Abstract
We describe the application of the NS3 cis-protease from hepatitis C virus (HCV) as a Ligand-Inducible Connection (LinC) that can be used to control the function and localization of engineered proteins in mammalian cells. The versatility of the approach is demonstrated in the design of drug-sensitive transcription factors (TFs) and transmembrane signaling proteins, the activities of which can be tightly and reversibly controlled using clinically-tested antiviral protease inhibitors.
A goal of synthetic biology is to program new functions into cells in ways that can be precisely manipulated for applications in medicine and basic research. Toward this end, researchers have recombined modular domains from natural signaling proteins and genetic control elements to produce new biological “parts” for cellular engineering.1 Among the parts that have been especially valuable in synthetic biology are a set of ligand-binding proteins that can be used to engineer drug-sensitivity into protein systems. These tools have been applied to render numerous cellular processes subject to drug-control, including transcription,2 cell signaling,3–4 genome-editing,5–6 and many others. However, as the goals and capabilities in synthetic biology continue to expand, there remains a need for new inducible platforms.
We set out to devise a new and modular platform that could not only be combined with the extant methods, but also generalized to many different systems. Given recent interest in developing drug-controllable cell-based therapies,7 we were especially interested in designing a strategy through which synthetic gene circuits and signaling pathways could be precisely gated using safe and well-characterized small molecules. Seeking to create an orthogonal system, we targeted inducer compounds that are typically absent from the environment, and in particular those with binding preference for non-human proteins. In order to identify such inducers, we turned our attention to antiviral drugs, especially those that have been previously tested (or are approved) for clinical use in humans.
Here, we describe use of the HCV NS3 protease domain and its various inhibitors as a protein-ligand pair that can be applied to install drug-sensitivity into both intracellular and cell-surface proteins. In its natural context, NS3 is a serine cis-protease that excises itself from the HCV polyprotein by cleaving recognition sites that flank it at either end.8 Because it is essential for HCV replication, numerous inhibitors targeting the viral protease have been developed. In previous work, cis-cleavage by NS3 has been used to conditionally link proteins to imaging tags and degradation sequences,9–11 and NS3 trans-proteolysis has been applied to control the activity of T7 RNA polymerase.12 Given its successful application in these non-natural contexts, we asked whether the viral protease could be used to design drug-sensitive systems and gain control over complex cellular processes such as protein localization, transcription, and intercellular communication.
We started by applying NS3 to design drug-sensitive TFs, using the protease as a Ligand-Inducible Connection (LInC) to control the association between modular DNA-binding (DB) and transcriptional activation (TA) domains. In an initial design, the protease was inserted between minimal DB and TA sequences sourced from the yeast TF Gal4, generating Gal4DB-NS3-Gal4TA, (Fig. 1a). In this configuration, we expected that the viral protease would serve as a self-immolating connection, excising itself from the fusion construct and, in doing so, separating the DB and TA elements. However, in the presence of an NS3 inhibitor, we anticipated that self-excision of the protease would be blocked, resulting in the preservation of full-length TF capable of activating the expression of targeted genes.
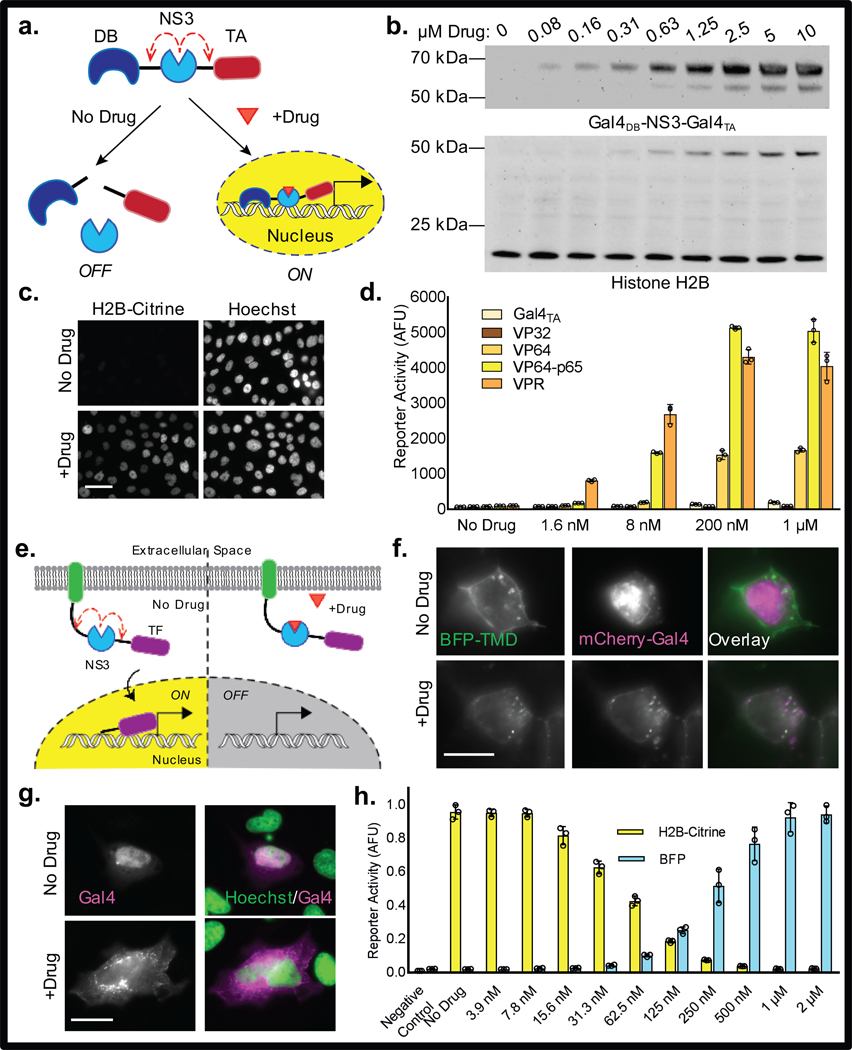
Figure 1. Drug-inducible “turn-on” and “turn-off” TFs.
(a) Design of the drug-inducible “turn-on” TFs. (b) Western blot showing accumulation of full-length Gal4DB-NS3-Gal4TA (anti-HA; 60.6 kDa) in response to BILN-2061, and corresponding H2B-Citrine expression (anti-H2B; 46.5 kDa). Endogenous histone H2B (13.8 kDa) served as a loading control. (c) Fluorescence images of CHO-derived reporter cells (UAS H2B-Citrine) stably expressing Gal4DB-NS3-Gal4TA in the absence and presence of 3 μM BILN-2061. Scale bar is 50 μm. (d) H2B-Citrine fluorescence as determined by flow cytometry in HEK 293FT cells transiently expressing TFs with the indicated TA domains and treated with varying concentrations of BILN-2061. (e) General design of the “turn-off” TFs containing a generic membrane-localizing element (green). (f) Fluorescence images of HEK 293A cells transiently expressing BFP-TMD-NS3-Gal4-mCherry. BFP is N-terminal to the TMD; Gal4 is tagged with mCherry. Scale bar is 10 μm. (g) Immunostained HeLa cells showing drug-induced membrane-targeting of Gal4min via myr-palm-NS3-Gal4min. Drug is 3 μM BILN-2061. Scale bar is 25 μm. (h) Fluorescence emission from HEK 293FT containing UAS H2B-Citrine and TRE BFP reporter constructs and transiently coexpressing rTetR-NS3-VP64-p65 and TMD-NS3-Gal4min. Cells were treated with 100 ng/mL doxycycline and varying concentrations of BILN-2061. Negative controls correspond to background activation of each reporter in cells containing mismatched TFs. Values plotted in (d) and (h) represent means +/− s.d., n = 3 biologically independent samples based on geometric mean fluorescence determined by flow cytometry.”
To determine whether Gal4DB-NS3-Gal4TA behaved in this manner, the protein was expressed in a mammalian cell line containing a stably-integrated Gal4-dependent reporter construct (UAS H2B-Citrine). Immunoblotting showed that, although full-length Gal4DB-NS3-Gal4TA copies were not detected in drug-untreated cells, formation of the intact TF was rapidly induced upon exposure to the NS3 inhibitor (Fig. 1b, Supplementary Figure 1). In addition to preserving newly made Gal4DB-NS3-Gal4TA copies, drug-treatment also induced expression of the H2B-Citrine reporter protein in a dose-dependent manner (Fig. 1b-c). Various inhibitors were tested, and multiple compounds capable of activating robust transcriptional responses were identified, including BILN-2061, asunaprevir, danoprevir, and grazoprevir (Supplementary Figure 2). Together, these results indicate that the LInC strategy can be used to precisely regulate the association of TF elements in order to achieve inducible control over gene expression.
Given the modular framework of our system, we predicted that substitution of the Gal4TA domain with more potent transcriptional effectors would yield TFs with enhanced drug sensitivity. Indeed, LInC-containing TFs possessing VP64, VP64-p65, or VPR13 as TA domains resulted in higher reporter activation levels at decreased drug concentrations as compared to the initial Gal4DB-NS3-Gal4TA (Fig. 1d, Supplementary Video 1). In addition, TFs with activity against alternative promoters were also designed, including one in which the reverse tetracycline repressor (rTetR) was used as the DB element (rTetR-NS3-VP64-p65). In transfected cells, rTetR-NS3-VP64-p65 exhibited “AND” gate activity, requiring both doxycycline and an NS3 inhibitor in order to activate transcription from the tetO-containing TRE promoter (Supplementary Figure 3). Notably, the effects of TF preservation and gene activation were reversed following inhibitor removal (Supplementary Figure 4).
In order to complement the “turn-on” systems described above, we also designed a strategy in which NS3 inhibitors could be applied to “turn-off” gene expression. In this approach, NS3 was used to conditionally tether an intact Gal4 (Gal4min) unit to a membrane-targeting domain, with the expectation that protease inhibition could be used to precisely control the amount of soluble versus membrane-bound TF.
Using a Type-l transmembrane protein as a targeting element, we generated a fusion construct containing NS3 and Gal4min as a C-terminal cytosolic domain (TMD-NS3-Gal4min) (Fig. 1e). Fluorescence imaging of cells expressing a dual-tagged version of the protein (BFP-TMD-NS3-Gal4min-mCherry) showed that Gal4min was released from its BFP-fused transmembrane domain in drug-untreated cells, resulting in a liberated TF unit that localized predominantly to the nucleus (Fig. 1f). However, in cells in which NS3 activity had been inhibited, the TF remained attached to its targeting element and thus trafficked to endoplasmic reticulum (ER) and plasma membrane (PM). A version in which an N-terminal myristoylation and palmitoylation substrate was used as the targeting sequence (myr-palm-NS3-Gal4min) exhibited similar behavior, becoming occluded from the nucleus in drug-treated cells (Fig. 1g).
Given that TFs localize to the nucleus in order to bind their DNA targets, we anticipated that TMD-NS3-Gal4min and myr-palm-NS3-Gal4min would facilitate target gene expression in a manner that could be down-regulated through NS3 inhibition. Consistent with this prediction were analyses showing that exposure to NS3 inhibitors led to the accumulation of membrane-tethered Gal4min, (Fig. 1f-g, Supplementary Figure 5), as well as the gradual depletion of previously-cleaved TF copies (Supplementary Figure 6). Measurements by flow cytometry confirmed that drug treatment suppressed reporter gene expression in a dose-dependent manner (Supplementary Figure 5), and live-cell imaging showed that the effect of downregulation could be reversed following inhibitor withdrawal (Supplementary Video 2). Together, these results demonstrate that TFs can be conditionally linked to localization signals to permit precise control over their subcellular distributions and activities.
Recognizing that natural gene expression often involves the synchronized control of multiple target genes, we next combined the “turn-on” and “turn-off” systems in order to create a platform for simultaneously regulating distinct promoters using drug. In cells that coexpressed both rTetR-NS3-VP64-p65 and TMD-NS3-Gal4min, we observed coinciding and inverse regulation of TRE- and UAS-controlled reporter constructs (Fig. 1h). Notably, “leaky” activation by each TF appeared to be low, comparing to that of control cells containing mismatched reporter-TF pairs (Fig. 1h). Together, these results demonstrate that multiple TFs can be controlled via LInC in order to achieve concurrent and opposing transcriptional changes in response to NS3 inhibition.
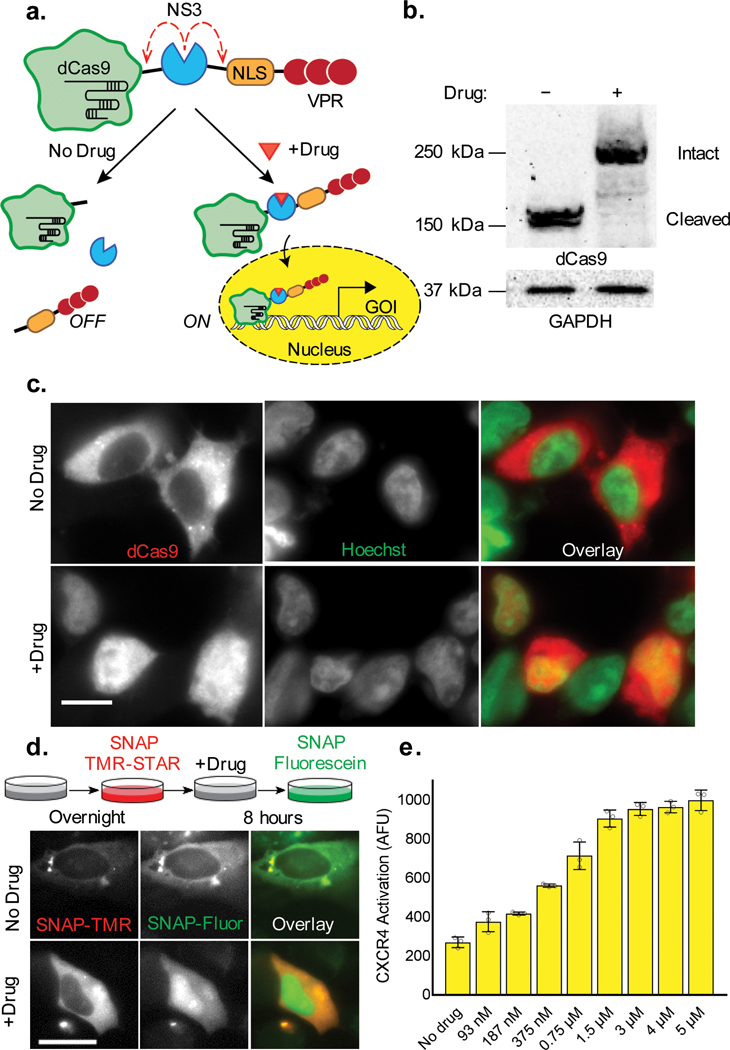
We also aimed to create a system that could be used to regulate expression from endogenous promoter sites. In order to achieve such control, we integrated the LInC module into dCas9-VPR13, positioning the viral protease in between the DB scaffold and a C-terminal region containing a nuclear localization sequence (NLS) and the VPR TA element (dCas9-NS3-NLS/VPR) (Fig. 2a). In this configuration, we anticipated that NS3 cleavage would not only inactivate the TF, but also generate a cytoplasmically contained dCas9 cleavage product. Indeed, immunoblotting showed that intact dCas9-NS3-NLS/VPR copies accumulated only drug-treated cells (Fig. 2b), and immunofluorescence confirmed that dCas9 localized to the nucleus in a drug-dependent manner (Fig. 2c, Supplementary Figure 7). In addition, time-dependent analyses using a SNAP-tag fusion (SNAP-dCas9-NS3-NLS/VPR) showed that only TF copies made in the presence of NS3 inhibitor were transported across the nuclear envelope (Fig. 2d). The cytoplasmic containment of cleaved dCas9 may serve to minimize any undesired repression in drug-untreated cells, as the unfused domain has been reported to suppress gene expression in certain cases through binding and occupying targeted DNA sites.16
Figure 2. Drug control over endogenous gene expression using dCas9-NS3-NLS/VPR.
(a) Schematic depicting the preservation and nuclear localization of dCas9-NS3-NLS/VPR upon NS3 inhibition. (b) Western blot showing the cleavage state of dCas9-NS3-NLS/VPR in untreated and drug-treated HEK 293FT cells (3 μM BILN-2061; dCas9-NS3-NLS/VPR, 249.4 kDa; cleaved dCas9, 160.6 kDa). (c) Immunolocalization of the dCas9 domain in untreated and drug-treated HEK 293FT. (d) Schematic of the time-dependent dye labeling strategy used to track SNAP-dCas9-NS3-NLS/VPR localization in live cells (top), and corresponding fluorescence images of temporally-defined protein populations in dye-labeled HeLa cells (bottom). (e) Drug-induced upregulation of CXCR-4 expression in HEK 293FT cells containing dCas9-NS3-NLS/VPR and an sgRNA targeting the endogenous CXCR4 promoter. Plotted values are mean + s.d. of n=3 biologically independent samples. All samples were analyzed 24 hours post-transfection. Scale bars are 25 μm.
To confirm that dCas9-NS3-NLS/VPR could be used to upregulate gene expression in a drug-inducible manner, the TF was co-expressed with sgRNAs targeting either a fluorescent reporter construct (UAS H2B-Citrine), or the promoter region of the endogenous CXCA4 gene. Analyses by flow cytometry and fluorescence imaging showed that the LInC-containing TF was able to upregulate each target in a dose-dependent and sgRNA-specified manner (Fig. 2e, Supplementary Figure 8, Supplementary Video 3). In addition, tests using separate sgRNAs targeting distinct CXCR4 promoter regions showed that under saturating drug concentrations, dCas9-NS3-NLS/VPR was able to upregulate CXCR-4 to a similar extent as was achieved using dCas9-VPR (Supplementary Figure 9). A system in which LInC was used to control TA domain recruitment to hairpin-modified sgRNA15 was also developed (Supplementary Figure 10). Together, these data indicate that NS3 can also be combined with dCas9 to achieve tunable control over the transcription of endogenous genes.
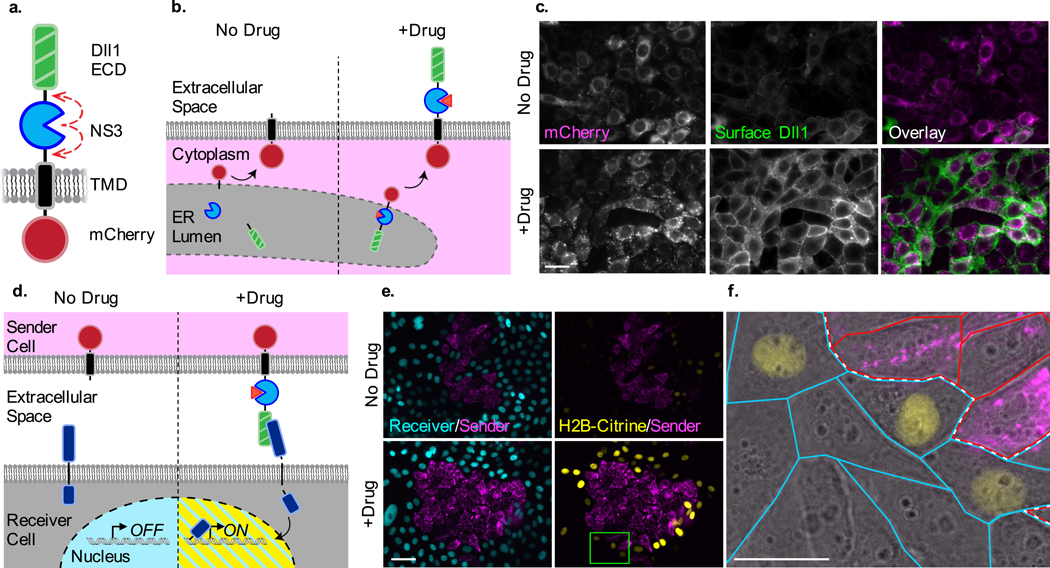
Certain transmembrane signaling proteins are also known to possess component-based architecture, including the Notch receptor, its ligands, and their synthetic derivatives.16–19 Thus, we predicted that LInC could also be used to regulate intercellular communication via the Notch/SynNotch pathway. We designed an NS3-containing version of the Notch ligand Delta-like 1 (DII1-NS3), positioning the cis-protease between the ligand’s ecto- and transmembrane-domains (Fig. 3a). In this configuration we anticipated that the self-excision of NS3 would yield a soluble ligand that, due to its lack of a membrane tether, would not be displayed on the plasma membrane (Fig. 3b). Indeed, immunostaining of cells stably expressing DII1-NS3 showed that the surface presentation of DII1-NS3 was induced upon NS3 inhibition (Fig. 3c).
Figure 3. Drug control over ligand presentation and intercellular Notch signaling.
(a) Design of Dll1-NS3-mCherry and (b) schematic of its drug-inducible preservation in response to NS3 inhibition. (c) Immunofluorescent detection of cell-surface localized Dll1-NS3-mCherry in untreated and drug-treated cells (1.5 μM BILN-2061). (d) Schematic depicting the drug-induced activation of Notch “receiver” cells by Dll1-NS3-mCherry expressing “sender” cells in trans. (e) Fluorescence images of cocultured sender and receiver cells in the absence or presence of drug (1.5 μM BILN-2061). Sender cells expressed the DII1-NS3-mCherry ligand (magenta). Receiver cells constitutively expressed H2B-Cerulean (cyan) and human Notch1, and conditionally expressed H2B-Citrine upon Notch activation (yellow). The interface between sender and receiver cell colonies is denoted by the white dotted line. (f) Magnified region from the inset shown in (e) shown as an overlay with transmitted light image. Scale bars are 25 μm.
In the prevailing model of Notch activation, the endocytosis of membrane-tethered ligand is thought to deliver a mechanical “pulling” energy that is required to trigger the “on” state of the receptor, in turn inducing the release of its intracellular domain (NICD, a transcriptional effector). Because Notch activation requires the endocytosis of membrane-tethered ligands,20 we anticipated only drug-preserved DII1-NS3 copies would be able to mediate trans-cellular signaling. To determine whether protease-containing ligand could activate Notch signaling in a drug-dependent manner, “sender” cells expressing DII1-NS3 were combined with Notch1-expressing “receiver” cells in a coculture assay (Fig. 3d). Using receiver cells containing a NICD-dependent fluorescent reporter gene (12xCSL H2B-Citrine), we observed Notch activation at sender cell-receiver cell interfaces only in drug-treated cocultures (Fig. 3e-f, Supplementary Figure 11). Thus, in addition to serving as a versatile module for controlling intracellular proteins, these results demonstrate that the LInC strategy can also be applied in luminal and cell-surface contexts in order to regulate cell-cell communication.
The applications described above demonstrate that the LInC strategy can be used to straightforwardly engineer drug-sensitivity into both intracellular and cell-surface proteins. A significant advantage of the method is the availability of highly-selective NS3 inhibitors, the pharmacological properties of which have previously been characterized (Supplementary Table 1). Finally, in addition to its potential applications in basic biology investigations, the LInC approach may also serve as a powerful strategy for regulating therapeutic cells in vivo using safe and clinically-approved drugs.
ONLINE METHODS
DNA constructs
Plasmid DNA and detailed sequence information for expression vectors encoding Gal4DB-NS3-Gal4TA, Gal4DB-NS3-VP64, Gal4DB-NS3-VP64-p65, rTetR-NS3-VP64-p65, myr-palm-NS3-Gal4min, dCas9-NS3-NLS/VPR, and SNAP-dCas9-NS3-NLS/VPR can be obtained via AddGene. Standard cloning procedures were used in the generation of all DNA constructs. The pEV-UAS-H2B-citrine reporter plasmid was a gift from Michael Elowitz (Caltech), the TRE-mTagBFP reporter plasmid was a gift from Wilson Wong (Boston University), the 5xGAL4-TATA-luciferase reporter plasmid (Addgene #46756) was a gift from Richard Maurer (Oregon Health Sciences University), the Tet-inducible mCherry reporter (Addgene #64128) and sgRNA1_Tet-inducible Luciferase plasmids (Addgene #64128) were gifts from Moritoshi Sato (University of Tokyo).
NS3 inhibitors
Asunaprevir, boceprevir, danoprevir, MK-5172 (a.k.a., grazoprevir), and simeprevir were from MedChemExpress. Telaprevir was from Selleck Chemicals. BILN-2061 was a gift from Roger Tsien and Stephen Adams (UC San Diego). Concentrated NS3 inhibitor stocks were dissolved in DMSO at concentrations between 3 – 10 mM and diluted into cell culture media at the indicated working concentrations.
Mammalian cell culture
All mammalian cell lines were cultured in a humidified incubator maintained at 37° C with 5% CO2. HEK 293FT cells (ThermoFisher) were cultured in Dulbecco’s modified Eagle medium (DMEM) containing with 10% FBS and supplemented with nonessential amino acids (Life Technologies), Glutamax (Life Technologies), and G418 (500 μg/mL, Invivogen). HeLa cells were obtained from ATCC and were cultured in DMEM containing 10% FBS and supplemented with Glutamax and penicillin/streptomycin. Stable cell lines based on CHO T-Rex (ThermoFisher) were were maintained in DMEM containing 10% FBS and supplemented with nonessential amino acids and glutamax.
DNA transfections
DNA transfections were performed using Lipofectamine 3000 Reagent (ThermoFisher) according to manufacturer’s instructions. For imaging experiments, cells were seeded in dishes or well plates containing coverslip bottoms either coated with bovine plasma fibronectin (Product #F 1141, Sigma-Aldrich) or treated for cell-adherence by the manufacturer (poly-D-lysine by MatTek, or ibiTreat by Ibidi).
Stable cell line generation
Stable cell lines were generated from previously reported CHO-K1 T-REx cells containing stably integrated Gal4- and Notch-dependent reporter constructs (UAS H2B-Citrine and 12xCSL H2B-Citrine, respectively; Sprinzak et al. 2010), which were gifts from Michael Elowitz (Caltech). Briefly, cells were transfected with linearized DNAs encoding the engineered protein of interest as well as antibiotic resistance gene for mammalian selection. Transfections were performed in 24-well plates containing 160,000 cells per well seeded the approximately 24 hours prior to transfection. At 48 hours post-transfection, the cells were transferred to 6-well plates and exposed to antibiotic selection using hygromycin (500 μg/mL). Upon elimination of non-transfected control cells (typically after 10 days of culture in the presence of antibiotic), surviving cells were transferred into 96-well plates using a limited dilution procedure in order to isolate single clones (see “Cell Cloning by Serial Dilution in 96 Well Plates” protocol by Corning).
Antibodies
The following primary antibodies were used: mouse anti-Cas9 (Santa Cruz Biotechnology, sc-517386, 1:500 dilution for western blotting, 1:50 for immunostaining), mouse anti-human CD184 (CXCR-4) APC conjugate (BioLegend, 306510, 1:200 dilution for flow cytometry), polyclonal sheep anti-mouse/rat Dll1 (R&D Systems, AF3970, 1:50 dilution for immunostaining), rabbit anti-Histone H2B (Cell Signalling, 12364, 1:1,000 dilution for western blotting), mouse anti-HA-HRP (Santa-Cruz, sc-7392, 1:1,000 dilution for blotting), rabbit anti-GAPDH (Sigma-Aldrich, G9545, 1:3,000 dilution for western blotting), and polyclonal rabbit anti-Gal4 (Santa Cruz Biotechnology, sc-577, diluted 1:500 for western blotting, 1:200 for immunostaining). The following secondary antibodies were used: goat anti-human AlexaFluor64/conjugate (ThermoFisher, A-21445, 1:1,000 dilution), donkey anti-sheep AlexaFluor64/conjugate (ThermoFisher, A-21448, 1:1,000 dilution), goat anti-rabbit CF647 conjugate (Sigma-Aldrich, SAB4600184, 1:300 dilution), anti-mouse HRP conjugate (Cell Signalling, 7076, 1:3,000 dilution), and anti-rabbit HRP conjugate (Bio-Rad, 170–6515, 1:3,000 dilution).
Preparation of cell lysates for immunoblotting
Cell lysates used in immunoblotting analyses were prepared by direct lysis of drug-treated and untreated cells in 1X LDS-PAGE loading buffer (ThermoFisher) following removal of cell culture media. Such procedure was applied in order to immediately denature proteins upon lysis, thus preventing undesired NS3 cis-cleavage in cell lysates. Viscous solutions were formed upon addition of the lysis reagent, which were clarified through sonication followed by centrifugation. The lysates were subsequently analyzed by standard immunoblotting procedures and probed using the antibodies listed above at the indicated dilutions. Detection of the labeled antigens was carried by chemiluminescence via the SuperSignal West Pico PLUS Chemiluminescent Substrate (Pierce).
Immunofluorescence staining of fixed cells
Cells were rinsed with PBS prior to fixation with formaldehyde (4% v/v, diluted into PBS from fresh vials containing 16% solutions purchased from ThermoFisher). Cells were fixed for 10 minutes at room temperature, followed by rinsing with PBS (three times) to remove residual fixative. When necessary, cells were then permeabilized with Triton-X 100 (0.2%, v/v, in PBS) for 10 minutes, following by rinsing with PBS. Cells were blocked with BSA solution (5%, v/v in PBS) for approximately 30 minutes at room temperature prior to staining with primary antibody solution (typically in PBS, or in the appropriate solution as suggested by the antibody supplier) at the dilutions indicated above for 1 hour at room temperature. Cells were stained with secondary antibody solution (in PBS at the solutions indicated above) for 1 hour at room temperature before imaging.
Time-dependent dye labeling of SNAP-tagged proteins
HeLa cells were transfected with DNA encoding SNAP-dCas9-NS3-NLS/VPR as described above. Approximately 24 hours later, cells were labeled with the red fluorescent dye SNAP-Cell TMR STAR (New England Biolabs) in complete culture media according to the manufacturer’s protocol. The dye was removed by gentle aspiration of the media, followed by rinsing with pre-warmed complete media (three times) to remove residual dye. Cells were subsequently incubated in culture media containing 3 μM BILN-2061. After 8 hours, cells were then labeled with the green fluorescent SNAP-Cell Fluorescein (New England Biolabs). Cells were fixed with 4% formaldehyde prior to imaging.
Luciferase assay
CHO-K1 cells stably expressing Gal4DB-NS3-VP64 were transfected with DNA encoding a UAS regulated firefly luciferase reporter construct (5xGAL4-TATA-luciferase). A constitutively transcribed NanoLuciferase construct (pNL1.1.TK[Nluc/TK], Promega) was used as a Co-transfection control. Approximately 16 hours after transfection, cells were treated with either BILN-2061, or Grazoprevir (both at 3 uM). Following a 12 hour period of drug treatment, a time-series was initiated during which the drug-containing media was removed from individual wells and replaced with drug-free media over the course of a 48 hour period. At the end of the series (approximately 56 hours after the initial drug exposure, and 72 hours after transfection), the amount of luciferase and NanoLuciferase present in cells was quantified using the Nano-Glo Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. The CHO-K1 cell line used in these analyses also contained a stably integrated UAS H2B-Citrine reporter construct, and fluorescence imaging confirmed the activation of the Gal4-dependent H2B-citrine gene in all drug treated-wells.
Image acquisition and analysis
Cells were imaged by epifluorescence microscopy in imaging-compatible vessels containing glass coverslip bottoms (MatTek) or optically clear plastic bottoms (ibidi). During imaging, cells were maintained in PBS, standard culture media, or FluoroBrite DMEM (ThermoFisher). For time-lapse analyses, cells were imaged in culture media supplemented with 30 mM HEPES diluted from a 1 M stock (pH 7.2–7.5, ThermoFisher) and maintained at 37° C in a heated imaging chamber throughout the duration of the analysis (typically 24 hours). Images were acquired using the ZEN imaging software (Zeiss). Image files were processed using the ImageJ-based image analysis package Fiji. The images were contrasted uniformly across experiments, and where applicable, pixel intensity profiles were plotted using the plot profile tool in Fiji. For the time-lapse analyses, movies were created using the ZEN imaging software.
Flow cytometry
Cells analyzed by flow cytometry were gated for living cells by scatter detection. The geometric mean measured reporter fluorescence levels were reported in arbitrary fluorescence units (AFU). Reporter activation analyses were performed using stable single-clones, or cells transiently transfected with DNA encoding the analyzed TF (as indicated in the figure captions). For analyses carried out using transiently expressing cells, plasmid DNA encoding a constitutively expressed fluorescent protein marker was co-delivered at the time of transfection and used to identify positively transfected cells populations (see Supplementary Figure 12). Briefly, transfected cells were were gated to the top 1% of marker fluorescence of non-transfected control cells under the same condition. Transient expression experiments carried out using the “turn-on” TFs (shown in Figure 1d and 1i, and Supplementary Figure 3) were gated via detection of an mCherry marker that was expressed via an IRES sequence on the TF-encoding plasmid. Transfected cells were incubated for 24–48 h after transfection either in the presence or absence of the indicated NS3 inhibitors before being analysed using an Attune NxT flow cytometer (ThermoFisher). For the analyses in which rTetR-NS3-VP64-p65 and TMD-NS3-Gal4min were combined, 125,000 HEK 293FT cells were transfected with 25 ng of DNA (per ~125,000 cells) with DNA mixtures containing a 3:3:2:2 molar ratio of rfetR-NS3-VP64-p65 to TMD-NS3-Gal4 to TRE mTagBFP to UAS H2B Citrine (as approximated by DNA size).
For expression analyses carried out using dCas9-based TFs, HEK 293FT cells were seeded in 48-well plates and transfected at a density of ~80,000 cells per well. A total 250 ng of DNA was delivered per well in each experiment; sgRNA- and dCas9-encoding constructs were transfected at a 1:1 molar ratio as approximated based on DNA size. The constructs encoding the sgRNAs targeting human CXCA4 promoter were acquired from AddGene and were previously reported (“sgC2” and “sgC3”, Zalatan et al. 2015). Expression of the chemokine receptor was analysed via flow cytometry using a fluorescently-conjugated CXCR-4 antibody; geometric means were recorded.
Statistics and Reproducibility
All flow cytometry and luminescence assay data were collected using 3 biologically independent samples. For fluorescence imaging analyses, ≥3 images per condition were recorded (encompassing hundreds of cells), and representative images are displayed in the figures. For immunoblotting, analyses were repeated ≥3 times and a representative blots were chosen for display.
Life Sciences Reporting Summary
Further information regarding the experimental designs and analytical procedures carried out in this work is available in the accompanying Life Sciences Reporting Summary.
DATA AVAILABILITY AND ACCESSION CODE AVAILABILITY STATEMENTS
Original data for the experiments can be obtained through requests sent to the corresponding author (jtngo@bu.edu).
Supplementary Material
ACKNOWLEDGEMENTS
EPT was supported through a T32 NIH Training Grant awarded to Boston University (EB006359).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Editor’s summary
Ligand-Inducible Connection (LinC) combines HCV protease and protease inhibitors to offer an orthogonal chemogenetic approach for controlling protein functions including protein localization, gene expression, and cell-cell communication.
REFERENCES
- 1.Lienert F, Lohmueller JJ, Garg A, & Silver PA (2014). Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nature Reviews Molecular Cell Biology, 15, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gossen M, & Bujard H. (1992). Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proceedings of the National Academy of Sciences, 89(12). 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spencer DM, Wandless TJ, Schreiber SL, & Crabtree GR (1993). Controlling signal transduction with synthetic ligands. Science, 262(5136). 1019–1024. [DOI] [PubMed] [Google Scholar]
- 4.Goreshnik I. & Maly DJ (2010) A Small Molecule-Regulated Guanine Nucleotide Exchange Factor. Journal of American Chemical Society. 132(3), 938–940. [DOI] [PubMed] [Google Scholar]
- 5.Davis KM, Pattanayak V, Thompson DB, Zuris JA. & Liu DR Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nature Chemical Biology 11, 316–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rose JC Stephany JJ, Valente WJ, Trevillian BM, Dang, Bielas JH, Maly DJ, & Fowler DM (2017). Rapidly inducible Cas9 and DSB-ddPCR to probe editing kinetics. Nat. Methods 14, 891–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu CY, Roybal KT, Puchner EM, Onuffer J, & Lim WA (2015) Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science 350, aab4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. (1993) Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. Journal of Virology, 67(7), 3835–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin MZ, Glenn JS, & Tsien RY (2008) A drug-controllable tag for visualizing newly synthesized proteins in cells and whole animals. Proceedings of the National Academy of Sciences, 105(22), 7744–7749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palida SF, Butko MT, Ngo JT, Mackey MR, Gross LA, Ellisman MH, & Tsien RY (2015). PKMζ,but not PKCλ, is rapidly synthesized and degraded at the neuronal synapse. Journal of Neuroscience, 35(20), 7736–7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung HK, Jacobs CL, Huo Y, Yang J, Krumm SA, Plemper RK, Tsien RY & Lin MZ (2015). Tunable and reversible drug control of protein production via a self-excising degron. Nature Chemical Biology, 11(9),713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pu J, Chronis I, Ahn D, Dickinson BC (2015) A Panel of Protease-Responsive RNA Polymerases Respond to Biochemical Signals by Production of Defined RNA Outputs in Live Cells. Journal of the American Chemical Society, 137(51), 15996–15999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, lyer E, Lin S, Kiani S, Guzman C, Wiegand DJ, Ter-Ovanesyan DJ, Braff JL, Davidsohn E, Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, & Church GM Highly efficient Cas9-mediated transcriptional programming. Nature Methods, 12(4), 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Pinera Kocak, D.D., Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, & Gersbach CA (2013) RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nature Methods, 10(10), 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, & Lim WA (2015). Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell, 160(1–2), 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell, 154(2), 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon WR, Zimmerman B, He L, Miles LJ, Huang J, Tiyanont K, McArthur DG, Aster JC, Perrimon N, Loparo JJ, & Blacklow SC (2015). Mechanical allostery: evidence for a force requirement in the proteolytic activation of Notch. Developmental Cell, 33(6), 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morsut L, Roybal KT, Xiong X, Gordley RM, Coyle SM, Thomson M, & Lim WA (2016). Engineering customized cell sensing and response behaviors using synthetic notch receptors. Cell, 164(4), 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roybal KT, Rupp LJ, Morsut L, Walker WJ, McNally KA, Park JS, & Lim WA (2016). Precision tumor recognition by T cells with combinatorial antigen-sensing circuits. Cell, 164(4), 770–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varnum-Finney B, Wu L, Yu M, Brashem-Stein C, Staats S, Flowers D, Griffin JD, & Bernstein ID (2000). Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. Journal of Cell Science, 113(23), 4313–4318. [DOI] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 21.McPhee Fiona, et al. “Preclinical profile and characterization of the hepatitis C virus NS3 protease inhibitor asunaprevir (BMS-650032).” Antimicrobial agents and chemotherapy 5610 (2012): 5387–5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamarre Daniel, et al. “An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus.” Nature 426.6963 (2003): 186. [DOI] [PubMed] [Google Scholar]
- 23.Seiwert Scott D., et al. “Preclinical characteristics of the hepatitis C virus NS3/4A protease inhibitor ITMN-191 (R7227).” Antimicrobial agents and chemotherapy 5212 (2008): 4432–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Summa Vincenzo, et al. “MK-5172, a selective inhibitor of hepatitis C virus NS3/4a protease with broad activity across genotypes and resistant variants.” Antimicrobial agents and chemotherapy (2012): AAC-00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis C. (2015) Pharmacology/Toxicology NDA Review and Evaluation (application number: 208,261), Food and Drug Administration, Center for Drug Evaluation and Research (https:/Awww.accessdata.fda.gov/drugsatfda_docs/nda/2016/208261Orig1s000PharmR.pdf)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data for the experiments can be obtained through requests sent to the corresponding author (jtngo@bu.edu).