Abstract
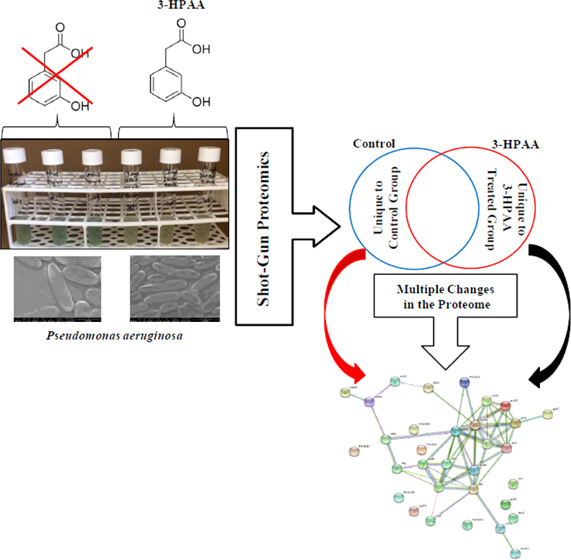
Pseudomonas aeruginosa, a widely distributed opportunistic pathogen, is an important threat to human health for causing serious infections worldwide. Due to its antibiotic resistance and virulence factors, it is so difficult to combat this bacterium; thus, new antimicrobial agents are in search. 3-Hydroxyphenylacetic acid (3-HPAA), which is a phenolic acid mostly found in olive oil wastewater, can be a promising candidate with its dose-dependent antimicrobial properties. Elucidating the molecular mechanism of action is crucial for future examinations and the presentation of 3-HPAA as a new agent. In this study, the antimicrobial activity of 3-HPAA on P. aeruginosa and its action mechanism was investigated via shot-gun proteomics. The data, which are available via ProteomeXchange with identifier PXD016243, were examined by STRING analysis to determine the interaction networks of proteins. KEGG Pathway enrichment analysis via the DAVID bioinformatics tool was also performed to investigate the metabolic pathways that undetected and newly detected groups of the proteins. The results displayed remarkable changes after 3-HPAA exposure in the protein profile of P. aeruginosa related to DNA replication and repair, RNA modifications, ribosomes and proteins, cell envelope, oxidative stress, as well as nutrient availability. 3-HPAA showed its antimicrobial action on P. aeruginosa by affecting multiple bacterial processes; hence, it could be categorized as a multitarget antimicrobial agent.
1. Introduction
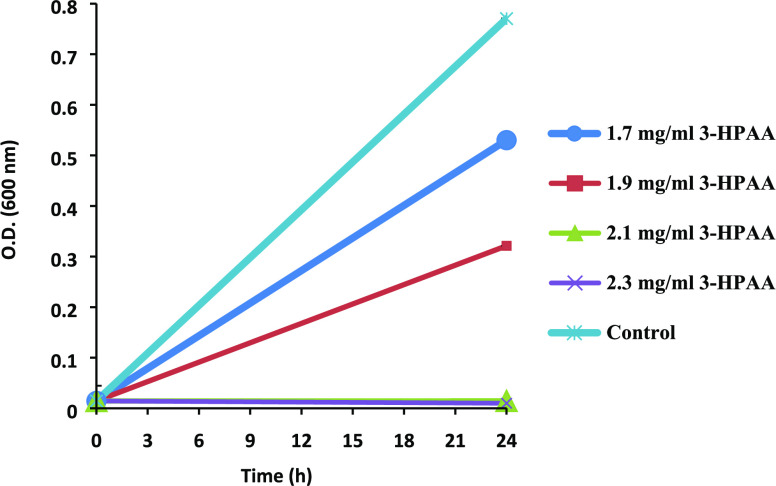
Pseudomonas aeruginosa, which is a Gram-negative bacterium, can colonize in soil, water, plant, and animal tissues in addition to the instruments and surfaces in the hospitals.1,2 Due to its highly diverse genotype and phenotype, it can switch the lifestyle according to the environmental conditions, leading to a rapid adaptation of broad changes in metabolism.3,4 These opportunistic pathogens are distributed extensively and accepted as one of the most critical agents in the nosocomial infections as well as the community-acquired ones, worldwide.5 They possess important virulence factors, including exotoxins and biofilm formation, in addition to their intrinsic and adaptive resistance abilities against various antibiotics.4,6 Their infections show high mortality and morbidity rates with a challenging treatment process, including extensive antibiotic therapy.7 Therefore, the effort for the development of new antimicrobial agents against P. aeruginosa is in progress. Phenolic acids, which can be accepted as promising antimicrobial agents, are secondary metabolites of the plants that function in various processes of the plant metabolism, including defense against predators and microorganisms.8,9 It is speculated that, because of their small size, phenolic acids can strongly interact with membranes or other targets in bacteria, such as easily cross through the outer membrane of Gram-negative bacteria,10 which can be an advantage in terms of antimicrobial effect. 3-Hydroxyphenylacetic acid (3-HPAA) is a phenolic acid that showed a dose-dependent antimicrobial effect on P. aeruginosa, either static or cidal, according to the studies of our group (Figure 1 and Table 1). 3-Hydroxyphenylacetic acid has a small and simple structure with only one hydroxy group on its benzene ring. It belongs to the hydroxycinnamic acid group of phenolic acids since the carboxylic group is attached to the benzene ring via an ethylene group.11 It is a water-soluble compound and mostly detected in olive oil wastewater, which is a byproduct of the olive oil extraction process.12 Although various studies demonstrated the antimicrobial effects of phenolic compounds on pathogens,13−16 studies focusing on the molecular mechanisms of antimicrobial effects of phenolic acids are limited in the literature. These mechanisms can be examined by omics-studies, to be able to present them as new antimicrobial agents.17 Since protein production is the final state of the molecular cell process under particular conditions, investigation of protein profile changes is a useful approach for obtaining potential action mechanisms in the field of antimicrobial discovery.18,19 Mass spectrometry is accepted as a reliable technique to examine bacterial processes such as the proteome comparison of different bacterial strains, determination of proteomic changes in the same bacterial strain under stress conditions such as a toxic compound exposure or determination of the proteomic basis of antibiotic resistance of the bacteria.20−26
Figure 1.
Growth of P. aeruginosa in the presence of various concentrations of 3-hydroxyphenylacetic acid (3-HPAA). The control group contains 0 mg/mL 3-HPAA. Optical density (OD) was measured at 600 nm.
Table 1. Percent Inhibition in Bacterial Growth and Cell Viability of P. aeruginosa in the Presence of 3-HPAA.
| 3-HPAA concentrations (mg/mL) | percent inhibitions (%) | cell viability (cfu/mL) |
|---|---|---|
| 1.7 | 31 | 5 × 106 |
| 1.9 | 58 | 3.4 × 106 |
| 2.1a | 98 | 1.9 × 105 |
| 2.3b | 99 | no survivors |
Minimum inhibitory concentration (MIC).
Minimum bacteriocidal concentration (MBC).
Herein, we report the antimicrobial effect of 3-HPAA and the changes in the protein profile of P. aeruginosa after treatment with 3-HPAA by liquid chromatography electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) accompanied with STRING and KEGG analyses to evaluate the protein–protein interactions and related metabolic pathways, respectively. To the best of our knowledge, this would be the first study to show proteomic changes of P. aeruginosa following 3-HPAA exposure. Thus, it would aid in improving the information about the antimicrobial effects of phenolic acids at the molecular level. Our study can be a starting point in the determination of the molecular action mechanism of 3-HPAA, which can be developed as an effective antimicrobial agent against important pathogenic bacteria in the future.
2. Results and Discussion
2.1. Antimicrobial Effect of 3-HPAA
The increasing concentrations of 3-HPAA resulted in a dose-dependent inhibition effect on the growth of P. aeruginosa (Figure 1). According to OD (600 nm) measurements, 1.9 mg/mL 3-HPAA showed a 58% inhibition, which is the subinhibitory concentration used in proteomic studies (Table 1). The minimum inhibitory concentration (MIC) was 2.1 mg/mL, with 98% inhibition. However, the enumeration studies demonstrated that 2.1 mg/mL had a bacteriostatic effect. Application of 2.3 mg/mL resulted in no survivors of P. aeruginosa, which was determined as minimum bacteriocidal concentration (MBC) (Table 1).
The concentration of 3-HPAA that resulted in 58% inhibition of bacterial growth (1.9 mg/mL) was determined as the 3-HPAA concentration used in the protein profile studies. The results demonstrate the dose-dependent antimicrobial effect of 3-HPAA on P. aeruginosa (Table 1).
2.2. Overall Protein Profile
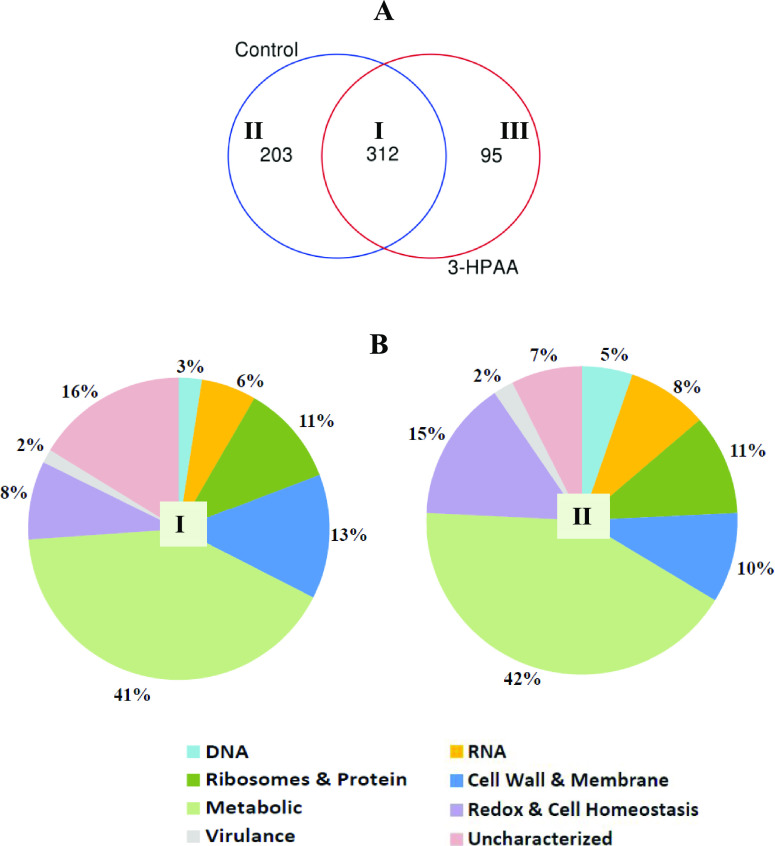
Bacterial response to changing environmental conditions such as exposure to toxic agents can be observed in their proteome.26 The application of 3-HPAA resulted in various changes in the protein profile of P. aeruginosa. When the MASCOT data (FDR < 1.3%) were subjected to Venn diagram analysis, 312 proteins were detected as mutually unchanged in both the control group and 3-HPAA-treated groups (Figure 2A, region I; Table S17). Besides, the proteins which showed changes depending on 3-HPAA exposure were also detected: 203 undetected proteins (unique to control group) (Figure 2A, region II) and 95 newly detected proteins (unique to 3-HPAA-treated group) (Figure 2A, region III). Depending on their functions reported in the UniProt database, these undetected and newly detected proteins were categorized into eight main groups: DNA-, RNA-, ribosomes and protein-, cell wall and membrane-, metabolism-, redox and cell homeostasis-, and virulence-related, as well as uncharacterized proteins. All proteins are presented in Tables S1–S17.
Figure 2.
Overall protein profile of P. aeruginosa after 3-HPAA exposure. (A) Venn diagram representation of protein profile after 3-HPAA exposure: (I) mutual proteins of the control and 3-HPAA-treated groups (unchanged proteins), (II) unique proteins of the control group (undetected proteins after 3-HPAA exposure), and (III) unique proteins of the 3-HPAA-treated group (newly detected proteins after 3-HPAA exposure). (B) Percentages of proteins categorized based on their functions after 3-HPAA exposure: (I) undetected proteins and (II) newly detected proteins.
The calculated percentage of the proteins in these groups (Figure 2B) showed that the lowest percentage of virulence-related proteins might demonstrate that 3-HPAA has a small effect on P. aeruginosa virulence. On the other hand, the highest percentage of metabolism-related proteins among these groups might be attributed to various changes in the protein profiles of general metabolism pathways for adaptation to the environment in the presence of 3-HPAA. Notable changes were recognized related to vital mechanisms of the bacteria after the exposure, which would be focused in this study as the basis of the antimicrobial effect of 3-HPAA (Tables 2 and 3).
Table 2. Significant Undetected Proteins of P. aeruginosa after 3-HPAA Exposure.
| protein IDaI | protein name | gene name | function | group of protein profileb |
|---|---|---|---|---|
| Q9HWG0 | UvrABC system protein A | uvrA PA4234 | nucleotide excision repair, SOS response | DNA |
| Q9HY08 | DNA mismatch repair protein | mutS PA3620 | mismatch repair | DNA |
| Q9HUJ8 | DNA topoisomerase 4 subunit B | parE PA4967 | DNA topological change | DNA |
| Q9HT76 | vitamin B12-dependent ribonucleotide reductase | nrdJA PA5497 | DNA biosynthetic process, DNA replication | DNA |
| P49988 | RNA polymerase sigma-54 factor | rpoN PA4462 | transcription initiation, sigma factor activity | RNA |
| Q9HU59 | transcriptional regulator NtrC | ntrC PA5125 | nitrogen fixation, regulation of nitrogen utilization, regulation of transcription | RNA |
| Q03456 | ferric uptake regulation protein | fur PA4764 | negative regulation of transcription | RNA |
| Q9HVY7 | stringent starvation protein A | sspA PA4428 | RNA polymerase core enzyme binding | RNA |
| Q9I382 | tRNA 2-selenouridine/geranyl-2-thiouridine synthase | selU PA1643 | tRNA seleno modification | RNA |
| Q9HY82 | ribonuclease T | rnt PA3528 | tRNA 3′-end processing | RNA |
| Q9I2U8 | glutamine-tRNA ligase | glnS PA1794 | glutamine-tRNA ligase activity | ribosomes and protein |
| Q9I2U7 | cysteine-tRNA ligase | cysS PA1795 | cysteine-tRNA ligase activity | ribosomes and protein |
| Q9HV58 | 30S ribosomal protein S15 | rpsO PA4741 | ribosome constituent, translation | ribosomes and protein |
| Q9HU56 | protein-export protein SecB | secB PA5128 | protein tetramerization, protein transport | ribosomes and protein |
| Q9HZC5 | aminopeptidase N | pepN PA3083 | peptide catabolic process, proteolysis | ribosomes and protein |
| O68822 | cytosol aminopeptidase | pepA phpA, PA3831 | release of an N-terminal amino acid, processing and regular turnover of intracellular proteins. | ribosomes and protein |
| Q9HT06 | membrane protein insertase YidC | yidC PA5568 | membrane insertase activity, insertion and/or proper folding and/or complex formation of integral membrane proteins into the membrane | cell wall and membrane |
| Q9I6C1 | signal recognition particle receptor FtsY | ftsY PA0373 | cotranslational protein targeting to membrane | cell wall and membrane |
| Q9HV48 | ATP-dependent zinc metalloprotease FtsH | ftsH PA4751 | cell division, response to antibiotic | cell wall and membrane |
| P50598 | Tol-Pal system protein TolQ | tolQ PA0969 | cell cycle, cell division, bacteriocin transport | cell wall and membrane |
| P23189 | glutathione reductase | gor PA2025 | cell redox homeostasis, response to oxygen radical | redox and cell homeostasis |
| Q9I6Z2 | alkyl hydroperoxide reductase | ahpF PA0140 | response to reactive oxygen species | redox and cell homeostasis |
| P53652 | superoxide dismutase [Mn] | sodA PA4468 | removal of superoxide radicals | redox and cell homeostasis |
| Q9I5R7 | S-adenosylmethionine decarboxylase proenzyme | speD PA0654 | S-adenosylmethioninamine biosynthetic process, spermidine biosynthetic process | redox and cell homeostasis |
| Q9I5F9 | lon protease | lon PA0779 | response to antibiotic, response to stress, nitric oxide metabolic process, peptidase activity | redox and cell homeostasis |
| Q9I2T9 | lon protease | lon PA1803 | response to antibiotics, response to drug, response to stress, pathogenesis, protein quality control for misfolded or incompletely synthesized proteins, single-species biofilm formation, type IV pilus-dependent motility, flagellum-dependent swarming motility | redox and cell homeostasis |
| Q9HTW6 | aminopeptidase P | pepP PA5224 | aminopeptidase activity | metabolism |
| Q9HWG9 | 5-methylphenazine-1-carboxylate 1-monooxygenase | phzS PA4217 | pyocyanine biosynthetic process | metabolism |
| O69753 | phenazine biosynthesis protein PhzB1 | phzB1 PA4211 | phenazine biosynthetic process | metabolism |
| Q7DC81 | phenazine biosynthesis protein PhzE | phzE1 phzE2, PA1903, PA4214 | glutamine metabolic process, tryptophan biosynthetic process, phenazine biosynthetic process | metabolism |
nformation of protein IDs, protein names, gene names, and functions were obtained from the UniProt database.
Group of protein profile was determined based on the function of the particular protein.
Table 3. Significant Newly Detected Proteins of P. aeruginosa after 3-HPAA Exposure.
| protein IDaI | protein name | gene name | function (UniProt) | protein profile groupb |
|---|---|---|---|---|
| Q9HXZ1 | DNA polymerase III subunit α | dnaE PA3640 | DNA replication | DNA |
| Q9I7C4 | β sliding clamp | dnaN PA0002 | DNA strand elongation involved in DNA replication, 3′–5′ exonuclease activity, DNA-directed DNA polymerase activity | DNA |
| P23620 | phosphate regulon transcriptional regulatory protein PhoB | phoB PA5360 | flagellum-dependent swarming motility, phosphate-ion transport, positive regulation of cellular response to phosphate starvation, transcription | RNA |
| Q9HTL4 | OxyR | oxyR PA5344 | RNA polymerase transcription activator, transcription regulatory region DNA binding source, negative regulation of secondary metabolite biosynthetic process, cell motility, lipid biosynthesis, response to reactive oxygen species | RNA |
| Q9HUN0 | 30S ribosomal protein S18 | rpsR PA4934 | structural constituent of ribosome, translation | ribosomes and protein |
| P33641 | outer membrane protein assembly factor BamD | bamD PA4545 | cell envelope organization, protein insertion into membrane | cell wall and membrane |
| G3XDB2 | cell division coordinator CpoB | cpoB PA0974 | FtsZ-dependent cytokinesis | cell wall and membrane |
| Q9HUF1 | peptide methionine sulfoxide reductase MsrA | msrA PA5018 | response to oxidative stress, response to hypochlorite | redox and cell homeostasis |
| Q9I6I9 | spermidine/putrescine import ATP-binding protein PotA | spuF potA, PA0302 | spermidine transport, putrescine transport | redox and cell homeostasis |
| Q9HUX1 | biosynthetic arginine decarboxylase | speA PA4839 | arginine catabolic process, putrescine biosynthetic process, spermidine biosynthetic process | redox and cell homeostasis |
| Q9X6R0 | polyamine aminopropyltransferase 1 | speE1 speE, PA1687 | spermidine biosynthetic process | redox and cell homeostasis |
nformation of protein IDs, protein names, gene names, and functions were obtained from the UniProt database.
Group of protein profile was determined based on the function of the particular protein.
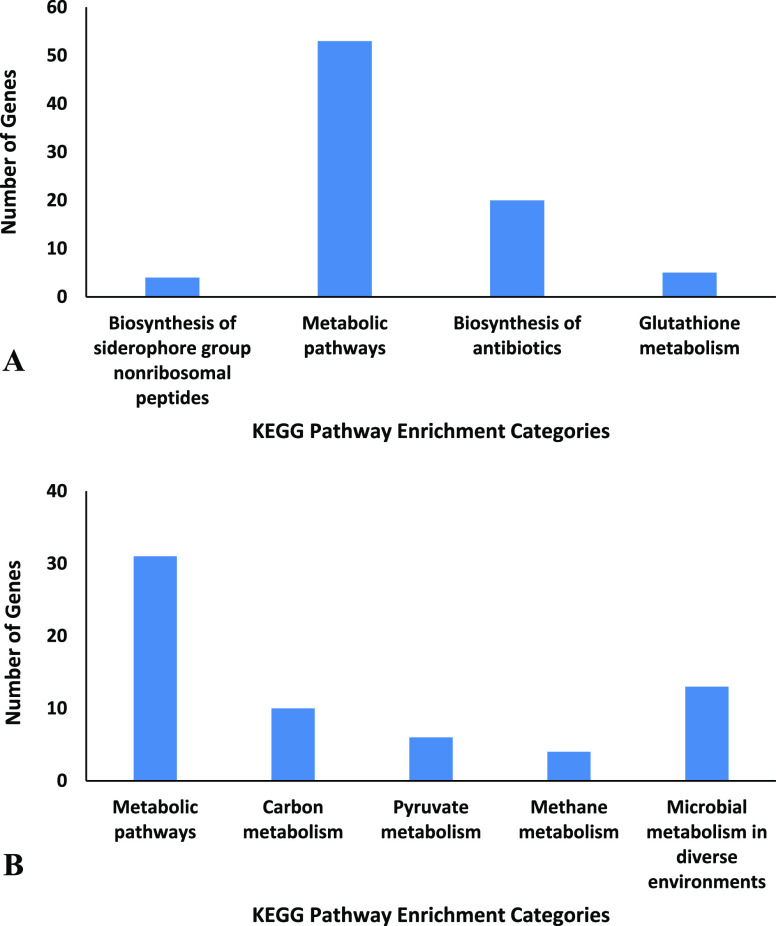
The pathway enrichment analysis of changed proteins in the presence of 3-HPAA (Table S19) demonstrated that the bacteria manage the adaptation to environmental changes by altering the proteins related to metabolic pathways (Figure 3). The application of 3-HPAA resulted in the nondetection of proteins related to the biosynthesis of antibiotics, which validates the reduction in the virulence of P. aeruginosa (Figure 3A).
Figure 3.
KEGG pathway enrichment analysis of the genes of changed proteins. (A) Genes of undetected proteins and (B) genes of newly detected proteins (for all categories, P value < 0.05).
The newly detected proteins related to the carbon, pyruvate, and methane metabolism might demonstrate the changes in the use of energy pathways to adapt to the nonoptimal conditions due to 3-HPAA treatment. The changes in the microbial metabolism in diverse environments support these changes that occurred in the metabolism of P. aeruginosa in the presence of 3-HPAA (Figure 3B).
2.3. DNA-Related Protein Profile
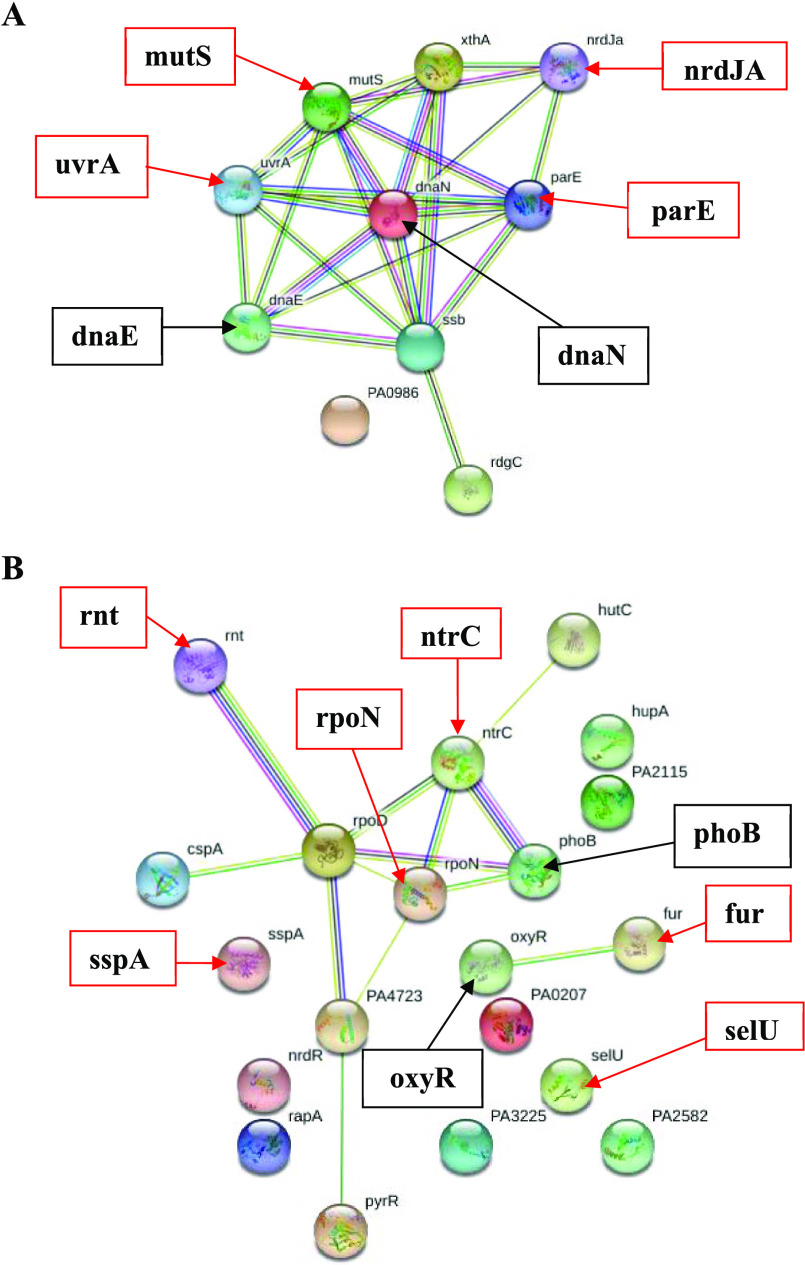
The changes in the protein profile of P. aeruginosa after 3-HPAA exposure were mostly observed in the proteins functioned in DNA replication and repair (Tables S1 and S2), which showed strong interactions with each other in STRING analysis (Figure 4A). One of the remarkable results of this study was the nondetection of UvrABC system protein A (uvrA) and DNA mismatch repair protein (mutS) after 3-HPAA exposure. These proteins play roles in nucleotide excision repair (NER) and mismatch repair (MMR), respectively (Table 2). Since these mechanisms are two of the primary DNA repair mechanisms in organisms,28−30 their impairment would cause the accumulation of mutations, leading to deterioration of the genome of P. aeruginosa, which eventually inhibits bacterial growth. Another significant result was the nondetection of DNA topoisomerase 4 subunit B (parE) and vitamin B12-dependent ribonucleotide reductase (nrdJA), which function in DNA replication (Table 2).
Figure 4.
Interaction networks of DNA-related and RNA-related proteins after 3-HPAA exposure by STRING. Proteins are represented as nodes. The undetected and newly detected significant proteins are shown with their names in bold by the red and black arrows, respectively. (A) DNA-related protein profile; (B) RNA-related protein profile.
On the other hand, newly detected proteins, such as DNA polymerase III subunit α (dnaE) and β sliding clamp (dnaN), are functioned in DNA replication and repair (Table 3) and showed strong interactions with undetected proteins in STRING analysis (Figure 4A). It might be speculated that the bacteria managed to compensate for the unperformed functions in DNA replication and repair. The permanence of cell survival requires proper DNA replication and repair mechanisms,29 and the results might display serious problems in the DNA metabolism of P. aeruginosa due to 3-HPAA exposure, which could be a reason for its antimicrobial activity.
2.4. RNA-Related Protein Profile
The undetected RNA-related proteins after 3-HPAA exposure were generally functioned in the regulation of transcription, RNA polymerase core enzyme binding, tRNA processing, and nutrient utilization (Table S3). The newly detected proteins were also functioned in transcriptional regulation in addition to transcription initiation and RNA polymerase transcriptional activation (Table S4). The nondetection of an important sigma factor, the RNA polymerase sigma-54 factor (σ54) (rpoN), would affect the function of σ54-dependent promoters in P. aeruginosa. Therefore, it leads to problems in the expression of nitrogen metabolism-related proteins due to the role of σ54 in the transcription of nitrogen-related genes under stress conditions.31 Accordingly, DNA binding transcriptional regulator NtrC (ntrC), which has a role in nitrogen utilization, was also undetected, which might show the reduction in nitrogen assimilation in bacteria. The phosphate regulon transcriptional regulatory protein PhoB (phoB), which is the major transcription factor that functions in the phosphate limitation of the bacteria,32 was newly detected (Table 3). Since rpoN, ntrC, and phoB show strong interactions (Figure 4B), the treatment of P. aeruginosa with 3-HPAA might cause the bacteria to experience nitrogen and phosphate starvation and, eventually, bacterial growth inhibition. Fur proteins, which are crucial for bacterial survival but not found in eukaryotic cells, would be useful targets for antimicrobial action.33 Therefore, 3-HPAA, which caused the nondetection of a Fur protein, ferric uptake regulation protein (fur), could be accepted as a promising antimicrobial agent. Depending on the metal chelation properties of the phenolic compounds,34,35 the presence of 3-HPAA in the growth environment of bacteria might result in the chelation of iron, which leads to iron limitation. Since this protein is functioned in iron limitation and oxidative stress adaptations of bacteria,36 nondetection of this protein after 3-HPAA treatment (Table 2) might indicate iron starvation accompanying with oxidative stress in P. aeruginosa. Relatedly, newly detected protein OxyR (oxyR), which showed an interaction with Fur in STRING analysis (Figure 4B), is one of the main transcriptional regulators of oxidative stress defense by regulation of oxidative stress response genes (katA, katB, ahpB, and ahpCF), and the genes function in iron homeostasis in P. aeruginosa.37−39 These results indicate that the antimicrobial effect of 3-HPAA could be due to the oxidative stress and the failure of bacteria to adapt to iron limitation in the environment containing 3-HPAA. Another critical result related to the problems of bacteria in adaptation was the nondetection of an essential protein of stringent stress response, stringent starvation protein A (sspA). Under stress conditions, stringent response proteins aid the performance of alternative sigma factors,38 which leads to the proper adaptation of bacteria to new environmental conditions for cell survival. The nondetection of this protein might show the deficiency of bacterial adaptation in the presence of 3-HPAA leading the bacterial inhibition.
After 3-HPAA exposure, tRNA 2-selenouridine/geranyl-2-thiouridine synthase (selU) and Ribonuclease T (rnt) were undetected as a vital result in terms of tRNA modification (Table 2). The protein SelU is the enzyme for geranylation of tRNA, which is a natural hydrophobic tRNA modification discovered in a few bacteria, including P. aeruginosa.40 The geranylation provides increase in codon recognition fidelity and reduction in frameshift reading by taking action at the wobble position (U34) in the anticodon of lysine, glutamine, and glutamic acid.41,42 Protein Rnt is an incumbent in tRNA 3′-end processing, which is very important for tRNA biogenesis for editing and repairing of tRNAs.43 The nondetection of these proteins might indicate the defects in tRNA biogenesis of the bacteria, which might lead to significant problems in bacterial RNA metabolism, depending on the 3-HPAA exposure.
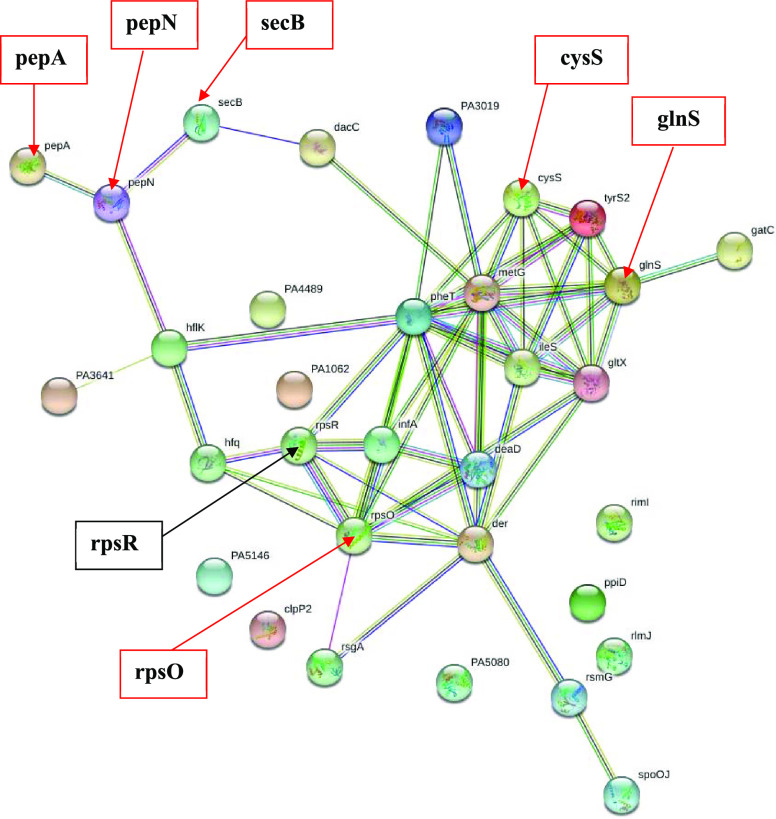
2.5. Ribosomes and Protein-Related Protein Profile
The nondetection of glutamine-tRNA ligase (glnS) and cysteine-tRNA ligase (cysS) (Table 2) could be indicated as important changes among the changes in tRNA ligase (aka aminoacyl-tRNA synthetase) profile due to 3-HPAA treatment. Since tRNA ligases are crucial for the growth of the peptide chain in protein translation by carrying the appropriate amino acids to the ribosomes,44 targeting the tRNA ligase processes is accepted as one of the potent antimicrobial drug development approaches.45,46 For instance, the topical antibiotic Mupirocin, which is still in use against Gram-positive bacteria, takes effect by reversible inhibition of isoleucyl-tRNAs.47 Likewise, the antibiotic purpuromycin inhibits the bacterial translation by inhibiting the tRNA aminoacylation.48 In this respect, it could be speculated that the nondetection of glutamine- and cysteine-tRNA ligases might be related to the antimicrobial effect of 3-HPAA on P. aeruginosa.
Besides the changes in the tRNA ligase profile, 3-HPAA resulted in changes in the structural components of ribosomal subunits, which might lead to an improper ribosome structure of the bacteria (Tables S5 and S6). One notable result on this subject was the nondetection of 30S ribosomal protein S15 (rpsO), which was also validated by our group via real-time quantitative PCR (Supporting Information 3, Figure S4 and Table S18).49,50 The subunits of prokaryotic ribosomes, 30S small subunit, and 50S large subunit, consist of ribosomal RNAs and various ribosomal proteins.51 In ribosomal assembly, small and large subunits are connected by intersubunit bridges.52 The 30S ribosomal protein is crucial for proper bacterial ribosome assembly and translation due to its location in one of these bridges: Bridge B4.52 It also plays a role in the binding of proteins S6, S18, S11, and S21 in vitro.53 When Bubunenko et al. examined in vivo function of the 30S ribosomal protein S15 in the binding of these proteins in E. coli, they have shown that the deletion of the rpsO gene did not keep them from binding in vivo.54 Nevertheless, they also pointed out that the mutant bacteria had a weaker 70S ribosomal structure, and it had a cold-sensitive phenotype. They concluded that under nonoptimal conditions, S15 protein is required for cell survival of Escherichia coli.54 Depending on the outcome of that study, it could be speculated that, for the survival of the bacteria, some other bacterial ribosomal assembly pathways might exist for use in the absence of S15 in vivo.53 According to STRING analysis, a newly detected protein, 30S ribosomal protein S18 (rpsR), showed strong interactions to protein S15 (Figure 4), which might be an alternative protein for the assembly of ribosomes in P. aeruginosa to compensate the function of undetected protein S15. All in all, 3-HPAA exposure formed an unideal condition for P. aeruginosa, and it might lead to detrimental effects by restraining the proper ribosomal assembly due to the nondetection of protein S15. Some antibiotics, such as gentamicin and kanamycin, show their antimicrobial effects by mainly targeting the small subunit of the bacterial ribosome.55 Since 3-HPAA displayed significant changes in the small ribosomal subunit of P. aeruginosa, its antimicrobial effect might be related to the aforementioned changes in small subunit. Besides the effects on the small subunit structure, 3-HPAA also resulted in changes in the large subunit assembly and RNA structure, translational initiation, and translational fidelity due to the nondetection of proteins that function in these processes (Table S5). These results demonstrate that the antimicrobial effect of 3-HPAA might be not only due to the defects in ribosomal structure and assembly but also due to the problems in protein translation. The protein modification processes were also affected by 3-HPAA exposure. For instance, protein-export protein SecB (secB), aminopeptidase N (pepN), and cytosol aminopeptidase (pepA) were undetected after the treatment (Table 2). According to the interaction network, aminopeptidase N interacted with cytosol aminopeptidase and protein-export protein SecB (Figure 5). The cleavage of the amino-terminal (N-terminal) of the peptides or proteins, which determine their structure and function, is carried out by the aminopeptidase enzymes.56 The role of the molecular chaperone SecB is to recognize the precursor proteins with the N-terminal signal sequences for their translocation during the bacterial growth.57,58 The nondetection of these proteins after 3-HPAA exposure caused defects in the modification and translocation of precursor proteins with N-terminal signals, which might eventually lead to serious consequences for the overall metabolism of P. aeruginosa. Thus, the inhibition of P. aeruginosa after 3-HPAA exposure might be attributed to the notable effects of this phenolic acid on tRNA loading, ribosomal structure, and assembly as well as protein translation and modification.
Figure 5.
Interaction network of ribosomes and protein-related undetected and newly detected proteins after 3-HPAA exposure by STRING. Proteins are represented as nodes. The undetected and newly detected significant proteins are shown with their names in bold by the red and black arrows, respectively.
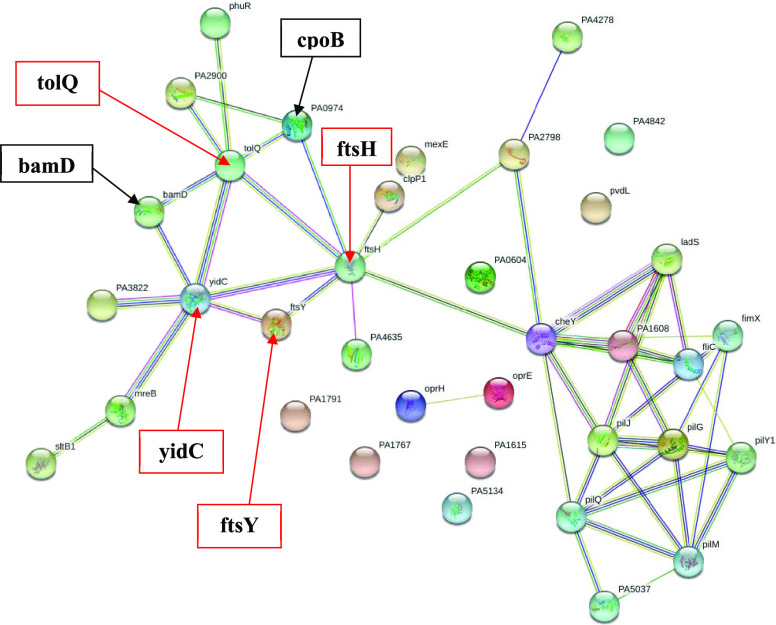
2.6. Cell Wall and Membrane-Related Protein Profile
The treatment of P. aeruginosa with 3-HPAA resulted in remarkable changes in the profile of cell wall and membrane proteins, which function in cell envelope structure composition and organization, cell division, motility, and chemotaxis (Tables S7 and S8). Since cell envelope functions as the protective component for bacterial cell,57 these vital changes in the proteome of cell envelope might be related to the mode of growth inhibition effect of 3-HPAA on P. aeruginosa. The proteins that showed strong interactions in STRING analysis, membrane protein insertase YidC (yidC), signal recognition particle receptor FtsY (ftsY), ATP-dependent zinc metalloprotease FtsH (ftsH), and Tol-Pal system protein TolQ (tolQ) were undetected due to 3-HPAA exposure (Figure 6). YidC protein is one of the major proteins for bacteria, which functions in the insertion/integration of small membrane proteins into the membrane by working together with Sec proteins.59 On the other hand, FtsY protein plays a role in the transfer of the newly formed proteins to the cytoplasmic region of the membrane since it targets the nascent polypeptide chains to the membrane.60 Therefore, 3-HPAA resulted in problems in the membrane protein targeting and insertion into the membrane, which might lead to membrane weakness and damage. The other undetected protein, FtsH, which is universally conserved among prokaryotes, is a protease for rapid control of the regulation of cell metabolism in the presence of environmental stress.27 It is the only one protease that is attached to the inner membrane, and it is responsible for quality control of membrane proteins as well as post-translation of transcription factors and enzymes.27 The nondetection of FtsH might cause defects in the regulation of these transcription factors, leading to problems in transcription processes of P. aeruginosa. Among the transcription factors, it also plays a role in the post-translational regulation of RpoN (σ54) in E. coli,61 which was also undetected after 3-HPAA exposure (Figure 4B). Therefore, we speculate that 3-HPAA treatment caused serious changes in the regulation of transcription in addition to the defects in the cell envelope structure of P. aeruginosa. Besides, the Tol-Pal system protein TolQ, which plays a role in cell cycle and division, was also undetected (Table 2). The Tol-Pal system is vital for Gram-negative bacteria for maintaining the outer membrane integrity.62,63 This system is structurally conserved among Gram-negative bacteria,62 which consists of seven genes in Pseudomonas: orf1, tolQ, tolR, tolA, tolB, oprL, and orf2.64 Since the generated mutations in the genes of the Tol-Pal system result in damages in the outer membrane,62 the nondetection of TolQ after 3-HPAA exposure might cause problems in the cell envelope integrity. The newly detected proteins, outer membrane protein assembly factor BamD (bamD), and cell division coordinator CpoB (PA0974) also showed interactions with YidC, FtsH, and TolQ (Figure 6). Since the lipoprotein BamD is crucial for outer membrane biogenesis64 and CpoB is important for peptidoglycan synthesis control and outer membrane construction,65,66 these might be used for the maintenance of the integrity of the cell envelope, especially the outer membrane, for survival. In our preliminary studies, we have determined the defects on P. aeruginosa cell morphology as the invagination lines throughout the surfaces of bacteria in the presence of 3-HPAA (TOC graphic). These morphological changes could be explained by the differences in the aforementioned proteome in the cell envelope. The cell envelope is the barrier against many unfavorable environmental conditions, such as the presence of antimicrobial agents. The notable results of our study could display that 3-HPAA seriously affected the structure and integrity of the P. aeruginosa cell wall and membrane, which might be a reason for the inhibition of bacterial growth.
Figure 6.
Interaction network of the cell wall and membrane-related undetected and newly detected proteins after 3-HPAA exposure by STRING. Proteins are represented as nodes. (The undetected and newly detected significant proteins are shown by the red and black arrows, respectively, with their names indicated in bold).
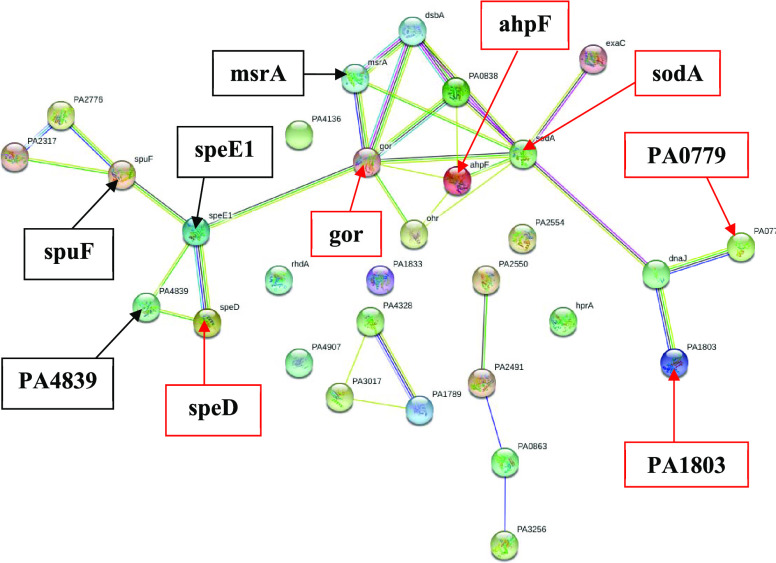
2.7. Redox and Cell Homeostasis-Related Protein Profile
Since reactive oxygen species (ROS) such as superoxide and hydrogen peroxide should be removed to maintain the cellular homeostasis and survival,67 the changes in the redox and cell homeostasis-related protein profile, which displayed significant stress of P. aeruginosa after 3-HPAA treatment (Tables S9 and S10), could be explanatory about bacterial inhibition. The most remarkable change in the redox and cell homeostasis-related protein profile was the nondetection of glutathione reductase (gor), which is an evolutionary conserved and highly homologous protein in both prokaryotes and eukaryotes.68 It plays a vital role in major response to oxidative stress and adaptation. This result might demonstrate that 3-HPAA caused major defects in the glutathione system of bacteria, leading to inadequate response to oxidative stress of 3-HPAA treatment. Gor protein protects the cell from oxidative stress with the detoxification of peroxidases using nicotinamide adenine dinucleotide phosphate (NADPH).69 It would not be surprising to think that the nondetection of Gor could be lethal due to restraining the NADPH recycling for the pentose phosphate pathway, thereby causing problems in energy metabolism.70 However, it was shown that E. coli with gor mutations could maintain the healthy growth and had enough reduced glutathione.71 Therefore, glutathione reduction could be performed by another Gor-independent pathway, providing NADPH for proper energy metabolism. Nevertheless, nondetection of Gor protein might be a strong indication of the problems in stress response of P. aeruginosa since it plays roles in the adaptation of bacteria to several stress conditions.68 Alkyl hydroperoxide reductase (ahpF) and superoxide dismutase [Mn] (sodA), which are the proteins that showed interactions to Gor (Figure 7), were also undetected (Table 2). Since AhpF and SodA function in the reduction of peroxidases and superoxidases, respectively,67,72 it could be concluded that 3-HPAA caused problems in oxidative stress response. Another incapability in stress response might be the nondetection of Lon proteases (PA1803, PA0779), which play roles in stress response and antibiotic response (Table 2).
Figure 7.
Interaction network of redox and cell homeostasis-related undetected and newly detected proteins after 3-HPAA exposure by STRING. Proteins are represented as nodes. (The undetected and newly detected significant proteins are shown by the red and black arrows, respectively, with their names indicated in bold).
Since RNA binding protein Hfq, which regulates Lon proteases,73 was also undetected after 3-HPAA treatment (Table S5), it might be speculated that 3-HPAA exposure damaged Lon protease-mediated processes of P. aeruginosa. The problems of P. aeruginosa related to 3-HPAA stress were also demonstrated by the newly detected proteins (Table 3). Under the oxidative stress conditions, the ROS cause oxidation of the methionine residues of proteins, resulting in the reduction of protein functions.74 The newly detected protein, methionine sulfoxide reductase MsrA (msrA), which exists in various tissues and organisms, is able to reduce the oxidized methionine back to methionine and aids the regaining of protein function.74 Since MsrA is one of the main enzymes that is induced under the oxidative stress, the bacteria might encounter serious problems via damages in the functional proteins due to 3-HPAA exposure. Besides, the newly detected proteins spermidine/putrescine import ATP-binding protein PotA (spuF), biosynthetic arginine decarboxylase (PA4839), and polyamine aminopropyltransferase 1 (speE1) are functioned in spermidine/putrescine metabolism and interacted with each other (Table 3 and Figure 7). However, the nondetection of S-adenosylmethionine decarboxylase proenzyme (speD), which also interacted with these proteins (Table 2 and Figure 7), might show that the spermidine/putrescine metabolism becomes defective in the presence of 3-HPAA. Spermidine and putrescine are main polyamines in bacteria that play roles in several processes, including cell viability and protecting the DNA from oxidative damage.75 Additionally, they have functions in signaling and regulation against stress conditions caused by ROS, heat, UV, acid, and osmotic pressure,76 which might validate various stresses of P. aeruginosa caused by 3-HPAA. Thus, either serious stress conditions or vital defects in stress responses of P. aeruginosa might be the reason for the antimicrobial effect of 3-HPAA.
2.8. Metabolism-Related Protein Profile
The profile of the metabolism-related proteins possessed a high quantity of undetected and newly detected proteins (Tables S11 and S12) after 3-HPAA exposure, which showed a complicated interaction network (Figure S1). These changes might be due to the necessity of the bacteria to change the metabolism for adapting the environment with nonoptimal conditions such as the presence of a toxic substance.27 On the other hand, the obtained profile changes of some particular metabolic systems of P. aeruginosa might have resulted in the inhibition of growth due to the antimicrobial effect of 3-HPAA. Among these, the changes in amino acid biosynthesis might be accepted as one of the notable results. The proteins functioned in the biosyntheses of alanine, lysine, leucine, cysteine, and aromatic amino acids, as well as the catabolic process of arginine, were undetected (Table S11). In contrast, lysine, alanine, histidine, glutamine, serine, and glycine metabolism-related proteins were newly detected (Table S12) that show interactions with each other (Figure S1). It could be speculated that P. aeruginosa was compelled to make regulations in the amino acid metabolism in the presence of 3-HPAA either due to the adaptation of the bacteria or the problems in amino acid and protein metabolism. Aminopeptidase P (pepP), which is a member of the aminopeptidases P (APPro) family and functions explicitly in the removal of the N-terminal residue of polypeptides containing the second residue of proline,77 was undetected (Table 2). It is accepted that PepP is a promising antimicrobial target since it is highly conserved among pseudomonads and played a role in the virulence of the bacteria.77 Therefore, the nondetection of this protein might be meaningful in terms of the 3-HPAA antimicrobial action mechanism. The profile of lipid metabolism-related proteins also changed 3-HPAA exposure.
The main change was the nondetection of proteins functioned in fatty acid β-oxidation, while the proteins functioned in lipid biosynthesis, phosphate transport, and phospholipase synthesis were newly detected (Tables S11 and S12). These changes demonstrated the increase in lipid biosynthesis and the decrease in fatty acid degradation of P. aeruginosa, which might indicate the effort of the bacteria to maintain the integrity of cell envelope in the presence of 3-HPAA. During the 3-HPAA exposure, the distinct greenish-blue color of P. aeruginosa culture was not observed (TOC graphic). The proteomic basis of this color change was due to the nondetection of the proteins functioned in phenazine biosynthesis (Table 2). The pyocyanin pigment, called phenazine produced by P. aeruginosa, is responsible for the specific blue color.78 Phenazine is an extracellular, water-soluble, heterocyclic pigment that can produce ROS and be toxic to other microorganisms and eukaryotes.78−80 It also elevates the virulence of P. aeruginosa by playing the leading role in the quorum sensing mechanism.80,81 Nowadays, the antimicrobial strategies which target quorum sensing, such as inhibiting the phenazine production, catch attention in combatting against pathogens.81,82 Thereby, the nondetection of 5-methylphenazine-1-carboxylate 1-monooxygenase (phzS), phenazine biosynthesis protein PhzB1 (phzB1), and phenazine biosynthesis protein PhzE (phzE1, phzE2) (Table 2) could make 3-HPAA a favorable antimicrobial agent candidate against P. aeruginosa in terms of reduction of phenazine synthesis.
3. Conclusions
Our study demonstrated that, when 3-HPAA, a dose-dependent antimicrobial agent, was present, notable changes in the protein profile of P. aeruginosa were determined. These might lead to serious problems to bacteria not only in maintaining the proper RNA and protein metabolism but also in shape, cell division, nutrient transport, motility, and chemotaxis, which might be stated as the basis of the antimicrobial effect. The other vital problems were in DNA repair mechanisms, which might cause deleterious results for bacteria due to the accumulation of mutations. Additionally, it could be stated that high oxidative stress and defected stress response of bacteria that 3-HPAA caused might be another reason for bacterial inhibition. Moreover, bacteria encountered iron, phosphate, and sulfate starvations in the presence of 3-HPAA, which might eventually damage the overall metabolism of the bacteria. Depending upon all of the results, we conclude that the antimicrobial activity of 3-HPAA on P. aeruginosa has occurred by more than one route, and it could be entitled as a multitarget antimicrobial agent. Our study could be elucidated as the starting point of further studies in examining the mechanisms of antimicrobial action of phenolic acids against pathogenic bacteria.
4. Experimental Section
4.1. Bacterial Growth Conditions
P. aeruginosa (ATCC 27853) were maintained on Mueller Hinton (Sigma-Aldrich) agar plates by overnight incubation at 37 °C. The stock cultures of bacteria were kept in glycerol at −80 °C. In each experiment, the overnight culture of P. aeruginosa was obtained by a single-colony inoculation in Mueller Hinton broth (MHB) following incubation at 37 °C for 18 h without shaking. The optical density (OD) of the overnight culture was determined at 600 nm.
4.2. Determination of MIC of 3-HPAA
3-HPAA was obtained commercially (Sigma-Aldrich). The solutions of 3-HPAA were prepared at the time of the experiment in sterile double-distilled water and added to MHB. The final bacterial load of 106 cfu/mL from overnight culture was inoculated into MHB, containing 3-HPAA, and incubated at 37 °C for 18 h without shaking. The OD of the treated and control cultures were measured at 600 nm at 0th and 24th hour time points of the growth. At the same time, the cell viabilities of the cultures grown for 24 h were tested by the enumeration method on MHA.
4.3. 3-HPAA Exposure for Protein Studies
The final bacterial load of 106 cfu/mL was inoculated from overnight culture into MHB (Sigma-Aldrich) containing subinhibitory concentration (1.9 mg/mL) of 3-HPAA for the treated group and incubated at 37 °C for 18 h without shaking. The same procedure was simultaneously applied for control bacteria without 3-HPAA addition.
4.4. Total Protein Isolation
The protocol of Sianglum et al. was used with minor modifications in the total protein isolation from the 3-HPAA-treated and the control bacteria.83 The cells were harvested at 10 000g for 20 min at 4 °C. They were washed twice with NaCl (0.85%) and centrifuged at 20 000g for 20 min at 4 °C. The pellet was dissolved in phosphate-buffered saline (PBS, pH 7.4) and sonicated for 9 s with 9 s intervals for 15 min. After centrifugation at 20 000g for 20 min at 4 °C, the supernatant was collected and kept at −80 °C until usage.
4.5. Peptide Sample Preparation
Acetone precipitation of the protein samples was performed: acetone (ice-cold) was added on the protein samples and mixed by a vortex. They were kept at −20 °C for overnight precipitation. The concentrations of the proteins were determined by Bradford assay (R2 > 0.98) and adjusted to 0.4 mg. The solution digestion protocol of Carrera et al. was used with minor modifications.23 It was initiated by a three-step procedure with dithiothreitol (DTT) (10/50 mM Tris–HCl, pH 7.8), iodoacetamide (20/50 mM Tris–HCl, pH 7.8) and DTT (20/50 mM Tris–HCl, pH 7.8) consecutively. The procedure was performed for 50 min each step in the dark at room temperature. The samples were cleaned by centrifugation at 14 000 rpm for 30 min in 10 K cutoff filters and washed with 50 mM Tris–HCl (pH 7.8). Each protein sample was treated with trypsin (in 50 mM Tris–HCl, pH 7.8) at the final concentration of 0.04 μg/mL and incubated overnight at 37 °C for trypsin digestion. The volume of each sample was decreased below 100 μL in a SpeedVac concentrator and kept at −20 °C.
4.6. LC-ESI-MS/MS
Fractionation of peptide samples was carried out by high-performance liquid chromatography (HPLC) (Shimadzu, LC20AD) via high-pH reversed-phase chromatography (reverse phase C18 Column, 25 cm × 0.46 cm, 5 μm) with fraction concatenation.84 The phases in fractionation were used as phase A (10 mM ammonium formate/double-distilled H2O (ddH2O), pH 10) and phase B (a mixture of 90% acetonitrile and 10 mM ammonium formate/ddH2O, pH 10). Peptide samples, which were adjusted to 100 μL with phase A, were fractionated by LC-solution software and collected by MALDI-Spotter (SunChrome, SunCollect System). Randomly collected fractions were completely dried in a SpeedVac concentrator, and zip-tip was applied to each sample. In the zip-tip process, each sample was prepared by the addition of an acetonitrile (20%) and formic acid (0.1%) mixture in ddH2O following ultrasonication in a bath (15 s) and vortexing (5 s) alternately for three times. After centrifugation at 14 000 rpm for 10 min, zip-tip was carried out for 10–15 cycles for each sample via the ZipTip column (C18), which was conditioned by acetonitrile (100%) and formic acid (1%) previously. The column was washed by formic acid (1%). Then, the peptides were collected by treatment with increasing concentrations of acetonitrile (twice with 50% and once with 75%). Each sample was mixed with 25 μL of formic acid (0.1%) and transferred into the insert tubes. These samples were analyzed by LC-ESI-MS/MS (Dionex Ultimate 3000) HPLC equipped with an LTQ XL mass spectrometer with electrospray ion source (Thermo Scientific). Chromeleon software (version 6.80) and low-pH column (Sigma Supelco Ascentis, 15 cm × 500 μm, 2.7 μm) were used for the analysis of peptides.84 The phases used were phase A (0.1% formic acid/ddH2O) and phase B (0.1% formic acid/acetonitrile) during the process (initiated by 98% phase A and 2% phase B; increasing phase B up to 90%). Ion trap electrospraying was carried out using helium gas, and the peaks of the peptide samples were collected twice as raw data by the LTQ Tune software.
4.7. Data Analysis
Proteome Discoverer software (version 1.4) and Mass Matrix MS Data File Conversion software were used for transferring the raw data into MGF format and merging the data of fractions, respectively. The reference protein database of P. aeruginosa was downloaded from UniProt proteomes (Proteome ID: UP000002438). It was used in search of merged data in MASCOT MS/MS ions software. Conditions in MASCOT (version 2.3) were: fixed modifications, carbamidomethyl; variable modifications, oxidation; peptide tolerance, +1.2 Da; MS/MS tolerance, +0.6 Da; peptide charge +2 and +3; instrument, ESI-TRAP; MASCOT percolator activated. Two biological repeats were analyzed, and the obtained protein data of two individual analyses were combined prior to comparison in the Draw Venn Diagram tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) to determine the mutual or unique proteins of the control and 3-HPAA-treated groups. The proteins that were unique to the control group (undetected proteins after 3-HPAA exposure) or unique to the 3-HPAA-treated group (newly detected proteins after 3-HPAA exposure) were investigated in terms of functions using UniProt Retrieve/I.D. Mapping with their protein IDs. The functions of uncharacterized proteins in UniProt were estimated by searching in the Protein BLAST tool. All proteins were grouped according to their functions in UniProt, and the interaction networks of the proteins in these groups were determined by STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) software version 10.5 (https://version-10-5.string-db.org/cgi/input.pl?sessionId=6mlAzREKIVnK&input_page_show_search=on).85 The significant metabolic pathways of the genes of these proteins were elucidated by the KEGG pathway enrichment analysis using the DAVID bioinformatics database (version 6.8). The gene IDs of the proteins which were determined in UNIPROT were converted to Entrez Gene IDs by Gene ID Conversion tool, and these were used for the functional annotation analysis tool. The determined categories that have a p-value below 0.05 were demonstrated in the bar chart.
Acknowledgments
The authors thank Prof. Dr. Talat Yalcin for generously providing his expertise and support for LC-ESI-MS/MS and MASCOT analyses. This work was funded by the Izmir Institute of Technology Research Fund (Project # 2014IYTE22) to F.S., and a Ph.D. grant to O.O.O. by the Izmir Institute of Technology. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE86,87 partner repository.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c00703.
All protein tables showing the proteins categorized based on functions in Uniprot and the list of unchanged mutual proteins (Supporting Information 1); validation of protein data of 30S ribosomal protein S15 (rpsO) by real-time quantitative polymerase chain reaction (qPCR) (Supporting Information 2); STRING representations of metabolism-related, virulence-related, and uncharacterized proteins (Supporting Information 3); and names of the genes in the categories of KEGG pathway enrichment analysis (Supporting Information 4) (PDF)
Author Contributions
O.O.O. and F.S. contributed to the design of this study, interpretation of data, and writing of the manuscript. O.O.O. performed the experiments and analyses.
The authors declare no competing financial interest.
Notes
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD016243.
Supplementary Material
References
- Valentini M.; Gonzalez D.; AI Mavridou D.; Filloux A. Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr. Opin. Microbiol. 2018, 41, 15–20. 10.1016/j.mib.2017.11.006. [DOI] [PubMed] [Google Scholar]
- Ha D.-G.; O’Toole G. A.. c-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review Microbiol. Spectrum 2015, 3 (2), 10.1128/microbiolspec.MB-0003-2014. [DOI] [PMC free article] [PubMed]
- Silby M. W.; Winstanley C.; Godfrey S. A. C.; Levy S. B.; Jackson R. W. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680. 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- Morita Y.; Tomida J.; Kawamura Y. Responses of Pseudomonas aeruginosa to antimicrobials. Front. Microbiol. 2014, 4, 422 10.3389/fmicb.2013.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klockgether J.; Tümmler B. Recent advances in understanding Pseudomonas aeruginosa as a pathogen [version 1; referees: 3 approved]. F1000Research 2017, 6, 1261 10.12688/f1000research.10506.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z.; Raudonis R.; Glick B. R.; Lin T.-J.; Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. 10.1016/j.biotechadv.2018.11.013. [DOI] [PubMed] [Google Scholar]
- Wagner S.; Sommer R.; Hinsberger S.; Lu C.; Hartmann R. W.; Empting M.; Titz A. Novel Strategies for the Treatment of Pseudomonas aeruginosa Infections. J. Med. Chem. 2016, 59, 5929–5969. 10.1021/acs.jmedchem.5b01698. [DOI] [PubMed] [Google Scholar]
- Silva L. N.; Zimmer K. R.; Macedo A. J.; Trentin D. S. Plant Natural Products Targeting Bacterial Virulence Factors. Chem. Rev. 2016, 116, 9162–9236. 10.1021/acs.chemrev.6b00184. [DOI] [PubMed] [Google Scholar]
- Kårlund A.; Salminen J.-P.; Koskinen P.; Ahern J. R.; Karonen M.; Tiilikkala K.; Karjalainen R. O. Polyphenols in Strawberry (Fragaria × ananassa) Leaves Induced by Plant Activators. J. Agric. Food Chem. 2014, 62, 4592–4600. 10.1021/jf405589f. [DOI] [PubMed] [Google Scholar]
- Lima M. C.; de Sousa C. P.; Fernandez-Prada C.; Harel J.; Dubreuil J. D.; de Souza E. L. A review of the current evidence of fruit phenolic compounds as potential antimicrobials against pathogenic bacteria. Microb. Pathog. 2019, 130, 259–270. 10.1016/j.micpath.2019.03.025. [DOI] [PubMed] [Google Scholar]
- Miklasińska-Majdanik M.; Kepa M.; Wojtyczka R. D.; Idzik D.; Wasik T. J. Phenolic Compounds Diminish Antibiotic Resistance of Staphylococcus aureus Clinical Strains. Int. J. Environ. Res. Public Health 2018, 15, 2321–2338. 10.3390/ijerph15102321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafra A.; Juarez M. J. B.; Blanc R.; Navalon A.; Gonzalez J.; Vilchez J. L. Determination of polyphenolic compounds in wastewater olive oil by gas chromatography-mass spectrometry. Talanta 2006, 70, 213–218. 10.1016/j.talanta.2005.12.038. [DOI] [PubMed] [Google Scholar]
- Albano M.; Crulhas B. P.; Alves F. C. B.; Pereira A. F. M.; Andrade B. F. M. T.; Barbosa L. N.; Furlanetto A.; da Silveira Lyra L. P.; Rall V. L. M.; Júnior A. F. Antibacterial and anti-biofilm activities of cinnamaldehyde against S. epidermidis. Microb. Pathog. 2019, 126, 231–238. 10.1016/j.micpath.2018.11.009. [DOI] [PubMed] [Google Scholar]
- Keman D.; Soyer F. Antibiotic-Resistant Staphylococcus aureus Does Not Develop Resistance to Vanillic Acid and 2-Hydroxycinnamic Acid after Continuous Exposure in Vitro. ACS Omega 2019, 4, 15393–15400. 10.1021/acsomega.9b01336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Larraínzar M.; Rúa J.; Caro I.; de Castro C.; de Arriaga D.; García-Armesto M. R.; del Valle P. Evaluation of antimicrobial and antioxidant activities of natural phenolic compounds against foodborne pathogens and spoilage bacteria. Food Control 2012, 26, 555–563. 10.1016/j.foodcont.2012.02.025. [DOI] [Google Scholar]
- Karaosmanoglu H.; Soyer F.; Ozen B.; Tokatlı F. Antimicrobial and antioxidant activities of Turkish extra virgin olive oils. J. Agric. Food Chem. 2010, 58, 8238–8245. 10.1021/jf1012105. [DOI] [PubMed] [Google Scholar]
- Gutierrez D. B.; Gant-Branum R. L.; Romer C. E.; Farrow M. A.; Allen J. L.; Dahal N.; Nei Y.-W.; Codreanu S. G.; Jordan A. T.; Palmer L. D.; Sherrod S. D.; McLean J. A.; Skaar E. P.; Norris J. L.; Caprioli R. M. An Integrated, High-Throughput Strategy for Multiomic Systems Level Analysis. J. Proteome Res. 2018, 17, 3396–3408. 10.1021/acs.jproteome.8b00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navare A. T.; Chavez J. D.; Zheng C.; Weisbrod C. R.; Eng J. K.; Siehnel R.; Singh P. K.; Manoil C.; Bruce J. E. Probing the protein interaction network of Pseudomonas aeruginosa cells by chemical cross-linking mass spectrometry. Structure 2015, 23, 762–773. 10.1016/j.str.2015.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François P.; Scherl A.; Hochstrasser D.; Schrenzel J. Proteomic approaches to study Staphylococcus aureus pathogenesis. J. Proteomics 2010, 73, 701–708. 10.1016/j.jprot.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Nilsson J. F.; Castellani L. G.; Draghi W. O.; Perez-Gimenez J.; Tejerizo G. A. T.; Pistorio M. Proteomic Analysis of Rhizobium favelukesii LPU83 in Response to Acid Stress. J. Proteome Res. 2019, 18, 3615–3629. 10.1021/acs.jproteome.9b00275. [DOI] [PubMed] [Google Scholar]
- Sharma D.; Garg A.; Kumar M.; Khan A. U. Proteome profiling of carbapenem-resistant K. pneumoniae clinical isolate (NDM-4): Exploring the mechanism of resistance and potential drug targets. J. Proteomics 2019, 200, 102–110. 10.1016/j.jprot.2019.04.003. [DOI] [PubMed] [Google Scholar]
- Li W.; Zhang S.; Wang X.; Yu J.; Li Z.; Lin W.; Lin X. Systematically integrated metabonomic-proteomic studies of Escherichia coli under ciprofloxacin stress. J. Proteomics 2018, 179, 61–70. 10.1016/j.jprot.2018.03.002. [DOI] [PubMed] [Google Scholar]
- Carrera M.; Böhme K.; Gallardo J. M.; Barros-Velazquez J.; Canas B.; Calo-Mata P. Characterization of foodborne strains of Staphylococcus aureus by shot-gun proteomics: Functional networks, virulence factors and species-specific peptide biomarkers. Front. Microbiol. 2017, 8, 2458 10.3389/fmicb.2017.02458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heunis T.; Deane S.; Smit S.; Dicks L. M. T. Proteomic Profiling of the Acid Stress Response in Lactobacillus plantarum 423. J. Proteome Res. 2014, 13, 4028–4039. 10.1021/pr500353x. [DOI] [PubMed] [Google Scholar]
- Lee J. Y.; Pajarillo E. A. B.; Kim M. J.; Chae J. P.; Kang D.-K. Proteomic and Transcriptional Analysis of Lactobacillus johnsonii PF01 during Bile Salt Exposure by iTRAQ Shotgun Proteomics and Quantitative RT-PCR. J. Proteome Res. 2013, 12, 432–443. 10.1021/pr300794y. [DOI] [PubMed] [Google Scholar]
- Santos P. M.; Benndorf D.; Sa-Correia I. Insights into Pseudomonas putida KT2440 response to phenol-induced stress by quantitative proteomics. Proteomics 2004, 4, 2640–2652. 10.1002/pmic.200300793. [DOI] [PubMed] [Google Scholar]
- Langklotz S.; Baumann U.; Narberhaus F. Structure and function of the bacterial AAA protease FtsH. Biochim. Biophys. Acta 2012, 1823, 40–48. 10.1016/j.bbamcr.2011.08.015. [DOI] [PubMed] [Google Scholar]
- Lakhani B.; Thayer K. M.; Hingorani M. M.; Beveridge D. L. Evolutionary Covariance Combined with Molecular Dynamics Predicts a Framework for Allostery in the MutS DNA Mismatch Repair Protein. J. Phys. Chem. B 2017, 121, 2049–2061. 10.1021/acs.jpcb.6b11976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X.; Ghosh A. K.; Van Houten B.; Greenberg M. M. Nucleotide Excision Repair of a DNA Interstrand Cross-Link Produces Single- and Double-Strand Breaks. Biochemistry 2010, 49, 11–19. 10.1021/bi901603h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko J.; Ukkivi K.; Kivisaar M. NER enzymes maintain genome integrity and suppress homologous recombination in the absence of exogenously induced DNA damage in Pseudomonas putida. DNA Repair 2015, 25, 15–26. 10.1016/j.dnarep.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Erickson K. E.; Winkler J. D.; Nguyen D. T.; Gill R. T.; Chatterjee A. The Tolerome: A Database of Transcriptome-Level Contributions to Diverse Escherichia coli Resistance and Tolerance Phenotypes. ACS Synth. Biol. 2017, 6, 2302–2315. 10.1021/acssynbio.7b00235. [DOI] [PubMed] [Google Scholar]
- Fontaine B. M.; Duggal Y.; Weinert E. E. Exploring the Links between Nucleotide Signaling and Quorum Sensing Pathways in Regulating Bacterial Virulence. ACS Infect. Dis. 2018, 4, 1645–1655. 10.1021/acsinfecdis.8b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu S.; Cisse C.; Vitale S.; Ahmadova A.; Degardin M.; Perard J.; Colas P.; Miras R.; Boturyn D.; Coves J.; Crouzy S.; Michaud-Soret I. From peptide aptamers to inhibitors of FUR, bacterial transcriptional regulator of iron homeostasis and virulence. ACS Chem. Biol. 2016, 11, 2519–2528. 10.1021/acschembio.6b00360. [DOI] [PubMed] [Google Scholar]
- Mazzone G. On the Inhibition of Hydroxyl Radical Formation by Hydroxycinnamic Acids: The Case of Caffeic Acid as a Promising Chelating Ligand of a Ferrous Ion. J. Phys. Chem. A 2019, 123, 9560–9566. 10.1021/acs.jpca.9b08384. [DOI] [PubMed] [Google Scholar]
- Andjelkovic M.; Camp J. V.; De Meulenaer B.; Depaemelaere G.; Socaciu C.; Verloo M.; Verhe R. Iron-chelation properties of phenolic acids bearing catechol and galloyl groups. Food Chem. 2006, 98, 23–31. 10.1016/j.foodchem.2005.05.044. [DOI] [Google Scholar]
- Pinochet-Barros A.; Helmann J. D. Redox sensing by Fe2+ in bacterial Fur family metalloregulators. Antioxid. Redox Signaling 2018, 29, 1858–1871. 10.1089/ars.2017.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y.; Park C.; Jang H.-J.; Kim B.-o.; Bae H.-W.; Chung I.-Y.; Kim E. S.; Cho Y.-H. Antibacterial strategies inspired by the oxidative stress and response Networks. J. Microbiol. 2019, 57, 203–212. 10.1007/s12275-019-8711-9. [DOI] [PubMed] [Google Scholar]
- Wei Q.; Le Minh P. N.; Dotsch A.; Hildebrand F.; Panmanee W.; Elfarash A.; Schulz S.; Plaisance S.; Charlier D.; Hassett D.; Haussler S.; Cornelis P. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res. 2012, 40, 4320–4333. 10.1093/nar/gks017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecilo A.; Zych-Wezyk I. Bacterial stress response as an adaptation to life in a soil environment. Pol. J. Environ. Stud. 2013, 22, 1577–1587. [Google Scholar]
- Wang R.; Vangaveti S.; Ranganathan S. V.; Basanta-Sanchez M.; Haruehanroengra P.; Chen A.; Sheng J. Synthesis, base pairing and structure studies of geranylated RNA. Nucleic Acids Res. 2016, 44, 6036–6045. 10.1093/nar/gkw544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierant M.; Leszczynska G.; Sadowska K.; Dziergowska A.; Rozanski M.; Sochacka E.; Nawrot B. S-Geranyl-2-thiouridine wobble nucleosides of bacterial tRNAs; chemical and enzymatic synthesis of S-geranylated-RNAs and their physicochemical characterization. Nucleic Acids Res. 2016, 44, 10986–10998. 10.1093/nar/gkw727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumelin C. E.; Chen Y.; Leconte A. M.; Chen Y. G.; Liu D. R. Discovery and biological characterization of geranylated RNA in bacteria. Nat. Chem. Biol. 2012, 8, 913–919. 10.1038/nchembio.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammelt C.; Rossmanith W. Repairing tRNA termini: News from the 3′ end. RNA Biol. 2016, 13, 1182–1188. 10.1080/15476286.2016.1239007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro I.; Chelysheva I.; Ignatova Z. Competition for amino acid flux among translation, growth and detoxification in bacteria. RNA Biol. 2018, 15, 991–994. 10.1080/15476286.2017.1306174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J. M.; Bakkalbasi E.; Söll D.; Miller C. A. Drugging tRNA aminoacylation. RNA Biol. 2018, 15, 667–677. 10.1080/15476286.2018.1429879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.-Y.; Kim S.; Kim M. H. Aminoacyl-tRNA synthetases, therapeutic targets for infectious diseases. Biochem. Pharmacol. 2018, 154, 424–434. 10.1016/j.bcp.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenti M. A.; Hatfield S. M.; Leyden J. J. Mupirocin: a topical antibiotic with a unique structure and mechanism of action. Clin. Pharm. 1987, 6, 761–70. [PubMed] [Google Scholar]
- Kirillov S.; Vitali L. A.; Goldstein B. P.; Monti F.; Semenkov Y.; Makhno V.; Ripa S.; Pon C. L.; Gualerzi C. O. Purpuromycin: An antibiotic inhibiting tRNA aminoacylation. RNA 1997, 3, 905–913. [PMC free article] [PubMed] [Google Scholar]
- Dumas J.-L.; van Delden C.; Perron K.; Köhler T. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol. Lett. 2006, 254, 217–225. 10.1111/j.1574-6968.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- Llanes C.; Hocquet D.; Vogne C.; Benali-Baitich D.; Neuwirth C.; Plesiat P. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 2004, 48, 1797–1802. 10.1128/AAC.48.5.1797-1802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Majdoub Z. M.; Carroll K. M.; Gaskell S. J.; Barber J. Quantification of the Proteins of the Bacterial Ribosome Using QconCAT Technology. J. Proteome Res. 2014, 13, 1211–1222. 10.1021/pr400667h. [DOI] [PubMed] [Google Scholar]
- Liu Q.; Fredrick K. Intersubunit bridges of the bacterial ribosome. J. Mol. Biol. 2016, 428, 2146–2164. 10.1016/j.jmb.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z.; Sykes M. T.; Williamson J. R. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 2011, 80, 501–26. 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- Bubunenko M.; Korepanov A.; Court D. L.; Jagannathan I.; Dickinson D.; Chaudhuri B. R.; Garber M. B.; Culver G. M. 30S ribosomal subunits can be assembled in vivo without primary binding ribosomal protein S15. RNA 2006, 12, 1229–1239. 10.1261/rna.2262106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. A. Improving on nature: antibiotics that target the ribosome. Curr. Opin. Microbiol. 2005, 8, 534–542. 10.1016/j.mib.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Nandan A.; Nampoothri K. Molecular advances in microbial aminopeptidases. Bioresour. Technol. 2017, 245, 1757–1765. 10.1016/j.biortech.2017.05.103. [DOI] [PubMed] [Google Scholar]
- Kedrov A.; Kusters I.; Driessen A. J. M. Single-Molecule Studies of Bacterial Protein Translocation. Biochemistry 2013, 52, 6740–6754. 10.1021/bi400913x. [DOI] [PubMed] [Google Scholar]
- Meyer K.; Addy C.; Akashi S.; Roper D. I.; Tame J. R. H. The crystal structure and oligomeric form of Escherichia coli L,D-carboxypeptidase A. Biochem. Biophys. Res. Commun. 2018, 499, 594–599. 10.1016/j.bbrc.2018.03.195. [DOI] [PubMed] [Google Scholar]
- Hennon S. W.; Dalbey R. E. Cross-Linking-Based Flexibility and Proximity Relationships between the T.M. Segments of the Escherichia coli YidC. Biochemistry 2014, 53, 3278–3286. 10.1021/bi500257u. [DOI] [PubMed] [Google Scholar]
- Dalbey R. E.; Wang P.; Kuhn A. Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 2011, 80, 161–87. 10.1146/annurev-biochem-060409-092524. [DOI] [PubMed] [Google Scholar]
- Carmona M.; de Lorenzo V. Involvement of the FtsH (HflB) protease in the activity of σ54 promoters. Mol. Microbiol. 1999, 31, 261–270. 10.1046/j.1365-2958.1999.01169.x. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C.; Sullivan C. J.; Kuehn M. J. Envelope Control of Outer Membrane Vesicle Production in GramNegative Bacteria. Biochemistry 2013, 52, 3031–3040. 10.1021/bi400164t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgis J. N. Organisation and evolution of the tol-pal gene cluster. J. Mol. Microbiol. Biotechnol. 2001, 3, 113–122. [PubMed] [Google Scholar]
- Llamas M. A.; Ramos J. L.; Rodriguez-Herva J. J. Transcriptional organization of the Pseudomonas putida tol-oprL genes. J. Bacteriol. 2003, 185, 184–195. 10.1128/JB.185.1.184-195.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori N.; Ishii Y.; Tateda K.; Kimura S.; Kouyama Y.; Inoko H.; Mitsunaga S.; Yamaguchi K.; Yoshihara E. A peptide based on homologous sequences of the b-barrel assembly machinery component BamD potentiates antibiotic susceptibility of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2012, 67, 2173–2181. 10.1093/jac/dks174. [DOI] [PubMed] [Google Scholar]
- Gray A. N.; Egan A. J. F.; van’t Veer I. L.; Verheul J.; Colavin A.; Koumoutsi A.; Biboy J.; Altelaar A. F. M.; Damen M. J.; Huang K. C.; Simorre J.-P.; Breukink E.; den Blaauwen T.; Typas A.; Gross C. A.; Vollmer W.. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division eLife 2015, 10.7554/eLife.07118. [DOI] [PMC free article] [PubMed]
- Kamariah N.; Manimekalai M. S. S.; Nartey W.; Eisenhaber F.; Eisenhaber B.; Gruber G. Crystallographic and solution studies of NAD(+)- and NADH-bound alkylhydroperoxide reductase subunit F (AhpF) from Escherichia coli provide insight into sequential enzymatic steps. Biochim. Biophys. Acta, Bioenerg. 2015, 1847, 1139–1152. 10.1016/j.bbabio.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Smirnova G. V.; Oktyabrsky O. N. Glutathione in bacteria. Biochemistry 2005, 70, 1199–1211. [DOI] [PubMed] [Google Scholar]
- Surya A.; Liu X.; Miller M. J. Glutathione Utilization in Lactobacillus fermentum CECT 5716. J. Agric. Food Chem. 2018, 66, 12651–12656. 10.1021/acs.jafc.8b06136. [DOI] [PubMed] [Google Scholar]
- Ogasawara Y.; Funakoshi M.; Ishii K. Determination of reduced nicotinamide adenine dinucleotide phosphate concentration using high-performance liquid chromatography with fluorescence detection: Ratio of the reduced form as a biomarker of oxidative stress. Biol. Pharm. Bull. 2009, 32, 1819–1823. 10.1248/bpb.32.1819. [DOI] [PubMed] [Google Scholar]
- Davis N. K.; Greer S.; Jones-Mortimer M. C.; Perham R. N. Isolation and Mapping of Glutathione Reductase-negative Mutants of Escherichia coli K 12. J. Gen. Microbiol. 1982, 128, 1631–1634. 10.1099/00221287-128-7-1631. [DOI] [PubMed] [Google Scholar]
- Iiyama K.; Chieda Y.; Lee J. M.; Kusakabe T.; Yasunaga-Aoki C.; Shimizu S. Effect of superoxide dismutase gene inactivation on virulence of Pseudomonas aeruginosa PAO1 toward the silkworm, Bombyx mori. Appl. Environ. Microbiol. 2007, 73, 1569–1575. 10.1128/AEM.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L.; Breidenstein E. B. M.; Taylor P. K.; Bains M.; de la Fuente-Nuñez C.; Fang Y.; Foster L. J.; Hancock R. E. W. Interconnection of posttranscriptional regulation: The RNA-binding protein Hfq is a novel target of the Lon protease in Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26811 10.1038/srep26811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A.-r.; Kim M.-J.; Kwak G.-H.; Son J.; Hwang K. Y.; Kim H.-Y. Essential Role of the Linker Region in the Higher Catalytic Efficiency of a Bifunctional MsrA-MsrB Fusion Protein. Biochemistry 2016, 55, 5117–5127. 10.1021/acs.biochem.6b00544. [DOI] [PubMed] [Google Scholar]
- Johnson L.; Mulcahy H.; Kanevets U.; Shi Y.; Lewenza S. Surface-localized spermidine protects the Pseudomonas aeruginosa outer membrane from antibiotic treatment and oxidative stress. J. Bacteriol. 2012, 194, 813–826. 10.1128/JB.05230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandounas L.; Ballerstedt H.; de Winde J. H.; Ruijssenaars H. J. Redundancy in putrescine catabolism in solvent tolerant Pseudomonas putida S12. J. Biotechnol. 2011, 154, 1–10. 10.1016/j.jbiotec.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Peng C.-T.; Liu L.; Li C.-C.; He L.-H.; Li T.; Shen Y.-L.; Gao C.; Wang N.-Y.; Xia Y.; Zhu Y.-B.; Song Y.-J.; Lei Q.; Yu L.-T.; Bao R. Structure-Function Relationship of Aminopeptidase P from Pseudomonas aeruginosa. Front. Microbiol. 2017, 8, 2385 10.3389/fmicb.2017.02385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada B.; Leto T. L. Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 2013, 21, 73. 10.1016/j.tim.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Fouly M. Z.; Sharaf A. M.; Shahin A. A. M.; El-Bialy H. A.; Omara A. M. A. Biosynthesis of pyocyanin pigment by Pseudomonas aeruginosa. J. Radiat. Res. Appl. Sci. 2015, 8, 36–48. 10.1016/j.jrras.2014.10.007. [DOI] [Google Scholar]
- Dietrich L. E. P.; Price-Whelan A.; Petersen A.; Whiteley M.; Newman D. K. The phenazine pyocyanin is a terminal signaling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 2006, 61, 1308–1321. 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- Ugurlu A.; Karahasan Yagci A.; Ulusoy S.; Aksu B.; Bosgelmez-Tinaz G. Phenolic compounds affect production of pyocyanin, swarming motility and biofilm formation of Pseudomonas aeruginosa. Asian Pac. J. Trop. Biomed. 2016, 6, 698–701. 10.1016/j.apjtb.2016.06.008. [DOI] [Google Scholar]
- Lu H. D.; Pearson E.; Ristroph K. D.; Duncan G. A.; Ensign L. M.; mSuk J. S.; Hanes J.; Prudhomme R. K. Pseudomonas aeruginosa pyocyanin production reduced by quorum-sensing inhibiting nanocarriers. Int. J. Pharm. 2018, 544, 75–82. 10.1016/j.ijpharm.2018.03.058. [DOI] [PubMed] [Google Scholar]
- Sianglum W.; Srimanote P.; Wonglumsom W.; Kittiniyom K.; Voravuthikunchai S. P. Proteome Analyses of Cellular Proteins in Methicillin-Resistant Staphylococcus aureus Treated with Rhodomyrtone, a Novel Antibiotic Candidate. PLoS One 2011, 6, e16628 10.1371/journal.pone.0016628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinç M.Proteomic Analyses of Biological Samples by Using Different Mass Spectrometric Strategies. Ph.D. Dissertation, Izmir Institute of Technology: Izmir, Turkey, 2018. [Google Scholar]
- Szklarczyk D.; Morris J. H.; Cook H.; Kuhn M.; Stefan Wyder S.; Simonovic M.; Santos A.; Doncheva N. T.; Roth A.; Bork P.; Jensen L. J.; von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y.; Csordas A.; Bai J.; Bernal-Llinares M.; Hewapathirana S.; Kundu D. J.; Inuganti A.; Griss J.; Mayer G.; Eisenacher M.; Pérez E.; Uszkoreit J.; Pfeuffer J.; Sachsenberg T.; Yilmaz S.; Tiwary S.; Cox J.; Audain E.; Walzer M.; Jarnuczak A. F.; Ternent T.; Brazma A.; Vizcaíno J. A. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch E. W.; Csordas A.; Sun Z.; Jarnuczak A.; Perez-Riverol Y.; Ternent T.; Campbell D. S.; Bernal-Llinares M.; Okuda S.; Kawano S.; Moritz R. L.; Carver J. J.; Wang M.; Ishihama Y.; Bandeira N.; Hermjakob H.; Vizcaíno J. A. The ProteomeXchange Consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017, 54, D1100–D1106. 10.1093/nar/gkw936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.