Cryo-EM structures reveal the ATP conduction and oligomeric assembly mechanisms of CALHM channels.
Abstract
Calcium homeostasis modulator (CALHM) family proteins are Ca2+-regulated adenosine triphosphate (ATP)–release channels involved in neural functions including neurotransmission in gustation. Here, we present the cryo–electron microscopy (EM) structures of killifish CALHM1, human CALHM2, and Caenorhabditis elegans CLHM-1 at resolutions of 2.66, 3.4, and 3.6 Å, respectively. The CALHM1 octamer structure reveals that the N-terminal helix forms the constriction site at the channel pore in the open state and modulates the ATP conductance. The CALHM2 undecamer and CLHM-1 nonamer structures show the different oligomeric stoichiometries among CALHM homologs. We further report the cryo-EM structures of the chimeric construct, revealing that the intersubunit interactions at the transmembrane domain (TMD) and the TMD–intracellular domain linker define the oligomeric stoichiometry. These findings advance our understanding of the ATP conduction and oligomerization mechanisms of CALHM channels.
INTRODUCTION
Adenosine triphosphate (ATP) as an extracellular ligand plays essential roles in various cellular functions, including neurotransmission (1,2). Although anionic ATP molecules cannot diffuse across the plasma membrane, ATP release from the cytosol is mediated by ATP-permeable channels, such as connexin hemichannels, pannexin 1 (PANX1), volume-regulated anion channels (VRACs), and calcium homeostasis modulator 1 (CALHM1) (3). CALHM1 was originally identified as a genetic risk factor for late-onset Alzheimer’s disease (4) and recently identified as an essential component of the neurotransmitter-release channel in taste bud cells that mediates the voltage-dependent release of ATP to afferent gustatory neurons, whereby mediating the perception of sweet, bitter, umami, and salty tastes (5–8). CALHMs are regulated by the membrane voltage and extracellular Ca2+. Strong depolarization and extracellular Ca2+ concentration ([Ca2+]o) reduction increase the open probability of CALHM channels (9,10). Six CALHM1 homologs were found in humans (11). CALHM genes are present throughout vertebrates, but they have no sequence homology to other known genes. CALHM2 modulates neural activity in the central nervous system (12). CALHM3 interacts with CALHM1, forming a CALHM1/CALHM3 heteromeric channel that confers fast activation (8, 13, 14). Outside of vertebrates, Caenorhabditis elegans has single CALHM ortholog, termed CLHM-1. The C. elegans CLHM-1 is expressed in sensory neurons and muscles and functions as an ion channel (15, 16).
Many ATP-permeable channels have been structurally characterized (17–20), but the molecular mechanism of CALHM channels has remained unclear. Here, we report the cryo–electron microscopy (EM) structures of three CALHM homologs (CALHM1, CALHM2, and C. elegans CLHM-1) and their chimeric construct in unexpectedly different oligomeric states, revealing the oligomerization mechanism of CALHM channels. Furthermore, the present cryo-EM structure of CALHM1 is the first to visualize the N-terminal helix (NTH), which forms the constriction gate at the channel pore in the open state, and the bioluminescence assay revealed that it modulates the ATP conductance. These findings provide insights into the ATP conduction and oligomerization mechanisms of CALHM channels.
RESULTS
Structure determination of CALHM1
For the structural analysis, we first screened the CALHM1 proteins from various vertebrates by fluorescence detection gel filtration chromatography (21) and identified the killifish Oryzias latipes CALHM1 (OlCALHM1) as a promising candidate (fig. S1A). OlCALHM1 and HsCALHM1 (“Hs” refers to Homo sapiens) share 62% sequence identity and 78% sequence similarity within the transmembrane (TM) region. Next, we analyzed the function of OlCALHM1. We transfected the DNA encoding the full-length OlCALHM1 into HeLa cells and measured the extracellular ATP concentration by the luciferin-luciferase reaction (fig. S2, A to C) (5). Lowering [Ca2+]o induced a larger increase in the extracellular ATP concentration than in mock-transfected cells (fig. S2A), and ruthenium red (RUR), a known blocker of human and mouse CALHM1 channels, abolished this ATP release (fig. S2B), indicating that OlCALHM1 forms an ATP-release channel activated by low [Ca2+]o, as also reported for HsCALHM1 (5).
For the cryo-EM analysis, we expressed the CALHM protein in human embryonic kidney (HEK) 293S GnTI− cells. To facilitate the expression and purification of OlCALHM1, we truncated the flexible C termini starting from Trp296 (OlCALHM1EM). OlCALHM1EM retains the ATP-release activity elicited by low [Ca2+]o when expressed in HeLa cells (fig. S2C). Moreover, we confirmed that the purified OlCALHM1EM forms an ATP-permeable channel by a liposome assay (fig. S2D). The OlCALHM1EM protein was purified in glyco-diosgenin (GDN) micelles without Ca2+ and vitrified on a grid. The samples were imaged using a Titan Krios EM equipped with a K3 camera. The two-dimensional (2D) class average showed obvious C8 symmetry (fig. S3, A and B). The 3D map was reconstructed from 102,109 particles with the imposed C8 symmetry, at the 2.66-Å resolution based on the Fourier shell correlation (FSC) gold-standard criteria (Fig. 1A), which was sufficient for de novo model building (figs. S3, C to E, and S4A and table S1). The processing summary is provided in fig. S3B. The N-terminal residues Met1 to Met6 and the C termini after Gln263 are disordered, consistent with the secondary structure prediction.
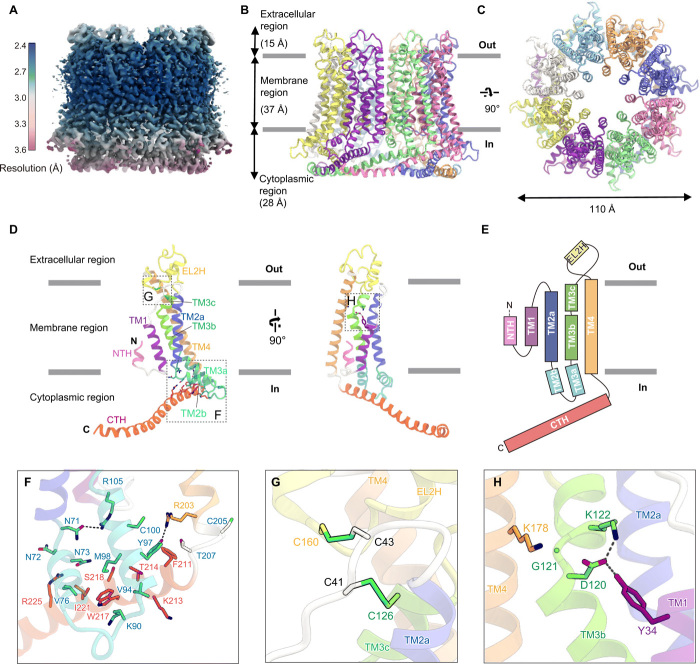
Fig. 1. Overall structure of OlCALHM1.
(A) Local resolution of the OlCALHM1EM structure, estimated by RELION. (B and C) Overall structure of the OlCALHM1EM octamer, viewed parallel to the membrane (B) and from the extracellular side (C). (D) Subunit structure of OlCALHM1EM, viewed parallel to the membrane. Each region is colored as follows: NTH, pink; TM1, purple; EL1, white; TM2a, light blue; TM2b, IL1, and TM3a, cyan; TM3b and 3c, light green; EL2, yellow; TM4, orange; CTH, red. The N and C termini are indicated by “N” and “C,” respectively. (E) Schematic representation of the OlCALHM1 membrane topology, colored as in (D). (F to H) Close-up views of the intracellular side (F), the two disulfide bonds in the extracellular region (G), and the interactions between TM1 and the other TMs around Asp120 (H).
Structure of OlCALHM1
OlCALHM1EM forms an octamer with a length of 80 Å and a width of 110 Å (Fig. 1, B and C). At the center of the octamer structure, a channel pore is formed along an axis perpendicular to the membrane. The extracellular region projects approximately 15 Å above the plasma membrane. The TM region is 37 Å thick, and the intracellular region protrudes about 28 Å from the plasma membrane. The subunit consists of extracellular loops 1 and 2 (EL1, Phe37-Tyr47 and EL2, Phe128-Asp166), an intracellular loop (IL; Arg83-Ala91), an NTH (Met7-Gln15), a TM domain (TMD; Gly25-Ser35, Tyr48-Leu70, Ile108-Ala127, and Asn167-Cys205), and a C-terminal helix (CTH; Phe206-Arg260), with the C terminus on the cytoplasmic side (Fig. 1, D and E). The TMD consists of four TM helices (TM1 to TM4), which are aligned counterclockwise (Fig. 1, D and E, and fig. S4B), as viewed from the extracellular side. This TM arrangement is quite different from those in the connexin (17), innexin (18), and LRRC8 (19, 20) structures (fig. S4B). TM1 faces the channel pore, while TM2 and TM4 face the phospholipid membrane. A clear density shows the NTH preceding TM1 and forming the channel pore, together with TM1 (fig. S4A). TM3 is located between TM1, TM2, and TM4, connecting these structural elements. TM4 is tilted by about 30° from the channel pore axis (perpendicular to the membrane plane) and extends from the membrane lipid. Notably, TM2 is kinked at Asn71 (TM2a and TM2b), and TM3 is kinked at Leu107 and Gly121 (TM3a to TM3c). Thereby, TM2b and TM3a protrude into the intracellular region and are stabilized by the hydrogen bonding interactions of Asn71-Arg105 and Tyr97-Arg203 (Fig. 1F). On the intracellular side, CTH adopts a long helix and extends toward the adjacent subunit. The protruded TM2b and TM3a form extensive van der Waals interactions with the CTH (Fig. 1F). On the extracellular side, EL2 between TM3 and TM4 forms a short helix (EL2H) and wraps around the short EL1 between TM1 and TM2, covering the extracellular side of the TM region. Between EL1 and EL2, two disulfide bonds are formed at the Cys41-Cys126 and Cys43-Cys160 pairs, which stabilize the EL structure (Fig. 1G). The residues constituting the TMD and the CTH, and the two cysteines in the ELs are highly conserved in the CALHM1 orthologs, including HsCALHM1 (fig. S1, A to C).
A previous study reported that two cysteines in mouse CALHM1 are palmitoylated and associated with the posttranslational regulation of the channel gating and localization (22). These two cysteines are completely conserved in the CALHM1 orthologs (fig. S1A). In the OlCALHM1EM structure, the corresponding cysteines Cys100 and Cys205 are located at TM3a and the intracellular end of TM4, respectively (Fig. 1F). The sulfhydryl groups of these Cys residues are directed toward the lipid bilayer and are likely to be palmitoylated, although we could not find the density of the palmitoylation in our EM map, probably due to the flexibility of the acyl chains.
In HsCALHM1, the negative charge of Asp121 reportedly plays a critical role in ion permeation property (9). This Asp residue is highly conserved in the CALHM1 orthologs, and the corresponding Asp120 in OlCALHM1 forms a salt bridge with Lys122 in TM3 (Fig. 1H), stabilizing its bent conformation at Gly121. Moreover, Asp120 forms a hydrogen bond with Tyr34 in TM1 and an electrostatic interaction with Lys178 in TM4. These residues are also conserved in HsCALHM1. Overall, Asp120 contributes to the structural organization of the TM helices inside the channel.
Subunit interface in OlCALHM1
The subunit interface is formed at the TM and intracellular regions (Fig. 2A). Unlike connexin (17), innexin (18), and LRRC8 (19, 20), the adjacent extracellular regions do not interact with each other. At the TM region (Fig. 2B), TM2 forms extensive hydrophobic interactions with TM4′ (apostrophe indicates the adjacent subunit). At the extracellular and intracellular ends of the interface, Tyr51 and Asn72 in TM2 form hydrogen bonds with Gln182 and Arg200 in TM4′, respectively (Fig. 2B). EL1 also forms hydrogen-bonding interactions with TM4′ (Fig. 2C). Moreover, the NTH regions interact with each other between adjacent subunits: Gln12 in the NTH forms a hydrogen bonding interaction with Ser13 in the NTH′ (Fig. 2C). At the intracellular region, the long CTH extends to the adjacent subunit and forms extensive interactions with it (Fig. 2D). Furthermore, Glu254 forms a salt bridge with Lys213 in the CTH between the two adjacent subunits (Fig. 2D).
Fig. 2. Subunit interfaces between two adjacent subunits.
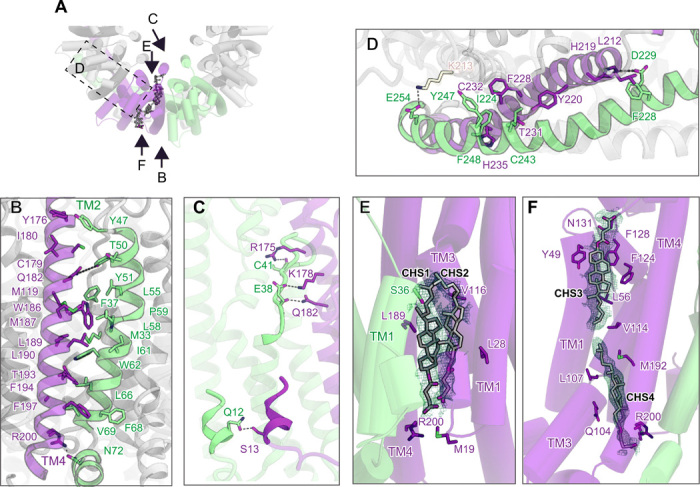
(A) Overview between two adjacent subunits, viewed from the extracellular side. (B to D) Close-up views between two adjacent subunits at the TM region (B and C) and intracellular region (D). (E and F) Close-up views of CHS1 and CHS2 (E) and CHS3 and CHS4 (F). CHS molecules are indicated by gray stick models. The cryo-EM density of each CHS is contoured at 4.0 σ.
In addition, there are extra densities at the subunit interface (Fig. 2, E and F, and fig. S4A). On the basis of their characteristic shape, we assigned them to the cholesteryl hemisuccinate (CHS) molecules used for membrane protein solubilization (CHS1 and CHS2) (Fig. 2E). These CHS molecules are surrounded by TM1 (Ser36) and TM1′ (Met19 and Leu28), TM3 (Val116), and TM4 (Arg200 and Leu189) and bridge the gap between the subunits behind the NTH, stabilizing the subunit interface and pore architecture. A lipid-mediated subunit interface has not been observed in connexin26, LRRC8, or other gap junction channels and is a unique structural feature of OlCALHM1. Moreover, two CHS molecules are observed on the membrane side (CHS3 and CHS4) (Fig. 2F). These CHS molecules form extensive hydrophobic interactions with the channel and fill the cleft between TM1, TM3, and TM4. The residues involved in the interaction with CHS are highly conserved in the CALHM1 orthologs (fig. S1), suggesting that lipids play an important role in the structural stabilization of the CALHM1 channels.
Pore architecture of OlCALHM1
OlCALHM1EM has a channel pore along a central axis perpendicular to the membrane. In the structures of gap junction channels (connexin and innexin) (17, 18), the NTH and extracellular loop form two constriction sites in the pore. In the OlCALHM1EM structure, only the NTH forms the single constriction site in the pore (Fig. 3A). At the constriction site, the neutral residues Gln9, Gln12, and Ser13 face the pore, with the narrowest diameter of 15.7 Å at Gln9 (Fig. 3B). Previous optical and electrophysiological analyses of the permeation of fluorescence dyes and ions with different sizes suggested that HsCALHM1 has a large pore greater than 14 Å (11), which is consistent with the diameter at the narrowest neck of OlCALHM1EM, allowing the permeation of all hydrated ions and ATP (~7 Å). HsCALHM1 is both cation and anion permeable at PNa:PK:PCa:PCl = 1:1.14:13.8:0.52 (9, 11). The pore-facing residues in the NTH are neutral (Gln9, Gln12, and Ser13), accounting for the weak charge selectivity. Together, the pore architecture in the OlCALHM1EM structure is in excellent agreement with the previous functional analysis of HsCALHM1.
Fig. 3. Pore architecture and ATP conduction of OlCALHM1.
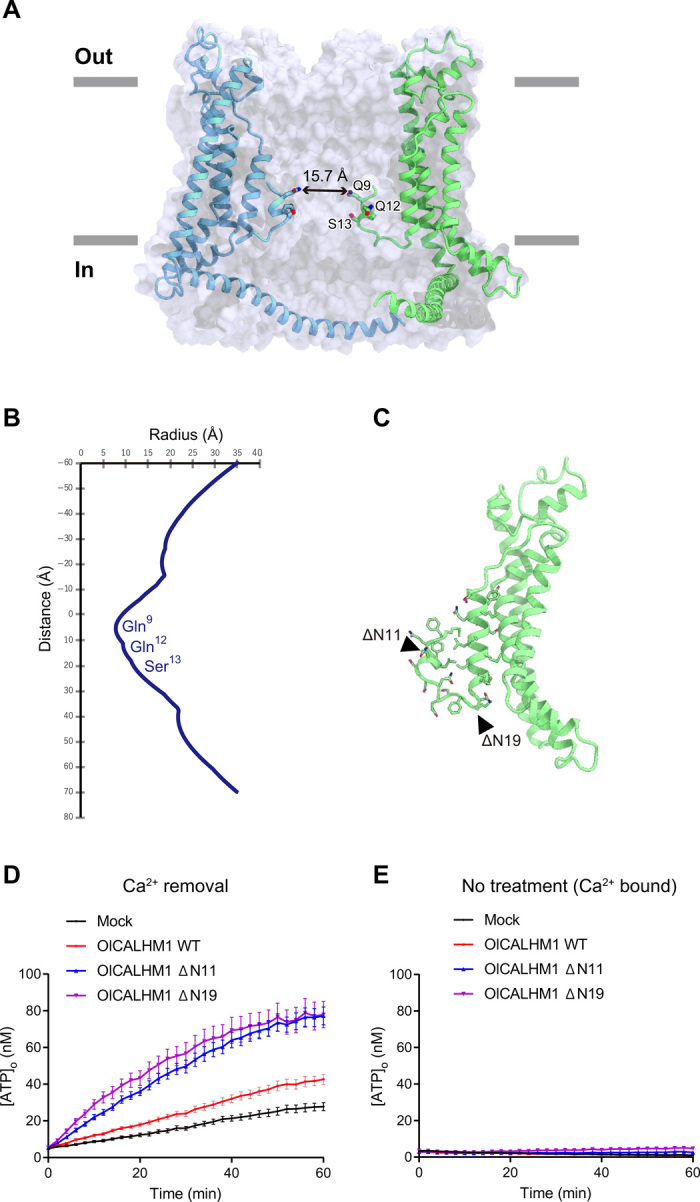
(A) Cross sections of the surface representation for the OlCALHM1EM subunits, showing the ion channel pore. (B) Pore radius for the OlCALHM1EM structure along the pore center axis. The pore size was calculated with the program HOLE (38). (C) Design of the ΔNTH constructs. The residues in the NTH and the loop between NTH and TM1 are shown as stick models. (D and E) Time courses of extracellular ATP levels due to release from HeLa cells transfected with the empty vector (mock), WT OlCALHM1, or NTH-truncated (ΔN11 or ΔN19) OlCALHM1 following exposure to essentially zero (17 nM) (D) or normal (1.9 mM) [Ca2+]o (E). Data are displayed as means ± SEM. n = 15 (mock), n = 16 (WT, ΔN11, and ΔN19) in (D). n = 16 in each group in (E).
To elucidate the functional role of the NTH, we truncated the NTH of OlCALHM1 (ΔN11 and ΔN19) (Fig. 3C) and measured the ATP-release activity by a cell-based assay. In the Ca2+-free open state, the ATP-release activities of the NTH-truncated mutant channels were markedly enhanced as compared to the wild type (WT), indicating that the NTH functions in the ATP release (Fig. 3D). However, in the Ca2+-bound closed state, the NTH-truncated mutants showed little ATP-release activities (Fig. 3E). These observations indicate that the NTH observed in our CALHM1 structure is involved in the channel conduction of ATP; however, the other structural components are associated with the Ca2+-dependent channel closing.
Structures of HsCALHM2 and CeCLHM-1 and structural differences among CALHM homologs
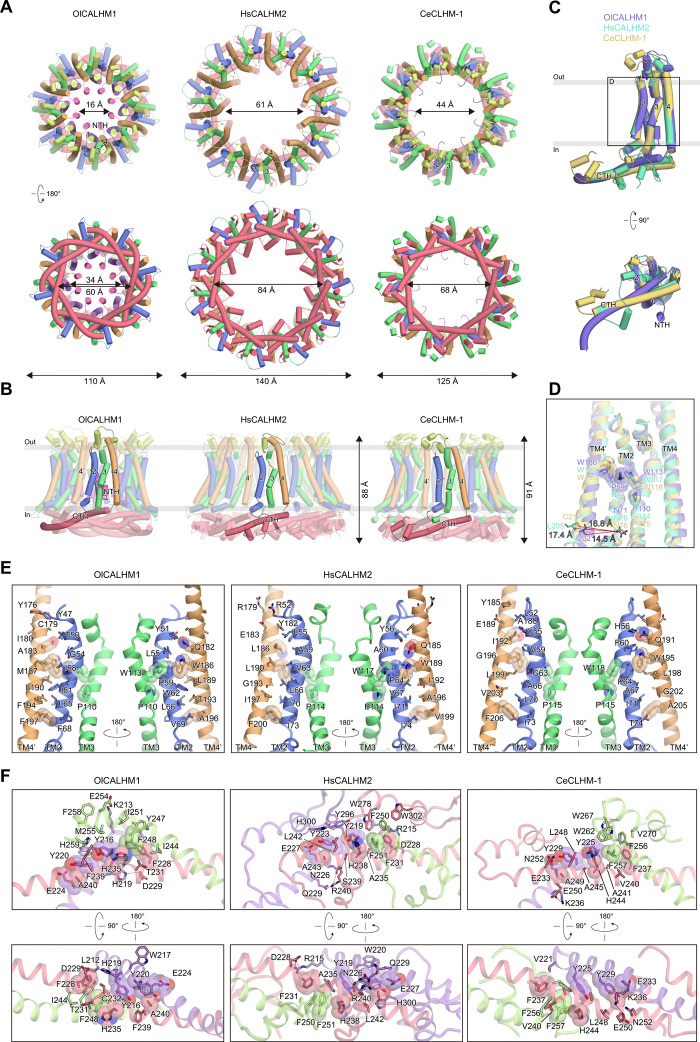
Next, we investigated the functional and structural diversity of the CALHM family. Although various members of the CALHM family have been identified (12, 15, 16), their ATP conductance activities remain elusive. Thus, we measured the ATP-release activities of HsCALHM2 and CeCLHM-1 by a cell-based assay. HsCALHM2- and CeCLHM-1–transfected HeLa cells exhibited greater RuR-sensitive increases in the extracellular ATP concentration in response to lowering [Ca2+]o compared to mock-transfected cells, indicating that HsCALHM2 and CeCLHM-1 form ATP-release channels activated by low [Ca2+]o, as in the CALHM1 channels (fig. S2, A and B). To further understand the ATP conduction mechanisms of the CALHM family members, we performed the cryo-EM analysis of HsCALHM2 and CeCLHM-1. HsCALHM2 and CeCLHM-1 were purified without Ca2+ and exchanged into the amphipol PMAL-C8. The cryo-EM images were acquired on a Titan Krios EM equipped with a Falcon 3 direct electron detector. The 2D class averages of HsCALHM2 and CeCLHM-1 showed the top views of 11- and 9-mer, respectively. We finally reconstructed the EM density maps of HsCALHM2 and CeCLHM-1 at resolutions of 3.4 and 3.6 Å, respectively, according to the gold-standard FSC = 0.143 criteria (figs. S5 and S6 and table S1).
Unlike the OlCALHM1 8-mer structure, HsCALHM2 and CeCLHM-1 form the 11-mer (approximate dimensions of 88 Å in height × 140 Å in intracellular width) and the 9-mer (approximate dimensions of 91 Å in height ×125 Å in intracellular width), respectively (Fig. 4A). The 11-mer structure of HsCALHM2 is similar to that of the recently reported structure in the open state (23). Each subunit of the three homologs, OlCALHM1, HsCALHM2, and CeCLHM-1, adopts almost similar architectures composed of the TMD and the C-terminal domain (CTD) (Fig. 4, B and C). At the TMD, both HsCALHM2 and CeCLHM-1 have the 4-TM helix topology, in which the NTH and TM1 of HsCALHM2 and CeCLHM-1 are also expected to face the pore, similar to OlCALHM1. However, the NTH and TM1 are not visualized in the density maps of HsCALHM2 and CeCLHM-1 (figs. S5G and S6G), indicating the conformational flexibility of these regions. Thus, we did not model the NTH and TM1 in both HsCALHM2 and CeCLHM-1. At the extracellular regions, two disulfide bonds between EL1 and EL2 are conserved among the three CALHM homologs (fig. S7). EL2, between TM3 and TM4, forms short helices that are arranged differently in the three structures. At the CTD, the three CALHM homologs adopt distinct architectures (Fig. 4, B and C). In contrast to OlCALHM1, HsCALHM2 and CeCLHM-1 have a CTH and additional helices, and their arrangements are different between the two homologs (Fig. 4, B and C): The CTD of HsCALHM2 consists of a CTH (Arg215-Phe250), which turns toward the pore at Phe251, and three short helices. The CTD of CeCLHM-1 consists of a CTH (Leu220-Arg254), which turns toward the membrane at Phe257, and at least one short helix. In addition to these structural observations, the low sequence identities within the CTDs among the three homologs (OlCALHM1 versus HsCALHM2, 23%; OlCALHM1 versus CeCLHM-1, 32%) suggest the structural divergence of the CTD among the CALHM family members.
Fig. 4. Structural differences among OlCALHM1, HsCALHM2, and CeCLHM-1.
(A) Structural comparisons among OlCALHM1, HsCALHM2, and CeCLHM-1. Overall structures are viewed from the extracellular side (top) and the intracellular side (bottom), colored as in Fig. 1D. (B) Subunit structures of the three CALHM channels, colored as in Fig. 1D. Adjacent subunits are shown in semitransparent colors. (C) Superimposition of the subunit structures of the three CALHM channels, viewed from the membrane side (top) and the intracellular side (bottom). OlCALHM1, HsCALHM2, and CeCLHM-1 are colored purple, cyan, and yellow, respectively. (D) Close-up view of the superimposition of the subunit structures of the three CALHM channels, colored as in Fig. 4C. Red arrows indicate the different orientations of TM4′ in the adjacent subunits among the three CALHM structures. (E) Subunit interfaces at the TMD of the three CALHM channels. TM2, TM3, and TM4 are colored blue, green, and orange, respectively. The conserved residues among the CALHM channels are shown in stick models and semitransparent CPK representations. Other residues involved in the interactions between TM2 and TM4′ in the adjacent subunit are shown in stick models. (F) Subunit interfaces at the CTDs of the three CALHM channels, viewed from the intracellular side (top) and the pore (bottom). The center CTDs are colored red, and the two adjacent CTDs are colored purple and green. The conserved residues among the CALHM channels are shown in stick models and semitransparent CPK representations. Other residues involved in interactions between adjacent CTDs are shown in stick models. Hydrogen bonds and salt bridges are shown as black dashed lines.
Similar to OlCALHM1, both the TMD and CTD are involved in the subunit interactions in HsCALHM2 and CeCLHM-1 (Fig. 4, D to F). At the TMD, TM2 of one subunit and TM4′ of the neighboring subunit form extensive hydrophobic interactions (Fig. 4, D and E). In particular, two Pro residues on TM2 and TM3 and two Trp residues on TM3 and TM4′ form van der Waals interactions, resulting in the tight contacts of the three TM helices (TM2, TM3, and TM4′), which are conserved among the three CALHM structures (Fig. 4, D and E, and fig. S7). Structural comparisons of the three CALHM homologs show the different relative positions and orientations of TM2 relative to TM4′ of the neighboring subunits (Fig. 4D). The subunit interfaces between TM2 and TM4′ in OlCALHM1 are formed by tighter hydrophobic interactions, relative to those in HsCALHM2 and CeCLHM-1: The distances between the Cα atoms of the intracellular ends in TM2 and TM4′ in OlCALHM1, HsCALHM2, and CeCLHM-1 are 14.5, 17.4, and 16.8 Å, respectively (Fig. 4D). These subtle differences in the subunit interfaces at the TMD are likely to be associated with the different oligomeric assemblies. At the CTD in the three CALHM structures, the neighboring CTHs entangle together and form stacking interactions between the conserved aromatic residues: Phe and His residues in one subunit, a Phe residue in the adjacent subunit, and two Tyr residues from the adjacent subunit on the opposite side (Fig. 4F and fig. S7). Together, the structural comparisons among the three CALHM homologs reveal both structural conservation (TM topology) and divergence (structures of CTD and oligomeric assembly).
Oligomeric assembly mechanism of CALHM channels
The three different oligomeric assembly structures of the CALHM homologs probably arise from the slight variations in the intersubunit interactions formed at the TMD and CTD. To investigate whether the TMD or CTD defines the oligomeric stoichiometry, we constructed a chimeric channel, consisting of the TMD from OlCALHM1 and the CTD from HsCALHM2 (named “OlCALHM1c”). We confirmed that OlCALHM1c also retains the ATP-release activity elicited by low [Ca2+]o by a cell-based assay (fig. S2E). The purified OlCALHM1c was exchanged into amphipol PMAL-C8 and subjected to the cryo-EM analysis. Without imposing any symmetry, the 3D classification provided cryo-EM maps of two different oligomeric states, the 8-mer and the 9-mer, but no 11-mer structure was obtained (Fig. 5A and figs. S8 and S9). We finally determined the cryo-EM structures of the 8-mer and the 9-mer, both at resolutions of 3.4 Å (Fig. 5, A and B; figs. S8 and S9; and table S1). The dimensions of the 8-mer and 9-mer structures of OlCALHM1c are consistent with the 8-mer and 9-mer structures of OlCALHM1 and CeCLHM-1, respectively (Fig. 5B). Because the densities of the CTDs are obscure in both the 8-mer and 9-mer EM maps (Fig. 5A), we did not model the entire CTD of the 8-mer and the C termini starting from Ile261 of the 9-mer (Fig. 5B). By contrast, the EM density maps corresponding to the TMD are resolved in both structures, allowing the identification of the residues in the TMD (Fig. 5B). The TMD structures of the respective subunits are almost identical between the 8-mer and the 9-mer [root mean square deviation (RMSD) of 0.61 Å over 184 Cα atoms] (Fig. 5C).
Fig. 5. Structures and oligomerization states of the chimeric CALHM proteins.
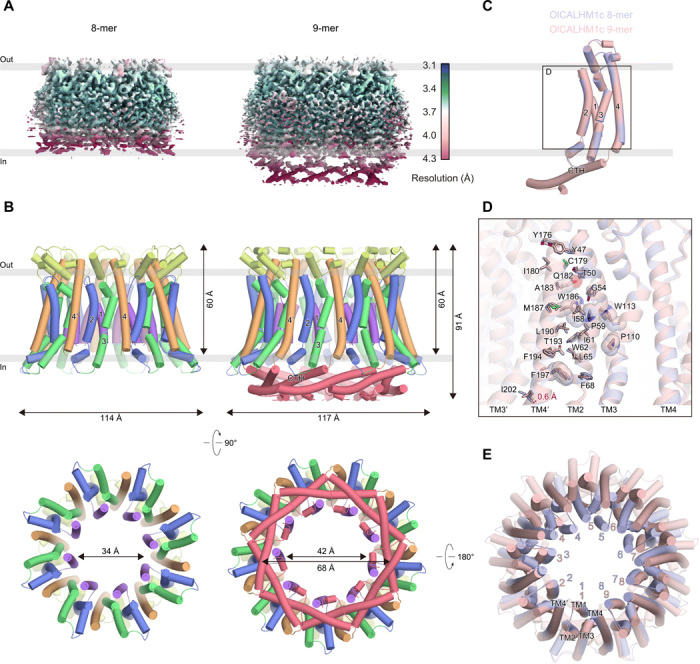
(A) EM density maps of the OlCALHM1c 8-mer (left) and 9-mer (right), colored according to the local resolution (blue: high resolution to red: low resolution), estimated by RELION. (B) Overall structures of the OlCALHM1c 8-mer (left) and 9-mer (right), viewed from the membrane side, colored as in Fig. 1D. (C) Superimposition of the OlCALHM1c 8-mer and 9-mer, viewed from the membrane side. The 8-mer and 9-mer structures are colored pale blue and pale pink, respectively. (D) Close-up view of the superimposition of the OlCALHM1c 8-mer and 9-mer, colored as in (C). The conserved residues among the CALHM channels are shown in stick models and semitransparent CPK representations. The red arrow indicates the different orientations of TM4′ in the adjacent subunit between the 8- and 9-mer structures. (E) Superimposition of the OlCALHM1c 8- and 9-mer, viewed from the extracellular side, colored as in (C). The two structures are superimposed based on subunit 1.
The subunit interfaces at the TMD of OlCALHM1c are almost the same as those of OlCALHM1EM, formed by hydrophobic interactions between TM2 of one subunit and TM4′ (Fig. 5D). Consistently, the 8-mer structure can be well superimposed with OlCALHM1EM, with the same TMD element. On the other hand, superimposition of the 8-mer and the 9-mer based on a subunit shows a 0.6-Å displacement of the Cα atoms of Ile202 on the intracellular end of TM4′ (Fig. 5D). The substitution of the CTD probably allowed this subtle difference of TM4′ at the TMD subunit interfaces, resulting in the 9-mer assembly (Fig. 5, B and E). These observations indicate that the assembly stoichiometry is mainly defined by the subunit interactions at the TMD (e.g., the TMD of OlCALHM1 forms the 8-mer or the 9-mer). Another chimeric structure of a CALHM1-CALHM2 channel was recently reported (24). Unlike our 8- and 9-mer structures, it was an 11-mer structure in which the CTD and the TM4-CTD linker define the relative orientation angle between the TMD and the CTD and thus determine the assembly number. The different oligomeric states between the two CALHM1-CALHM2 chimeric channels are probably due to the sequence diversities within the TMDs and the TM4-CTD linker. The sequences of the TM4-CTD linkers are different between the chimeric channels [derived from OlCALHM1 (Arg203-Ala209) in OlCALHM1c and from HsCALHM2 (Lys206-Leu212) in the recent report; fig. S10A]. The TM4-CTD linkers in the chimeric channels adopt distinct conformations (fig. S10B), thereby affecting the orientation of the CTD and its interactions between the adjacent subunits. As shown in the current OlCALHM1c structures, the substitution of the CTD could slightly modify the interaction mode at the TMD. However, the oligomeric state is probably affected and limited by the tolerance at the TMD interfaces, because the two chimeric channels, our OlCALHM1c (OlCALHM1-TMD) and the reported chimera (chicken CALHM1-TMD), show different TMD assemblies. The TMD of OlCALHM1 also has low sequence identities with those of HsCALHM2 (29%) and CeCLHM-1 (23%), although the three CALHM homologs adopt similar TM topologies. These sequence differences within the TMDs and TM4-CTD linkers among the CALHM family members are likely to be reflected in their diverse oligomeric assemblies and eventually in the different diameters of the channel pore, which may be associated with the different specificities of the released substrates at distinct expression sites (5, 12, 15).
DISCUSSION
In this study, we present the cryo-EM structures of three CALHM homologs and the two oligomeric states of the chimeric construct, revealing the ATP conduction and oligomeric assembly mechanisms. Notably, the present 2.66-Å resolution OlCALHM1 structure allowed the accurate modeling of the NTH and TM1 (fig. S4A), which form the pore architecture in the Ca2+-free open state (Fig. 3, A and B). Consistent with the previous report, OlCALHM1 has the constriction site with a pore diameter of 15.7 Å, which is narrower than that in the recently reported chicken CALHM1 structure (24), in which the NTH is not visualized. Thus, the present OlCALHM1 structure provides the most detailed information on the pore architecture. The analysis of the ΔNTH mutants indicates that the NTH is involved in the channel conduction of the ATP (Fig. 3D). Although the NTH and TM1 are disordered in our HsCALHM2 and CeCLHM-1 structures, the comparable arrangement of the TM helices among OlCALHM1, HsCALHM2, and CeCLHM-1 suggests that the NTH would similarly constitute the channel pores in the CALHM family members. The pore constriction by the NTH is also observed in gap junction channels (17, 18), indicating that this is a conserved structural feature of these oligomeric channels, despite the lack of sequence similarity among them.
The ΔNTH mutant showed almost no ATP-release activity upon Ca2+ binding, similar to the WT (Fig. 3E), suggesting that other structural components are associated with the channel gating in OlCALHM1 (Fig. 6, A and B). In the OlCALHM1EM structure, TM1 protrudes toward the channel pore, as compared to the other TMs (Fig. 1D). Recently, the cryo-EM structures of HsCALHM2 were reported in the open and inhibited states at 3.3 and 2.7 Å resolutions, respectively (23). These structures showed channel pore closing by the swing motion of TM1 by about 60° toward the pore axis, driven by the binding of the inhibitor RUR at the base of the TM segment. Therefore, these structures suggest the large rearrangement of TM1 upon the channel gating (23). Given the structural and sequence similarities between OlCALHM1 and HsCALHM2 (Fig. 4, B and C, and fig. S7), TM1 may undergo a large structural movement and block the pore upon Ca2+ binding in OlCALHM1 (Fig. 6, A and B).
Fig. 6. Insight into channel gating.
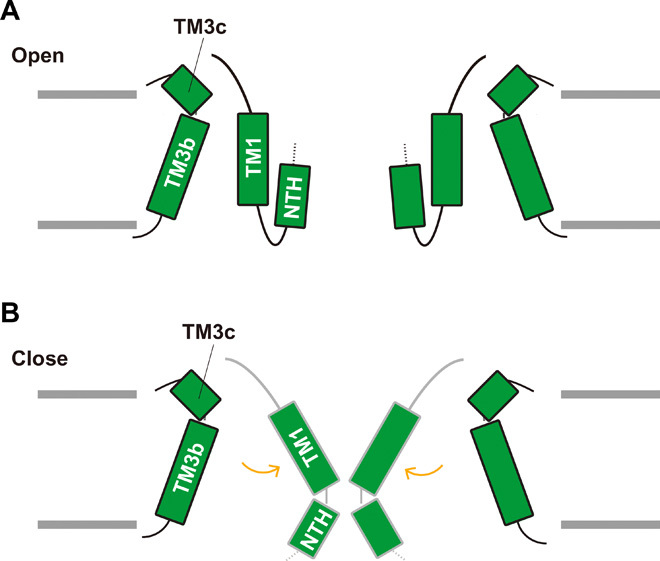
(A and B) Schematic model of the Ca2+-dependent gating mechanism. In the Ca2+-free open state, TM1 forms the stable interaction with TM3 and moves perpendicularly to the membrane. The NTH forms the constriction site in the pore (A). In the Ca2+-bound closed state, TM1 blocks the pore (B).
For the subunit assembly of CALHM family proteins, the interactions at the TMD and the TM4-CTD linker sequences are important for the oligomeric stoichiometry. The structural comparisons of the present structures with the recently reported structures (24) revealed that the respective TM4-CTD linker regions adopt different conformations in the structures, affecting the orientations of the CTD (fig. S10B). The 8- and 9-mer structures of OlCALHM1c also suggested that the CTD may affect the assembly, but the CTD alone could not necessarily govern the oligomeric numbers. The hydrophobic residues involved in the subunit interactions between TM2 and TM4′ at the TMD are conserved in the CALHM1 orthologs, except for Met187 and Phe194 (fig. S1, A to C), implying a similar assembly number of HsCALHM1. In taste bud cells, CALHM1 and CALHM3 form a functionally heteromeric channel with fast activation (13). HsCALHM1 and HsCALHM3 have sequence identities of 47% overall and 54% within the TMD (fig. S11), which are relatively higher as compared to the sequence identity between HsCALHM1 and HsCALHM2 (30% over the full length and 33% within the TMD). Moreover, the residues that are probably involved in the subunit assembly and the TM4-CTD linker sequence are conserved in both HsCALHM1 and HsCALHM3, implying that they can form a heterooctamer, although the different residues of the CTD could slightly modify this assembly. Overall, our current study provides the ATP conduction and diverse assembly mechanisms of the CALHM family proteins.
MATERIALS AND METHODS
Expression and purification of OlCALHM1, HsCALHM2, and the chimeric construct OlCALHM1c
The DNAs encoding the OlCALHM1 channel (UniProt ID: H2MCM1) and the HsCALHM2 (UniProt ID: Q9HA72) were codon-optimized for eukaryotic expression systems, synthesized (Genewiz Inc.), and served as the templates for subcloning. To improve the expression and thermostability, the C-terminal residues starting from Trp296 were truncated (OlCALHM1EM). The chimeric construct consists of the TMD in OlCALHM1 (Met1-Ala209) followed by the CTD in HsCALHM2 (Ser213-Ser323). These DNA fragments were ligated into a modified pEG BacMam vector, with a C-terminal green fluorescent protein (GFP)–His8 tag and a tobacco etch virus (TEV) cleavage site, and were expressed in HEK293S GnTI− (N-acetylglucosaminyl-transferase I–negative) cells (American Type Culture Collection, catalog no. CRL-3022) (25). Cells were collected by centrifugation (8000g, 10 min, 4°C). For the cells expressing OlCALHM1c, the cells were disrupted by probe sonication and the membrane fraction was collected by ultracentrifugation (186,000g, 1 hour, 4°C). The cells or the membrane fraction was solubilized for 1 hour in buffer containing 50 mM tris, pH 8.0, 150 mM NaCl, 2 mM dithiothreitol, 1% N-dodecyl β-d-maltoside (DDM; Calbiochem), and 0.2% CHS. The mixture was centrifuged at 40,000g for 20 min, and the supernatant was incubated with CNBr-Activated Sepharose 4 Fast Flow beads (GE Healthcare) coupled with an anti-GFP nanobody (GFP enhancer or GFP minimizer) (26) for 2 hours at 4°C. The protein-bound resins were washed with gel filtration buffer (50 mM tris, pH 8.0, 150 mM NaCl, and 2 mM dithiothreitol) containing 0.05% GDN for OlCALHM1EM and OlCALHM1c or 0.06% digitonin for HsCALHM2 and further incubated overnight with TEV protease. The flow through was concentrated using a centrifugal filter unit (Merck Millipore, 100-kDa molecular weight cutoff) and separated on a Superose 6 Increase 10/300 GL column (GE Healthcare) equilibrated with gel filtration buffer. The peak fractions of the protein were pooled.
Expression and purification of CeCLHM-1
The C. elegans calcium homeostasis modulator protein (CeCLHM-1, UniProt: Q18593) complementary DNA (cDNA) was synthesized, codon-optimized for expression in human cell lines, and cloned into the pcDNA3.4 vector. The CeCLHM-1 sequence was fused with a C-terminal His8 tag and enhanced GFP (EGFP), followed by a TEV protease site. HEK293S GnTI− cells were transiently transfected at a density of 2.0 × 106 cells ml−1, with the plasmids and FectoPRO (Polyplus). Approximately 360 μg of the CeCLHM-1 plasmid was premixed with 360 μl of FectoPRO reagent in 80 ml of fresh FreeStyle 293 medium for 10 to 20 min before transfection. For transfection, 80 ml of the mixture was added to 0.8 liters of cell culture and incubated at 37°C in the presence of 8% CO2. After 18 to 20 hours, 2.2 mM valproic acid was added, and the cells were further incubated at 30°C in the presence of 8% CO2 for 48 hours. The cells were collected by centrifugation (3000g, 10 min, 4°C) and disrupted by Dounce homogenization in hypotonic buffer [50 mM Hepes-NaOH (pH 7.5), 10 mM KCl, deoxyribonuclease (DNase) I (0.04 mg ml−1), and protease inhibitor cocktail]. The membrane fraction was collected by ultracentrifugation (138,000g, 1 hour, 4°C) and solubilized for 1 hour at 4°C in buffer [50 mM Hepes-NaOH (pH 7.5), 300 mM NaCl, 1.5% (w/v) DDM, and 0.15% (w/v) CHS]. After ultracentrifugation (138,000g, 30 min, 4°C), the supernatant was incubated with AffiGel 10 (Bio-Rad) coupled with a GFP-binding nanobody for 2 hours at 4°C. The resin was washed five times with three column volumes of wash buffer [50 mM Hepes-NaOH (pH 7.5), 300 mM NaCl, and 0.06% GDN] and gently suspended overnight with TEV protease to cleave the His8-EGFP tag. After the TEV protease digestion, the flow through was pooled, concentrated, and purified by size-exclusion chromatography on a Superose 6 Increase 10/300 GL column (GE Healthcare), equilibrated with gel filtration buffer [20 mM Hepes-NaOH (pH 7.5), 150 mM NaCl, and 0.06% GDN]. The peak fractions of the protein were pooled.
Grid preparation
The purified OlCALHM1EM protein in GDN was concentrated to 5 mg ml−1. For cryo-EM analyses of HsCALHM2, CeCLHM-1, and OlCALHM1c, the detergents were exchanged with amphipols. The purified HsCALHM2 in digitonin, CeCLHM-1 in GDN, and OlCALHM1c in GDN were mixed with amphipol PMAL-C8 (Anatrace) at a 1:100 ratio (w/w) for 2 to 2.5 hours at 4°C. Detergents were removed using Bio-Beads SM-2 (Bio-Rad, 100 mg per 1 ml of mixture) for 2 to 2.5 hours at 4°C. After removal of the Bio-Beads, the samples were concentrated and subjected to size-exclusion chromatography on a Superose 6 Increase 10/300 GL column (GE Healthcare), equilibrated with buffer (20 mM tris-HCl, pH 8.0, 150 mM NaCl, and 2 mM dithiothreitol). The peak fractions containing HsCALHM2, CeCLHM-1, and OlCALHM1c in PMAL-C8 were concentrated to 1.6, 2.2, and 1 mg ml−1, respectively. Portions (3 μl) of the concentrated samples were applied to glow-discharged R1.2/1.3 Cu/Rh 300 mesh grids (Quantifoil). The grids were subsequently blotted (4 s for OlCALHM1EM, 8 s for HsCALHM2, 4 s for CeCLHM-1, and 4 s for OlCALHM1c) and vitrified using Vitrobot Mark IV (FEI) under 4°C and 100% humidity conditions.
EM image acquisition and data processing
For OlCALHM1EM and OlCALHM1c, data collections were performed on an FEI Titan Krios (FEI) EM, operating at an acceleration voltage of 300 kV, and equipped with a BioQuantum K3 imaging filter and a K3 direct electron detector (Gatan). EM images were acquired at a nominal magnification of ×105,000, corresponding to a physical pixel size of 0.83 Å pixel−1, using the SerialEM software (27). For OlCALHM1EM, movies were dose-fractionated to 54 frames at a dose rate of 14.9 e− pixel−1 per second, resulting in a total accumulated dose of 59.4 e− Å−2. For OlCALHM1c, movies were dose-fractionated to 48 frames at a dose rate of 14.0 e− pixel−1 per second, resulting in a total accumulated dose of 50 e− Å−2. For HsCALHM2 and CeCLHM-1, data collections were performed on a 300-kV Titan Krios EM equipped with a Falcon III direct electron detector. EM images were acquired at a nominal magnification of ×96,000, corresponding to a calibrated pixel size of 0.8346 Å pixel−1, using the EPU software. For HsCALHM2, movies were dose-fractionated to 60 frames at a dose rate of 0.95 e− pixel−1 per second, resulting in a total accumulated dose of 60 e− Å−2. For CeCLHM-1, movies were dose-fractionated to 48 frames at a dose rate of 0.7 e− pixel−1 per second, resulting in a total accumulated dose of 50 e− Å−2. For all datasets, the movie frames were aligned in 4 × 4 patches and dose-weighted using RELION (28). CTF estimation was performed by CTFFIND 4.1 (29).
For the OlCALHM1EM dataset, a total of 1,220,980 particles were extracted from 3075 micrographs in 3.55 Å pixel−1 using AutoPick in RELION-3.0 (28), applying the 2D class averages of reference-free autopicked particles based on a Laplacian-of-Gaussian filter as templates. The initial model was generated in RELION. The particles were passed to RELION to calculate the 2D classification, refinement, and postprocessing. Because some classes of the 2D classification exhibited the density for eight subunits, 137,146 particles in three good classes were re-extracted in the original pixel size of 0.83 Å pixel−1 and refined in C8 symmetry. The particles were further cleaned using 3D classification, and 102,109 good particles were subsequently subjected to micelle subtraction, polishing (30), and CTF refinement. The overall gold-standard resolution calculated at the FSC (FSC = 0.143) (31) was 2.66 Å.
For the HsCALHM2 dataset, a total of 946,353 particles were initially extracted from 2110 micrographs, with a binned pixel size of 3.52 Å pixel−1, as described above. These particles were subjected to several rounds of 2D and 3D classifications, resulting in the best class from the 3D classification, which contained 365,116 particles. These particles were then re-extracted with 1.36 Å pixel−1 and subsequently subjected to 3D refinement with C11 symmetry, polishing, CTF refinement, and micelle subtraction. The final 3D refinement with C11 symmetry and postprocessing yielded a density map with an overall resolution of 3.4 Å at a gold-standard FSC = 0.143. The image processing workflow is described in fig. S5.
For the CeCLHM-1 dataset, a total of 832,750 particles were initially extracted from 2906 micrographs, with a binned pixel size of 3.01 Å pixel−1, as described above. These particles were subjected to several rounds of 2D classification, resulting in the best classes containing 261,169 particles. These particles were re-extracted with 1.5 Å pixel−1 and subjected to 3D classification, resulting in the best classes containing 65,887 particles. The final 3D refinement with C9 symmetry and postprocessing yielded a density map with an overall resolution of 3.6 Å at a gold-standard FSC = 0.143. The image processing workflow is described in fig. S6.
For the OlCALHM1c dataset, a total of 1,689,949 particles were initially extracted from 2808 micrographs, with a binned pixel size of 3.39 Å pixel−1, as described above. These particles were subjected to several rounds of 2D and 3D classifications, resulting in the two distinct classes from the 3D classification, which represented the 8-mer and 9-mer structures. The 8-mer and 9-mer classes contained 233,566 and 227,781 particles, respectively. These particles were separately re-extracted with 1.41 Å pixel−1 and subsequently subjected to 3D refinement with C8 or C9 symmetry, respectively. The final 3D refinement and postprocessing of the two classes yielded density maps with overall resolutions of 3.4 Å in both cases at a gold-standard FSC = 0.143. The image processing workflow is described in fig. S8.
Model building and refinement
The models of OlCALHM1EM, HsCALHM2, and CeCLHM-1 were built de novo into the density map using COOT (32) and rebuilt using Rosetta (33). The model was again rebuilt using COOT with multiple rounds of real-space refinement in PHENIX (34, 35), with secondary structure restraints. For the OlCALHM1c 8-mer and 9-mer, the TMD of OlCALHM1EM (8-mer and 9-mer) and the CTD of HsCALHM2 (9-mer) were modeled into the density map by rigid-body fitting and modified using COOT and Rosetta with multiple rounds of real-space refinement in PHENIX, with secondary structure restraints.
For cross-validation, the final models were randomly shaken and refined against one of the half maps generated in RELION. FSC curves were calculated between the refined model and half map 1 and between the refined model and half map 2 (figs. S3, S5, S6, and S8) (36). The statistics of the model refinement and validation are summarized in table S1. Figures of the density maps and molecular graphics were prepared using CueMol (http://www.cuemol.org) and UCSF Chimera (37).
Bioluminescent ATP-release assay
The extracellular ATP concentration was measured by the luciferin-luciferase reaction, as previously reported (22). HeLa cells seeded at a density of 1.2 × 104 cells well−1 on 96-well cell culture plates (Corning Costar, Corning, NY, USA) were transfected with 0.6 μg of pIRES2.AcGFP1 encoding WT or mutant OlCALHM1 cDNA or the empty vector using Lipofectamine 3000 (Thermo Fisher Scientific), according to the manufacturer’s instruction, and incubated for 4 hours at 37°C. Subsequently, transfection media were replaced with fresh culture media, and cells were incubated at 27°C for 4 hours before being tested. In experiments shown in fig. S2 (A to C), cells were transfected with 0.2 μg of pIRES2.AcGFP1 encoding HsCALHM1, HsCALHM2, OlCALHM1, or CeCLHM-1 cDNA or the empty vector and incubated in the transfection media for 4 hours at 37°C and then in fresh culture media for 20 hours at 27°C. Cells were then washed twice with the standard bath solution and incubated in 100 μl of the standard bath solution for another 1 hour at room temperature. Immediately after 75 μl of the bath solution was replaced with an equal volume of the Ca2+-free bath solution to make final [Ca2+]o ~ 17 nM, 10 μl of the luciferin-luciferase solution (FL-AAM and FL-AAB, Sigma, St. Louis, MO, USA) was added to each well and the plate was placed in a microplate luminometer (Centro LB960, Berthold Technologies, Bad Wildbad, Germany). Luminescence was measured every 2 min at 25°C. The extracellular ATP concentration was calculated from a standard curve created in each plate. The standard bath solution contained 150 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM Hepes, and 10 mM glucose, pH 7.4, adjusted with NaOH. The Ca2+-free bath solution contained 150 mM NaCl, 5 mM KCl, 5 mM EGTA, 1 mM MgCl2, 10 mM Hepes, and 10 mM glucose, pH 7.4, adjusted with NaOH.
Preparation of proteoliposomes
Lipids from chloroform stocks, mixed at a 2:1 weight ratio of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (Avanti) with 12% egg phosphatidylcholine:1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (Avanti), were dried and solubilized in a solution containing 100 mM KCl, 0.1 mM EGTA, 2.3% n-octyl-β-d-glucopyranoside, and 25 mM Hepes, pH 7.6. Solubilized OlCALHM1EM was added to the lipid-detergent mixture at a 50:1 lipid to protein ratio. After a short incubation, Bio-Beads SM-2 (Bio-Rad) were added and mixed at 4°C overnight. To confirm the reconstitution, after ultracentrifugation (125,000g, 15 min, 4°C) and solubilization in buffer (50 mM tris-HCl, pH 8.0, 150 mM NaCl, 1.0% DDM, and 0.2% CHS) at 4°C for 1 hour, the proteoliposomal solutions were subjected to SDS–polyacrylamide gel electrophoresis.
ATP transport assay
Liposomes and proteoliposomes were loaded with 1 mM ATP and sonicated for 30 s using a Bioruptor (CosmoBio). After removal of the extraliposomal ATP by chromatography on Sephadex G-50 fine resin (GE Healthcare), 50-μl portions of the samples were combined with 50 μl of a mixture containing 45 nM luciferase (QuantiLum Photinus pyralis recombinant luciferase; Promega, Madison, WI), 1.2 nM luciferin (Nacalai Tesque), 1.0 mM EDTA, and 10 mM MgSO4. After 1-hour incubation with or without 0.4% Triton X-100, the luminescence was measured on an ARVO-X3 microplate reader (PerkinElmer). The amount of ATP retained in the liposomes was determined as the difference in the luminescence between total ATP and extraliposomal ATP that was accessible to luciferase without Triton X-100.
Supplementary Material
Acknowledgments
We thank T. Nakane for assistance with the single-particle analysis. We also thank the staff scientists at the University of Tokyo’s cryo-EM facility, especially K. Kobayashi, H. Yanagisawa, A. Tsutsumi, M. Kikkawa, and R. Danev. Funding: This work was supported by a MEXT Grant-in-Aid for Specially Promoted Research (grant 16H06294) to O.N. and JST PRESTO (grant JPMJPR1886) to A.T. This research was supported by the Platform Project for Supporting Drug Discovery and Life Science Research [Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)] from AMED under grant number JP19am01011115 (support number 1111). Author contributions: K.D. prepared the cryo-EM sample of OlCALHM1 and performed the liposome assay. K.D. and T.K. collected and processed the cryo-EM data and built the structure of OlCALHM1. T.K. prepared the cryo-EM sample of the CALHM chimera, collected and processed the cryo-EM data, and built the structures of HsCALHM2, CeCLHM-1, and the CALHM chimera. W.S. initially screened the CALHM homologs, established the expression and purification procedure, and prepared the cryo-EM sample of HsCALHM2. H.S. assisted the purification and grid preparation of HsCALHM2. M.H. screened and prepared the cryo-EM sample of CeCLHM-1, collected and processed the cryo-EM data, and built the structure of CeCLHM-1. T.N. assisted with the data collection and processing. K.Y. assisted with the data processing and structure refinement. K.N. and A.T. measured the ATP-release activities of the CALHM proteins. K.D., T.K., W.S., and O.N. mainly wrote the manuscript. A.T. and O.N. supervised the research. Competing interests: M.H. is a graduate student at Mitsubishi Tanabe Pharma Corporation and is supported by the company with nonresearch funds. The company has no financial or other interest in this research. O.N. is a co-founder and scientific advisor for Curreio. All other authors declare no competing interests. Data and materials availability: The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank, under the accession codes EMD-0919 (OlCALHM1EM), EMD-0920 (HsCALHM2), EMD-0921 (CeCLHM-1), EMD-0922 (OlCALHM1c 8-mer), and EMD-0923 (OlCALHM1c 9-mer). The atomic coordinates have been deposited in the Protein Data Bank, under IDs 6LMT (OlCALHM1EM), 6LMU (HsCALHM2), 6LMV (CeCLHM-1), 6LMW (OlCALHM1c 8-mer), and 6LMX (OlCALHM1c 9-mer). The raw images have been deposited in the Electron Microscopy Public Image Archive, under the accession code EMPIAR-10444.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/29/eaba8105/DC1
REFERENCES AND NOTES
- 1.Burnstock G., Historical review: ATP as a neurotransmitter. Trends Pharmacol. Sci. 27, 166–176 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Abbracchio M. P., Burnstock G., Verkhratsky A., Zimmermann H., Purinergic signalling in the nervous system: An overview. Trends Neurosci. 32, 19–29 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Taruno A., ATP release channels. Int. J. Mol. Sci. 19, 808 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dreses-Werringloer U., Lambert J.-C., Vingtdeux V., Zhao H., Vais H., Siebert A., Jain A., Koppel J., Rovelet-Lecrux A., Hannequin D., Pasquier F., Galimberti D., Scarpini E., Mann D., Lendon C., Campion D., Amouyel P., Davies P., Foskett J. K., Campagne F., Marambaud P., A polymorphism in CALHM1 influences Ca2+ homeostasis, Aβ levels, and Alzheimer’s disease risk. Cell 133, 1149–1161 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taruno A., Vingtdeux V., Ohmoto M., Ma Z., Dvoryanchikov G., Li A., Adrien L., Zhao H., Leung S., Abernethy M., Koppel J., Davies P., Civan M. M., Chaudhari N., Matsumoto I., Hellekant G., Tordoff M. G., Marambaud P., Foskett J. K., CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes. Nature 495, 223–226 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma Z., Saung W. T., Foskett J. K., Action potentials and ion conductances in wild-type and CALHM1-knockout type II taste cells. J. Neurophysiol. 117, 1865–1876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taruno A., Matsumoto I., Ma Z., Marambaud P., Foskett J. K., How do taste cells lacking synapses mediate neurotransmission? CALHM1, a voltage-gated ATP channel. Bioessays 35, 1111–1118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nomura K., Nakanishi M., Ishidate F., Iwata K., Taruno A., All-electrical Ca2+-independent signal transduction mediates attractive sodium taste in taste buds. Neuron 106, 816–829.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Ma Z., Siebert A. P., Cheung K.-H., Lee R. J., Johnson B., Cohen A. S., Vingtdeux V., Marambaud P., Foskett J. K., Calcium homeostasis modulator 1 (CALHM1) is the pore-forming subunit of an ion channel that mediates extracellular Ca2+ regulation of neuronal excitability. Proc. Natl. Acad. Sci. U.S.A. 109, E1963–E1971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Z., Tanis J. E., Taruno A., Foskett J. K., Calcium homeostasis modulator (CALHM) ion channels. Pflugers Arch. 468, 395–403 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siebert A. P., Ma Z., Grevet J. D., Demuro A., Parker I., Foskett J. K., Structural and functional similarities of calcium homeostasis modulator 1 (CALHM1) ion channel with connexins, pannexins, and innexins. J. Biol. Chem. 288, 6140–6153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma J., Qi X., Yang C., Pan R., Wang S., Wu J., Huang L., Chen H., Cheng J., Wu R., Liao Y., Mao L., Wang F., Wu Z., An J., Wang Y., Zhang X., Zhang C., Yuan Z., Calhm2 governs astrocytic ATP releasing in the development of depression-like behaviors. Mol. Psychiatry 23, 883–891 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Ma Z., Taruno A., Ohmoto M., Jyotaki M., Lim J. C., Miyazaki H., Niisato N., Marunaka Y., Lee R. J., Hoff H., Payne R., Demuro A., Parker I., Mitchell C. H., Henao-Mejia J., Tanis J. E., Matsumoto I., Tordoff M. G., Foskett J. K., CALHM3 is essential for rapid ion channel-mediated purinergic neurotransmission of GPCR-mediated tastes. Neuron 98, 547–561.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kashio M., Wei-qi G., Ohsaki Y., Kido M. A., Taruno A., CALHM1/CALHM3 channel is intrinsically sorted to the basolateral membrane of epithelial cells including taste cells. Sci. Rep. 9, 2681 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanis J. E., Ma Z., Krajacic P., He L., Foskett K. J., Lamitina T., CLHM-1 is a functionally conserved and conditionally toxic Ca2+-permeable ion channel in Caenorhabditis elegans. J. Neurosci. 33, 12275–12286 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanis J. E., Ma Z., Foskett J. K., The NH2 terminus regulates voltage-dependent gating of CALHM ion channels. Am. J. Physiol. Cell Physiol. 313, C173–C186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeda S., Nakagawa S., Suga M., Yamashita E., Oshima A., Fujiyoshi Y., Tsukihara T., Structure of the connexin 26 gap junction channel at 3.5 Å resolution. Nature 458, 597–602 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Oshima A., Tani K., Fujiyoshi Y., Atomic structure of the innexin-6 gap junction channel determined by cryo-EM. Nat. Commun. 7, 13681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasuya G., Nakane T., Yokoyama T., Jia Y., Inoue M., Watanabe K., Nakamura R., Nishizawa T., Kusakizako T., Tsutsumi A., Yanagisawa H., Dohmae N., Hattori M., Ichijo H., Yan Z., Kikkawa M., Shirouzu M., Ishitani R., Nureki O., Cryo-EM structures of the human volume-regulated anion channel LRRC8. Nat. Struct. Mol. Biol. 25, 797–804 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Deneka D., Sawicka M., Lam A. K. M., Paulino C., Dutzler R., Structure of a volume-regulated anion channel of the LRRC8 family. Nature 558, 254–259 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Kawate T., Gouaux E., Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Taruno A., Sun H., Nakajo K., Murakami T., Ohsaki Y., Kido M. A., Ono F., Marunaka Y., Post-translational palmitoylation controls the voltage gating and lipid raft association of the CALHM1 channel. J. Physiol. 595, 6121–6145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi W., Clemente N., Sun W., Du J., Lü W., The structures and gating mechanism of human calcium homeostasis modulator 2. Nature 576, 163–167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syrjanen J. L., Michalski K., Chou T.-H., Grant T., Rao S., Simorowski N., Tucker S. J., Grigorieff N., Furukawa H., Structure and assembly of calcium homeostasis modulator proteins. Nat. Struct. Mol. Biol. 27, 150–159 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goehring A., Lee C.-H., Wang K. H., Michel J. C., Claxton D. P., Baconguis I., Althoff T., Fischer S., Garcia K. C., Gouaux E., Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirchhofer A., Helma J., Schmidthals K., Frauer C., Cui S., Karcher A., Pellis M., Muyldermans S., Casas-Delucchi C. S., Cardoso M. C., Leonhardt H., Hopfner K.-P., Rothbauer U., Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17, 133–139 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J. H., Lindahl E., Scheres S. H. W., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zivanov J., Nakane T., Scheres S. H. W., A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenthal P. B., Henderson R., Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R. Y.-R., Song Y., Barad B. A., Cheng Y., Fraser J. S., DiMaio F., Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. eLife 5, e17219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afonine P. V., Poon B. K., Read R. J., Sobolev O. V., Terwilliger T. C., Urzhumtsev A., Adams P. D., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown A., Long F., Nicholls R. A., Toots J., Emsley P., Murshudov G., Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 71, 136–153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S. P., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/29/eaba8105/DC1