Abstract
Lysine ubiquitination, a widely studied posttranslational modification, plays vital roles in various biological processes in eukaryotic cells. Although several studies have examined the plant ubiquitylome, no such research has been performed in tobacco, a model plant for molecular biology. Here, we comprehensively analyzed lysine ubiquitination in tobacco (Nicotiana tabacum) using LC–MS/MS along with highly sensitive immune-affinity purification. In total, 964 lysine-ubiquitinated (Kub) sites were identified in 572 proteins. Extensive bioinformatics studies revealed the distribution of these proteins in various cellular locations, including the cytoplasm, chloroplast, nucleus, and plasma membrane. Notably, 25% of the Kub proteins were located in the chloroplast of which 21 were enzymatically involved in important pathways, that is, photosynthesis and carbon fixation. Western blot analysis indicated that TMV infection can cause changes in ubiquitination levels. This is the first comprehensive proteomic analysis of lysine ubiquitination in tobacco, illustrating the vital role of ubiquitination in various physiological and biochemical processes and representing a valuable addition to the existing landscape of lysine ubiquitination.
1. Introduction
Posttranslational modifications (PTMs) play vital roles in modulating the ultimate functions of newly synthesized proteins in the cellular process by adding and/or removing functional groups such as methyl, acetyl, phospho, and ubiquityl groups.1,2 More than 300 PTMs have been characterized and additional modifications are still being discovered. These modifications show great potential for functional regulation by changing protein structures and modulating the stability, activities, and interactions with other proteins and molecules.3 Among these different types of PTMs, lysine ubiquitination is widely involved in regulating cell cycle progression and gene expression via covalent conjugation of ubiquitin (Ub) with lysine (Lys) residues of the proteasome.4,5 Ub is a highly conserved small protein, consisting of 76 amino acids(aa) found in various eukaryotic cells6 and is known as a signal for protein degradation through the proteasome pathway that degrades and/or regulates target proteins via three distinct enzymatic activities involving E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin-ligase enzymes.7−9 The process is activated by E1 through a thioester bond, which is formed between the ubiquitin C-terminal carboxyl group of Lys and a cysteinyl sulfhydryl residue of cysteine (Cys) on E1.10 Activated Ub then gets attached to a c sulfhydryl of E2 by a thioester bond. Finally, E3s bring the activated Ub and substrate together via an isopeptide linkage between the Ub C-terminal carboxyl group and the ε-amino group of the Lys residue on target proteins.11 Unlike the single group addition that occurs during methylation, acetylation, or phosphorylation, the linkage may occur as monoubiquitination, that is, the addition of one Ub molecule, or as polyubiquitination, that is, the addition of a chain of Ub.12 Due to the strong influence of ubiquitination on the ultimate fate of the substrate, this modification regulates a wide variety of biological processes, including but not limited to immune and stress responses, homeostasis, photosynthesis, DNA and hormone synthesis, embryogenesis regulation, transcription, and signal transduction.3,8,13−15 Owing to the tremendous sophistication of mass spectrometry (MS) and affinity purification, numerous ubiquitylation modifications have been reported on a proteomic scale.16−18 For instance, in the past few years, a massive number of lysine-ubiquitylated proteins have been reported in yeasts19,20 and mammalian cells.21−23 However, the ubiquitylome of plants has rarely been studied.6,16 Therefore, the study of the ubiquitination profile of plants may play a vital role in explaining the important functional characteristics of these modified proteins for many applications, including potential biotechnological and pathological applications. In this study, we analyzed lysine ubiquitination in tobacco (Nicotiana tabacum), which is both an important economic leaf crop and a model plant in molecular biological research.24 As one of the most valuable research biomaterials, common tobacco is widely used in the study of various aspects of botany, including genetic, cytological, physiological, and phytopathological studies. Because lysine ubiquitination has been well studied in the regulation of cellular and physiological processes in plants, animals, and humans in the past three decades,25 elucidation of the ubiquitination sites in tobacco may provide an important breakthrough for understanding the potential role of this PTM in virus resistance in plants. In the present research, for the first time, we performed a comprehensive proteomic analysis of lysine-ubiquitinated (Kub) sites in N. tabacum seedlings using high-resolution liquid chromatography-tandem mass spectrometry (LC–MS/MS) accompanied by highly sensitive immune-affinity purification and heuristic bioinformatics tools. In total, 964 ubiquitination sites distributed on 572 proteins were identified as being present in various cellular compartments including the cytoplasm, chloroplast, nucleus, plasma membrane, mitochondria, and cytoskeleton, which are mainly involved in photosynthesis, carbon fixation metabolism, and protein metabolism. Western blot results indicated that tobacco mosaic virus (TMV) infection can cause changes in ubiquitination levels. To our knowledge, this is the first global qualitative lysine ubiquitylome of N. tabacum, providing a rich set of data for exploration of functions of ubiquitinated proteins in tobacco. The results are a valuable addition in the existing landscape of lysine ubiquitination.
2. Results and Discussion
2.1. Proteome-Wide Identification of Kub Sites in Tobacco
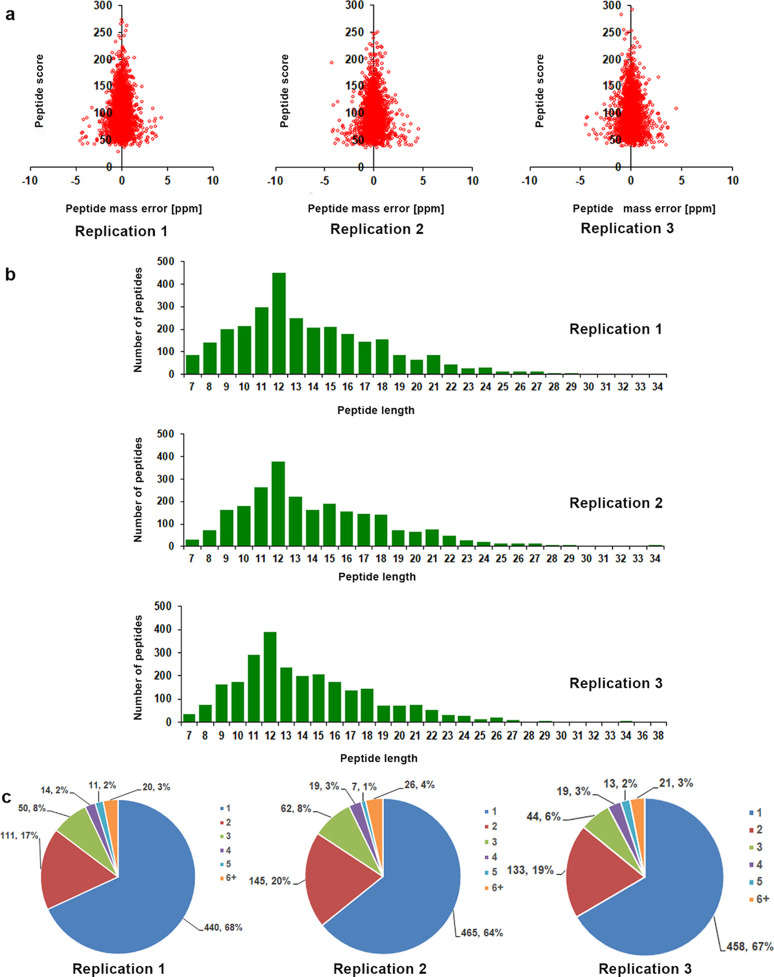
Ubiquitination is an extensively studied conserved PTM that plays an important role in many cellular and biological processes in eukaryotes. In recent years, the ubiquitylome of important plant species, including wheat (Triticum aestivum L.), rice (Oryza sativa), soybean (Glycine max), Arabidopsis, and Pyrus spp. has been reported.6,16,25−28 In this study, we analyzed the global lysine ubiquitylome of N. tabacum, which is a widely used model plant in molecular research. To identify Kub peptides in tobacco, LC–MS/MS along with highly specific enrichment analysis was used (Figure 1). A total of 1963 ubiquitinated sites located on 1106 proteins were identified. Further analysis was conducted for only one selected replication because the trend of Kub proteins was similar in all three repeats (Figure 2a–c and Supporting Information Table S1). The results showed the high accuracy of the MS data (Figure 2a). The length of most Kub peptides ranged from 7 to 27 aa (Figure 2b). A systematic bioinformatics analysis of the identified Kub proteins revealed that 964 ubiquitinated sites were located on 572 proteins (Supporting Information Table S2). Most of the ubiquitinated sites were identified between positions 1 and 11 (Supporting Information Table S1). Eighty-five percent of the identified proteins had one or two ubiquitination sites, whereas 15% of the Kub proteins showed three or more modification sites (Figure 2c).
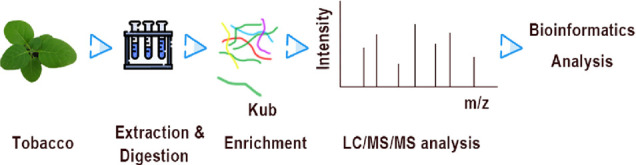
Figure 1.
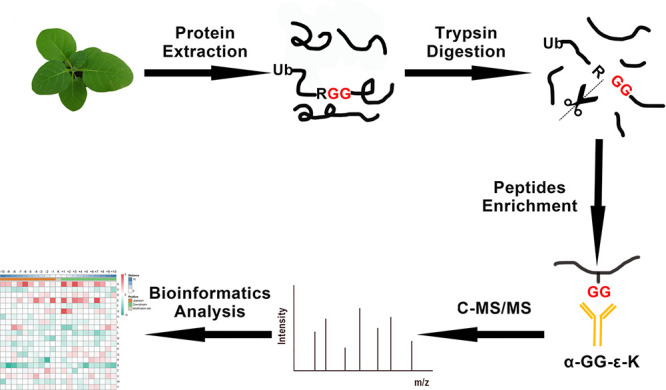
Overview of the experimental procedures used in this study.
Figure 2.
Proteome-wide identification of lysine ubiquitination sites in Nicotiana tabacum. (a) Mass error distribution of identified ubiquitinated peptides. (b) Peptide length distribution of ubiquitinated proteins. (c) Pie chart illustration of the number and percentage of lysine ubiquitination sites per protein. All the data were analyzed for three replications.
2.2. Motif Analysis and Secondary Structures of Kub Proteins
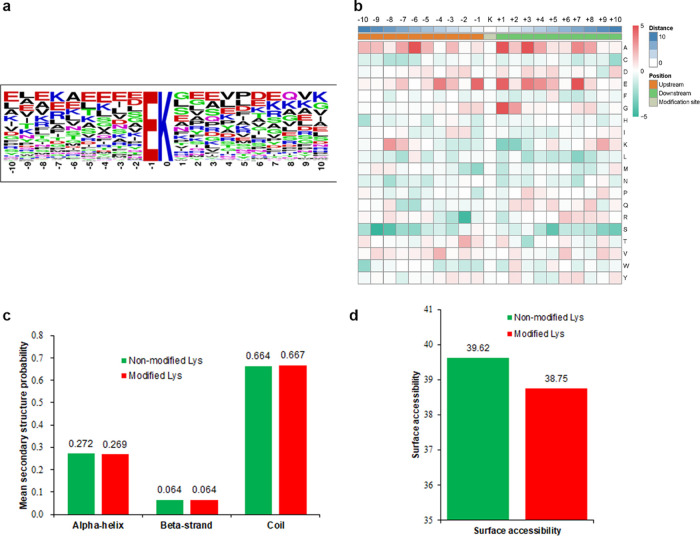
To characterize the features of Kub sites in tobacco, the sequence motifs were analyzed using Motif-X software, which showed one conserved motif, that is, EKub (Kub indicates the ubiquitinated lysine), in 143 peptides (Figure 3a and Supporting Information Table S3). Only glutamic acid (E) was found upstream of Kub (Figure 3a). The enriched Kub motif (EKub) in tobacco has been reported previously in rice16 and petunia,29 indicating the relationship among these plants at the proteomic level, which is consistent with the fact that Kub is a highly conserved PTM. The heat map (Figure 3b and Supporting Information Table S4) showed the enrichment of lysine (K) in the −8 to −7 and +10 to +8 and +5 positions, while the occurrence of glutamic acid (E) was found to be enriched in the −7 to +8 position. We used NetSurfP software to reveal the relationship between Kub and protein secondary structures (Figure 3c). The results showed that 26.9% of the ubiquitinated sites were located in α-helices, and the percentage of the sites located in β-strands was 6.4%. The remaining 66.7% sites were located in disordered regions of the proteins. There seemed to be no tendency of lysine ubiquitination in tobacco leaves according to the similarity of distribution patterns between Kub and nonmodified K. Moreover, the data for surface accessibility showed that 38.8% of Kub sites were exposed to the protein surface, close to the 39.6% of nonmodified lysine observed (Figure 3d). Therefore, lysine ubiquitination could barely change the surface properties of proteins.
Figure 3.
Properties of the Kub sites. (a) Sequence probability logos of significantly enriched ubiquitination motifs around the Kub sites. (b) Heat map of amino acid compositions showing the frequency of different types of amino acids around the lysine ubiquitination sites. Capital letters on the right side of the figure represent amino acid abbreviations. Red indicates enrichment, while green indicates depletion. (c) Distribution of secondary structures containing Kub sites. (d) Predicted surface accessibility of ubiquitinated sites.
2.3. Functional Annotation and Cellular Localization of Ubiquitinated Proteins
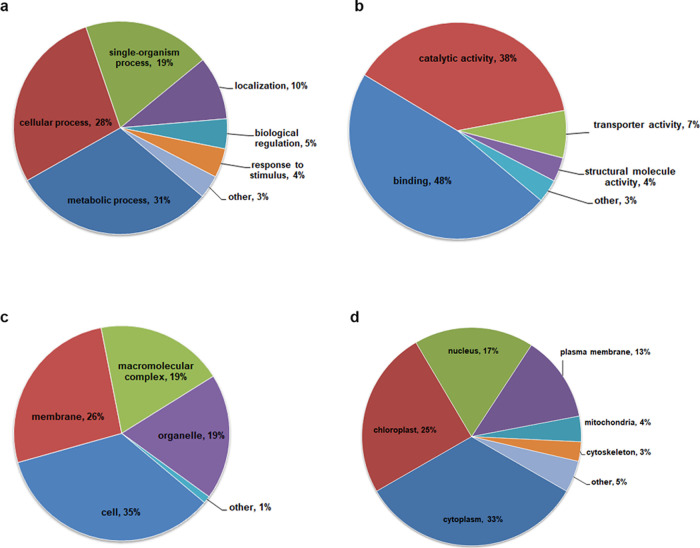
To thoroughly understand the ubiquitylome in tobacco, GO functional annotation of all Kub proteins was performed under the biological process, cellular component, and molecular function categories (Figure 4a–c and Supporting Information Table S5). The biological process analysis showed that a majority of the Kub proteins were related to metabolic processes (236; 31%), cellular processes (215; 28%), single-organism processes (148; 19%), and localization (74; 10%) (Figure 4a). The identified proteins were involved in multiple cellular processes, suggesting that lysine ubiquitination plays an important role in the regulation of different metabolic and cellular processes in tobacco. For the cellular component category, the modified proteins were distributed within the cell (126; 35%), membrane (96; 26%), macromolecular complex (70; 19%), and organelle (69; 19%) (Figure 4b). According to the molecular function classification, 48, 38, and 7% of the Kub proteins were associated with binding function, catalytic activity, and transporter activity, respectively (Figure 4c). Subcellular localization analysis indicated that a large number of Kub proteins were distributed in the cytoplasm (185; 33%), chloroplast (138; 25%), nucleus (98; 17%), and plasma membrane (71; 13%) (Figure 4d and Supporting Information Table S6). The results mentioned above suggest that the newly identified Kub proteins are involved in various important biological processes in tobacco, especially involved in photosynthesis, the conversion of light energy to chemical energy in the form of sugars and ATP30 that are readily available to plants.
Figure 4.
Functional classification of ubiquitinated proteins in tobacco leaves. (a) Classification of ubiquitinated proteins based on biological processes. (b) Molecular function classification. (c) Cellular component classification. (d) Subcellular localization.
2.4. Functional Enrichment Analysis
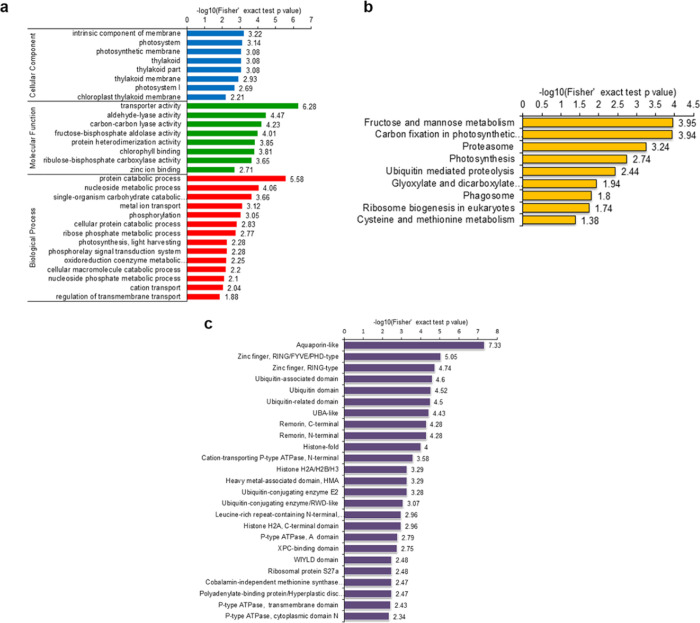
Functional enrichment of identified Kub proteins was investigated through Gene Ontology (GO), the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and protein domain analyses (Supporting Information Tables S7–S9). The results of biological process enrichment revealed that Kub proteins were mainly related to the protein catabolic process, nucleoside metabolic process, single-organism carbohydrate catabolic process, and metal ion transport (Figure 5a, red bars). Consistent with these results, many ubiquitinated proteins were revealed to be linked with transporter activity, aldehyde-lyase activity, carbon–carbon lyase activity, and fructose-bisphosphate aldolase activity in the molecular function enrichment analysis (Figure 5a, green bars). The cellular component enrichment results indicated that proteins located in the intrinsic components of the membrane, photosystem, photosynthetic membrane, and thylakoid were highly likely to be ubiquitinated (Figure 5a, blue bars). Furthermore, based on KEGG pathway enrichment analysis, most of the ubiquitinated proteins were involved in fructose and mannose metabolism, carbon fixation in photosynthetic organisms, the proteasome, and photosynthesis (Figure 5b).The analysis of protein domain enrichment revealed that proteins with aquaporin-like, zinc finger, ubiquitin-associated remorin and histone-fold domains were highly prone to ubiquitylation (Figure 5c), suggesting an important role of ubiquitylation in cellular metabolic pathways.
Figure 5.
Enrichment analysis of ubiquitinated proteins. (a) GO-based enrichment analysis in terms of biological process (red bars), molecular function (green bars), and cell component (blue bars). (b) KEGG pathway-based enrichment analysis. (c) Protein domain enrichment analysis. The numbers on the X axes represent significant values. Data were considered statistically significant when the value was greater than 1.3 and p-value <0.05.
2.5. Analysis of Ubiquitinated Proteins Involved in Metabolic Pathways
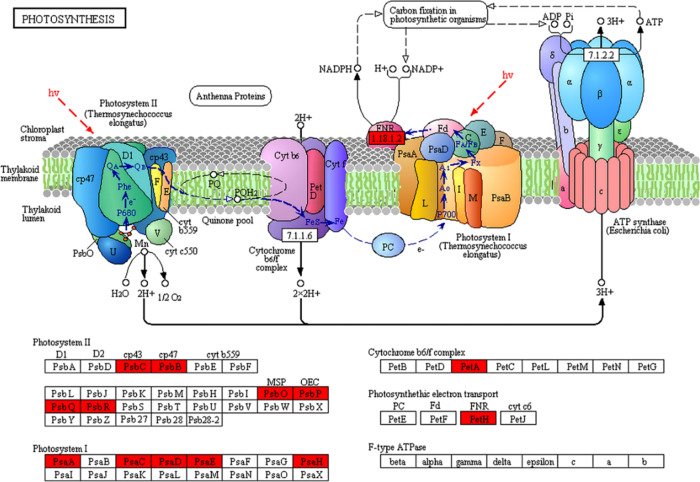
Lysine ubiquitination plays essential roles in metabolic regulation,26 cellular signaling events,31 and cellular process regulation.32 In this study, GO annotation (Figure 5a) and KEGG pathway analysis (Figure 5b) showed that 25% of the Kub proteins in tobacco were positioned in the chloroplast (Figure 4c), suggesting that lysine ubiquitination may play a vital role in carbon fixation in photosynthetic organisms. Several Kub proteins were identified in photosystems I and II, the cytochrome b6/f complex, and photosynthetic electron transport (Figure 6). The results of LC–MS/MS analysis showed that five protein subunits of photosystem I (PSI) (Psa A, Psa C, Psa D, Psa E, and Psa H), six subunits of photosystem II (PSII) (Psb B, Psb C, Psb O, Psb P, Psb Q, and Psb R), and one subunit of the cytochrome b6/f complex (Pet A) were among the important ubiquitinated proteins (Figure 6). Our results also showed that Kub occurred in ferredoxin-NADP+ oxidoreductase (FNR) (Figure 6), which catalyzes electron transfer from ferredoxin (Fd) to NADP+ for the production of NADPH.33,34
Figure 6.
Kub proteins involved in photosynthesis. The newly identified proteins are highlighted in red.
Photosynthesis is accomplished by two light reaction systems, that is, PSI and PSII. As important subunits, Psa A and Psa B are located in the center of PSI monomers, which contain the majority of the chlorophyll, carotenoids, and cofactors of the electron transport chain from P700 to the first FeS cluster, FX.35 Psa C, Psa D, and Psa E are the subunits present outside the membrane and constitute the stromal hump. All three subunits provide the docking point for Fd.35 As the proximal antennae for PSII, CP47 and CP43, encoded by the Psb B and Psb C genes, are also located in chlorophyll and offer a channel for excitation of energy from the exterior antennae of the photosystem to the reaction center core.36 Furthermore, PSI and PSII are connected by a complex consisting of cytochrome b6/f and a ferrosulfur protein,37 wherein the cytochrome complex may affect the production of ATP and NADPH by transferring electrons from PSII to PSI.38,39 Additionally, FNR catalyzes the last step of electron transfer in photosynthesis, from reduced Fd to NADPH.40 All the abovementioned findings strongly support the importance of the identified Kub proteins in tobacco and the potential role of ubiquitination in the ultimate functional profile of these proteins during photosynthesis. Proteins participated in photosynthesis were identified in the tobacco ubiquitome. PSI catalyzes electron transfer from the plastocyanin (PC) to the flavodoxin (FD) through a series of electron transfer components.41 The thylakoid-bound [4Fe-4S] clusters X on Psa A (A0A140G1R3) or clusters A/B on Psa C (A0A140G1X0) of the PSI complex involve electron transfer. PSII catalyzes electron transfer from water to plastoquinone driven by light.42 We found 3 ubiquitination sites on Psb P (A0A1S3XWU9), which plays a vital role in maintaining photosynthetic electron transfer.43 Additionally, other components involved in electron transfer such as FNR (A0A1S3X0Y5) and PetA (A0A140G1S8) were identified as ubiquitinated proteins in tobacco. Plants can connect light-driven electron transport with various metabolic processes. Chloroplasts with an intense rate of electron flow are a major source of reactive oxygen species (ROS) production in plant cells.44 ROS is closely related to plant pathogen defense and abiotic stress response.45 It has been proved that higher ROS production levels could enhance disease resistance in tobacco.46 These findings demonstrate that lysine ubiquitylation plays an important role in the process of photosynthesis and a potential role in disease resistance.
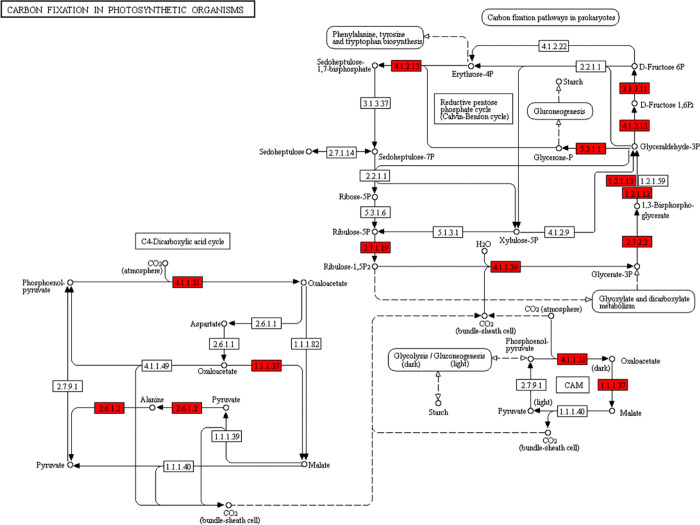
It has been reported that the ubiquitin-proteasome system is closely involved in carbon and nitrogen metabolism.47 We further studied the Kub proteins associated with carbon fixation in tobacco. Eleven ubiquitinated enzymes were determined to be involved in the carbon fixation pathway in photosynthesis (Figure 7). Among these enzymes, 10 were related to the Calvin cycle: fructose-bisphosphate aldolase, fructose-1,6- bisphosphatase, triosephosphate isomerase, glyceraldehyde-3-phosphate dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase (NADP+) (phosphorylating), phosphoglycerate kinase, phosphoribulokinase, ribulose bisphosphate carboxylase, phosphoenolpyruvate carboxylase, and malate dehydrogenase. In addition, three enzymes participating in the C4-dicarboxylic acid cycle, namely, phosphoenolpyruvate carboxylase, alanine transaminase, and malate dehydrogenase, were also found to be ubiquitinated. Collectively, these results illustrated that tobacco Kub proteins share close interactions in photosynthesis and carbon metabolism.
Figure 7.
Kub proteins involved in carbon fixation. The newly identified proteins are highlighted in red.
Moreover, carbon fixation is usually driven by products of photosynthesis, converting CO2 and other compounds to glucose. There are three biochemical pathways for carbon fixation in higher plants: the Calvin–Benson cycle (CBC), the C4-dicarboxylic acid cycle, and crassulacean acid metabolism (CAM). We found that ubiquitination occurred in 11 enzymes that were involved in carbon fixation in tobacco (Figure 7). Among these enzymes, eight were involved in the CBC, while three were relevant to the C4-dicarboxylic acid cycle and the CAM pathway, namely, phosphoenolpyruvate carboxylase (PEPC) [EC:4.1.1.31], malate dehydrogenase (MDH) [EC:1.1.1.37], and alanine transaminase (ALT) [EC:2.6.1.2]. MDH catalyzes the reduction of NAD+ to NADH48 to facilitate the oxidation of malate to oxaloacetate. This process is part of some crucial metabolic pathways, for example, the citric acid cycle. PEPC plays a key role in plant carbon metabolism.49 Many PEPC and PEPC isoforms were found to be ubiquitinated in Arabidopsis, harsh hakea, lily, and castor oil plants.16 We found that PEPC (A0A1S4DHV3, A0A1S4DJ63, and P27154) was also modified by ubiquitination in tobacco. Furthermore, ribulose bisphosphate carboxylase (rubisco), which mediates the fixation of inorganic CO2 in photosynthesis by catalyzing the carboxylation of ribulose-1,5- bisphosphate (RuBP) as the first step of the CBC,50,51 was also identified to be ubiquitinated in this study. Rubisco with accession nos. A0A140G1S3, P69249, and Q84QE5 contained 11, 2, and 1 sites, respectively (Supporting Information Table S2). Taken together, it is clearly indicated that lysine ubiquitination greatly modified the important enzymes associated with the regulation of carbon-fixation pathways in tobacco.
2.6. Protein Interaction Network Analysis
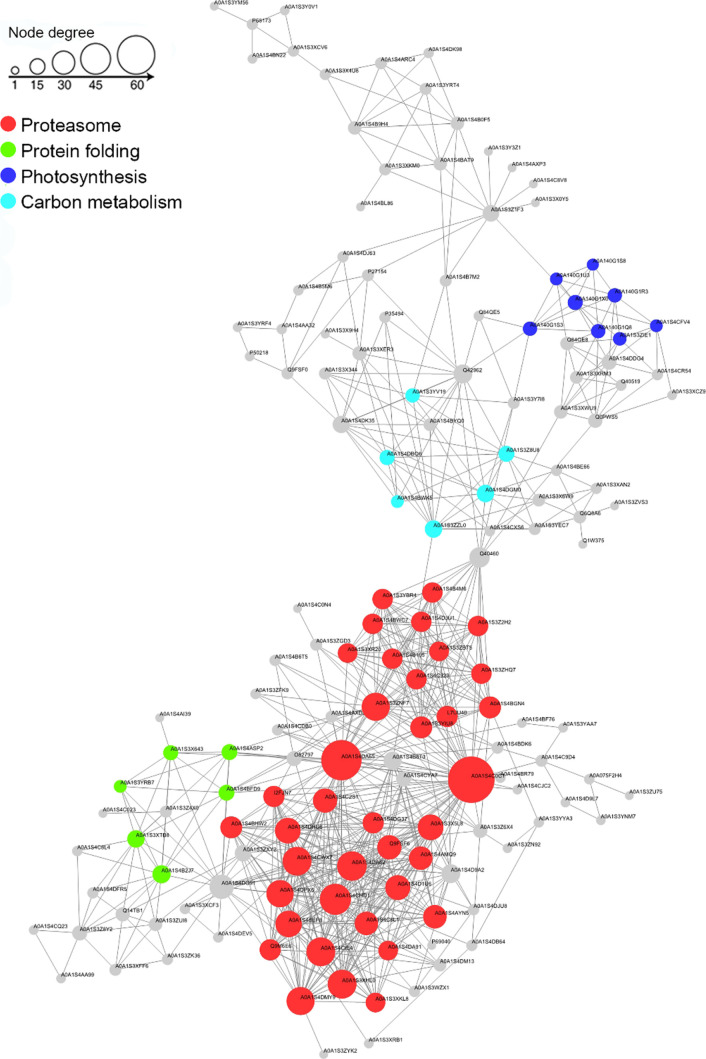
To analyze how ubiquitination regulates the cellular processes in tobacco, Cytoscape software and the STRING database were used to construct the PPI network. A total of 157 ubiquitinated proteins were mapped through the MCODE plug-in tool kit, which showed that the top four clusters were the proteasome (Cluster I), protein folding (Cluster II), photosynthesis (Cluster III), and carbon metabolism (Cluster IV) (Figure 8, Suppoting Information Table S10), presenting an overall view of how Kub proteins participate in multiple pathways in tobacco. These complex interactions among Kub proteins possibly indicate the strong coordination among these proteins in tobacco.
Figure 8.
Protein interaction network of newly identified ubiquitinated proteins in tobacco.
2.7. Detection of Kub Proteins under TMV Inoculation Treatment
The relationship between TMV infection and tobacco ubiquitination was verified by western blots. Proteins were extracted from the treated samples, which were gathered at 2, 4, 6, 8 days past inoculation (dpi). The results of the western blot were measured by Image J. The expression of TMV coat protein (cp) showed an upward trend over time and was highest at 6 dpi (Figure 9). A strong immunoblot signal was observed at 6 dpi, and no changes were evident in mock samples (Figure 10). This indicated that TMV infection can cause changes in ubiquitination levels. The increasing trend of TMV replication expression in tobacco was consistent with the trend of the ubiquitination level. Therefore, the sixth day could be the quantitative analysis time point of ubiquitination. It has been reported that the ubiquitin variant induces the development of necrotic lesions and influences plant responses to TMV infection.52 Marino53 indicated that ubiquitination has a close relationship with the regulation of plant immunity and plant defense responses to abiotic or biotic stresses. We identified ubiquitination of the abscisic acid receptor (A0A1S3X1P6), phenylalanine ammonia-lyase (C6ZIA5), and electron transport-associated proteins (A0A140G1R3, A0A140G1X0, A0A1S3XWU9, A0A1S3X0Y5, and A0A140G1S8) in Nicotiana tabacum, which were associated with plant defense responses. The western blot analysis suggested that there is a relevance between TMV infection and tobacco ubiquitination.
Figure 9.
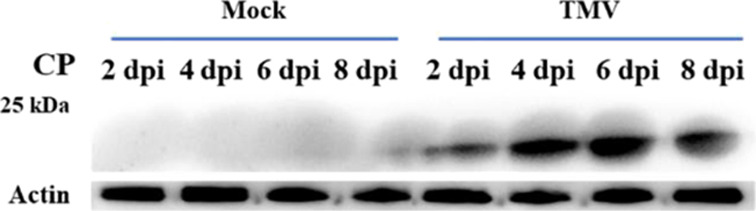
Western blot analysis with the TMV cp antibody. A β-actin antibody was used as a loading control. Tobacco leaves inoculated with PBS was used as mock. Dpi represents day past inoculation.
Figure 10.
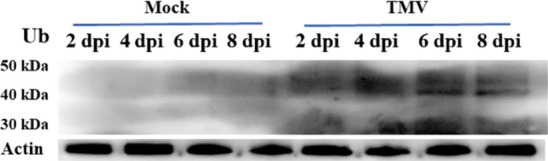
Western blot analysis with an antiubiquitin antibody. A β-actin antibody was used as a loading control. Tobacco leaves inoculated with PBS was used as mock. Dpi represents day past inoculation.
3. Conclusions
The present study presented the first comprehensive lysine ubiquitination analysis in tobacco. We identified 964 Kub sites in 572 proteins from tobacco seedling leaves using LC–MS/MS along with highly sensitive immune-affinity purification and bioinformatics tools. The characterization of ubiquitinated proteins indicates that the ubiquitome plays a vital role in diverse cellular processes, metabolic pathways and protein interaction networks. Our results reveal that many different proteins are involved in diverse cellular processes, especially photosynthesis and carbon fixation. These findings could provide a valuable reference for further research on the functions of lysine ubiquitination in tobacco and other plants under different environmental stresses or pathogenic infections. In the future, we will conduct LC–MS/MS quantitative analysis and verification experiments to clarify the relationship between ubiquitination modification and TMV resistance in tobacco.
4. Experimental Section
4.1. Sample Preparation
Tobacco plants (N. tabacum) were grown in a greenhouse with a photoperiod of 16/8 h (light/dark) at 25 °C. The leaves of three replications were collected from 4 week-old tobacco seedlings for downstream experiments. The Qin54 method was used for TMV solution preparation. Tobacco seedling leaves were inoculated with TMV solution by the rub-inoculation method. Tobacco leaves were collected at 2, 4, 6, and 8 dpi. PBS-treated tobacco seedlings were used as controls. Three biological replicates were used for assessment.
4.2. Protein Extraction
Tobacco leaves were ground in liquid nitrogen and resuspended in extraction buffer comprising 8 M urea, 1% Triton X-100, 10 mM dithiothreitol (DTT), 50 μM deubiquitinase inhibitor MPR-619, and 1% protease inhibitor cocktail. Next, the debris was removed by centrifugation at 20,000× g at 4 °C for 10 min. Finally, the proteins were precipitated using cold 20% TCA for 2 h at 4 °C. The supernatant was discarded after centrifugation at 4 °C and 12,000× g for 3 min, and the precipitate was washed thrice with cold acetone. The protein was redissolved in buffer (100 mM NH4CO3, 8 M urea, pH 8.0), and the concentration was measured by a BCA kit (Beyotime, Shanghai, China) following the manufacturer’s protocol.
4.3. Trypsin Digestion
The obtained protein concentrate was reduced using 5 mM DTT at 56 °C for 30 min and alkylated with 11 mM iodoacetamide at room temperature for 15 min in darkness. The protein sample was then diluted by adding 100 mM NH4CO3 to adjust the urea concentration to less than 2 M. Finally, trypsin (1:50 trypsin-to-protein mass ratio) was added for the first digestion at 37 °C for 12 h, while a 1:100 trypsin-to-protein mass ratio was used for the second digestion for 4 h.
4.4. Affinity Enrichment
Tryptic peptides dissolved in IP buffer (100 mM NaCl, 0.5% NP-40, 50 mM Tris–HCl, 1 mM EDTA, pH 8.0) were incubated with prewashed antibody beads (PTM Biolabs, Hangzhou, China) at 4 °C for 12 h to analyze the affinity enrichment. The beads were then washed four times with IP buffer and twice with ddH2O. The lysine-ubiquitinated peptides were eluted from the agarose beads with 0.1% trifluoroacetic acid (TFA) followed by mixing and vacuum drying.
4.5. Liquid Chromatography (LC)-Tandem Mass Spectrometry (MS/MS) Analysis
A mass spectrometer (Thermo Scientific Q Exactive Plus) was used to analyze the enriched peptides, which were cleaned using C18 Zip Tips (Millipore, Bedford, MA, USA). The peptides were loaded onto a column (Acclaim PepMap 100, 100 μm × 2 cm, nanoViper C18, Thermo Fisher Scientific Inc., Waltham, MA, USA) connected to a reversed-phase analytical column (Acclaim PepMap 100 C18, 75 μm, 150 mm, 3 μm, Thermo Fisher Scientific Inc., Waltham, MA, USA) in 0.1% formic acid (solvent A) and then isolated with a linear gradient of 0.1% formic acid and 90% acetonitrile (solvent B) at a flow rate of 350 nL/min on an EASY-nLC 1000 UPLC system (Thermo Fisher Scientific Inc). The gradient was as follows: 0–60 min, 6–24% solvent B; 60–82 min, 24–36% solvent B; 82–86 min, 36–80% solvent B; and 86–90 min, solvent B at 80%. The peptides were analyzed using MS/MS in an Orbitrap Exactive Plus (Thermo Fisher Scientific Inc) coupled with an online UPLC system. An electrospray voltage of 2.0 kV was applied. To read MS scans, the m/z scan range was set at 350 to 1550. The intact peptides were detected at a resolution of 60,000, whereas the resolution for higher-energy collisional dissociation (HCD) spectra was 17,500. The automatic gain control (AGC) target was set to 1E4, and the maximum injection time was 100 ms with a 15 s dynamic exclusion duration.
4.6. Database Analysis
The proteins with ubiquitination sites were identified using MaxQuant with an integrated Andromeda search engine (v.1.5.2.8). MS/MS data were searched against the UniProt N. tabacum database concatenated with the reverse decoy database.55 The fixed modification was carbamidomethyl on Cys, and the variable modifications were Gly-Gly modification on lysine (K) and oxidation on methionine. The maximal missed cleavage of trypsin/P was 4. For precursor ions, mass error was set as 5 ppm in the main search and 20 ppm in the first search. The mass tolerance for fragment ions was set as 0.02 Da. The minimal peptide length was 7, and maximal number of modifications per peptide was 5. The maximal number of labeled aa was 5, and the minimum score for modified peptides was set above 40. The false discovery rate (FDR) thresholds for proteins, peptides, and modification sites were adjusted below 1%.
4.7. Bioinformatics Analysis
Motif-X software (http://motif-x.med.harvard.edu/) was used to determine the model of the sequences with aa in specific positions of ubiquityl-21-mers (10 aa upstream and downstream of the ubiquitination site).16 We used all protein sequences as background parameters, and other parameters were set as default values. The “heatmap.2” function in the “gplot2” R-package was used to describe the cluster membership in the form of a heat map. Secondary structures were predicted by NetSurfP software.56 Further hierarchical clustering was performed according to categories, and the categories of the ubiquitinated substances and their p-values were compared after enrichment. Then, these categories were enriched in one of the clusters with a p-value <0.05. Gene Ontology (GO) classification was performed using the UniProt-GOA database (http://www.ebi.ac.uk/GOA/),57 which classified Kub proteins into three categories: biological processes, cellular components, and molecular functions. Protein pathways were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database58 using the online service tool KAAS, and KEGG Mapper was used to map the annotation results. GO term and KEGG pathway analyses were performed using DAVID bioinformatics resources 6.7.59 Functional descriptions of the identified domains were annotated by the InterProScan online service tool (http://www.ebi.ac.uk/interpro/beta/). Fisher’s exact test (two-tailed test) was used to detect the enrichment of the differentially expressed proteins against all ubiquitinated proteins. Any term with adjusted p-values <0.05 in any cluster was considered significant. WoLF PSORT software (http://wolfpsort.org/) was used to predict subcellular localization.60 Protein–protein interaction (PPI) information for the identified proteins was acquired from STRING software (http://string-db.org/). The PPI network map was created using Cytoscape (version 3.7.0) software (http://www.cytoscape.org/),61 with molecular complex detection (MCODE) analyzing densely connected regions.62
4.8. Western Blot Analysis
To examine the TMV coat protein (cp) and tobacco ubiquitination levels, proteins were extracted from tobacco seedlings that had been treated with PBS and TMV solution. Samples were collected at different time points. We performed the western blot analysis using a commercially available antibody against ubiquitination as previously described.32,63 Proteins were separated by SDS-PAGE and transferred to a PVDF (Immobilon-P, Merck Millipore, United States) membrane. The TMV cp antibody (Youlong, Shanghai, China), anti-rabbit secondary antibody (CWBIO, Beijing, China), and β-actin (CWBIO, Beijing, China) antibody were used for this assay. Ubiquitination levels were then detected using antiubiquitin as a primary antibody (PTM Biolabs, Hangzhou, China) and anti-rabbit secondary antibody conjugated to HRP (CWBIO, Beijing, China). The results of the western blot were analyzed by Image J (Version 1.52a).
Acknowledgments
This work was supported by the State Tobacco Monopoly Administration major special projects (110201901041 [LS-04], 110101601024 [LS-04]), Sichuan Tobacco Company projects (SCYC201804), and Hunan Tobacco Company projects (201743010020088).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c01741.
Identifed lysine-ubiquitinated protein information for three replicates; identifed lysine-ubiquitinated sites in tobacco; motif retrieved from lysine-ubiquitinated peptides; frequency of the different types of amino acids around the ubiquitinated lysines; GO annotation of the ubiquitinated proteins in tobacco; subcellular location of the ubiquitinated proteins in tobacco; GO-based enrichment analysis of ubiquitinated proteins in terms of cellular component, molecular function, and biological process; KEGG pathway analysis of the ubiquitinated proteins; protein domain enrichment analysis of ubiquitinated proteins; and information on protein interaction networks (XLSX)
Author Contributions
# H. Zhan., L. Song., and A. Kamran are co-first authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Sun H.; Liu X.; Li F.; Li W.; Zhang J.; Xiao Z.; Shen L.; Li Y.; Wang F.; Yang J. First comprehensive proteome analysis of lysine crotonylation in seedling leaves of Nicotiana tabacum. Sci. Rep. 2017, 7, 3013. 10.1038/s41598-017-03369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Guo L.; Liang W.; Chi Z.; Liu L. Systematic analysis of the lysine acetylome reveals diverse functions of lysine acetylation in the oleaginous yeast Yarrowia lipolytica. AMB Express. 2017, 7, 94. 10.1186/s13568-017-0393-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S. J.; Choi E. Y.; Choi Y. J.; Ahn J. H.; Park O. K. Proteomics studies of post-translational modifications in plants. J. Exp. Bot. 2006, 57, 1547–1551. 10.1093/jxb/erj137. [DOI] [PubMed] [Google Scholar]
- Joo H. Y.; Zhai L.; Yang C.; Nie S.; Erdjument-Bromage H.; Tempst P.; Chang C.; Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature 2007, 449, 1068–1072. 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- Glickman M. H.; Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Zhang N.; Zhang L.; Shi C.; Tian Q.; Lv G.; Wang Y.; Cui D.; Chen F. Comprehensive profiling of lysine ubiquitome reveals diverse functions of lysine ubiquitination in common wheat. Sci. Rep. 2017, 7, 13601. 10.1038/s41598-017-13992-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A.; Novatchkova M.; Potuschak T.; Eisenhaber F. Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends Plant Sci. 2001, 6, 463–470. 10.1016/S1360-1385(01)02080-5. [DOI] [PubMed] [Google Scholar]
- Sadanandom A.; Bailey M.; Ewan R.; Lee J.; Nelis S. The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol. 2012, 196, 13–28. 10.1111/j.1469-8137.2012.04266.x. [DOI] [PubMed] [Google Scholar]
- Dreher K.; Callis J. Ubiquitin, hormones and biotic stress in plants. Ann. Bot. 2007, 99, 787–822. 10.1093/aob/mcl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M.; Eddins M. J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 2004, 1695, 55–72. 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Deshaies R. J.; Joazeiro C. A. P. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Polo S.; Sigismund S.; Faretta M.; Guidi M.; Capua M. R.; Bossi G.; Chen H.; De Camilli P.; Di Fiore P. P. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 2002, 416, 451. 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- Yates G.; Sadanandom A. Ubiquitination in plant nutrient utilization. Front Plant Sci. 2013, 4, 452. 10.3389/fpls.2013.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X.; Chen Z. J. The role of ubiquitylation in immune defence and pathogen evasion. Nat. Rev. Immunol. 2011, 12, 35–48. 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchman R. L.; Gordon C.; Mayer R. J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 2005, 6, 599. 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Xie X.; Kang H.; Liu W.; Wang G. L. Comprehensive profiling of the rice ubiquitome reveals the significance of lysine ubiquitination in young leaves. J. Proteome Res. 2015, 14, 2017–2025. 10.1021/pr5009724. [DOI] [PubMed] [Google Scholar]
- Xu G.; Jaffrey S. R. Proteomic identification of protein ubiquitination events. Biotechnol. Genet. Eng. Rev. 2013, 29, 73–109. 10.1080/02648725.2013.801232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P.; Huang L. Global approaches to understanding ubiquitination. Genome Biol. 2005, 6, 233. 10.1186/gb-2005-6-10-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J.; Schwartz D.; Elias J. E.; Thoreen C. C.; Cheng D.; Marsischky G.; Roelofs J.; Finley D.; Gygi S. P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003, 21, 921–926. 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Simonetti F.; Candelli T.; Leon S.; Libri D.; Rougemaille M. Ubiquitination-dependent control of sexual differentiation in fission yeast. eLife. 2017, 6, e28046 10.7554/eLife.28046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wal L.; Bezstarosti K.; Sap K. A.; Dekkers D. H. W.; Rijkers E.; Mientjes E.; Elgersma Y.; Demmers J. A. A. Improvement of ubiquitylation site detection by Orbitrap mass spectrometry. J. Proteomics 2018, 172, 49–56. 10.1016/j.jprot.2017.10.014. [DOI] [PubMed] [Google Scholar]
- Schwertman P.; Bezstarosti K.; Laffeber C.; Vermeulen W.; Demmers J. A.; Marteijn J. A. An immunoaffinity purification method for the proteomic analysis of ubiquitinated protein complexes. Anal. Biochem. 2013, 440, 227–236. 10.1016/j.ab.2013.05.020. [DOI] [PubMed] [Google Scholar]
- Danielsen J. M.; Sylvestersen K. B.; Bekker-Jensen S.; Szklarczyk D.; Poulsen J. W.; Horn H.; Jensen L. J.; Mailand N.; Nielsen M. L. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics 2011, 10, M110.003590. 10.1074/mcp.M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai S.-S.; Liu G.-S.; Sun Y.-H.; Chen J. Cloning and Expression of Calcium-Dependent Protein Kinase (CDPK) Gene Family in Common Tobacco (Nicotiana tabacum). Agric. Sci. China. 2009, 8, 1448–1457. 10.1016/S1671-2927(08)60358-2. [DOI] [Google Scholar]
- Zhang C.; Song L.; Choudhary M. K.; Zhou B.; Sun G.; Broderick K.; Giesler L.; Zeng L. Genome-wide analysis of genes encoding core components of the ubiquitin system in soybean (Glycine max) reveals a potential role for ubiquitination in host immunity against soybean cyst nematode. BMC Plant Biol. 2018, 18, 149. 10.1186/s12870-018-1365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Y.; Scalf M.; Smith L. M.; Vierstra R. D. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 2013, 25, 1523–1540. 10.1105/tpc.112.108613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.; Lv Y.; Mujahid H.; Edelmann M. J.; Zhao H.; Peng X.; Peng Z. Proteome-wide lysine acetylation identification in developing rice (Oryza sativa) seeds and protein co-modification by acetylation, succinylation, ubiquitination, and phosphorylation. Biochim. Biophys. Acta. Proteins Proteomics. 2018, 1866, 451–463. 10.1016/j.bbapap.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Cao Y.; Meng D.; Chen Y.; Abdullah M.; Jin Q.; Lin Y.; Cai Y. Comparative and Expression Analysis of Ubiquitin Conjugating Domain-Containing Genes in Two Pyrus Species. Cell 2018, 7, 77. 10.3390/cells7070077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.; Liu J.; Wei Q.; Wang R.; Yang W.; Ma Y.; Chen G.; Yu Y. Proteomes and Ubiquitylomes Analysis Reveals the Involvement of Ubiquitination in Protein Degradation in Petunias. Plant Physiol. 2017, 173, 668–687. 10.1104/pp.16.00795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gest H. History of the word photosynthesis and evolution of its definition. Photosynth. Res. 2002, 73, 7–10. 10.1023/A:1020419417954. [DOI] [PubMed] [Google Scholar]
- Vu L. D.; Gevaert K.; De Smet I. Protein Language: Post-Translational Modifications Talking to Each Other. Trends Plant Sci. 2018, 23, 1068–1080. 10.1016/j.tplants.2018.09.004. [DOI] [PubMed] [Google Scholar]
- Chen X. L.; Xie X.; Wu L.; Liu C.; Zeng L.; Zhou X.; Luo F.; Wang G. L.; Liu W. Proteomic Analysis of Ubiquitinated Proteins in Rice (Oryza sativa) After Treatment With Pathogen-Associated Molecular Pattern (PAMP) Elicitors. Front. Plant Sci. 2018, 9, 1064. 10.3389/fpls.2018.01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulo P. Chloroplast-targeted ferredoxin-NADP(+) oxidoreductase (FNR): structure, function and location. Biochim. Biophys. Acta 2011, 1807, 927–934. 10.1016/j.bbabio.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Batie C. J.; Kamin H. Electron transfer by ferredoxin:NADP+ reductase. Rapid-reaction evidence for participation of a ternary complex. J. Biol. Chem. 1984, 259, 11976–11985. [PubMed] [Google Scholar]
- Grotjohann I.; Fromme P. Structure of cyanobacterial photosystem I. Photosynth. Res. 2005, 85, 51–72. 10.1007/s11120-005-1440-4. [DOI] [PubMed] [Google Scholar]
- Bricker T. M.; Frankel L. K. The structure and function of CP47 and CP43 in Photosystem II. Photosynth. Res. 2002, 72, 131–146. 10.1023/A:1016128715865. [DOI] [PubMed] [Google Scholar]
- Campbell A. D.Photosynthesis. In New Studies in Biology Series, fourth ed; Hall D. O.; Rao K. K. Ed. ; Cambridge University Press: Cambridge, 1992; pp. 122. [Google Scholar]
- Cramer W.; Yamashita E.; Baniulis D.; Hasan S. S., Cytochrome b 6 f Complex. In Encyclopedia of Biophysics, first ed.; Springer: Berlin Heidelberg, 2013; pp. 417–422. [Google Scholar]
- Rowland J. G.; Simon W. J.; Nishiyama Y.; Slabas A. R. Differential proteomic analysis using iTRAQ reveals changes in thylakoids associated with Photosystem II-acquired thermotolerance in Synechocystis sp. PCC 6803. Proteomics 2010, 10, 1917–1929. 10.1002/pmic.200900337. [DOI] [PubMed] [Google Scholar]
- Moolna A.; Bowsher C. G. The physiological importance of photosynthetic ferredoxin NADP+ oxidoreductase (FNR) isoforms in wheat. J. Exp. Bot. 2010, 61, 2669–2681. 10.1093/jxb/erq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q.; Armbrust T.; Guikema J.; Chitnis P. R. Organization of Photosystem I Polypeptides (A Structural Interaction between the PsaD and PsaL Subunits). Plant Physiol. 1994, 106, 1057–1063. 10.1104/pp.106.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L. B.; Maderia M.; Ouellette A. J. A.; Putnam-Evans C.; Higgins L.; Krick T.; MacCoss M. J.; Lim H.; Yates J. R.; Barry B. A. Posttranslational modifications in the CP43 subunit of photosystem II. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 14676. 10.1073/pnas.232591599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifuku K.; Ishihara S.; Shimamoto R.; Ido K.; Sato F. Structure, function, and evolution of the PsbP protein family in higher plants. Photosynth. Res. 2008, 98, 427–437. 10.1007/s11120-008-9359-1. [DOI] [PubMed] [Google Scholar]
- Mittler R.; Vanderauwera S.; Gollery M.; Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Apel K.; Hirt H. REACTIVE OXYGEN SPECIES: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Chamnogpol S.; Willeekens H.; Moeder W.; van Camp W. Defense activation and enhanced pathogen tolerance induced by H2O2 in transgenic tobacco. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 5818–5823. 10.1073/pnas.95.10.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T.; Maekawa S.; Yasuda S.; Yamaguchi J. Carbon and nitrogen metabolism regulated by the ubiquitin-proteasome system. Plant Signal. Behav. 2011, 6, 1465–1468. 10.4161/psb.6.10.17343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. M. Moonlighting NAD(+) Malate Dehydrogenase Is Essential for Chloroplast Biogenesis. Plant Cell 2018, 30, 1663–1664. 10.1105/tpc.18.00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet L.; Zaffagnini M.; Morisse S.; Sparla F.; Perez-Perez M. E.; Francia F.; Danon A.; Marchand C. H.; Fermani S.; Trost P.; Lemaire S. D. Redox regulation of the Calvin-Benson cycle: something old, something new. Front. Plant Sci. 2013, 4, 470. 10.3389/fpls.2013.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Cajar O.; Stotz M.; Bracher A. Maintaining photosynthetic CO2 fixation via protein remodelling: the Rubisco activases. Photosynth. Res. 2014, 119, 191–201. 10.1007/s11120-013-9819-0. [DOI] [PubMed] [Google Scholar]
- Pichersky E.; Bernatzky R.; Tanksley S.; Cashmore A. R. Evidence for Selection as a Mechanism in the Concerted Evolution of Lycopersicon esculentum (Tomato) Genes Encoding the Small Subunit of Ribulose1,5-bisphosphate Carboxylase/Oxygenase. Proc. Natl. Acad. Sci. U. S. A. 1986, 83, 3880–3884. 10.1073/pnas.83.11.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker F.; Buschfeld E.; Schell J.; Bachmair A. Altered response to viral infection by tobacco plants perturbed in ubiquitin system. Plant J. 2005, 3, 875–881. 10.1111/j.1365-313X.1993.00875.x. [DOI] [Google Scholar]
- Marino D.; Peeters N.; Rivas S. Ubiquitination during Plant Immune Signaling. Plant Physiol. 2012, 160, 15. 10.1104/pp.112.199281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y.; Wang J.; Wang F.; Shen L.; Zhou H.; Sun H.; Hao K.; Song L.; Zhou Z.; Zhang C.; Wu Y.; Yang J. Purification and Characterization of a Secretory Alkaline Metalloprotease with Highly Potent Antiviral Activity from Serratia marcescens Strain S3. J. Agric. Food Chem. 2019, 67, 3168–3178. 10.1021/acs.jafc.8b06909. [DOI] [PubMed] [Google Scholar]
- Xu X.; Liu T.; Yang J.; Chen L.; Liu B.; Wang L.; Jin Q. The First Whole-Cell Proteome- and Lysine-Acetylome-Based Comparison between Trichophyton rubrum Conidial and Mycelial Stages. J. Proteome Res. 2018, 17, 1436–1451. 10.1021/acs.jproteome.7b00793. [DOI] [PubMed] [Google Scholar]
- Petersen B.; Petersen T. N.; Andersen P.; Nielsen M.; Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 2009, 9, 51. 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell D.; Dimmer E.; Huntley R. P.; Binns D.; O’Donovan C.; Apweiler R. The GOA database in 2009--an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009, 37, D396–D403. 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M.; Furumichi M.; Tanabe M.; Sato Y.; Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X.; Sherman B. T.; da Huang W.; Stephens R.; Baseler M. W.; Lane H. C.; Lempicki R. A. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012, 28, 1805–1806. 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P.; Park K. J.; Obayashi T.; Fujita N.; Harada H.; Adams-Collier C. J.; Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T.; Copley R. R.; Schultz J.; Ponting C. P.; Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002, 12, 47–56. 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G. D.; Hogue C. W. V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003, 4, 2. 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.-Q.; Chen Z.-R.; Chen D.-Z.; Hao X.-J.; Li S.-L. Anti-TMV Effects of Amaryllidaceae Alkaloids Isolated from the Bulbs of Lycoris radiata and Lycoricidine Derivatives. Nat. Prod. Bioprospect. 2018, 8, 189–197. 10.1007/s13659-018-0163-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.