Human oxytocin receptor structure reveals the basis for ligand recognition and allosteric modulation by lipids and ions.
Abstract
The peptide hormone oxytocin modulates socioemotional behavior and sexual reproduction via the centrally expressed oxytocin receptor (OTR) across several species. Here, we report the crystal structure of human OTR in complex with retosiban, a nonpeptidic antagonist developed as an oral drug for the prevention of preterm labor. Our structure reveals insights into the detailed interactions between the G protein–coupled receptor (GPCR) and an OTR-selective antagonist. The observation of an extrahelical cholesterol molecule, binding in an unexpected location between helices IV and V, provides a structural rationale for its allosteric effect and critical influence on OTR function. Furthermore, our structure in combination with experimental data allows the identification of a conserved neurohypophyseal receptor-specific coordination site for Mg2+ that acts as potent, positive allosteric modulator for agonist binding. Together, these results further our molecular understanding of the oxytocin/vasopressin receptor family and will facilitate structure-guided development of new therapeutics.
INTRODUCTION
Across many species, signaling mediated by oxytocin- and vasopressin-related neuropeptides provides a global, biologically conserved organizing role in sexual reproduction and social behavior (1). In mammals, oxytocin and vasopressin orchestrate their biological role through binding and activation of closely related G protein–coupled receptors (GPCRs), called the oxytocin receptor (OTR) and the vasopressin receptors (V1aR, V1bR and V2R) (2). The neurohypophyseal peptide hormones oxytocin and vasopressin differ only in two of their nine amino acids, but they are structurally distinct from other neuropeptides in being cyclized through a disulfide bond between two cysteines at positions 1 and 6 of the peptide chain (3).
Oxytocin has numerous functions, and its central neurotransmitter-like action has been shown to widely modulate cognitive effects, regulating the establishment and maintenance of complex social and bonding behavior (4), while peripheral activation of OTR is intimately linked to parturition and lactation (5, 6). Translational neuroscience research suggests that targeted activation of the OTR may have therapeutic potential in the treatment of mental health disorders including autism, Asperger’s syndrome, social anxiety disorder, and schizophrenia (7–10), while OTR antagonism can be of value in the treatment of male sexual disorders, assisted reproductive technologies, and spontaneous premature labor (11–13). However, to date, the only clinically approved OTR-targeted treatments are peptide-based ligands in obstetrics that have to be delivered through intravenous administration. To induce labor, the endogenous agonist oxytocin is administered (14), while conversely for the prevention of preterm labor, which is still the leading cause of morbidity and mortality of children under the age of five (www.unicef.org/publications/index_103264.html), the antagonist atosiban is clinically approved (15).
Despite the clear therapeutic potential of targeted intervention within this evolutionarily ancient signaling system, it has, so far, remained challenging to identify suitable orally available small-molecule ligands that display high specificity and potency toward the OTR. Certainly, these issues can be attributed to the lack of any detailed structural information on the OTR (or the closely related vasopressin receptors) and, therefore, the absence of a precise understanding of the structural determinants for ligand interaction and selectivity. Furthermore, drug discovery efforts are complicated by the OTR’s strong and incompletely understood dependence on allosteric modulators such as cholesterol and magnesium (Mg2+), which were shown to critically influence the interaction between the receptor and extracellular ligands (16, 17).
Here, we report the first crystal structure of OTR, bound to the orally available antagonist retosiban that has been clinically developed as a tocolytic agent to efficiently block the oxytocin-mediated contraction of the smooth muscle in the uterus that occurs during the initiation of preterm labor.
In comparison to other peptidergic GPCRs, the antagonist-bound OTR structure displays an enlarged binding pocket, which is exposed to the extracellular solvent. Specific contacts with the cocrystallized antagonist retosiban are mediated through both polar and hydrophobic interacting residues that are located on opposing hemispheres of the binding cavity. Furthermore, the crystal structure allowed the identification of the binding site for the physiologically essential allosteric cholesterol molecule, which we find to be located in a pocket formed by transmembrane helices IV and V.
Last, the identification and functional verification of two highly conserved, negatively charged residues at the extracellular tips of transmembrane helices I and II, exclusive within the neurohypophyseal oxytocin/vasopressin receptor family, allow us to postulate a structural rationale for the strong allosteric effect that Mg2+ exerts on agonist binding affinity. Thus, the OTR structure reported here provides the first detailed structural information on this peculiar neuropeptide-binding GPCR and provides answers to long-standing questions with regard to its allosteric modulation by both a sterol and ions.
RESULTS
Structure determination
To achieve sufficient purification yields of functional OTR, two consecutive rounds of directed evolution in Saccharomyces cerevisiae were performed on the human wild-type OTR (wtOTR), introducing a total of eight mutations (18). Furthermore, to allow for crystallization in lipidic cubic phase (LCP), 34 residues (residues R232 to L265) of the third intracellular loop (ICL3) were replaced with the thermostable Pyrococcus abysii glycogen synthase (PGS) domain, 30 residues of the flexible C terminus (residues 360 to 389) were truncated, and residues D1534.42 and S2245.62 [Ballesteros-Weinstein numbering denoted in superscript (19)] were substituted with alanine to increase thermostability. These were identified through a limited alanine screen, using the previously reported 7-diethylamino-3-(4-maleimidophenyl)-4-methylcoumarin (CPM)–based thermostability assay as a readout (20). In combination, these modifications resulted in almost unaltered binding affinity for the co-crystallized antagonist retosiban (table S2). From crystals grown in LCP, the OTR:retosiban complex structure was determined at 3.2-Å resolution with strong and unambiguous electron density for retosiban, cholesterol, and key interaction residues of the receptor (Table 1 and figs. S1 and S2).
Table 1. Data collection and structure refinement statistics.
Values in parenthesis indicate highest-resolution shell. RMSD, root mean square deviations.
| Data collection | |
| Number of crystals | 9 |
| Space group | C2221 |
| Cell dimensions | |
| a, b, c (Å) | 76.99, 78.86, 283.49 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Number of reflections measured | 472,505 |
| Number of unique reflections | 14,739 |
| Resolution (Å) | 47.59–3.20 (3.42–3.20) |
| Rpim | 0.119 (1.309) |
| CC1/2 | 0.998 (0.578) |
| Mean I/s(I) | 7.5 (1.1) |
| Completeness (%) | 99.9 (99.7) |
| Redundancy | 32.1 (32.9) |
| Refinement | |
| Resolution (Å) | 29.843–3.20 |
| Number of reflections (test set) Reflection test set (%) |
14,602 (632) 4.33 |
| Rwork/Rfree | 0.252/0.266 |
| Number of atoms | |
| All | 3,718 |
| Protein | 3,647 |
| Ligand | 36 |
| Others (cholesterol, PEG) | 35 |
| Average B factors (Å2) | |
| All | 100.89 |
| Protein | 100.64 |
| Ligand | 120.71 |
| Others (cholesterol, PEG) | 105.82 |
| RMSD | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.725 |
| Ramachandran statistics | |
| Favored regions (%) | 96.47 |
| Allowed regions (%) | 3.53 |
| Outliers (%) | 0.00 |
| MolProbity overall score | 10.34 |
Overall, OTR displays the canonical GPCR topology consisting of a seven-transmembrane helical bundle (helices I to VII), two ICLs and three extracellular loops (ECLs), and a C-terminal amphipathic helix VIII of which only the N-terminal part is resolved (Fig. 1, A and B). Similar to other class A peptide GPCRs, the ECL2 of OTR forms an extended β-hairpin structure that is anchored to the extracellular tip of helix III by the conserved disulfide bridge between C1123.25 and C187 of ECL2.
Fig. 1. Overall structure of retosiban-bound OTR.
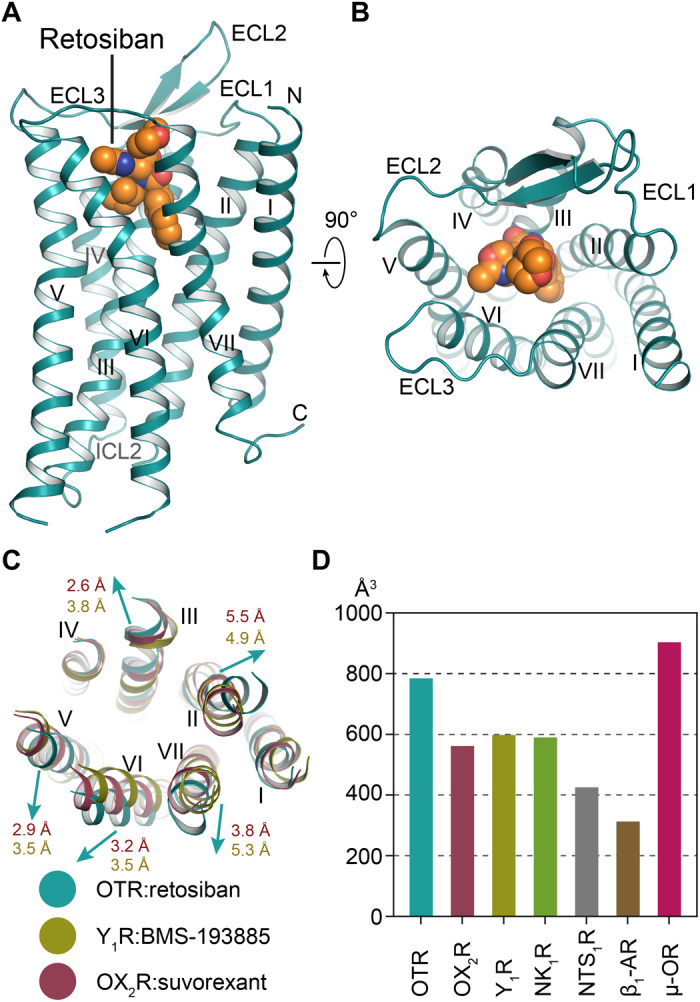
(A) OTR (turquoise) in complex with retosiban (orange) viewed parallel to the membrane plane. OTR is depicted as ribbon, and retosiban is shown in sphere representation, with oxygen and nitrogen atoms highlighted in red and blue, respectively. (B) OTR:retosiban complex as viewed from the extracellular space. (C) Structural superposition of OTR with Y1R (yellow; PDB ID: 5ZBH) and OX2R (raspberry; PDB ID: 4S0V). Arrows indicate shifts of the OTR extracellular helix tips relative to the reference receptors. Helix tip movement in Å is indicated in the respective receptor colors (raspberry and yellow, respectively). (D) Calculated extracellular ligand-binding pocket volumes of selected neuropeptide GPCRs and one adrenergic receptor (β1-AR).
Ligand-binding pocket of OTR
Structural superpositions with other small-molecule bound GPCRs belonging to the β-branch of class A GPCRs reveal notable conformational differences at the extracellular helical ends of OTR, even though their overall root-mean-square deviations for backbone atoms are as low as 1.6 Å (fig. S3 and table S1). For the OTR, we find that the extracellular tips of helices II, III, V, VI, and VII are moved away from the central axis of the heptahelical bundle, with the most marked outward shifts observed for the extracellular ends of helices II, VI, and VII (Fig. 1C and fig. S3). However, these extracellular conformational differences are not transferred to the intracellular side of the helical bundle since, on this part of the receptor, the helical conformations coincide with those of previously determined inactive peptidergic GPCR structures, consistent with the expectation that the OTR structure in complex with the antagonist retosiban is captured in the inactive state.
To quantify the consequences of the observed extracellular helical bundle expansion on the size of the solvent-accessible ligand-binding pocket, we used an analogous strategy as recently reported for the detailed analysis of the β1-adrenoceptor (β1-AR) (21). We calculated the accessible volumes of the ligand-binding sites of the different receptors (table S1), and from this analysis, we find the OTR binding pocket volume to be enlarged by approximately 25% compared with other non–peptide antagonist-bound neuropeptide GPCRs. Moreover, in comparison with the neurotensin receptor (NTS1R) in complex with its peptide agonist, the pocket volume of retosiban-bound OTR is increased by almost 50%, in agreement with the reported contraction of the orthosteric pocket in the agonist-bound state of NTS1R (22). Overall, we find that the OTR-binding pocket volume is most similar in size and shape to the previously reported exceptionally large binding site of the μ-opioid receptor (μ-OR; Fig. 1, fig. S4, and table S1).
The increased size of the extracellular binding pocket of OTR may indicate the necessity to accommodate a cyclic peptide, a feature that is shared within the closely related oxytocin/vasopressin receptor family. Thus, this feature may provide an opportunity in future drug discovery programs targeting these receptors.
Retosiban binding mode
Retosiban is a potent, nonpeptidic antagonist of OTR with >1400-fold selectivity over the closely related vasopressin receptors (V1aR, V1bR, V2R) (23). Chemically, retosiban (Fig. 2A) is composed of a central 2,5-diketopiperazine (2,5-DKP) core motif with three substituents at its 3, 6, and 7 positions (butan-2-yl, indanyl, and 2′-methyl-1′,3′-oxazol-4′-yl morpholine amide, respectively) that were found to provide the best balance between potency and selectivity of all tested compounds (23, 24).
Fig. 2. The OTR-binding pocket for retosiban.
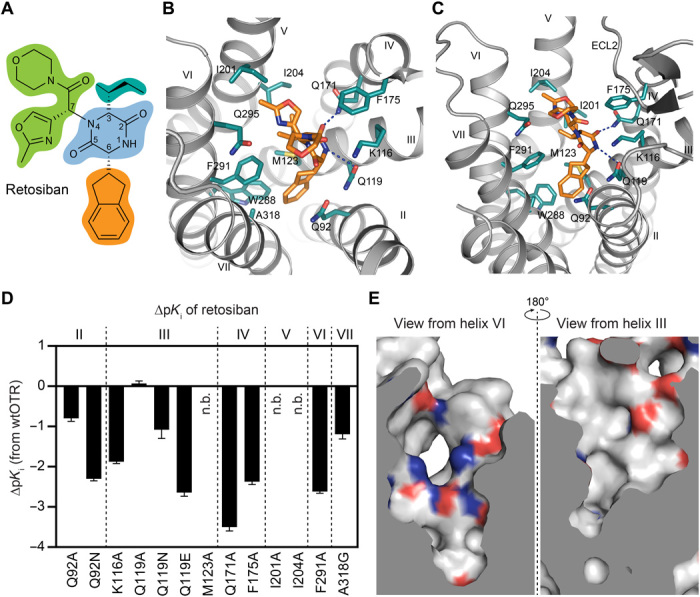
(A) Chemical structure of retosiban with structural topology highlighted by colored circles (2,5-diketopiperazine core in blue, indanyl group in orange, sec-butyl group in cyan, and oxazol-morpholine amide moiety in green). (B) Detailed interactions of retosiban with OTR as viewed from the extracellular space from a position above helices I and II. The receptor backbone is shown in gray. Retosiban and key interaction residues within 4 Å of the ligand are shown as sticks and are colored as in Fig. 1. Hydrogen bonds are indicated by dashed, blue lines. (C) Interactions of retosiban with OTR as viewed from the membrane plane. (D) Retosiban affinity profiles of selected mutants in comparison to wtOTR. Bars represent differences in calculated affinity (pKi values) for each mutant relative to the wild-type receptor. pKi values were derived from competition ligand-binding experiments (table S2). Data are shown as means ± SEM from three independent experiments performed in quadruplicates. n.b., no binding. (E) The two chemically distinct interaction surfaces (left: polar, charged; right: hydrophobic) of the extracellular ligand-binding pocket of OTR, depicted as electrostatic surfaces, with negatively and positively charged amino acids highlighted in red and blue, respectively.
Retosiban adopts an upright, elongated conformation along the central axis of the helical bundle with the indanyl located at the bottom, the 2,5-DKP core in a central position, the sec-butyl substituent oriented toward the helix V, and the oxazol-morpholine amide moiety closest to the extracellular surface (Fig. 2, B and C). Overall, the binding pocket can be divided into a polar interaction surface, created by a network of residues located on helices II to IV, and a largely hydrophobic hemisphere stretching from transmembrane helices V to VII (Fig. 2E).
The central 2,5-DKP core is found in a slightly kinked conformation with its 2-keto group pointing toward helix IV and the 5-keto group oriented toward helix VII. The 2,5-DKP core specifically interacts with the receptor through a polar interaction interface, which is formed by two glutamine (Q1193.32 and Q1714.60) and one lysine (K1163.29) residue on the surface of helices III and IV on this side of the ligand-binding pocket (Fig. 2, A, B, and E). Q1714.60 forms a strong hydrogen bond (2.5-Å distance between side-chain nitrogen and ligand oxygen) with the 2-keto oxygen, while Q1193.32 forms a weaker hydrogen bond with N1 of the 2,5-DKP core of retosiban (3.5-Å distance between side-chain oxygen and ligand nitrogen; Fig. 2, A and B). The critical contributions of these polar interactions for the high-affinity binding of retosiban to the OTR are highlighted by a severe loss in binding affinity when K1163.29 is mutated to alanine (75-fold), Q1714.60 is mutated to alanine (1600-fold), and by a notable loss in binding affinity if either the length of the side chain is varied (Q1193.32N, 12-fold) or if a negative charge (Q1193.32E, 440-fold) is introduced at position Q1193.32 (Fig. 2D, fig. S6, and table S2).
The hydrophobic indanyl substituent attached at position 6 of the central 2,5-DKP core, which during initial structure-activity relationship (SAR) studies was shown to increase potency ∼15-fold (25), optimally penetrates into a deep, mainly hydrophobic crevice at the bottom of the binding pocket, formed by side-chain residues of helices II, III, VI, and VII. Within this pocket extension, the indanyl is laterally sandwiched between Q922.57 and M1233.36 and forms additional hydrophobic interactions with F2916.51, A3187.42, and W2886.48 (Fig. 2, A and B).
Additional interactions between OTR and retosiban are formed by the butan-2-yl substituent and the hydrophobic side-chains I2015.39, I2045.42, and F2916.51 on helices V and VI. The importance of this hydrophobic interaction surface opposite to the polar network is underscored by a 214-fold loss in binding affinity of retosiban if F2916.51 is mutated to alanine (Fig. 2D, fig. S6, and table S2). Higher up in the binding pocket, as part of the third, most solvent-exposed substituent, the oxazol moiety is oriented toward I2015.39 and is in close proximity to Q2956.55, while F1754.64 provides an additional hydrophobic interaction with the amide linker to the morpholine ring. In line with the extensive SAR studies performed during the development of retosiban, the morpholine ring itself has no direct interactions with the receptor (24).
Distinct extrahelical cholesterol binding site
Cholesterol is an abundant constituent of animal cell plasma membranes and has been shown to be intimately involved in the regulation of membrane proteins. This regulatory role of cholesterol can be achieved either indirectly by its ability to modulate the physical properties of the lipid bilayer (e.g., modulation of membrane fluidity and maintenance of lipid microdomains) or directly through specific protein-cholesterol interactions (26, 27). Modulation of protein function through cholesterol has previously been reported for a number of GPCRs including the OTR (16, 28), and multiple class A GPCR crystal structures have revealed direct structural evidence for specific cholesterol-GPCR interactions (29, 30), albeit at different locations in the structures.
In particular, the physiological function and the thermostability of the OTR have been shown to be critically dependent on the presence of cholesterol. These findings provided evidence that cholesterol or its analogs act as allosteric modulators of the OTR by shifting the receptor to a high-affinity state for both agonist and antagonist binding (16, 28, 31). However, despite extensive efforts, no specific cholesterol binding site has been identified for the OTR to date.
The structure of the OTR:retosiban complex displays unambiguous extrahelical electron density for one molecule of cholesterol in a distinct extrahelical surface groove that is formed by hydrophobic residues of helices IV and V and is capped toward the extracellular space by residues of ECL2 (Fig. 3A). The receptor was crystallized from a monoolein:cholesterol mixture.
Fig. 3. Cholesterol binding site of OTR.
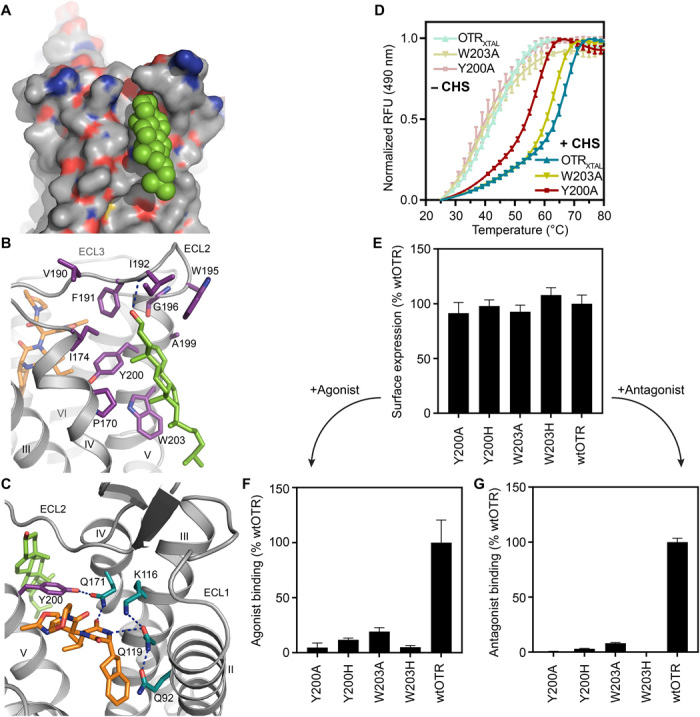
(A) Surface representation of cholesterol binding site between helices IV and V of OTR, with cholesterol shown in green spheres. (B) Detailed interactions of cholesterol with OTR, as viewed from the membrane. Residues of OTR within 4 Å of cholesterol and retosiban are depicted as sticks colored in violet and orange, respectively. Hydrogen bonds are indicated by dashed blue lines. (C) Cholesterol modulating the polar network of the ligand-binding pocket through the positioning of Y2005.38, which establishes a hydrogen bond to Q1714.60. Through its interaction with Q1714.60, Y2005.38 coordinates the formation of a hydrogen bonding network including residues from helices II to IV. (D) Thermostability assay (CPM) of two selected mutants in the OTRXTAL background and OTRXTAL performed in DDM in the presence (saturated colors) or absence (pale colors) of the cholesterol analog cholesteryl hemisuccinate (CHS). For each experiment, a representative curve is shown. RFU, relative fluorescence units. Data are shown as means ± SEM from three independent experiments. (E) Receptor surface expression levels of selected mutants in wtOTR background compared to wtOTR. Data are shown as means ± SD from three independent experiments with 64 data points each (table S3). (F and G) Maximal binding (Bmax) of agonist oxytocin (F) and antagonist PVA (G) on the OTR variants from (E) depicted relative to wtOTR. Data are shown as means ± SEM from three independent experiments in triplicates (table S3)
The steroid core of cholesterol specifically contacts the receptor through main hydrophobic interactions with the highly conserved (within the oxytocin/vasopressin receptor family) residues Y2005.38 and W2035.41 (fig. S5) of helix V, and further with the more distal residues P1704.59 and I1744.63 on helix IV, residues V190, F191, and W295, all of ECL2, as well as residues G1965.43 and A1995.37 on helix V (Fig. 3B). In addition to these hydrophobic interactions, cholesterol forms a hydrogen bond to the backbone amide of residue I192 of ECL2 with its β3-hydroxyl group.
To probe whether the observed cholesterol interaction is critical for the integrity and stability of OTR, we characterized the thermal stability of purified, detergent-solubilized receptor and several mutants in the presence or absence of the cholesterol analog cholesteryl hemisuccinate (CHS). We monitored protein unfolding using the previously described CPM-based thermal shift assay (20) (Fig. 3D). Since wtOTR could not be purified, because of very low expression yields and insufficient thermostability, the thermal stability analyses were performed with OTRXTAL or mutants thereof.
In accordance with previous findings (31), the stability of OTRXTAL is significantly impaired in n-dodecyl-β-d-maltopyranoside (DDM) in the absence of CHS, as evidenced by a decrease of 12°C in thermostability, when compared with the same construct measured in DDM supplemented with CHS (Fig. 3D and table S4). A similar effect has also been reported for several other GPCRs (32–34). To test the individual contributions of residues within the observed cholesterol binding site, we next expressed and purified OTRXTAL mutants where the two most prominent aromatic residues within the cholesterol binding site, Y2005.38 and W2035.41, were each mutated to alanine.
In the absence of CHS, OTRXTAL and the two mutants Y2005.38A and W2035.41A display comparable melting temperatures (43.7°, 44.5°, and 41.9°C, respectively), indicating a similar limited stability of all three receptor variants (Fig. 3D and table S4). This similarity of melting temperatures of OTRXTAL mutants and OTRXTAL shows that there is no strongly destabilizing effect of the introduced mutations themselves. In the presence of CHS, in contrast, all three variants are stabilized as evidenced by higher melting temperatures, albeit to varying extents (Fig. 3D). The two tested OTRXTAL mutants, where key cholesterol-interacting residues were exchanged to alanine, display a significant decrease in thermostability of 9°C (Y2005.38A) and 3°C (W2035.41A) in comparison to OTRXTAL, when CHS is present. This observed decrease in thermal stability of the mutants can be best rationalized by the perturbation and weakening of the binding of CHS to the cholesterol binding pocket.
To further validate the biological relevance of the observed cholesterol binding site and delineate its influence on antagonist and agonist binding in a native lipid environment, we generated point mutants of Y2005.38 and W2035.41 in the background of wtOTR and probed their involvement in cholesterol binding to the OTR in a cell-based assay. Y2005.38 and W2035.41 were individually mutated to either alanine or histidine, and ligand binding was assessed using a homogeneous time-resolved fluorescence (HTRF)–based assay (35). This experimental setup allowed us to measure simultaneously total receptor surface expression levels through the donor-only signal (E620) originating from the fluorescently labeled SNAP tag and the maximum attainable response Bmax (E665/E620; as a measure of functional protein) at saturating concentrations of a fluorescently labeled agonist or antagonist (see Methods for details).
While all tested mutants displayed total surface expression levels similar to the wtOTR (Fig. 3E), the measured Bmax values of the respective mutants were markedly reduced in comparison with the wt receptor (Fig. 3, F and G). The strong reduction in the Bmax values suggests that, although wtOTR and the respective mutants are expressed and localized in equivalent amounts on the cellular surface, upon mutation of residues involved in the cholesterol interaction, only a fraction of receptors remain functional with regard to both agonist and antagonist binding. Therefore, also in a native environment, the mutations introduced in the cholesterol binding site appear to have a detrimental effect on the structural integrity of the extracellular binding pocket itself, with direct consequences for agonist and antagonist binding.
Mechanistically, the receptor modulation by the presence of cholesterol in this particular binding site might occur through the direct interaction with ECL2, which has been reported to be intimately involved in agonist binding (36). However, cholesterol appears to also directly influence the polar network of the ligand-binding pocket through the positioning of Y2005.38, which can then form a hydrogen bond with the carbonyl side chain of Q1714.60 and thereby optimally orient this residue within the extracellular binding cavity (Fig. 3C), thereby rendering the OTR in a state for optimal ligand recognition and interaction—the aforementioned high-affinity state.
Allosteric divalent cation coordination site
GPCRs are intrinsically allosteric molecules, with many of them being negatively modulated by the monovalent cation Na+. High-resolution structures of several receptors have provided a structural basis for this observation: An interhelical Na+ binding site within the conserved water-mediated hydrogen bond network connecting helices II, III, VI, and VII stabilizes the inactive receptor conformation in the presence of Na+ and leads to a measurable decrease in agonist affinity (37). Likewise, for the OTR, Na+ has been shown to act as a negative allosteric modulator with the sodium binding pocket located in the aforementioned network in the core of the receptor (38). In contrast to the negative allosteric effect of monovalent sodium ions, divalent cations such as Mg2+ (but also Ni2+, Zn2+, Mn2+, and Co2+) were shown to act as positive allosteric modulators of the OTR (17, 39).
In the antagonist-bound OTR structure, we found two acidic amino acids, E421.35 and D1002.65, located at the extracellular tips of helices I and II, which are found in unusually close proximity to each other (Fig. 4A). Both residues have previously been described to be involved in the binding of endogenous cyclic peptide agonists [E421.35 in OTR for oxytocin binding, and both corresponding residues, E371.35 and D952.65 in V1AR, for vasopressin binding (40, 41)]. The short distance of only 2.8 Å between the closest oxygens of the carboxyl head groups of these two amino acids in the antagonist-bound state suggests that in this arrangement in the determined structure, the two residues share a proton. We hypothesized that, on the basis of the charged nature and close proximity of these residues to the presumed orthosteric binding site of the OTR, this region might represent the elusive coordination site for divalent cations, known to be involved in modulating neurohypophyseal hormone binding. We note, however, that in the captured antagonist-bound state, no divalent cation could be observed.
Fig. 4. Divalent cation binding site of OTR.
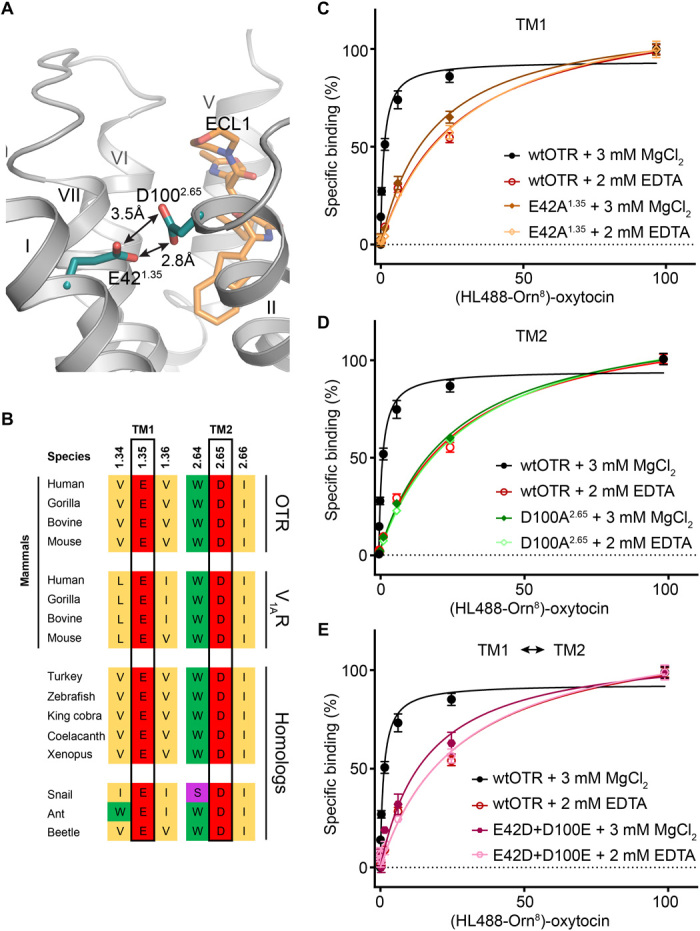
(A) Location of the allosteric divalent cation binding site created by E421.35 and D1002.65 shown in cyan sticks with head group oxygen-oxygen distances indicated by arrows. (B) Amino acid sequence alignment of specific residues comprising the human OTR divalent cation binding site (black boxes). The divalent cation binding site of human OTR is highly conserved in homologs from different taxa including mammals, nonmammalian vertebrates, and invertebrates. Furthermore, the divalent cation binding site of OTR is also conserved in mammalian homologs (human, gorilla, bovine, and mouse) of the closely related V1AR of the oxytocin/vasopressin system. Accession codes are provided in fig. S5A. (C) Specific oxytocin binding to wtOTR and wtOTR with E421.35 of the divalent cation binding site mutated to alanine (E421.35A) in the presence (3 mM MgCl2) or absence (2 mM EDTA) of Mg2+. Data are shown as means ± SEM from three independent experiments performed in triplicates (table S5). (D) As in (C) with the mutant D1002.65A. (E) As in (C) with a double mutant, in which both acidic residues of the divalent cation binding site are interchanged (E421.35D and D1002.65E).
Initial amino acid sequence alignments across both the OTR and the related V1AR sequences revealed that this region of transmembrane helices I and II is highly conserved in both receptors across mammalian species (Fig. 4B and table S4). Since the evolutionary lineage of disulfide-linked oxytocin- and vasopressin-related hormones, and thus possibly their associated GPCRs, existed for at least 700 million years (1), we next expanded our search to receptor homologs identified in more distant taxa such as nonmammalian vertebrates and invertebrates. We find that the glutamate in helix I and the aspartate in helix II as well as surrounding residues are strictly conserved throughout taxa and might, thus, represent a mechanistically critical motif for the function of these GPCRs. Moreover, sequence comparisons with closely related class A GPCRs binding to linear peptides (fig. S5) did not reveal the presence of these acidic side chains at homologous positions, thereby lending further evidence for a specific functional role of this arrangement within the neurohypophyseal hormone receptors and their related homologs.
To test whether the acidic residues of helices I and II are involved in the positive allosteric modulation of agonist binding through divalent cation coordination, we measured the binding affinity of fluorescently labeled oxytocin (HiLyte Fluor 488-Orn8 oxytocin) in the presence or absence of Mg2+, which is most likely the physiologically relevant cation (35), with an HTRF-based assay. We used human embryonic kidney (HEK) 293T cells expressing either the wtOTR, one of the single point mutants E421.35A and D1002.65A, or the double mutant E421.35D/D1002.65E (Fig. 4C and table S5). In accordance with previous studies, which reported a pronounced influence of magnesium on the binding of different agonists to OTR (38, 39), we observed a 30-fold increase in affinity for oxytocin to the wt receptor in the presence of 3 mM Mg2+, compared with the same experiment performed in its absence. When divalent cations were depleted through addition of EDTA, the affinity for oxytocin to all three evaluated mutants and wtOTR was found to be very similar (table S5).
In contrast, in the presence of 3 mM Mg2+, the individual substitution of either E421.35 or D1002.65 by alanine abolishes the positive modulatory effect of Mg2+ on oxytocin binding, thus highlighting the importance of these two specific residues in divalent cation binding. Under the same experimental conditions, the positive allosteric effect of Mg2+ can only be partly restored by interchanging residues, E421.35D and D1002.65E, suggesting a significant influence of the precise spatial orientation of these two acidic residues and participation of further Mg2+-liganding atoms from the agonist and receptor, which is supported by the strong conservation of surrounding residues.
Together, these results strongly suggest that E421.35 and D1002.65 constitute part of the physiologically important coordination site of the OTR for divalent cations and presumably also in the closely related vasopressin receptors, and that the precise spatial orientation of these residues is critical for high-affinity binding of the cyclic peptide agonists of these receptors.
DISCUSSION
Because of their distinct roles in social behavior, cognition, and reproduction (42), the receptors of the oxytocin and vasopressin family represent attractive drug targets. The crystal structure of the OTR in complex with the small-molecule antagonist retosiban presented in this study provides the first structural insights into these distinct neuropeptide GPCRs; it furthermore allows us to provide answers to several long-standing questions regarding the structural basis of the essential modulatory effects by both cholesterol and divalent cations. Both of these can likely be extrapolated to the closely related vasopressin receptors due to their high-sequence conservation.
The solvent-exposed binding pocket of retosiban-bound OTR is significantly larger in comparison to other antagonist-bound peptidergic GPCRs, which is possibly a direct consequence of the requirement to accommodate a cyclic nonapeptide agonist. Within the binding pocket, the ligand-accessible interface, ranging from helix II to helix IV, is dominated by a polar network, while the opposing side of the cavity is largely hydrophobic. By virtue of its 2,5-DKP core and its chemically diverse substituents, the cocrystallized nonpeptidic oral antagonist retosiban optimally addresses this challenging environment, lending credit to decades of SAR studies to optimize these compounds.
Retosiban occupies an overlapping binding site at the bottom of the predicted orthosteric binding pocket that is at least partially overlapping with previously proposed agonist binding modes. Retosiban directly interacts with residues Q922.57, K1163.29, Q1193.32, Q1714.60, and Q2956.55, all of which were previously described to be part of the presumed orthosteric pocket of OTR, based on SAR studies performed on V1AR (43). Of the 11 contact residues between retosiban and OTR, 8 are conserved in the V1AR receptor (except I2015.39, I2045.42, and A3187.42 in OTR, which are V2175.39, M2205.42, and G3377.42 in V1AR, respectively). Mutating A3187.42 to glycine, which takes this position in V1AR, has already previously been reported to influence the selectivity profile of other nonpeptidic OTR antagonists (L-371,257 and L-372,622) (44). Thus, the high selectivity of retosiban for the OTR over the V1AR might at least be partially rationalized by these three nonconserved interaction residues in the OTR-retosiban binding pocket.
In addition to the structural basis for antagonist recognition, the OTR structure provides new structural insights into the binding location of the modulatory cholesterol molecule. We find that the cholesterol molecule binds to an extrahelical crevice (formed between helices IV, V, and ECL2) that is distal from previously reported cholesterol interaction sites on other GPCRs, consistent with nonconserved sequences in these regions (30). Only in the P2Y1 receptor, cholesterol binds to a similar location, albeit with different contact residues and a different orientation (fig. S2) (45). Through mutagenesis experiments using orthogonal assays, we established that binding of cholesterol in OTR is critically governed by the conserved aromatic residues Y2005.38 and W2035.41 on helix V. The presence of cholesterol within this binding site has a direct effect on the integrity of the binding pocket as evidenced by an almost complete loss in measurable ligand binding for the antagonist as well as the agonist, if single-point mutations are introduced.
This finding, obtained both from purified protein preparations in vitro and from cellular in vivo experiments, makes it conceivable to now specifically target this extrahelical binding site for the development of potent, positive or negative allosteric modulators for the OTR.
Finally, a mechanistic basis of the allosteric modulation of OTR function through interaction with Mg2+ is provided by the identification of an evolutionarily highly conserved Mg2+ binding site, which is formed by two acidic residues at the extracellular tips of helices I and II, likely involving other coordinating atoms in agonist and receptor. To our knowledge, the identification and verification of this distinct, solvent-exposed coordination site in the OTR represent the first example for a positive allosteric modulatory mechanism of GPCRs by divalent cations and, thus, further expands our knowledge of this intrinsically allosteric receptor superfamily.
Despite their diverse physiological roles, the receptors for the cyclic peptide hormones oxytocin and vasopressin are very closely related. Thus, not only do these studies presented here provide answers concerning the OTR itself, but these findings will support research advances within the vasopressin receptors, or possibly can even be directly extrapolated. We anticipate that the inaugural structure from this medically highly relevant receptor family branch opens up attractive possibilities to deploy structure-based drug design methods to not only the OTR but also, eventually, the vasopressin receptors in an effort to develop new therapeutics for the efficient treatments of a diverse and important array of diseases.
METHODS
Generation of OTR crystallization construct
The human OTR gene was codon-optimized for Spodoptera frugiperda (Sf9) expression, synthesized (Integrated DNA Technologies) and cloned into a modified pFL vector (MultiBac system, Geneva Biotech), resulting in an expression construct with an N-terminal melittin signal sequence followed by a FLAG tag, His10 tag, and a human rhinovirus 3C protease cleavage site.
In the crystallization construct, several modifications were introduced to the receptor to increase the functional expression yield, stability, and crystallization: To aid crystal contact formation in LCP, 34 residues (R232-L265) of the ICL3 were replaced by the thermostable PGS domain, and 30 residues (360 to 389) were truncated from the C-terminus of the receptor. Analogous to previously reported structures, the gene was further modified by introducing mutations to improve protein yield and thermostability: A19T, R65H, V120L, A167V, Q193R, S322C, N325K, and T333M were selected through two consecutive rounds of directed evolution in yeast (18) on the wtOTR using the fluorescently labeled peptide antagonist phenylpropionyl linear vasopressin antagonist (PVA) [HiLyte Fluor 647-Lys8 PVA (Eurogentec)] (46). To further increase the thermostability of the evolved OTR mutant, we subsequently substituted selected amino acids within the transmembrane helical bundle with either alanine or leucine (if the amino acid was already an alanine). These single-point mutants were then individually expressed, purified, and evaluated on the basis of their influence on apparent thermostability as evidenced by a change in melting temperature (Tm) when assessed in the CPM assay (20). The two-point mutants D153A and S224A that individually displayed the highest gain in Tm were combined and, together with the eight mutations stemming from directed evolution, yielded the stabilized receptor variant OTRSTAB.
For ligand-binding experiments, the codon-optimized wtOTR gene was subcloned into a mammalian expression vector [pcDNA3.1(+)] containing an N-terminal SNAP tag (Cisbio). Point mutations were derived from this construct using SLIC cloning (47).
Expression and purification of OTR
Recombinant baculovirus was generated using the MultiBac expression system. The receptor expression was performed as previously described (48). For crystallization experiments, 4 liters of Sf9 insect cells was infected with high-titer P1 or P2 baculovirus stocks (multiplicity of infection ≥5) at a cell density of 3 × 106 cells/ml in Sf-900 II SFM medium (Thermo Fisher Scientific). Cells were harvested 72 hours post-infection by centrifugation, washed with phosphate-buffered saline, flash frozen in liquid nitrogen, and stored at −80°C until further use.
Receptor-containing insect cell membranes were isolated by repeated Dounce homogenization in hypotonic [10 mM Hepes (pH 7.5), 20 mM KCl, 10 mM MgCl2, Pefabloc SC (50 μg/ml; Carl Roth), and pepstatin A (1 μg/ml; Carl Roth)] and hypertonic buffer [10 mM Hepes (pH 7.5), 20 mM KCl, 10 mM MgCl2, 1.0 M NaCl, Pefabloc SC (50 μg/ml), pepstatin A (1 μg/ml)] followed by a final wash in hypotonic buffer to reduce the NaCl concentration. The purified membrane fraction was resuspended in hypotonic buffer supplemented with 20 μM of retosiban, incubated for 30 min at 4°C, frozen in liquid nitrogen, and stored at −80°C until further use.
Before solubilization, membranes were thawed on ice, retosiban was added to a final concentration of 100 μM, and the suspension was incubated for 35 min in the presence of iodoacetamide (2 mg/ml; Sigma-Aldrich) while turning. The membranes were then solubilized in 30 mM Hepes (pH 7.5), 500 mM NaCl, 10 mM KCl, 5 mM MgCl2, Pefabloc SC (50 μg/ml), pepstatin A (1 μg/ml), 1% (w/v) DDM (Anatrace), and 0.2% (w/v) CHS (Sigma-Aldrich) at 4°C for 2.5 hours. Insoluble material was removed by ultracentrifugation, and the cleared supernatant was incubated with TALON IMAC resin (GE Healthcare) overnight at 4°C.
The receptor-bound resin was washed with 30 column volumes (CV) of Wash Buffer I [50 mM Hepes (pH 7.5), 500 mM NaCl, 10 mM MgCl2, 5 mM imidazole, 10% (v/v) glycerol, 1.0% (w/v) DDM, 0.2% (w/v) CHS, 8 mM ATP, and 50 μM retosiban] followed by 30 CV of Wash Buffer II [50 mM Hepes (pH 7.5), 500 mM NaCl, 15 mM imidazole, 10% (v/v) glycerol, 0.05% (w/v) DDM, 0.01% (w/v) CHS, and 50 μM retosiban]. Retosiban-bound OTR was eluted in fractions using a total of 4 CV of elution buffer [50 mM Hepes (pH 7.5), 500 mM NaCl, 250 mM imidazole, 10% (v/v) glycerol, 0.05% (w/v) DDM, 0.01% (w/v) CHS, and 100 μM retosiban]. Those fractions that contained purified OTR were concentrated to 0.5 ml using a 100-kDa molecular weight cutoff (MWCO) Vivaspin 2 concentrator (Sartorius Stedim) and applied to a PD MiniTrap G-25 column (GE Healthcare) equilibrated with G25 buffer [50 mM Hepes (pH 7.5), 500 mM NaCl, 10% (v/v) glycerol, 0.03% (w/v) DDM, 0.006% (w/v) CHS, and 100 μM retosiban] to remove imidazole. The eluted receptor:antagonist complex was treated overnight with His-tagged 3C protease and PNGase F (both prepared in-house) to remove the N-terminal affinity tags and deglycosylate the receptor. The mixture was then incubated with Ni-NTA resin (GE Healthcare) for 1 hour, and cleaved receptor was collected as the flow-through. Typically, we obtained 0.8 mg of purified, tag-free receptor per liter of expression volume. Pure OTR:retosiban complex was concentrated to ~70 mg/ml with a 100-kDa MWCO Vivaspin 2 concentrator. All protein concentrations were determined by measuring absorbance at 280 nm on a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). For protein concentrations, ≥10 mg/ml 1:10 dilutions were prepared using the respective buffer to ensure measurement in the linear range of the instrument. Protein purity and monodispersity were assessed by SDS–polyacrylamide gel electrophoresis and analytical size-exclusion chromatography using a Sepax Nanofilm SEC-250 column.
Crystallization in LCP
The OTR:retosiban complex was crystallized using the in meso method, however, at a reduced temperature of 16°C. For reconstitution, the concentrated OTR:retosiban complex (∼70 mg/ml) was mixed with molten monoolein (Sigma-Aldrich) supplemented with 10% (w/w) cholesterol (Sigma-Aldrich) using the twin-syringe method (49). The final protein:lipid ratio was 40:60 (v/v). Boli (40 nl) were dispensed onto 96-well glass bases with a 120-μm spacer (SWISSCI), overlaid with 800 nl of precipitant solution using a Gryphon LCP crystallization robot (Art Robbins Instruments), and sealed with a coverglass. All initial steps were performed at 20°C; however, for optimal crystal growth and diffraction, the screening plates were subsequently transferred to a humidified incubator set to 16°C. Optimized crystals used for data collection of retosiban-bound OTR were obtained in a condition consisting of 100 mM N-(2-acetamido)iminodiacetic acid (pH 5.8 to 6.0), 27 to 29% (v/v) PEG400, 320 to 350 mM NaH2PO4, 1 mM TCEP, and 50 μM retosiban. Single crystals were mounted with 20- or 30-μm Dual Thickness MicroMounts (MiTeGen) for data collection and cryo-cooled in liquid nitrogen without the addition of further cryo-protectant.
Data collection, structure determination, and refinement
X-ray diffraction data were collected using an EIGER 16M detector and a beam size of 10 μm × 10 μm at the Swiss Light Source of the Paul Scherrer Institute (Villigen, Switzerland). Datasets were collected using a beam attenuated to 30%, using 0.1° of oscillation and 0.05-s exposure time per frame. Data from individual crystals were integrated using XDS. Data from the nine best diffracting crystals were merged and scaled using the program AIMLESS from the CCP4 suite (50, 51). Data collection statistics are reported in Table 1.
Initial phase information was obtained by molecular replacement with the program Phaser (52) using a truncated polyalanine model of the OX1R transmembrane domain [Protein Data Bank (PDB) ID 4ZJ8] and the separated PGS fusion protein (53) as independent search models looking for one copy of each domain. Manual model building was performed in COOT (54) using sigma-A weighted 2 m|Fo|-|DFc| and m|Fo|-D|Fc| maps together with simulated-annealing and simple composite omit maps calculated using Phenix (55). Initial refinement was carried out with REFMAC5 (56) using maximum-likelihood restrained refinement in combination with the jelly-body protocol. Further and final stages of refinement were performed with Phenix.refine (57) with positional, individual isotropic B-factor refinement and Translation/Liberation/Screw (TLS). The final refinement statistics are presented in Table 1. Coordinates and structure factors have been deposited in the PDB under accession code 6TPK.
Whole-cell ligand-binding assay
HEK293T/17 cells (American Type Culture Collection) were cultivated in Dulbecco’s modified Eagle’s medium (Sigma) supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml; Sigma), and 10% (v/v) fetal calf serum (BioConcept). Cells were maintained at 37°C in a humidified atmosphere of 5% CO2, 95% air. Transient transfections were performed with TransIT-293 (Mirus Bio) according to the manufacturer’s instructions.
Ligand-binding experiments were performed on whole HEK293T cells for comparison of affinities for wt and receptor mutants using an HTRF-binding assay. HEK293T cells were seeded and transfected in poly-d-lysine–coated 384-well plates (Greiner) at a cell density of 7500 cells per well. Forty-eight hours after transfection, cells were labeled with 50 nM SNAP-Lumi4-Tb (Cisbio) in assay buffer [20 mM Hepes (pH 7.5), 100 mM NaCl, 3 mM MgCl2, and 0.05% (w/v) bovine serum albumin (BSA)] for 1.5 hours at 37°C. Cells were washed four times with assay buffer and were then incubated for 1 hour at RT in assay buffer containing fluorescently labeled tracer peptide (HiLyte Fluor 647-Lys8 PVA). Depending on the determined dissociation constant (Kd) from saturation binding at each mutant, tracer peptide for competition binding was used at concentrations of 5, 25, or 150 nM together with a concentration range of unlabeled retosiban as competitor. For characterization of the Mg2+-binding site and its influence on oxytocin binding, fluorescently labeled oxytocin [HiLyte Fluor 488-Orn8 oxytocin (Eurogentec)] was used. Cells were washed in wash buffer [20 mM Hepes (pH 7.5), 100 mM KCl, and 0.05% (w/v) BSA)], and assay buffer [20 mM Hepes (pH 7.5), 100 mM KCl, 3 mM MgCl2 or 2 mM EDTA, and 0.05% (w/v) BSA] was adjusted to contain either divalent magnesium or the chelator EDTA. Fluorescence intensities were measured on an Infinite M1000 fluorescence plate reader (Tecan) with an excitation wavelength of 340 nm. Acceptor emission was measured at 520 or 665 nm for the HiLyte Fluor 488 or 647, respectively. The Tb3+ donor emission was recorded at 620 nm. The ratio of FRET (fluorescence resonance energy transfer) donor and acceptor fluorescence intensities was calculated (F665 nm or F520 nm/F620 nm). Total binding was obtained in the absence of competitor, and nonspecific binding was determined in the presence of 1000-fold excess of unlabeled competitor. Data were normalized to the specific binding for each individual experiment and were analyzed by global fitting to a one-site heterologous competition equation with the GraphPad Prism software (version 8.1.1, GraphPad Prism). To obtain Ki values, data were corrected for fluorescent ligand occupancy of each mutant with the Cheng-Prusoff equation as Ki = IC50/(1 + [fl. ligand]/Kd).
Supplementary Material
Acknowledgments
We thank B. Blattmann of the Protein Crystallization Center at the University of Zürich for support with crystallization, the staff of the X06SA beamline at the Paul Scherrer Institute for support during data collection, and I. Berger at the European Molecular Biology Laboratory for providing us with baculovirus transfer vectors. We further thank C. Thom for help with protein expression, L. Wiedmer for helpful discussions during structure analysis, and GlaxoSmithKline for the generous gift of retosiban. Funding: This work was supported by Schweizerischer Nationalfonds Grants 31003A_153143 and 31003A_182334, both to A.P. Author contributions: Y.W. and L.K. created libraries and performed directed evolution in yeast. Y.W. and J.S. carried out the mutagenesis and thermostabilization of the receptor and designed and characterized crystallization constructs. Y.W. and J.E. expressed the protein. Y.W. and J.S. purified and crystallized the OTR-PGS fusion protein and harvested crystals. J.E. supported cloning and expression. Y.W., J.S., and J.E collected and processed the data, and solved and refined the structures. Y.W. performed ligand-binding experiments and analyzed the data. Project management was carried out by Y.W. and A.P. The manuscript was prepared by Y.W., J.S., J.E., and A.P. All authors contributed to the final editing and approval of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Atomic coordinates and structure factors of the OTR:retosiban complex structure have been deposited in the PDB under accession code 6TPK. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/29/eabb5419/DC1
REFERENCES AND NOTES
- 1.Donaldson Z. R., Young L. J., Oxytocin, vasopressin, and the neurogenetics of sociality. Science 322, 900–904 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Barberis C., Mouillac B., Durroux T., Structural bases of vasopressin/oxytocin receptor function. J. Endocrinol. 156, 223–229 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Duvigneaud V., Ressler C., Trippett S., The sequence of amino acids in oxytocin, with a proposal for the structure of oxytocin. J. Biol. Chem. 205, 949–957 (1953). [PubMed] [Google Scholar]
- 4.Carter C. S., Neuroendocrine perspectives on social attachment and love. Psychoneuroendocrinology 23, 779–818 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Fuchs A. R., Fuchs F., Husslein P., Soloff M. S., Fernström M. J., Oxytocin receptors and human parturition: A dual role for oxytocin in the initiation of labor. Science 215, 1396–1398 (1982). [DOI] [PubMed] [Google Scholar]
- 6.Uvnäs-Moberg K., Widström A.-M., Werner S., Matthiesen A.-S., Winberg J., Oxytocin and prolactin levels in breast-feeding women. Correlation with milk yield and duration of breast-feeding. Acta Obstet. Gynecol. Scand. 69, 301–306 (1990). [DOI] [PubMed] [Google Scholar]
- 7.Modahl C., Fein D., Waterhouse L., Newton N., Does oxytocin deficiency mediate social deficits in autism? J. Autism Dev. Disord. 22, 449–451 (1992). [DOI] [PubMed] [Google Scholar]
- 8.Hollander E., Novotny S., Hanratty M., Yaffe R., De Caria C. M., Aronowitz B. R., Mosovich S., Oxytocin infusion reduces repetitive behaviors in adults with autistic and Asperger's disorders. Neuropsychopharmacology 28, 193–198 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Pedersen C. A., Gibson C. M., Rau S. W., Salimi K., Smedley K. L., Casey R. L., Leserman J., Fredrik Jarskog L., Penn D. L., Intranasal oxytocin reduces psychotic symptoms and improves Theory of Mind and social perception in schizophrenia. Schizophr. Res. 132, 50–53 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Windle R. J., Shanks N., Lightman S. L., Ingram C. D., Central oxytocin administration reduces stress-induced corticosterone release and anxiety behavior in rats. Endocrinology 138, 2829–2834 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Giuliano F., Clèment P., Pharmacology for the treatment of premature ejaculation. Pharmacol. Rev. 64, 621–644 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Moraloglu O., Tonguc E., Var T., Zeyrek T., Batıoglu S., Treatment with oxytocin antagonists before embryo transfer may increase implantation rates after IVF. Reprod. Biomed. Online 21, 338–343 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Andersen L. F., Lyndrup J., Åkerlund M., Melin P., Oxytocin receptor blockade: A new principle in the treatment of preterm labor? Am. J. Perinatol. 6, 196–199 (1989). [DOI] [PubMed] [Google Scholar]
- 14.Theobald G. W., Graham A., Campbell J., Gange P. D., Driscoll W. J., The use of post-pituitary extract in physiological amounts in obstetrics; a preliminary report. Br. Med. J. 2, 123–127 (1948). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsatsaris V., Carbonne B., Cabrol D., Atosiban for preterm labour. Drugs 64, 375–382 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Klein U., Gimpl G., Fahrenholz F., Alteration of the myometrial plasma membrane cholesterol content with beta-cyclodextrin modulates the binding affinity of the oxytocin receptor. Biochemistry 34, 13784–13793 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Pearlmutter A. F., Soloff M. S., Characterization of the metal ion requirement for oxytocin-receptor interaction in rat mammary gland membranes. J. Biol. Chem. 254, 3899–3906 (1979). [PubMed] [Google Scholar]
- 18.Schütz M., Schöppe J., Sedlák E., Hillenbrand M., Nagy-Davidescu G., Ehrenmann J., Klenk C., Egloff P., Kummer L., Plückthun A., Directed evolution of G protein-coupled receptors in yeast for higher functional production in eukaryotic expression hosts. Sci. Rep. 6, 21508 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.J. A. Ballesteros, H. Weinstein, Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors, in Methods in Neurosciences, S. C. Sealfon, Ed. (Academic Press, 1995), vol. 25, pp. 366–428. [Google Scholar]
- 20.Alexandrov A. I., Mileni M., Chien E. Y. T., Hanson M. A., Stevens R. C., Microscale fluorescent thermal stability assay for membrane proteins. Structure 16, 351–359 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Warne T., Edwards P. C., Doré A. S., Leslie A. G. W., Tate C. G., Molecular basis for high-affinity agonist binding in GPCRs. Science 364, 775–778 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Draper-Joyce C. J., Khoshouei M., Thal D. M., Liang Y.-L., Nguyen A. T. N., Furness S. G. B., Venugopal H., Baltos J.-A., Plitzko J. M., Danev R., Baumeister W., May L. T., Wootten D., Sexton P. M., Glukhova A., Christopoulos A., Structure of the adenosine-bound human adenosine A1 receptor-Gi complex. Nature 558, 559–563 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Borthwick A. D., Liddle J., The design of orally bioavailable 2, 5-diketopiperazine oxytocin antagonists: From concept to clinical candidate for premature labor. Med. Res. Rev. 31, 576–604 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Liddle J., Allen M. J., Borthwick A. D., Brooks D. P., Davies D. E., Edwards R. M., Exall A. M., Hamlett C., Irving W. R., Mason A. M., McCafferty G. P., Nerozzi F., Peace S., Philp J., Pollard D., Pullen M. A., Shabbir S. S., Sollis S. L., Westfall T. D., Woollard P. M., Wu C., Hickey D. M. B., The discovery of GSK221149A: A potent and selective oxytocin antagonist. Bioorg. Med. Chem. Lett. 18, 90–94 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Wyatt P. G., Allen M. J., Borthwick A. D., Davies D. E., Exall A. M., Hatley R. J. D., Irving W. R., Livermore D. G., Miller N. D., Nerozzi F., Sollis S. L., Szardenings A. K., 2,5-Diketopiperazines as potent and selective oxytocin antagonists 1: Identification, stereochemistry and initial SAR. Bioorg. Med. Chem. Lett. 15, 2579–2582 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Hulce J. J., Cognetta A. B., Niphakis M. J., Tully S. E., Cravatt B. F., Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat. Methods 10, 259–264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lingwood D., Simons K., Lipid rafts as a membrane-organizing principle. Science 327, 46–50 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Gimpl G., Burger K., Fahrenholz F., Cholesterol as modulator of receptor function. Biochemistry 36, 10959–10974 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G. F., Thian F. S., Kobilka T. S., Choi H.-J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C., High-resolution crystal structure of an engineered human β2-adrenergic G protein–coupled receptor. Science 318, 1258–1265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gimpl G., Interaction of G protein coupled receptors and cholesterol. Chem. Phys. Lipids 199, 61–73 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Gimpl G., Fahrenholz F., Cholesterol as stabilizer of the oxytocin receptor. BBA-Biomembranes 1564, 384–392 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Yao Z., Kobilka B., Using synthetic lipids to stabilize purified β2 adrenoceptor in detergent micelles. Anal. Biochem. 343, 344–346 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Hanson M. A., Cherezov V., Griffith M. T., Roth C. B., Jaakola V.-P., Chien E. Y. T., Velasquez J., Kuhn P., Stevens R. C., A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure 16, 897–905 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson A. A., Liu J. J., Chun E., Wacker D., Wu H., Cherezov V., Stevens R. C., GPCR stabilization using the bicelle-like architecture of mixed sterol-detergent micelles. Methods 55, 310–317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Degorce F., Card A., Soh S., Trinquet E., Knapik G. P., Xie B., HTRF: A technology tailored for drug discovery –a review of theoretical aspects and recent applications. Curr. Chem. Genomics 3, 22–32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Postina R., Kojro E., Fahrenholz F., Separate agonist and peptide antagonist binding sites of the oxytocin receptor defined by their transfer into the V2 vasopressin receptor. J. Biol. Chem. 271, 31593–31601 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Katritch V., Fenalti G., Abola E. E., Roth B. L., Cherezov V., Stevens R. C., Allosteric sodium in class A GPCR signaling. Trends Biochem. Sci. 39, 233–244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schiffmann A., Gimpl G., Sodium functions as a negative allosteric modulator of the oxytocin receptor. Biochim. Biophys. Acta Biomembr. 1860, 1301–1308 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Antoni F. A., Chadio S. E., Essential role of magnesium in oxytocin-receptor affinity and ligand specificity. Biochem. J. 257, 611–614 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wootten D. L., Simms J., Massoura A. J., Trim J. E., Wheatley M., Agonist-specific requirement for a glutamate in transmembrane helix 1 of the oxytocin receptor. Mol. Cell. Endocrinol. 333, 20–27 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Rodrigo J., Pena A., Murat B., Trueba M., Durroux T., Guillon G., Rognan D., Mapping the binding site of arginine vasopressin to V1a and V1b vasopressin receptors. Mol. Endocrinol. 21, 512–523 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Insel T. R., A neurobiological basis of social attachment. Am. J. Psychiatry 154, 726–735 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Mouillac B., Chini B., Balestre M.-N., Elands J., Trumpp-Kallmeyer S., Hoflack J., Hibert M., Jard S., Barberis C., The binding site of neuropeptide vasopressin V1a receptor: Evidence for a major localization within transmembrane regions. J. Biol. Chem. 270, 25771–25777 (1995). [DOI] [PubMed] [Google Scholar]
- 44.Hawtin S. R., Ha S. N., Pettibone D. J., Wheatley M., A Gly/Ala switch contributes to high affinity binding of benzoxazinone-based non-peptide oxytocin receptor antagonists. FEBS Lett. 579, 349–356 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Zhang K., Zhang J., Gao Z.-G., Zhang D., Zhu L., Han G. W., Moss S. M., Paoletta S., Kiselev E., Lu W., Fenalti G., Zhang W., Müller C. E., Yang H., Jiang H., Cherezov V., Katritch V., Jacobson K. A., Stevens R. C., Wu B., Zhao Q., Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature 509, 115–118 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mouillac B., Manning M., Durroux T., Fluorescent agonists and antagonists for vasopressin/oxytocin G protein-coupled receptors: Usefulness in ligand screening assays and receptor studies. Mini Rev. Med. Chem. 8, 996–1005 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.M. Z. Li, S. J. Elledge, in Gene Synthesis: Methods and Protocols, J. Peccoud, Ed. (Humana Press, Totowa, NJ, 2012), pp. 51–59. [Google Scholar]
- 48.Schöppe J., Ehrenmann J., Klenk C., Rucktooa P., Schütz M., Doré A. S., Plückthun A., Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists. Nat. Commun. 10, 17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caffrey M., Cherezov V., Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collaborative Computational Project, Number 4 , The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 51.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin J., Mobarec J. C., Kolb P., Rosenbaum D. M., Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant. Nature 519, 247–250 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/29/eabb5419/DC1


