Abstract
Objective
This study aimed to investigate the abilities of long non‐coding RNA PVT1 (lnc‐PVT1) and microRNA‐146a (miR‐146a) in predicting chronic obstructive pulmonary disease (COPD) susceptibility and acute exacerbation risk, moreover, to explore the association of lnc‐PVT1 with disease severity, inflammation, and miR‐146a in patients with COPD.
Methods
A total of 80 acute exacerbation of COPD (AECOPD) patients, 80 stable COPD patients, and 80 healthy controls (HCs) were consecutively recruited. Peripheral blood samples of all participants were collected to isolate peripheral blood mononuclear cells (PBMCs), and serum: PBMCs were used to detect lnc‐PVT1 and miR‐146a by RT‐qPCR; serum was used to detect inflammatory cytokines by ELISA. Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage of COPD was assessed.
Results
Lnc‐PVT1 expression was highest in AECOPD patients, followed by stable COPD patients and HCs. Receiver operating characteristic curves disclosed that lnc‐PVT1 distinguished AECOPD patients and stable COPD patients from HCs, also distinguished AECOPD patients from stable COPD patients. In AECOPD patients and stable COPD patients, lnc‐PVT1 expression positively correlated with GOLD stage and levels of TNF‐α, IL‐6, IL‐8, and IL‐17. Moreover, lnc‐PVT1 was negatively correlated with miR‐146a. For miR‐146a, its expression was lowest in AECOPD patients, followed by stable COPD patients and HCs, and it predicted reduced COPD susceptibility and decreased acute exacerbation risk; meanwhile, it negatively correlated with GOLD stage and inflammatory cytokine levels in AECOPD patients and stable COPD patients.
Conclusion
Lnc‐PVT1 assists the disease management and acute exacerbation risk monitoring of COPD via interaction with miR‐146a.
Keywords: acute exacerbation risk, chronic obstructive pulmonary disease, disease severity, inflammation, long non‐coding RNAPVT1, miRNA‐146a
1. INTRODUCTION
Chronic obstructive pulmonary disease (COPD) is characterized by irreversible airflow limitation and persistent respiratory symptoms, which is becoming a major burden on healthcare systems worldwide. 1 Notably, acute exacerbation of COPD (AECOPD) refers to an acute event featured by a worsening of the patient's respiratory symptoms, which is by far the most devastating aspect of the disease management, and contributes to the increasing morbidity and mortality in COPD patients. 2 Despite managements (such as bronchodilators, corticosteroids, and antibiotics) are available for AECOPD patients to minimize the influence of the current exacerbation, these patients still face increasing social burden, poor life quality, and huge medical expenditures. 3 Therefore, it is imperative to exploring strategies preventing AECOPD and improving prognosis of COPD patients.
Long non‐coding RNAs (lncRNAs) have been confirmed to be involved in a myriad of biologic processes, and their dysregulations are implicated in various disease pathologies. 4 As one of most critical lncRNAs, lncRNA plasmacytoma variant translocation 1 (lnc‐PVT1) functions as a regulator in inflammatory responses and possesses pro‐inflammatory impact in acute kidney injury in vitro and sepsis in vivo. 5 , 6 Regarding clinical practices, lnc‐PVT1 has excellent ability to distinguish severe asthmatics from non‐severe asthmatics, and its single nucleotide polymorphism (SNP) associates with increased Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage and worse lung function in smokers with COPD, suggesting that lnc‐PVT1 might participate in COPD progression. 7 , 8 MicroRNA‐146a (miR‐146a), as a target of lnc‐PVT1, has been recognized as an anti‐inflammatory mediator in chronic airway respiratory diseases, which not only negatively correlates with inflammatory cytokines but also distinguishes AECOPD patients from stable COPD patients. 9 , 10 , 11
Considering that lnc‐PVT1 might be involved in COPD pathology and showed the potential in enhancing COPD progression, meanwhile, its target miR‐146a was downregulated and predicted decreased acute exacerbation risk in patients with COPD, we hypothesized that lnc‐PVT1 might participate in the disease development of COPD via the inter‐correlation with miR‐146a, and if so, lnc‐PVT1 might facilitate the COPD susceptibility and acute exacerbation risk as well as surveillance of disease progression; however, limited evidence was reported. In this study, we enrolled AECOPD patients, stable COPD patients, and healthy controls (HCs), respectively, to investigate the abilities of lnc‐PVT and miR‐146a in predicting COPD susceptibility and acute exacerbation risk, moreover, to explore the association of lnc‐PVT1 with disease severity, inflammation and miR‐146a in COPD patients, which might help explore strategies preventing AECOPD and improving prognosis of COPD patients.
2. MATERIALS AND METHODS
2.1. Subjects
In this case‐control study, a total of 80 AECOPD patients, 80 stable COPD patients, and 80 HCs (who were from the individuals performed health examination in our hospitals) were consecutively recruited from our hospitals between September 2018 and August 2019. The eligible criteria for AECOPD patients were (a) diagnosed as COPD according to the criteria of GOLD 12 ; (b) presenting with acute exacerbation of symptoms in accordance with the definitions of the GOLD criteria; (c) age ≥ 40 years; (d) not complicated with asthma, lung cancer, or other relevant lung diseases; (e) no history of severe infection; and (f) no history of tumors, autoimmune diseases or malignant hematologic diseases. The eligible criteria for stable COPD patients were (a) diagnosed as COPD according to the criteria of the GOLD; (b) clinically stable for at least 3 months without acute exacerbations; (c) age ≥ 40 years; (d) not complicated with asthma, lung cancer, or other relevant lung diseases; (e) no history of severe infection; and (f) no history of tumors, autoimmune diseases, or malignant hematologic diseases. The eligible criteria for HCs were (a) no history of COPD, asthma or other respiratory diseases (such as interstitial lung disease, bronchiectasis, pneumonia, pulmonary thromboembolism, and so on); (b) no history of severe infection or malignancies; and (c) not complicated with active inflammatory diseases, hematological diseases, or autoimmune diseases. In addition, it needed to be emphasized that patients were assigned to AECOPD cohort or stable COPD cohort depending on first disease status after study initiation, and the assignment of patients was not changed as patients' disease status change. (That is, if patients were initially assigned to the stable COPD cohort, the patients would not assign to AECOPD cohort even though they later experienced the acute exacerbations; correspondingly, if patients were initially assigned to the AECOPD cohort, the patients would not assign to stable COPD cohort even though their disease status were later under effective control.)
2.2. Ethic approval
This study was approved by the Ethic Committee of our hospitals and conducted in accordance with the provisions of the Declaration of Helsinki as well as the Good Clinical Practice guidelines by the International Conference on Harmonisation. The informed consents were signed by all subjects.
2.3. Data collection
Clinical characteristics of enrolled subjects were documented after enrollment, which included age, gender, body mass index (BMI), family history of COPD, history of smoke, forced expiratory volume in 1 second (FEV1), and forced vital capacity (FVC). Then, FEV1 (% predicted) and FEV1/FVC ratio were calculated. The FEV1 (% predicted) represented the percentage of FEV1 in predicted FEV1. Furthermore, as for the AECOPD patients and stable COPD patients, severity of airflow obstruction was graded in accordance with the GOLD guidelines as follows: GOLD 1: FEV1 (% predicted) ≥ 80%; GOLD 2: FEV1 (% predicted) within 50%‐79%; GOLD 3: FEV1 (% predicted) within 30%‐49%; and GOLD 4: FEV1 (% predicted) < 30%.
2.4. Sample collection and detection
Peripheral blood samples of AECOPD patients (within 24 hours after admission) and stable COPD patients were collected to isolate the serum and peripheral blood mononuclear cells (PBMCs) by centrifugal separation and density gradient separation, which were then stored at −80°C. Besides, the peripheral blood samples of HCs were collected from physical examinations and then were subjected to separate serum and PBMCs as well. The lnc‐PVT1 and miR‐146a relative expressions in the PBMCs of all subjects were determined by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) assay. And the inflammatory cytokines including tumor necrosis factor‐α (TNF‐α), interleukin‐6 (IL‐6), interleukin‐8 (IL‐8), and interleukin‐17 (IL‐17) in serum of all subjects were detected by human enzyme‐linked immunosorbent assay (ELISA) kits (R&D Systems Inc, USA) according to the manufacturer's instructions: In brief, samples were added to the 96‐well plates that coated with antibodies and then were blocked with the sealing solution, followed by the addition of the second antibody diluent; after color reaction by tetramethylbenzidine (TMB), the results could be read by micro‐plate reader (Biotek) at 450 nm.
2.5. RT‐qPCR assay
Total RNA was extracted from PBMCs using TRIzol™ Reagent (Thermo Fisher Scientific), and then, reverse transcription to cDNA was conducted using PrimeScript™ RT reagent Kit (Perfect Real Time) (Takara). qPCR process was performed by SYBR® Premix DimerEraser™ (Takara). Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) was set as the internal reference for lnc‐PVT1, and U6 was applied as the internal reference for miR‐146a (the selection of different reference gene was according to the different sedimentation coefficients between lnc‐PVT1 and miR‐146a). The results of lnc‐PVT1 expression and miR‐146a expression were calculated by the 2−ΔΔCt formula. Information of primers used for RT‐qPCR were as follows: lnc‐PVT1, forward (5′→3′): CGAGCTGCGAGCAAAGATGT, reverse (5′→3′): CCGTGTCTCCACAGGTCACA; miR‐146a, forward (5′→3′): ACACTCCAGCTGGGTGAGAACTGAATTCCA, reverse (5′→3′): TGTCGTGGAGTCGGCAATTC; GAPDH, forward (5′→3′): GGAGCGAGATCCCTCCAAAAT, reverse (5′→3′): GGCTGTTGTCATACTTCTCATGG; U6, forward (5′→3′): CTCGCTTCGGCAGCACATATACTA, reverse (5′→3′): ACGAATTTGCGTGTCATCCTTGC.
2.6. Statistical analysis
Statistical analysis was performed by SPSS 20.0 software (SPSS Inc, USA), and all figures were plotted by GraphPad Prism 7.01 software (GraphPad Software Inc, USA). Data were described as mean value ± standard deviation (SD), median and interquartile range (IQR), or count (percentage). Comparison among three groups was determined by one‐way analysis of variance (ANOVA), Kruskal‐Wallis test, or chi‐square test. Comparison between two groups was determined by Wilcoxon rank‐sum test. Correlation analysis was performed using Spearman's rank correlation test. Receiver operating characteristic (ROC) curves and areas under the curve (AUC) were used to assess the performance of the variables in distinguishing different subjects. P value <.05 was considered significant.
3. RESULTS
3.1. Comparison of clinical characteristics among AECOPD patients, stable COPD patients, and HCs
No difference of age (P = .718), gender (P = .170), or BMI (P = .381) was observed among AECOPD patients, stable COPD patients, and HCs (Table 1). However, the percentages of family history of COPD (P = .017) and history of smoke (P = .002) were increased in AECOPD patients and stable COPD patients compared to HCs. For FEV1/FVC (P < .001) and FEV1 (% predicted), (P < .001), their values were lowest in AECOPD patients, followed by stable COPD patients, and were highest in HCs. For inflammatory cytokine levels, TNF‐α (P < .001), IL‐6 (P < .001), IL‐8 (P < .001), and IL‐17 (P < .001) values were all highest in AECOPD, followed by stable COPD, and were lowest in HCs. Additionally, GOLD stage was raised to some extent in AECOPD patients compared to stable COPD patients (P = .025).
TABLE 1.
Comparison of clinical characteristics
| Items |
HCs (N = 80) |
Stable COPD (N = 80) |
AECPOD (N = 80) |
P value |
|---|---|---|---|---|
| Age (y), mean ± SD | 66.0 ± 7.6 | 66.3 ± 6.8 | 66.9 ± 7.0 | .718 |
| Gender, No. (%) | .170 | |||
| Female | 24 (30.0) | 18 (22.5) | 14 (17.5) | |
| Male | 56 (70.0) | 62 (77.5) | 66 (82.5) | |
| BMI (kg/m2), mean ± SD | 22.4 ± 2.5 | 21.8 ± 2.7 | 22.3 ± 3.0 | .381 |
| Family history of COPD, No. (%) | 11 (13.8) | 26 (32.5) | 22 (27.5) | .017 |
| History of smoke, No. (%) | 22 (27.5) | 43 (53.8) | 39 (48.8) | .002 |
| FEV1/FVC (%), mean ± SD | 81.9 ± 4.0 | 60.3 ± 6.1 | 58.3 ± 8.1 | <.001 |
| FEV1 (% predicted), mean ± SD | 98.8 ± 4.0 | 68.3 ± 18.4 | 59.3 ± 18.7 | <.001 |
| GOLD stage, No. (%) | .025 | |||
| 1 | ‐ | 38 (47.5) | 25 (31.3) | |
| 2 | ‐ | 27 (33.7) | 31 (38.7) | |
| 3 | ‐ | 14 (17.5) | 22 (27.5) | |
| 4 | ‐ | 1 (1.3) | 2 (2.5) | |
| TNF‐α (pg/mL), median (IQR) | 13.6 (7.8‐21.3) | 17.9 (9.4‐34.1) | 58.2 (32.9‐84.3) | <.001 |
| IL‐6 (pg/mL), median (IQR) | 7.9 (4.1‐11.2) | 9.4 (4.6‐18.4) | 39.4 (17.0‐60.8) | <.001 |
| IL‐8 (pg/mL), median (IQR) | 12.2 (7.0‐21.8) | 20.0 (7.8‐45.5) | 47.1 (27.6‐134.9) | <.001 |
| IL‐17 (pg/mL), median (IQR) | 10.8 (6.6‐17.1) | 14.1 (6.0‐31.7) | 50.5 (22.1‐100.5) | <.001 |
Comparison was determined by one‐way analysis of variance (ANOVA), chi‐square test, Wilcoxon rank‐sum test or Kruskal‐Wallis H rank‐sum test.
Abbreviations: AECOPD, acute exacerbation of chronic obstructive pulmonary disease; BMI, body mass index; COPD, chronic obstructive pulmonary disease; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; GOLD, Global Initiative for Chronic Obstructive Lung Disease; HCs, healthy controls; IL, interleukin; IQR, interquartile range; SD, standard deviation; TNF‐α, tumor necrosis factor‐α.
3.2. Comparison of lnc‐PVT1 and miR‐146a expressions among AECOPD patients, stable COPD patients and HCs
Lnc‐PVT1 expression was highest in AECOPD patients, followed by stable COPD patients, and was lowest in HCs (P < .001) (Figure 1A). Moreover, miR‐146a expression was lowest in AECOPD patients, followed by stable COPD patients, and was highest in HCs (P < .001) (Figure 1B).
FIGURE 1.
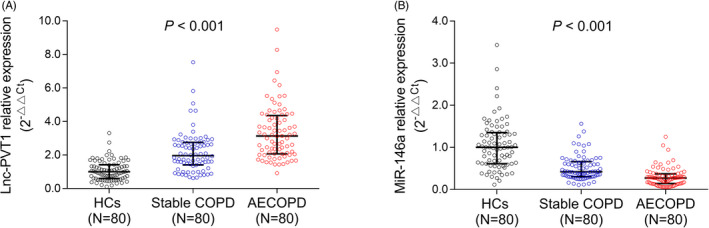
Lnc‐PVT1 and miR‐146a expressions. Comparison of lnc‐PVT1 (A) and miR‐146a (B) expressions among AECOPD patients, stable COPD patients, and HCs. AECOPD, acute exacerbation of chronic obstructive pulmonary disease; COPD, chronic obstructive pulmonary disease; HCs, healthy controls; Lnc‐PVT1, long non‐coding RNA plasmacytoma variant translocation 1; miR‐146a, microRNA 146a
3.3. The abilities of lnc‐PVT1 and miR‐146a to distinguish AECOPD patients, stable COPD patients, and HCs
Lnc‐PVT1 was able to distinguish AECOPD patients from stable COPD patients (AUC: 0.739, 95% CI: 0.663‐0.815) (Figure 2A), AECOPD patients from HCs (AUC: 0.953, 95% CI: 0.925‐0.982) (Figure 2B), and stable COPD patients from HCs (AUC: 0.813, 95% CI: 0.748‐0.878) (Figure 2C). Regarding miR‐146a, it could also distinguish AECOPD patients from stable COPD patients (AUC: 0.765, 95% CI: 0.691‐0.838) (Figure 2D), AECOPD patients from HCs (AUC: 0.927, 95% CI: 0.887‐0.967) (Figure 2E), and stable COPD patients from HCs (AUC: 0.812, 95% CI: 0.745‐0.879) (Figure 2F).
FIGURE 2.
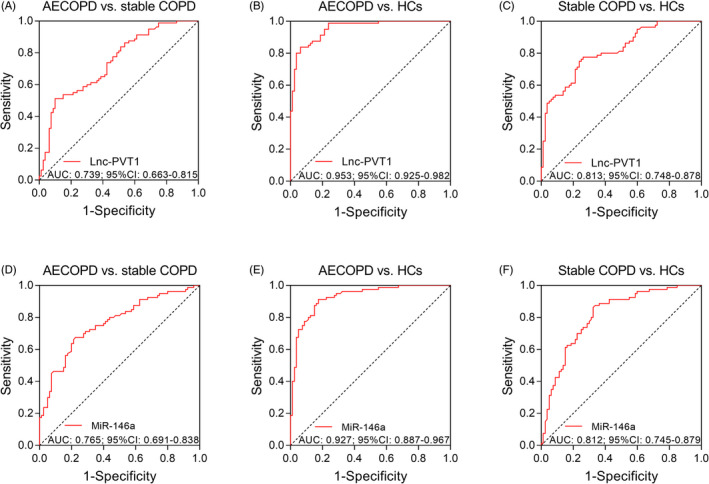
ROC curves. ROC curves showed the ability of lnc‐PVT1 for distinguishing AECOPD patients from stable COPD patients (A), AECOPD patients from HCs (B) and stable COPD patients from HCs (C), as well as the ability of miR‐146a for distinguishing AECOPD patients from stable COPD patients (D), AECOPD patients from HCs (E) and stable COPD patients from HCs (F). AECOPD, acute exacerbation of chronic obstructive pulmonary disease; AUC, areas under the curve; COPD, chronic obstructive pulmonary disease; HCs, healthy controls; lnc‐PVT1, long non‐coding RNA plasmacytoma variant translocation 1; miR‐146a, microRNA 146a; ROC, receiver operating characteristic
3.4. Correlation between lnc‐PVT1 and miR‐146a expressions in AECOPD patients and stable COPD patients
Both in AECOPD patients (P < .001, r = −.384) (Figure 3A) and stable COPD patients (P = .001, r = −.367) (Figure 3B), lnc‐PVT1 expression was negatively correlated with miR‐146a expression.
FIGURE 3.
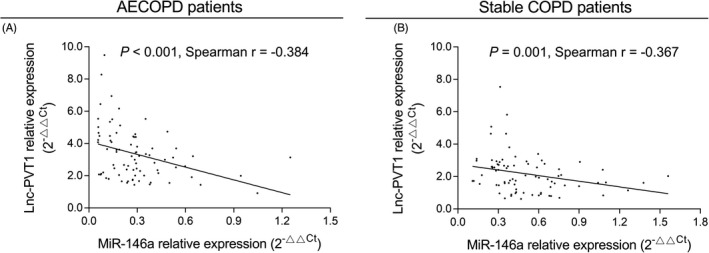
Association of lnc‐PVT1 expression with miR‐146a expression. The negative correlation of lnc‐PVT1 expression with miR‐146a expression in AECOPD patients (A) and stable COPD patients (B). AECOPD, acute exacerbation of chronic obstructive pulmonary disease; COPD, chronic obstructive pulmonary disease; Lnc‐PVT1, long non‐coding RNA plasmacytoma variant translocation 1; miR‐146a, microRNA 146a
3.5. Correlation of lnc‐PVT1 and miR‐146a expressions with GOLD stage in AECOPD patients and stable COPD patients
In AECOPD patients, lnc‐PVT1 expression was positively correlated with GOLD stage (P < .001, r = .501) (Figure 4A). Whereas miR‐146a expression was negatively associated with GOLD stage (P < .001, r = −.562) (Figure 4B). In stable COPD patients, lnc‐PVT1 expression was positively associated with GOLD stage (P = .018, r = .264) (Figure 4C), while miR‐146a expression was negatively correlated with GOLD stage (P = .045, r = −.225) (Figure 4D).
FIGURE 4.
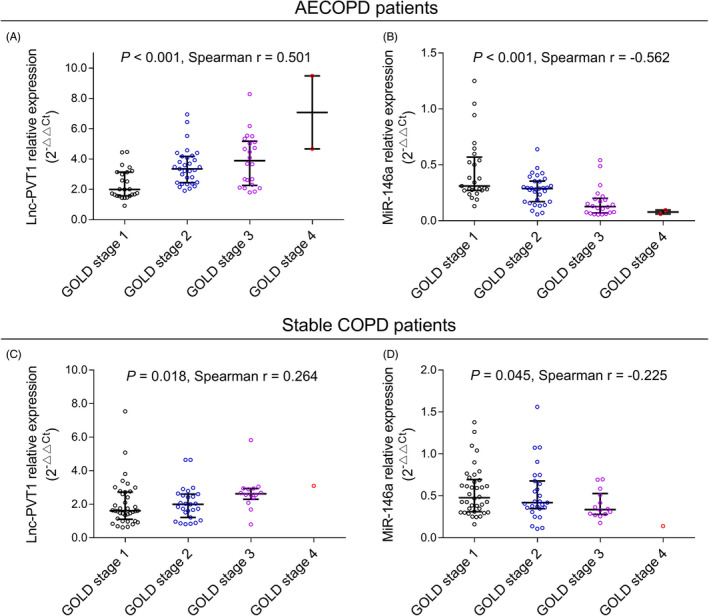
Association of lnc‐PVT1 and miR‐146a expressions with GOLD stage. A positive correlation of lnc‐PVT1 expression with GOLD stage (A) and a negative correlation of miR‐146a expression with GOLD stage (B) in AECOPD patients. A positive correlation of lnc‐PVT1 expression with GOLD stage (C) and a negative correlation of miR‐146a expression with GOLD stage (D) in stable COPD patients. AECOPD, acute exacerbation of chronic obstructive pulmonary disease; COPD, chronic obstructive pulmonary disease; GOLD, Global Initiative for Chronic Obstructive Lung Disease; Lnc‐PVT1, long non‐coding RNA plasmacytoma variant translocation 1; miR‐146a, microRNA 146a
3.6. Correlation of lnc‐PVT1 and miR‐146a expressions with inflammatory cytokine levels in AECOPD patients and stable COPD patients
In AECOPD patients, lnc‐PVT1 expression was positively correlated with levels of TNF‐α (P < .001, r = .479) (Figure 5A), IL‐6 (P = .010, r = .287) (Figure 5B), IL‐8 (P = .011, r = .282) (Figure 5C), and IL‐17 (P < .001, r = .473) (Figure 5D), while miR‐146a expression was negatively associated with levels of TNF‐α (P < .001, r = −.455) (Figure 5E), IL‐6 (P = .002, r = −.335) (Figure 5F), IL‐8 (P = .006, r = −.306) (Figure 5G), and IL‐17 (P = .003, r = −.327) (Figure 5H). In stable COPD patients, lnc‐PVT1 expression was positively associated with levels of TNF‐α (P = .001, r = .359) (Figure 5I), IL‐6 (P = .012, r = .280) (Figure 5J), IL‐8 (P < .001, r = .388) (Figure 5K), and IL‐17 (P < .001, r = .421) (Figure 5L), but miR‐146a was negatively associated with levels of TNF‐α (P = .005, r = −.310) (Figure 5M), IL‐6 (P < .001, r = −.382) (Figure 5N), IL‐8 (P = .024, r = −.252) (Figure 5O), and IL‐17 (P = .007, r = −.298) (Figure 5P).
FIGURE 5.
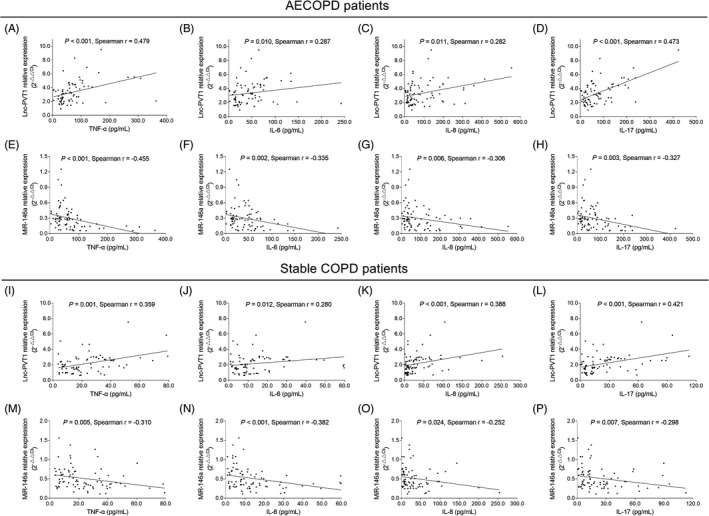
Association of lnc‐PVT1 and miR‐146a expressions with inflammatory cytokines. Positive correlations of lnc‐PVT1 expression with TNF‐α (A), IL‐6 (B), IL‐8 (C), and IL‐17 (D) levels; as well as negative correlations of miR‐146a expression with TNF‐α (E), IL‐6 (F), IL‐8 (G), and IL‐17 (H) levels in AECOPD patients. Positive correlations of lnc‐PVT1 expression with TNF‐α (I), IL‐6 (J), IL‐8 (K), and IL‐17 (L) levels; as well as negative correlations of miR‐146a expression with TNF‐α (M), IL‐6 (N), IL‐8 (O), and IL‐17 (P) levels in stable COPD patients. AECOPD, acute exacerbation of chronic obstructive pulmonary disease; COPD, chronic obstructive pulmonary disease; IL‐17, interleukin‐17; IL‐6, interleukin‐6; IL‐8, interleukin‐8; Lnc‐PVT1, long non‐coding RNA plasmacytoma variant translocation 1; miR‐146a, microRNA 146a; TNF‐α, tumor necrosis factor‐α
4. DISCUSSION
Lnc‐PVT1 is a novel cancer diagnostic biomarker with properties of high specificity and easy detection in the serum, plasma and saliva, which is shown overexpressed in several cancer cells while downregulated in normal cells. 13 Moreover, lnc‐PVT1 not only exerts regulatory effects mainly through modulation of miRNAs at the cytoplasmic level, but also regulates gene transcription and protein levels in the cell nucleus and extracellular fluid. 14 Although lnc‐PVT1 is mainly known as an oncogene in various cancers, while its importance in inflammatory responses has just recently been elucidated. 6 , 15 , 16 For example, lnc‐PVT1 is overexpressed in myocardial tissues, and it inhibits cardiac function as well as enhances inflammatory cytokine levels via activating mitogen‐activated protein kinase (MAPK)/nuclear factor (NF)‐κB pathway in sepsis rats. 6 Also, lnc‐PVT1 knockdown inhibits rheumatoid arthritis (RA)‐fibroblast‐like synoviocyte proliferation and reduces inflammation in RA rat models. 16 For chronic airway respiratory diseases, lnc‐PVT1 is found downregulated in respiratory syncytial virus (RSV) infected‐airway smooth muscle (ASM) cells after α‐asarone treatment, and it is involved in the regulation of viability, proliferation and migration of ASM cells via miR‐1207‐5p/p53 or miR‐203a/E2F3 pathways. 17 , 18 These data suggest that lnc‐PVT1 may act as an important factor contributing to the occurrence of several inflammatory diseases.
Some evidences in clinical practices also uncover the implication of lnc‐PVT1 in inflammatory diseases. 7 , 19 , 20 , 21 For instance, lnc‐PVT1 expression is elevated in osteoarthritis (OA) patients, diabetic nephropathy (DN) patients, and atrial fibrillation (AF) patients compared to HCs. 19 , 20 , 21 In addition, one study shows that lnc‐PVT1 has excellent ability to distinguish corticosteroid‐insensitive severe asthmatics from corticosteroid sensitive non‐severe asthmatics, 7 whereas the role of lnc‐PVT1 in COPD occurrence remains poorly elucidated. To assess the value of lnc‐PVT1 in predicting COPD susceptibility and acute exacerbation risk, we detected the lnc‐PVT1 expression in AECOPD patients, stable COPD patients, and HCs and performed ROC curves to evaluate the distinguishing ability of lnc‐PVT1 among them. We observed that lnc‐PVT1 expression was highest in AECOPD patients, followed by stable COPD patients, and was lowest in HCs. Furthermore, ROC curves displayed that lnc‐PVT1 not only distinguished AECOPD patients and stable COPD patients from HCs, but also distinguished AECOPD patients from stable COPD patients. The possible reasons might be as follows: lnc‐PVT1 might induce the oxidative stress condition and the activated inflammatory cascades, which led to sever damage in ADM cells or lung bronchial epithelial cells and eventually impaired lung function, thus highly expressed lnc‐PVT1 predicted increased COPD susceptibility and acute exacerbation risk. 17 , 18
In addition, lnc‐PVT1 is shown to be correlated with disease severity and inflammation level in some inflammatory diseases. 8 , 20 For example, the expression of lnc‐PVT1 is gradually increased with the elevation of cardiac function classification, and is positively correlated with collagen I and collagen III (two major proteins contributing to atrial fibrosis) in AF patients 20 ; its SNP is positively associated with GOLD stage, FVC, FEV1, and diffusing capacity of carbon monoxide in COPD smoker patients. 8 In this study, we observed that lnc‐PVT1 expression was positively correlated with GOLD stage and inflammatory cytokine levels (TNF‐α, IL‐6, IL‐8, and IL‐17, which are well known as pro‐inflammatory cytokines in COPD 22 , 23 ) in both AECOPD patients and stable COPD patients, which might be explained as follows: (a) upregulated lnc‐PVT1 might make the ASM cells in an over‐activated state via promoting the viability, proliferation, and migration of ASM cells, resulting in airway narrowing (due to the contractile properties of ASM cells) and further enhanced airflow obstruction; therefore, lnc‐PVT1 expression was positively correlated with GOLD stage in AECOPD patients and stable COPD patients 17 ; and (b) lnc‐PVT1 might promote inflammatory responses via activating MAPK/NF‐κB pathway or via sponging its certain targeted miRNAs, thereby facilitated the production and release of inflammatory cytokines, and lnc‐PVT1 expression was positively associated with inflammation level in AECOPD patients and stable COPD patients. 6 , 17 , 18
As one of the frequently investigated miRNAs, miR‐146a is reported to take a crucial role in chronic airway respiratory diseases. MiR‐146a decreases IL‐1α–induced pro‐inflammatory responses in lung fibroblasts and reduces particulate matter‐induced inflammation via NF‐κB signaling pathway in human lung bronchial epithelial cells. 9 , 10 Moreover, clinical practices show that miR‐146a is downregulated in primary human lung fibroblasts from COPD patients. 9 Given that miR‐146a may exert anti‐inflammatory role in chronic airway respiratory diseases, especially COPD, and miR‐146a (or its SNP) has been confirmed as a target miRNA of lnc‐PVT1 in multiple diseases (prostate cancer, colon cancer), we speculated that miR‐146a might also be correlated with lnc‐PVT1 and accordingly be involved in the COPD progression. 15 , 24 Hence, we explored the association of lnc‐PVT1 with miR‐146a expressions and the correlation of miR‐146a with disease severity in COPD patients. We observed that in both AECOPD patients and stable COPD patients, lnc‐PVT1 presented with a negative correlation with miR‐146a; meanwhile, miR‐146a low expression correlated with elevated COPD susceptibility and acute exacerbation risk, as well as raised COPD severity and inflammation. These data suggested that the predictive ability of lnc‐PVT1 for COPD susceptibility and acute exacerbation risk as well as its surveillance of disease progression might work through its inter‐correlation with miR‐146a.
There were some limitations in our study. Firstly, the sample size in this study was relatively small, which might lead to the relatively insufficient statistical power, and so further study with larger sample size was needed; secondly, the detailed regulation between lnc‐PVT1 and miR‐146a, and the underlying mechanisms of lnc‐PVT1 in COPD remained unclear, which required further exploration; thirdly, there were differences of clinical characteristics among AECOPD patients, stable COPD patients, and HCs, which might affect the patients' prognosis, while this study mainly aimed to investigate the abilities of lnc‐PVT and miR‐146a in predicting COPD susceptibility and acute exacerbation risk, and also to explore the association of lnc‐PVT1 with disease severity, inflammation, and miR‐146a in COPD patients, thus further study exploring factors affecting the treatment outcomes in AECOPD and stable COPD patients was needed to facilitate the treatment strategy in these patients; Fourthly, this preliminary study mainly aimed to investigate the abilities of lnc‐PVT1 and miR‐146a in predicting COPD susceptibility and acute exacerbation risk, and it was worth noting that the lnc‐PVT1 and miR‐146a expressions of patients were restricted to the levels within 24 hours after admission in this study; thus, patients were defined as AECOPD or stable COPD only at study initiation and disease progress during study was not taken into account to reach the accurate results.
To conclude, lnc‐PVT1 not only distinguishes AECOPD patients and stable COPD patients from HCs but also distinguishes AECOPD patients from stable COPD patients; meanwhile, lnc‐PVT1 expression positively correlates while miR‐146a expression negatively correlates with GOLD stage and inflammatory cytokine levels in both AECOPD patients and stable COPD patients, implying that lnc‐PVT1 assists the disease management and acute exacerbation risk monitoring of COPD, which may work through interaction with miR‐146a.
Wang Y, Lyu X, Wu X, Yu L, Hu K. Long non‐coding RNA PVT1, a novel biomarker for chronic obstructive pulmonary disease progression surveillance and acute exacerbation prediction potentially through interaction with microRNA‐146a. J Clin Lab Anal. 2020;34:e23346 10.1002/jcla.23346
Contributor Information
Li Yu, Email: yuli641006@sina.com.
Ke Hu, Email: huke_rmhospital@163.com.
REFERENCES
- 1. Huang X, Mu X, Deng L, et al. The etiologic origins for chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2019;14:1139‐1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sun Y, Zhou J. New insights into early intervention of chronic obstructive pulmonary disease with mild airflow limitation. Int J Chron Obstruct Pulmon Dis. 2019;14:1119‐1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cai BQ, Cai SX, Chen RC, et al. Expert consensus on acute exacerbation of chronic obstructive pulmonary disease in the People's Republic of China. Int J Chron Obstruct Pulmon Dis. 2014;9:381‐395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang Z, Tang Y, Lu H, et al. Long non‐coding RNA reprogramming (lncRNA‐ROR) regulates cell apoptosis and autophagy in chondrocytes. J Cell Biochem. 2018;119(10):8432‐8440. [DOI] [PubMed] [Google Scholar]
- 5. Huang W, Lan X, Li X, et al. Long non‐coding RNA PVT1 promote LPS‐induced septic acute kidney injury by regulating TNFalpha and JNK/NF‐kappaB pathways in HK‐2 cells. Int Immunopharmacol. 2017;47:134‐140. [DOI] [PubMed] [Google Scholar]
- 6. Feng F, Qi Y, Dong C, Yang C. PVT1 regulates inflammation and cardiac function via the MAPK/NF‐kappaB pathway in a sepsis model. Exp Ther Med. 2018;16(6):4471‐4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Austin PJ, Tsitsiou E, Boardman C, et al. Transcriptional profiling identifies the long noncoding RNA plasmacytoma variant translocation (PVT1) as a novel regulator of the asthmatic phenotype in human airway smooth muscle. J Allergy Clin Immunol. 2017;139(3):780‐789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou S, Liu Y, Li M, et al. Combined effects of PVT1 and MiR‐146a single nucleotide polymorphism on the lung function of smokers with chronic obstructive pulmonary disease. Int J Biol Sci. 2018;14(10):1153‐1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Osei ET, Florez‐Sampedro L, Tasena H, et al. miR‐146a‐5p plays an essential role in the aberrant epithelial‐fibroblast cross‐talk in COPD. Eur Respir J. 2017;49(5):1602538. [DOI] [PubMed] [Google Scholar]
- 10. Liu L, Wan C, Zhang W, et al. MiR‐146a regulates PM1 ‐induced inflammation via NF‐kappaB signaling pathway in BEAS‐2B cells. Environ Toxicol. 2018;33(7):743‐751. [DOI] [PubMed] [Google Scholar]
- 11. Chen BB, Li ZH, Gao S. Circulating miR‐146a/b correlates with inflammatory cytokines in COPD and could predict the risk of acute exacerbation COPD. Medicine (Baltimore). 2018;97(7):e9820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pauwels RA, Buist AS, Calverley PM, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med. 2001;163(5):1256‐1276. [DOI] [PubMed] [Google Scholar]
- 13. Li MY, Tang XH, Fu Y, Wang T‐J, Zhu J‐M. Regulatory mechanisms and clinical applications of the long non‐coding RNA PVT1 in cancer treatment. Front Oncol. 2019;9:787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jin K, Wang S, Zhang Y, et al. Long non‐coding RNA PVT1 interacts with MYC and its downstream molecules to synergistically promote tumorigenesis. Cell Mol Life Sci. 2019;76(21):4275‐4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu HT, Fang L, Cheng YX, Sun Q. LncRNA PVT1 regulates prostate cancer cell growth by inducing the methylation of miR‐146a. Cancer Med. 2016;5(12):3512‐3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang CW, Wu X, Liu D et al. Long non‐coding RNA PVT1 knockdown suppresses fibroblast‐like synoviocyte inflammation and induces apoptosis in rheumatoid arthritis through demethylation of sirt6. J Biol Eng. 2019;13:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perry MM, Tsitsiou E, Austin PJ, et al. Role of non‐coding RNAs in maintaining primary airway smooth muscle cells. Respir Res. 2014;15:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu X, Zhe Z, Tang B, et al. alpha‐Asarone suppresses the proliferation and migration of ASMCs through targeting the lncRNA‐PVT1/miR‐203a/E2F3 signal pathway in RSV‐infected rats. Acta Biochim Biophys Sin (Shanghai). 2017;49(7):598‐608. [DOI] [PubMed] [Google Scholar]
- 19. Zhao Y, Zhao J, Guo X, She J, Liu Y. Long non‐coding RNA PVT1, a molecular sponge for miR‐149, contributes aberrant metabolic dysfunction and inflammation in IL‐1beta‐simulated osteoarthritic chondrocytes. Biosci Rep. 2018;38(5). 10.1042/BSR20180576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu DW, Zhang JH, Liu FX, et al. Silencing of long noncoding RNA PVT1 inhibits podocyte damage and apoptosis in diabetic nephropathy by upregulating FOXA1. Exp Mol Med. 2019;51(8):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cao F, Li Z, Ding WM, Yan L, Zhao QY. LncRNA PVT1 regulates atrial fibrosis via miR‐128‐3p‐SP1‐TGF‐beta1‐Smad axis in atrial fibrillation. Mol Med. 2019;25(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iqbal H, Rhee DK. Ginseng alleviates microbial infections of the respiratory tract: a review. J Ginseng Res. 2020;44(2):194‐204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao N, Xu W, Ji J, et al. Lung function and systemic inflammation associated with short‐term air pollution exposure in chronic obstructive pulmonary disease patients in Beijing, China. Environ Health. 2020;19(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang W, Xiao J, Lu X, et al. PVT1 (rs13281615) and miR‐146a (rs2910164) polymorphisms affect the prognosis of colon cancer by regulating COX2 expression and cell apoptosis. J Cell Physiol. 2019;234(10):17538‐17548. [DOI] [PubMed] [Google Scholar]


