical specimens (porcine small intestine, feces, and serum) were obtained from 10 pig farms in China. Field strains and clinical samples were obtained from a commercial company (Yongshun Biological Pharmaceutical). Total RNA/DNA was extracted (RaPure viral RNA/DNA kit; Magen Technologies), and the RNA derived from PRRSV, CSFV, PEDV, PDCoV, TGEV, PRCV, SADS-CoV, and clinical specimens was reverse transcribed (Primer Script RT reagent kit; TaKaRa) according to the kit manufacturer’s instructions. In the repeatability tests of the ddPCR reaction system, 10-fold diluted PEDV plasmids (PUC57-N) were simultaneously tested in triplicate. The coefficients of variation (CV) and the concentrations (copies/μL) were calculated. These experiments revealed that the intra-assay CV for concentration was 2.8–7.44% and that the CV of the inter-assay was 8.5–14.4%, which indicated that the repeatability of the ddPCR reaction system was high (Table 1).
Table 1.
Robustness and repeatability of the droplet digital PCR assay for porcine epidemic diarrhea virus.
| Concentration of PUC57-N plasmid (copies/μL) | Intra-assay |
Inter-assay |
||||||
|---|---|---|---|---|---|---|---|---|
| Viral load of 3 tests (copies/μL) | CV% | Viral load of 3 tests (copies/μL) | CV% | |||||
| 4 × 103 | 23.3 | 22.6 | 23.9 | 2.80 | 20 | 22 | 18 | 10.7 |
| 4 × 104 | 226 | 241 | 262 | 7.44 | 230 | 239 | 297 | 14.4 |
| 4 × 105 | 2,370 | 2,510 | 2,380 | 3.29 | 2,120 | 2,250 | 2,500 | 8.5 |
To compare the sensitivity, linearity, and quantification agreement between ddPCR and rtPCR, 10-fold diluted templates of plasmid PUC57-N (4 × 108–4 × 101 copies/μL) were used to conduct parallel tests (Table 2). The detection limit of ddPCR was 0.26 copies/μL; the detection limit of rtPCR was 1.47 copies/μL. Thus, the sensitivity of the ddPCR assay was 5.7 times higher than that of rtPCR. In this experiment, both rtPCR (R2 = 0.999) and ddPCR (R2 = 0.992) exhibited excellent linearity according to the regression analysis (Fig. 3A, 3B). The Pearson correlation coefficient was used to assess the correlation between ddPCR and rtPCR assays at each concentration of PEDV. The correlation coefficient between rtPCR and ddPCR was 1, indicating that the 2 methods were positively correlated (Fig. 3C).
Table 2.
Comparison of real-time PCR (rtPCR) and droplet digital PCR (ddPCR) assays using serially diluted porcine epidemic diarrhea virus plasmids.
| Concentration of PUC57-N plasmid (copies/μL) | rtPCR (mean Cq value) | ddPCR (mean concentration,* copies/μL) |
|---|---|---|
| 4 × 108 | 11.0 | ND |
| 4 × 107 | 14.7 | ND |
| 4 × 106 | 18.2 | 100,000 |
| 4 × 105 | 21.7 | 2,420 |
| 4 × 104 | 25.2 | 243 |
| 4 × 103 | 28.6 | 23.3 |
| 4 × 102 | 32.1 | 1.47 |
| 4 × 101 | Neg | 0.26 |
Cq = quantification cycle; ND = not detected; Neg = 1 replicate of 3 was negative.
Concentration based on ddPCR detection.
Figure 3.
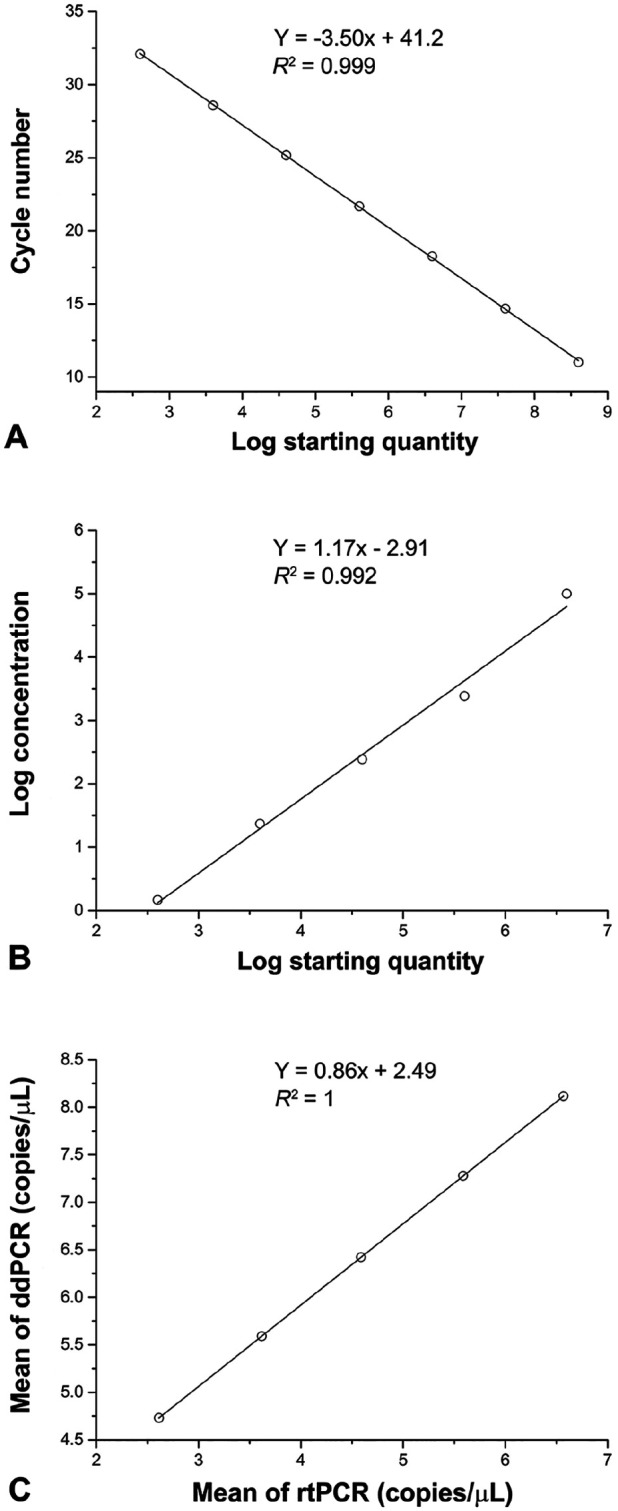
Sensitivity comparison between droplet digital PCR (ddPCR) and real-time PCR (rtPCR) using serially diluted porcine epidemic diarrhea virus (PEDV) plasmids. A. rtPCR standard curve generated from a 10-fold dilution series of PEDV plasmids. The quantification correlation was obtained by plotting the quantification cycle value against the log starting concentration. B. Standard curve generated by ddPCR. The quantification correlation was obtained by plotting the log absolute concentration against the log starting concentration. C. Quantitative correlation of mean values for each concentration of PEDV plasmids between ddPCR and rtPCR methods.
The 147 clinical specimens were tested simultaneously using ddPCR and rtPCR to compare the sensitivities of the assays. PEDV was detected with a positive rate of 14.3% (21 of 147) by ddPCR and 12.9% (19 of 147) by rtPCR. The overall coincidence rate was 98.6%, the kappa value was 0.942, and 95% CI was 0.863~1.022. Two of the samples identified as negative by rtPCR were positive by ddPCR (Table 3). The conflicting results detected by ddPCR and rtPCR were subsequently sequenced, and the sequencing results verified that these samples were positive for PEDV. The conflicting results from these samples could be a result of the higher sensitivity provided by the ddPCR assay compared to that of the rtPCR method.
Table 3.
Comparison of droplet digital PCR (ddPCR) and real-time PCR (rtPCR) assays for porcine epidemic diarrhea virus using 147 clinical specimens.
| rtPCR | ddPCR | Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 19 | 0 | 19 |
| Negative | 2 | 126 | 128 |
| Total | 21 | 126 | 147 |
Our results indicated that the 2 methods had a high coincidence and that the ddPCR assay provided a more sensitive method for the precise quantification of PEDV compared to that of the rtPCR system, particularly for detecting lower concentrations of PEDV.9 Additionally, the ddPCR system quantified the DNA in a highly reproducible manner without relying on a standard curve.2,8 However, based on analyzing 96 samples, the ddPCR took 3.3 times longer to complete than the rtPCR, the operation of ddPCR was more complicated than the rtPCR, and the overall cost (consumables and labor) of ddPCR was 2 times higher than rtPCR.2,18 In terms of high-throughput detection, the ddPCR instrument has only 2 fluorescence channels, testing a maximum of 4 target genes; the rtPCR instrument has 5 fluorescence channels, testing a maximum of 5 target genes.3,10 In general, the ddPCR system was verified as a sensitive and accurate method for detecting PEDV in clinical molecular virology.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest in regard to the research, authorship, and/or publication of this article.
Funding: This work was supported by the National Key Research and Development Program of China (grant 2016YFD0500600) and the Natural Science Foundation of Guangdong Province of China (grant 2017A030310175).
ORCID iD: Lei Ye  https://orcid.org/0000-0003-1696-3701
https://orcid.org/0000-0003-1696-3701
References
- 1. Alexandra BK, et al. Droplet volume variability as a critical factor for accuracy of absolute quantification using droplet digital PCR. Anal Bioanal Chem 2017;409:6689–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cao W, et al. Species identification and quantification of silver pomfret using the droplet digital PCR assay. Food Chem 2020;302:125331. [DOI] [PubMed] [Google Scholar]
- 3. Dobnik D, et al. Multiplex quantification of four DNA targets in one reaction with Bio-Rad droplet digital PCR system for GMO detection. Sci Rep 2016;6:35451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Floren C, et al. Species identification and quantification in meat and meat products using droplet digital PCR (ddPCR). Food Chem 2015;173:1054–1058. [DOI] [PubMed] [Google Scholar]
- 5. Huang Y-W, et al. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 2013;4:e00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kiselinova M, et al. Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA. PLoS One 2014;9:e85999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee C. Erratum to: Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol J 2015;12:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lievens A, et al. Measuring digital PCR quality: performance parameters and their optimization. PLoS One 2016;11:e0153317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu Y, et al. Development of a droplet digital polymerase chain reaction for sensitive and simultaneous identification of porcine circovirus type 2 and 3. J Virol Methods 2019;270:34–37. [DOI] [PubMed] [Google Scholar]
- 10. Mokany E, Todd AV. MNAzyme qPCR: a superior tool for multiplex qPCR. Methods Mol Biol 2013;1039:31–49. [DOI] [PubMed] [Google Scholar]
- 11. Notomi T, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 2000;28:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ogawa H, et al. Multiplex PCR and multiplex RT-PCR for inclusive detection of major swine DNA and RNA viruses in pigs with multiple infections. J Virol Methods 2009;160:210–214. [DOI] [PubMed] [Google Scholar]
- 13. Qian S, et al. Isolation and identification of porcine epidemic diarrhea virus and its effect on host natural immune response. Front Microbiol 2019;10:2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ren X, et al. Development of reverse transcription loop-mediated isothermal amplification for rapid detection of porcine epidemic diarrhea virus. Virus Genes 2011;42:229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Song D, et al. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin Exp Vaccine Res 2015;4:166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun P, et al. Sequence and phylogenetic analyses of the M and N genes of porcine epidemic diarrhea virus (PEDV) strains in Anhui Province, China. Genet Mol Res 2015;14:13403–13413. [DOI] [PubMed] [Google Scholar]
- 17. Wang Q, et al. Droplet digital PCR (ddPCR) method for the detection and quantification of goat and sheep derivatives in commercial meat products. Eur Food Res Technol 2017;244:1–8. [Google Scholar]
- 18. Yang R, et al. Comparison of next-generation droplet digital PCR (ddPCR) with quantitative PCR (qPCR) for enumeration of Cryptosporidium oocysts in faecal samples. Int J Parasitol 2014;44:1105–1113. [DOI] [PubMed] [Google Scholar]
- 19. Zhang X, et al. Identification and pathogenicity of a variant porcine epidemic diarrhea virus field strain with reduced virulence. Virol J 2015;12:88. [DOI] [PMC free article] [PubMed] [Google Scholar]


