Co-delivery of IKBKE siRNA and cabazitaxel inhibits invasiveness and growth of triple-negative breast cancer.
Abstract
IKBKE is an oncogene in triple-negative breast cancer (TNBC), and we demonstrate that IKBKE small interfering RNA (siRNA) inhibits the proliferation, migration, and invasion of TNBC cells. Despite the recent success of siRNA therapeutics targeting to the liver, there still remains a great challenge to deliver siRNAs to solid tumors. Here, we report a hybrid nanocomplex to co-deliver the IKBKE siRNA and cabazitaxel to TNBC to achieve an optimal antitumor effect. The nanocomplex is modified with hyaluronic acid to target CD44 on TNBC cells. The nanocomplex shows higher cellular uptake and better tumor penetration of the encapsulated cargos. The nanocomplex also exhibits high tumor accumulation and antitumor activity in an orthotopic TNBC mouse model. Encapsulation of cabazitaxel in the nanocomplex enhances the activity of the IKBKE siRNA. The hybrid nanocomplex provides a novel and versatile platform for combination therapies using siRNAs and chemotherapy.
INTRODUCTION
Triple-negative breast cancer (TNBC), defined as lacking the expression of estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 (HER2), accounts for approximately 12 to 17% of all breast cancers. TNBC is more aggressive, proliferative, and has poorer prognoses and survival rates than non-TNBC (1). Nuclear factor κB (NF-κB) is a key regulator in TNBC. In normal cells, NF-κB is bound to the inhibitor of κB (IκB) and stays in the cytoplasm. In cancer cells, the inhibitory κB kinases (IKKs) are activated to phosphorylate the IκBs and induce their degradation, leading to the translocation of NF-κB to the nucleus. The activated NF-κB signaling pathways enhance cellular proliferation, invasion, and metastasis and decrease cellular apoptosis in breast cancer, especially in ER-negative breast cancers such as TNBC (2).
The IKK family contains five members including IKKα, IKKβ, IKK𝛾, TBK-1 (TANK-binding kinase 1), and IKKε (3). IKBKE (also known as IKKε) was originally identified as an oncogene in breast cancer and is overexpressed in approximately 30% of breast carcinomas (4). IKBKE is also overexpressed in a variety of human cancers and plays important roles in migration, invasion, malignant transformation, poor prognosis, and chemoresistance of cancer cells (5, 6). For example, IKBKE was found to be overexpressed in human glioma cells, and down-regulation of IKBKE reduced invasion and migration of the cells (6). Recently, there is growing evidence showing the important role of IKBKE in TNBC (7). IKBKE overexpression is associated with signal transducer and activator of transcription 3 (STAT3) activation and contributes to the proliferation and survival of TNBC (7). Meanwhile, IKBKE induces the production of the cytokines chemokine (C-C motif) ligand 5 (CCL5) and interleukin-6 (IL-6), which stimulates the migration of TNBC cells and endothelial cells through autocrine and paracrine signaling processes (7). Moreover, activation of NF-κB is linked to chemoresistance of breast cancer cells. In one study, nuclear staining of NF-κB was characterized in tumor specimen from patients with breast cancer who received anthracycline- and/or taxane-based chemotherapy. Patients with negative nuclear staining of NF-κB in pretreatment specimen had a 91% response rate to chemotherapy, but patients with positive NF-κB staining only showed a 20% response rate (8). Similarly, down-regulation of NF-κB with sulforaphane sensitizes breast cancer cells to paclitaxel-induced apoptosis (9). Therefore, down-regulation of IKBKE inhibits the nuclear translocation of NF-κB and subsequently sensitizes breast cancer cells to chemotherapy. Several small molecular inhibitors have been developed to suppress IKBKE. However, none of them is specific for IKBKE, which may lead to unexpected potential side effects (5, 10, 11). In our previous report, we found IKBKE small interfering RNAs (siRNAs) and demonstrated their inhibitory effect on the proliferation and invasion of HER2-positive breast cancer MCF7 and SK-BR-3 cells in vitro. However, the siRNAs have negligible effect on the apoptosis of the tumor cells (3).
Taxanes, including paclitaxel, docetaxel, and cabazitaxel, are a group of microtubule stabilizers that induce cellular apoptosis of cancer cells. Taxanes are more effective in patients with TNBC than in non-TNBC patients (12, 13). Cabazitaxel is a novel second-generation semisynthetic taxane that was approved by the U.S. Food and Drug Administration (FDA) for the treatment of hormone-refractory prostate cancer (14). As a microtubule stabilizer, cabazitaxel has similar activity as docetaxel but is more potent against taxane-resistant tumors because of its poor affinity to P-glycoprotein, which is responsible for taxane resistance (14, 15). As a result, cabazitaxel provides a promising opportunity for the treatment of metastatic prostate cancer and breast cancer (16, 17). However, cabazitaxel has high systemic toxicity, which limits its applications for TNBC treatment. Innovative delivery strategies have been developed to improve cabazitaxel’s antitumor efficacy with alleviated toxicity in metastatic TNBC (17, 18).
Because of the heterogeneity and complexity of cancers, combinational therapy has become the mainstay in cancer therapy. We hypothesize that co-delivery of the IKBKE siRNA and cabazitaxel to TNBC cells will achieve optimal therapeutic outcomes because they target different pathways in tumorigenesis. We have previously reported a cholesterol-modified peptide, which can self-assemble into a micelle-like structure with a high siRNA condensation capability (19). In this study, cabazitaxel was encapsulated into the cholesterol-peptide micelle without affecting its condensation with the IKBKE siRNA to form a hybrid siRNA nanocomplex. The nanocomplex was modified with hyaluronic acid (HA) to target CD44, which is overexpressed on TNBC cells. To improve the release of the IKBKE siRNA and cabazitaxel in tumor cells, a cathepsin B–liable dipeptide sequence (Val-Cit) was inserted between cholesterol and the peptide (20). Cathepsin B is a type of lysosomal cysteine protease that localizes in the lysosomes of cancer cells including TNBC (21). The Val-Cit dipeptide linker has been widely used in antibody-drug conjugates and targeted drug delivery systems to release active agents inside tumor cells (20, 22). The hybrid siRNA nanocomplex represents a promising strategy to co-deliver cabazitaxel and IKBKE siRNA for TNBC therapy.
RESULTS
Antitumor activities of the IKBKE siRNA in TNBC cells
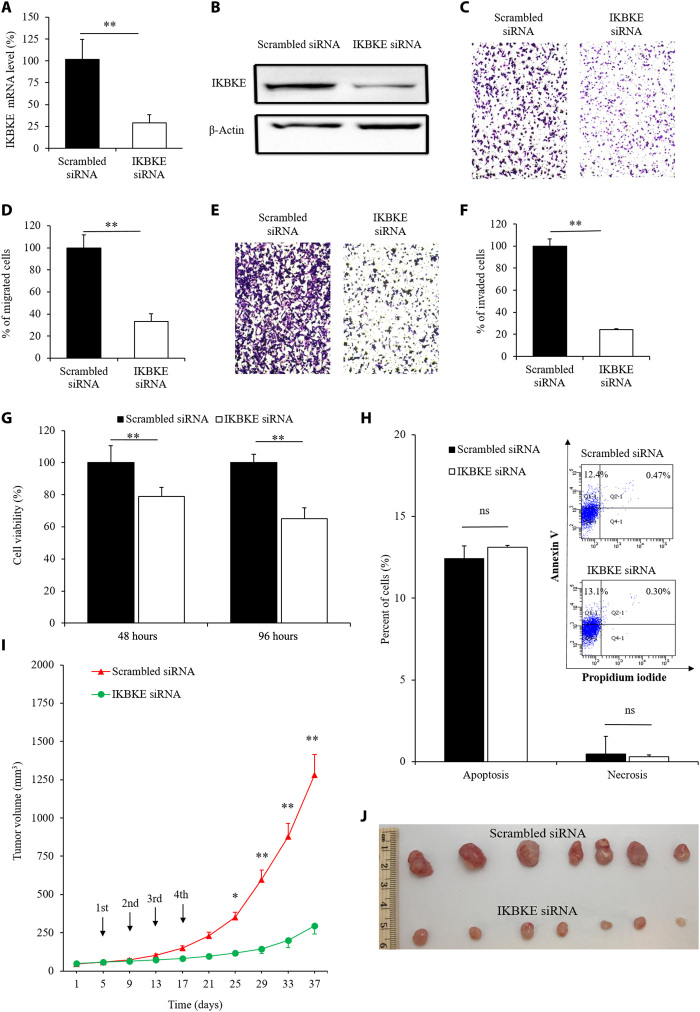
We previously designed an IKBKE siRNA that efficiently silences the expression of IKBKE in HER2-positive breast cancer cells MCF7 and SK-BR-3 (3). Here, we further explored whether the IKBKE siRNA has similar antitumor activities in TNBC MDA-MB-231 cells. As shown in Fig. 1A, the IKBKE siRNA silenced 71% of the expression of IKBKE in MDA-MB-231 cells 24 hours after transfection, and the silencing effect was confirmed by a Western blot assay 48 hours after transfection (Fig. 1B). We next evaluated whether the siRNA inhibits the migration and invasion of TNBC cells. Compared with the scrambled siRNA, only 33% of MDA-MB-231 cells treated with the IKBKE siRNA migrated from fetal bovine serum (FBS)–free medium to 10% FBS-containing medium in 6 hours (Fig. 1, C and D). Similar trends were observed in the invasion assay, as the invasion of the cells treated with the IKBKE siRNA was inhibited by approximately 76% compared with the control group (Fig. 1, E and F).
Fig. 1. Biological activities of the IKBKE siRNA in MDA-MB-231 cells.
(A) Silencing effect of the IKBKE siRNA at the mRNA level. (B) Silencing activity of the IKBKE siRNA at the protein level. Inhibitory effect of the IKBKE siRNA on the migration (C and D), invasion (E and F), and proliferation (G) of MDA-MB-231 cells. Cell migration and invasion were evaluated at 6 and 24 hours after transfection, respectively. Quantitative analysis was performed by counting the number of migrated cells in six random microscope fields from three independent samples. Cell proliferation was determined at 48 and 96 hours after transfection using the CellTiter-Glo Luminescent Cell Viability Assay Kit. All results are presented as the means ± SD (n = 3). (H) Effect of the IKBKE siRNA on the apoptosis and necrosis of MDA-MB-231 cells. All results are presented as the means ± SD (n = 3). (I and J) In vivo antitumor efficacy of the IKBKE siRNA in a subcutaneous xenograft TNBC mouse model. The IKBKE siRNA was peritumorally injected at a dose of 0.3 mg/kg. All results are presented as the means ± SEM (n = 7). *P < 0.05 and **P < 0.01. ns, not significant. Photo credit: Z.Z., School of Pharmacy, University of Missouri-Kansas City.
A CellTiter-Glo assay was performed to measure the inhibitory effect of the IKBKE siRNA on the proliferation of TNBC cells. As shown in Fig. 1G, 48 and 96 hours after transfection, the IKBKE siRNA significantly inhibited the proliferation of MDA-MB-231 cells by 21 and 35%, respectively. We also assessed the apoptosis of TNBC cells treated with the IKBKE siRNA. As illustrated in Fig. 1H, the IKBKE siRNA induced the apoptosis of 13.1% of MDA-MB-231 cells, while the scrambled siRNA induced the apoptosis of 12.4% of cells, suggesting that knockdown of the IKBKE gene alone in TNBC cells may not induce apoptosis, which is consistent with our previous finding in HER2-positive breast cancer cells (3).
Next, we studied the antitumor effect of the IKBKE siRNA in an MDA-MB-231 subcutaneous xenograft tumor model. The IKBKE siRNA was condensed into nanocomplexes using a cholesterol-modified peptide (cholesterol-HHHKKHHHKK), which was previously developed by our group and exhibited high transfection efficiency and low cytotoxicity in various cancer cell lines (19). The siRNA nanocomplex was injected peritumorally at a dose of 0.3 mg siRNA/kg for four injections. As illustrated in Fig. 1 (I and J), tumor growth was significantly inhibited in the IKBKE siRNA–treated group, as compared with the scrambled siRNA group, indicating that the IKBKE siRNA efficiently inhibits TNBC tumor growth in vivo after localized delivery to the tumor microenvironment. This is in accordance with our previous finding that silencing the expression of IKBKE inhibits the proliferation of breast cancer cells by blocking cell cycle progression at the G0/G1 phase (3). A recent study also reported that IKBKE-associated cytokine signaling promotes tumorigenicity of TNBC. Inhibition of the signaling restrains the growth of TNBC cells and patient-derived xenografts (7). In summary, the proof-of-concept studies in the xenograft mouse model (Fig. 1) clearly demonstrated that targeting IKBKE with siRNA is a potential therapeutic strategy for TNBC.
Fabrication and characterization of the HA-modified siRNA nanocomplex
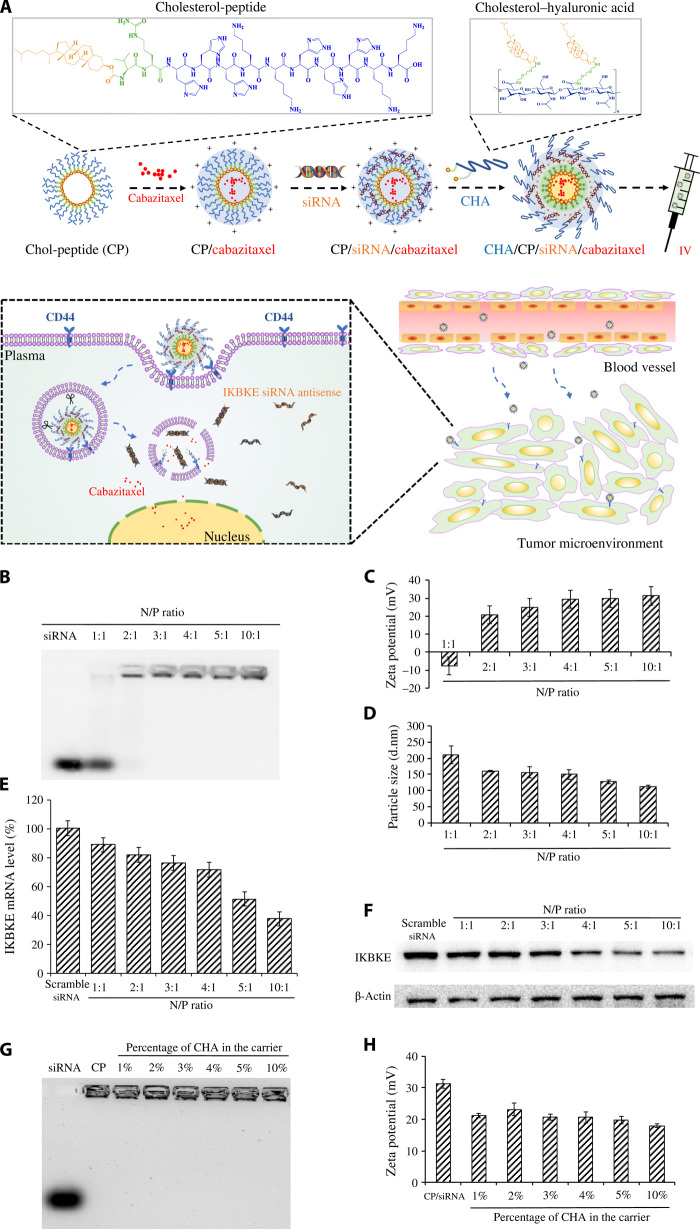
Because of the limited applications of localized drug delivery in cancer therapy, we aimed to develop a targeted siRNA nanocomplex for systemic administration of the siRNA. We previously conjugated cholesterol to a small peptide (HHHKKHHHKK) to increase the charge density of lysine by forming a micellar structure, thus avoiding the use of a long chain of positively charged amino acids, which is highly toxic in the body (19). Here, we improved the platform to achieve active targeting to TNBC cells. As illustrated in Fig. 2A, cholesterol-peptide and cholesteryl-HA were mixed to form positively charged micelles, which condense siRNAs to form nanocomplexes. HA was coated to the surface of the nanocomplex to target CD44, which is overexpressed on TNBC cells. To improve the release of siRNA from nanocomplex in tumor cells, the cathepsin B–labile dipeptide (Val-Cit) was introduced into the cholesterol-peptide. Synthetic schemes of the cholesterol-peptide and cholesteryl-HA are illustrated in fig. S1.
Fig. 2. Schematics, characterization, and silencing activity of the HA-modified siRNA nanocomplex.
(A) Cholesterol-peptide (CP) is self-assembled into a micelle-like structure, followed by loading with cabazitaxel, condensing with the IKBKE siRNA, and coating with cholesteryl-HA (CHA) to form the hybrid siRNA nanocomplex. The nanocomplex will accumulate in TNBC cells by recognizing CD44 on the tumor cells. The nanocomplex will dissociate inside the tumor cells to release the IKBKE siRNA and cabazitaxel to exert antitumor activities. IV, intravenous. (B) Gel retardation assay of the siRNA nanocomplexes at different N/P ratios. Zeta potential (C), particle size (D), silencing activity at the mRNA level (E), and silencing activity at the protein level (F) of the siRNA nanocomplexes at different N/P ratios. Gel retardation assay (G) and zeta potential (H) of the HA-modified siRNA nanocomplexes at different percentages of cholesteryl-HA (CHA) in the carrier. All results are presented as means ± SD (n = 3).
The modified cholesterol-peptide was used to condense the siRNA at different N/P ratios and examined on a 2% agarose gel. As shown in Fig. 2B, the cholesterol-peptide completely condensed the siRNA at an N/P ratio of 2:1, and this result was confirmed by the zeta potential result in Fig. 2C. The surface charge of the siRNA nanocomplexes changed from negative (N/P ratio of 1:1) to positive (N/P ratio of 2:1) and increased as the N/P ratio increased. In Fig. 2D, the particle size decreased as the N/P ratio increased, indicating an increase in the interaction between siRNA and cholesterol-peptide as the N/P ratio increased. We next coated the complex with cholesterol-modified HA to target CD44, which is a polymorphic transmembrane glycoprotein and highly expressed on several types of cancers including TNBC (23). HA is a natural, biodegradable, nontoxic polysaccharide that specifically binds to the extracellular domain of CD44. HA has been used as a tumor-targeting ligand for numerous TNBC therapeutics (24).
Because HA has a slightly negative charge, it can be coated on the surface of the positively charged siRNA nanocomplex. Conjugation of cholesterol to HA further enhances the condensation of HA into the siRNA nanocomplex. We performed a gel retardation assay to determine whether the negatively charged HA displaces siRNA from the nanocomplex. As illustrated in Fig. 2G, no free siRNA was detected when the percentage of the cholesterol-HA in the carrier increased from 1 to 10%. The zeta potential of the siRNA nanocomplexes was reduced as the amount of cholesterol-HA increased (Fig. 2H).
Silencing activity of the HA-modified siRNA nanocomplex
Real-time polymerase chain reaction (PCR) was performed to evaluate the silencing activity of the siRNA nanocomplexes at different N/P ratios 24 hours after transfection. As illustrated in Fig. 2E, the expression of the IKBKE gene in MDA-MB-231 cells was reduced as the N/P ratio increased. Similar silencing effects at the protein level were observed in the Western blot (Fig. 2F). After endocytosis, encapsulated siRNAs have to be released from the nanocomplex to exert their silencing activity. We, therefore, investigated whether incorporation of the cathepsin B–labile dipeptide (Val-Cit) in the cholesterol-peptide improves the silencing activity of the IKBKE siRNA. As shown in fig. S2, the siRNA nanocomplex prepared from the cholesterol-peptide containing Val-Cit exhibited higher silencing activity compared with the siRNA nanocomplex made from the cholesterol-peptide without Val-Cit, suggesting the importance of releasing encapsulated siRNA in the cells.
Cellular uptake of the HA-modified siRNA nanocomplex
Flow cytometry was used to study the cellular uptake of the nanocomplexes at different time intervals. Compared with free siRNA, both unmodified and HA-modified siRNA nanocomplexes exhibited a higher cellular uptake (Fig. 3, A and B). The fluorescence intensity of the HA-modified siRNA nanocomplex was 3.06-, 2.80-, and 2.18-fold higher than that of the unmodified siRNA nanocomplex after 2, 4, and 6 hours of incubation, respectively, indicating that HA significantly enhanced the cellular uptake of the siRNA nanocomplex (Fig. 3C). Meanwhile, the HA-mediated increase in fluorescence intensity was reduced from 3.06-fold at 4 hours to 2.18-fold at 6 hours, potentially due to the nonspecific accumulation of the nanocomplex in the cells after the long incubation period. Cellular uptake of the HA-modified siRNA nanocomplex was confirmed with confocal microscopy after 2 and 6 hours of incubation. As illustrated in Fig. 3 (D and E), the cellular uptake of the HA-modified siRNA was higher than that of the unmodified siRNA 2 and 6 hours after transfection. By contrast, preincubation of the cells with free HA decreased the cellular uptake of the HA-modified siRNA nanocomplex, indicating the specific targeting effect of HA in the cellular uptake of the siRNA nanocomplex. Moreover, Pearson’s correlation coefficient (PCC) was calculated using ImageJ to study the colocalization of Cy5-siRNA and lysosomes. In general, PCC values between 0 and 0.5 indicate a low colocalization of siRNA and lysosomes, which suggests efficient endosomal release of the siRNA (25). As shown in fig. S6, both HA-modified and unmodified siRNA nanocomplexes have low PCC values (<0.5) after 2 and 6 hours of transfection, suggesting that the cholesterol-peptide–based siRNA delivery system can facilitate the endosomal escape of siRNA.
Fig. 3. Cellular uptake of the HA-modified siRNA nanocomplex.
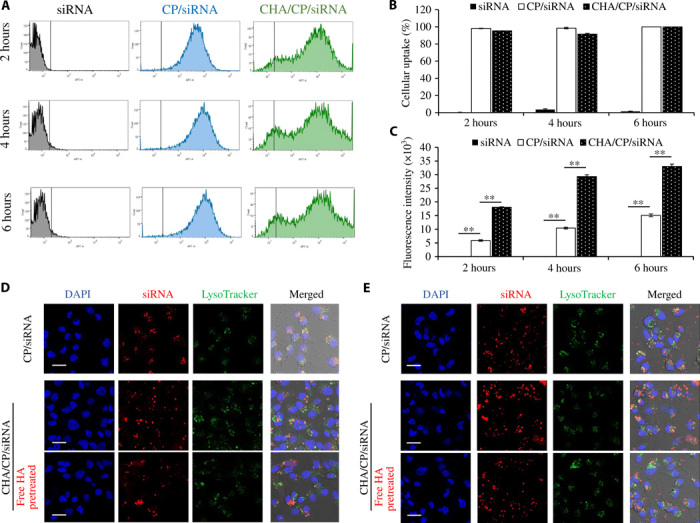
The IKBKE siRNA was labeled with Cy5 for fluorescence analysis using flow cytometry and confocal microscopy. (A) Cellular uptake of free siRNA, CP/siRNA nanocomplex, and CHA/CP/siRNA nanocomplex after 2, 4, and 6 hours of transfection. (B) Percent of the labeled cells. (C) Fluorescence intensity of the labeled cells. Representative confocal images of the cells treated with the siRNA nanocomplexes after 2 hours (D) and 6 hours (E) of transfection. HA (5 mg/ml) was used to pretreat the cells to block the cellular uptake of the nanocomplexes. **P < 0.01. Scale bars, 20 μm. CP, cholesterol-peptide; CHA, cholesteryl-HA.
Preparation of and characterization of cabazitaxel-loaded siRNA nanocomplexes
Because the IKBKE siRNA mainly suppresses the migration and proliferation of TNBC cells but does not induce apoptosis of TNBC cells (Fig. 1G), we, therefore, encapsulated cabazitaxel into the HA-modified siRNA nanocomplex to improve its antitumor effect by inducing apoptosis of TNBC cells (Fig. 2A). Cabazitaxel was encapsulated into the hydrophobic core of the cholesterol-peptide micelle, followed by condensing with the IKBKE siRNA and coating with cholesterol-HA to form the cabazitaxel-loaded HA-modified siRNA nanocomplexes. The drug loading and entrapment efficiency of cabazitaxel were 18.3% (w/w) and 19.8%, respectively. The drug loading efficiency of siRNA was 14.9% (w/w) at an N/P ratio of 10:1. The critical micelle concentrations (CMCs) of the siRNA nanocomplexes were measured with an iodine probe to determine whether siRNA condensation and HA coating affect the stability of the micellar structure. As shown in Fig. 4A, CMCs of cholesterol-peptide, siRNA nanocomplex, and HA-coated siRNA nanocomplex were 72, 76, and 82 μg/ml, respectively, suggesting that siRNA condensation and HA coating have negligible effect on the stability of the cholesterol-peptide–based micellar structure. The particle sizes of the HA-coated nanocomplexes with and without cabazitaxel encapsulation were 160 and 165 nm, respectively (Fig. 4B). The particle size and structure of the nanocomplexes were confirmed by transmission electron microscopy (TEM) (Fig. 4C). Meanwhile, encapsulation of cabazitaxel into the HA-coated siRNA nanocomplex does not comprise the silencing activity of the siRNA (Fig. 4D). The nanocomplex effectively protected the encapsulated siRNA from degradation in human and mouse serum for up to 24 hours (Fig. 4, E and F). Moreover, premature release of siRNA was not observed after incubation with human serum (fig. S5).
Fig. 4. Characterization of the cabazitaxel-loaded siRNA nanocomplexes.
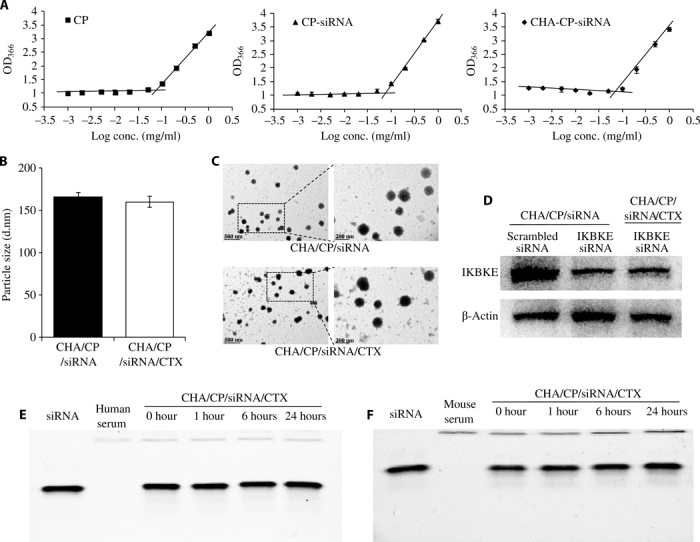
(A) Critical micelle concentrations of CP, CP/siRNA nanocomplex, and CHA/CP/siRNA nanocomplex. Particle size (B) and TEM images (C) of the CHA/CP/siRNA and CHA/CP/siRNA/CTX nanocomplexes. (D) Silencing activity of the nanocomplexes at the protein level. Serum stability of the CHA/CP/siRNA/CTX nanocomplex in 50% human serum (E) and mouse serum (F). Heparin was used to dissociate the siRNA from the nanocomplex after incubation with the serum. All results are presented as means ± SD (n = 3). CP, cholesterol-peptide; CHA, cholesteryl-HA; cabazitaxel, CTX.
Three-dimensional tumor spheroid penetration study
Three-dimensional (3D) tumor spheroid model has been used in cancer research to mimic the complex cell-cell and cell–extracellular matrix (ECM) interactions in the tumor microenvironment (26, 27). The presence of targeting ligand has been proved to improve the penetration of siRNA into tumor spheroids (28). Here, we evaluated the tumor penetration capability of the HA-modified siRNA nanocomplex in MDA-MB-231 spheroids with a diameter of approximately 500 μm, as previously reported (29).
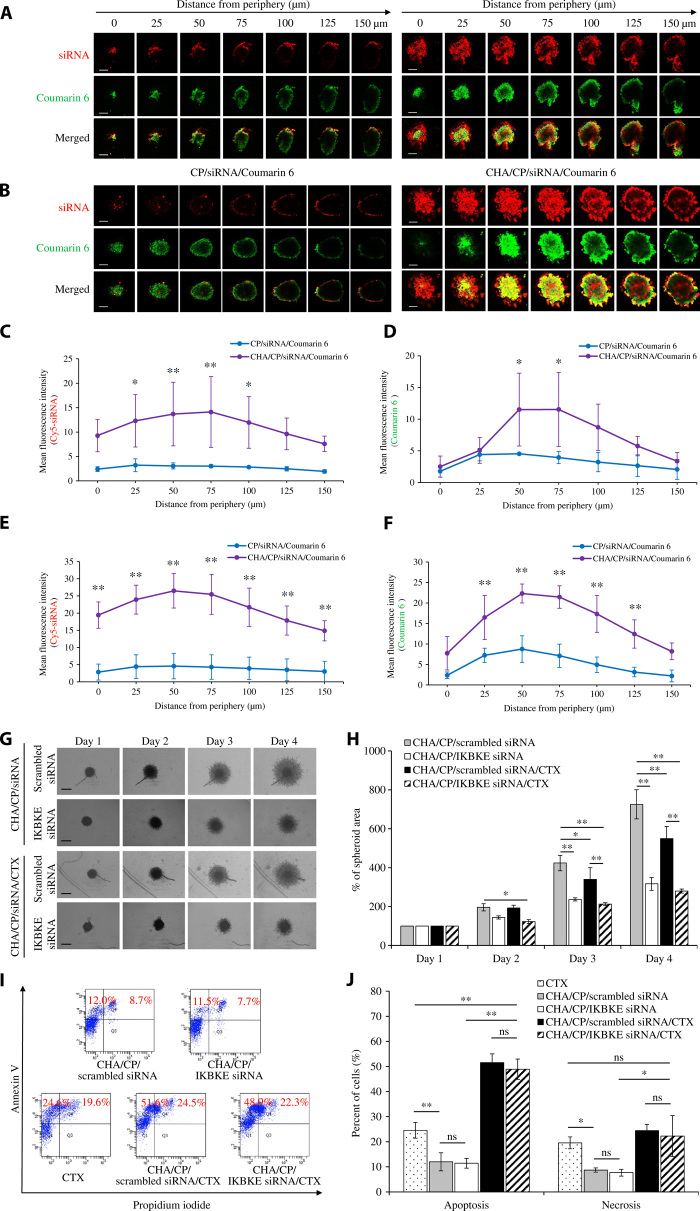
The IKBKE siRNA was labeled with Cy5, and a hydrophobic dye (Coumarin 6) was used to mimic cabazitaxel in this study. After 1 and 2 hours of incubation with the MDA-MB-231 spheroids, the HA-modified siRNA nanocomplex demonstrated a much deeper penetration and higher fluorescence intensity of the Cy5-siRNA (red) and Coumarin 6 (green) in the center of the spheroids (Fig. 5, A to F). Compared with the unmodified siRNA nanocomplex, the HA-modified siRNA nanocomplex increased the penetration of both Cy5-siRNA (Fig. 5, C and E) and Coumarin 6 (Fig. 5, D and F). Overall, Cy5-siRNA and Coumarin 6 exhibited significant colocalization in the spheroids, suggesting co-delivery of these molecules by the nanocomplex. However, we also observed a slight difference in the distribution of Cy5-siRNA and Coumarin 6 inside the spheroids. During the penetration of the nanocomplex into the spheroids, a small fraction of Cy5-siRNA and Coumarin 6 could be released from nanocomplexes and subsequently exhibited different distribution because of their very different physical and chemical properties. By contrast, the unmodified siRNA nanocomplex was mainly observed in the peripheral areas of the spheroids. The results clearly demonstrated that the HA-coated nanocomplex can efficiently deliver its siRNA and small-molecule drug into the tumor spheroids.
Fig. 5. 3D tumor spheroid penetration, invasion, and apoptosis study of the HA-modified siRNA nanocomplex.
Representative z-stack confocal images of the spheroids with a z-step of 25 μm for CP/Cy5-siRNA/Coumarin 6 and CHA/CP/Cy5-siRNA/Coumarin 6 nanocomplexes after 1 hour (A) and 2 hours (B) of incubation. Red for Cy5-siRNA, green for Coumarin 6, and yellow for merged area. Scale bars, 200 μm. Mean fluorescence intensity of Cy5-siRNA after 1 hour (C) and 2 hours (E) of incubation and Coumarin 6 after 1 hour (D) and 2 hours (F) of incubation in the z-stacked confocal images versus the distance from the periphery of the spheroids (n = 3). (G) Representative images of 3D spheroid invasion of MDA-MB-231 cells (n = 3). Scale bars, 500 μm. (H) Quantification of spheroid invasion based on the change of the total areas of the spheroids. The spheroid areas of each day were normalized to the areas measured at day 1. The data are presented as means ± SD (n = 3). (I) Apoptosis study of the cells using the Alexa Fluor 488 Annexin V/Dead Cell Apoptosis Kit and flow cytometry. (J) Percentage of apoptotic and necrotic cells after 48 hours of transfection. All results are presented as means ± SD (n = 3). *P < 0.05; **P < 0.01; ns, not significant. CP, cholesterol-peptide; CHA, cholesteryl-HA; cabazitaxel, CTX.
3D tumor spheroid invasion study
After showing the inhibitory effect of the IKBKE siRNA on the migration and invasion of monolayer MDA-MB-231 cells (Fig. 1), we further evaluated whether the HA-modified siRNA nanocomplex suppresses the invasion of the MDA-MB-231 spheroid, which more closely mimics the complex scenario in vivo. Compared with the scrambled siRNA group, the spheroids treated with the IKBKE siRNA exhibited markedly reduced invasion (Fig. 5G). The IKBKE siRNA alone inhibited the invasion of MDA-MB-231 spheroids by 26.6, 44.1, and 56.3% after 24, 48, and 72 hours, respectively (Fig. 5H). Cabazitaxel was also found to moderately inhibit the invasion of the spheroids (19.8% for 48 hours and 24.3% for 72 hours) treated with the scrambled siRNA, but not the spheroids treated with the IKBKE siRNA.
Apoptosis study of the HA-modified siRNA nanocomplex
We next assessed the apoptosis effect of the HA-modified siRNA nanocomplexes on MDA-MB-231 cells (Fig. 5, I and J). Cabazitaxel induced significant apoptosis, while the nanocomplexes containing either scrambled or IKBKE siRNA showed very limited apoptosis, which is possibly due to the transfection procedure. Moreover, the cabazitaxel-loaded siRNA nanocomplexes exhibited markedly higher apoptosis than cabazitaxel alone, indicating that encapsulation of cabazitaxel into the nanocomplex increased its cellular uptake and subsequently enhanced its activity. Similar results were observed in cell necrosis. These findings suggested that the IKBKE siRNA has a negligible effect on apoptosis and necrosis of MDA-MB-231 cells, which is consistent with our previous findings in SK-BR-3 and MCF7 breast cancer cells (3).
Biodistribution and in vivo pharmacokinetics of the HA-modified siRNA nanocomplex
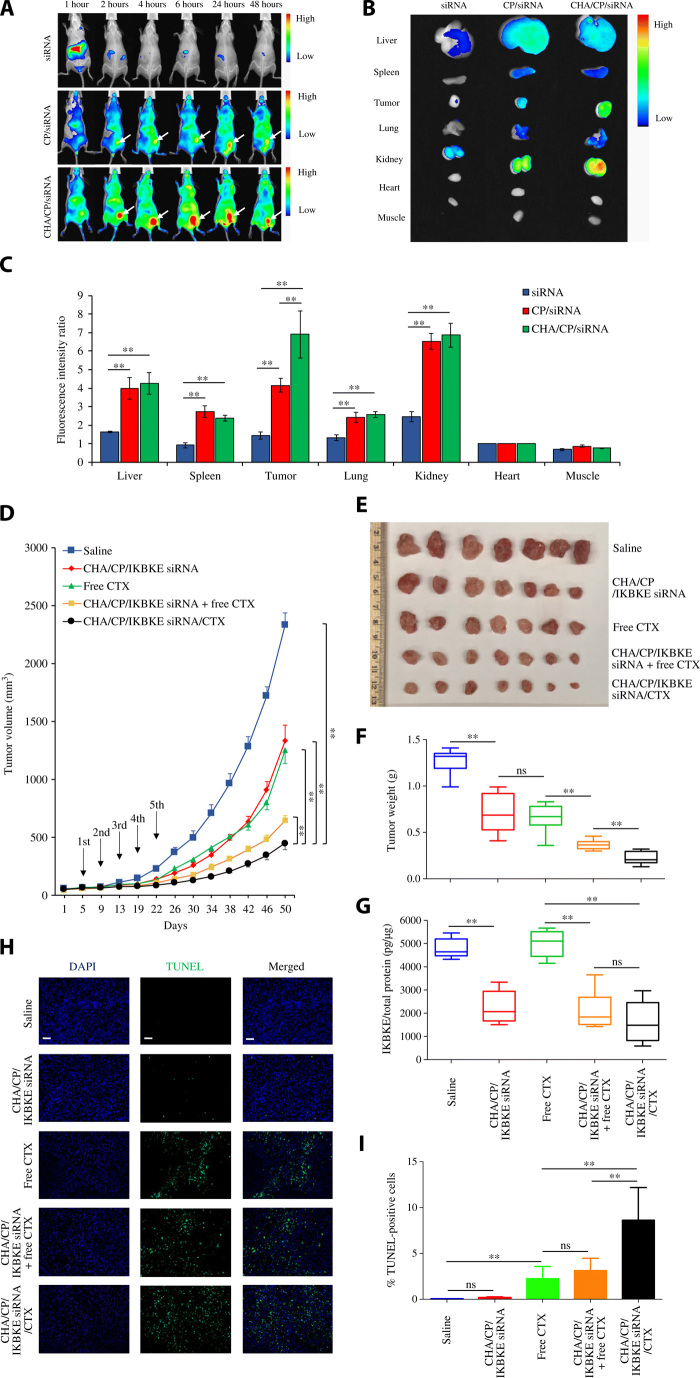
In this study, an orthotopic xenograft mouse model of TNBC was established to closely mimic the tumor microenvironment in human cancer. Live animal imaging of mice at different time intervals is presented in Fig. 6A. After 48 hours, the major organs were harvested for imaging and quantitation of the fluorescence intensity (Fig. 6, B and C). After intravenous administration, free siRNA quickly accumulated in the liver and then eliminated from the body by 2 hours. By contrast, the unmodified siRNA nanocomplex showed high fluorescence intensity across the body for up to 48 hours, which is due to the protection of the siRNA and longer circulation time of the nanocomplex. Meanwhile, the unmodified nanocomplex also showed accumulation of the siRNA in the tumor. This is consistent with our previous finding that nanoscale particles can accumulate in the tumor via the enhanced permeability and retention effect (29). Compared with the unmodified siRNA nanocomplex, the HA-modified siRNA nanocomplex exhibited markedly higher uptake in the tumor but similar distribution in other organs, including the liver, spleen, lung, kidney, heart, and muscle. This result clearly demonstrated the tumor-targeting effect of HA in vivo.
Fig. 6. Biodistribution and antitumor activity of the HA-modified siRNA nanocomplex in an orthotopic TNBC mouse model.
For biodistribution study, the siRNA was labeled with Cy5 and injected via tail vein at a dose of 1.5 mg siRNA/kg. (A) Live animal imaging of the mice at different time intervals. n = 3. (B) Fluorescence images of the liver, spleen, tumor, lung, kidney, heart, and muscle were taken at 48 hours after injection. (C) Mean fluorescence intensity of each organ was normalized to that of the heart. The results are presented as means ± SD (n = 3). **P < 0.01. For antitumor activity study, saline, CHA/CP/IKBKE siRNA nanocomplex, CTX, CHA/CP/IKBKE siRNA nanocomplex plus free CTX, and CHA/CP/IKBKE siRNA/CTX nanocomplex were administered via the tail vein every 4 days for a total of five times at a dose of 1.5 mg siRNA/kg and 5 mg cabazitaxel/kg. (D) Tumor volumes are presented as means ± SEM. (E) Images of harvested tumors. Photo credit: Z.Z., School of Pharmacy, University of Missouri-Kansas City). (F) Tumor weight. The results are presented as means ± SD (n = 7). (G) Expression of IKBKE protein in tumor tissues (n = 6). Fluorescence images (H) and quantitative analysis (I) of apoptotic cells in tumor specimen using a TUNEL assay. Scale bars, 100 μm. Quantitative analysis was performed by counting the fluorescence intensity of apoptotic cells in three random microscope fields from three independent samples. The results are presented as means ± SD. **P < 0.01. ns, not significant. CP, ns, cholesterol-peptide; CHA, cholesteryl-HA; CTX, cabazitaxel.
Next, we evaluated the pharmacokinetic profiles of free siRNA and HA-modified siRNA nanocomplex. As shown in fig. S4, the concentration of free siRNA in the blood decreased to less than 10% after 10 min, and the concentration was not detectable after 2 hours. By contrast, the HA-modified siRNA nanocomplex prolongs the circulation of encapsulated siRNA to more than 24 hours. The areas under the curve (AUCs) for free siRNA and HA-modified siRNA nanocomplex are 95.2 and 1104.0 μg⋅min/ml, respectively. The HA-modified siRNA nanocomplex increased the AUC of the siRNA by approximately 11.6-fold.
In vivo antitumor activity of the HA-modified IKBKE siRNA nanocomplex
Antitumor activities of the siRNA nanocomplexes were evaluated using the same orthotopic xenograft mouse model, as described above. Thirty-five mice were randomly divided into five groups and treated with saline, HA-modified IKBKE siRNA nanocomplex, cabazitaxel, HA-modified IKBKE siRNA nanocomplex plus cabazitaxel, and cabazitaxel-loaded HA-modified IKBKE siRNA nanocomplex. The nanocomplexes and cabazitaxel were administered via the tail vein every 4 days for a total of five injections at a dose of 1.5 mg IKBKE siRNA/kg and 5 mg cabazitaxel/kg.
As illustrated in Fig. 6 (D to F), the IKBKE siRNA nanocomplex alone exhibited a similar inhibition effect on tumor growth as cabazitaxel, demonstrating that the nanocomplex can efficiently deliver the siRNA into the tumor to exert its antitumor effect. While combination of free cabazitaxel with the siRNA nanocomplex enhanced siRNA’s antitumor effect, coencapsulation of cabazitaxel and the siRNA in the nanocomplex exhibited the highest activity. This is in accordance with previous findings that activation of NF-κB is linked to chemoresistance of breast cancer cells and down-regulation of NF-κB sensitizes breast cancer cells to chemotherapy-induced apoptosis (8, 9). Our results also demonstrated the importance of combinational therapy targeting different pathways in TNBC.
To evaluate the silencing effect of the siRNA nanocomplex in vivo, we isolated proteins from the tumor specimen and measured the protein expression of IKBKE using enzyme-linked immunosorbent assay (ELISA). As shown in Fig. 6G, all the tumors treated with the IKBKE siRNA nanocomplex exhibited the down-regulation of the IKBKE protein, whereas cabazitaxel alone did not affect the expression of the protein. This result clearly demonstrated that the nanocomplex efficiently delivered the siRNA into the tumor and down-regulated the expression of IKBKE, leading to the suppression of tumor growth.
A TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) assay was performed to detect cell apoptosis in tumor tissues (Fig. 6, H and I). No difference was observed between the saline group and the IKBKE siRNA nanocomplex group, which is in accordance with the in vitro apoptosis study in Fig. 1H and 5I. While treatment with free cabazitaxel induced significant apoptosis of tumor cells, encapsulation of cabazitaxel in the siRNA nanocomplex markedly increased its apoptotic effect on tumor cells. All these results indicated that this multifunctional co-delivery system for cabazitaxel and IKBKE siRNA may offer a promising option for TNBC therapy.
DISCUSSION
Recently, there is growing evidence showing the important role of IKBKE in TNBC. IKBKE overexpression is associated with STAT3 activation and contributes to the proliferation and survival of TNBC (7). IKBKE also induces the production of the cytokines CCL5 and IL-6, which stimulates the migration of TNBC cells and endothelial cells through autocrine and paracrine signaling processes (7). As a result, IKBKE is a promising therapeutic target for TNBC. Several small molecular inhibitors, such as BX-795, MRT67307, and MRT68921, have been developed to suppress IKBKE. However, none of them is specific for IKBKE, which may lead to unexpected potential side effects (5, 10, 11). By contrast, RNA interference (RNAi) can specifically down-regulate a target gene with high potency. Meanwhile, there has been growing interest in RNAi-based therapeutics since the approval of the first-ever RNAi therapy by the FDA in 2018.
We have developed IKBKE siRNAs and demonstrated their in vitro antitumor activities in HER2-positive breast cancer cells (3). Here, we explored the potential application of the IKBKE siRNA in TNBC. As shown in Fig. 1, the IKBKE siRNA silenced the expression of IKBKE in TNBC cells and subsequently suppressed the proliferation, migration, and invasion of TNBC cells. Moreover, peritumoral injection of the siRNA markedly inhibited tumor growth in an MDA-MB-231 subcutaneous xenograft tumor model. These proof-of-concept studies clearly demonstrated that targeting IKBKE with siRNA is a potential therapeutic strategy for TNBC.
Metastasis is a major challenge for TNBC treatment. Compared with non-TNBC patients, patients with TNBC have a fourfold higher risk to develop visceral metastasis (30). The metastatic cascade is a complex multistep process containing invasion, intravasation, and extravasation (31). Inhibition of tumor invasion is, therefore, critical to reducing metastasis in TNBC. The IKK/NF-κB signaling pathway plays important roles in TNBC metastasis (32). In this study, we demonstrated that knockdown of the expression of IKBKE with siRNA markedly decreases the migration and invasion of TNBCs (Fig. 1), and the hybrid siRNA nanocomplex inhibited the invasion of TNBC in a 3D tumor spheroid model (Fig. 5, G and H).
Combination therapy has attracted wide attention in cancer therapy because of the heterogeneity and complexity of cancers (33). While the IKBKE siRNA exhibited potent antitumor activities in TNBC, it has a negligible effect on cell apoptosis. We, therefore, combined cabazitaxel, a microtubule stabilizer, with the IKBKE siRNA to achieve optimal antitumor effect against TNBC. Although cabazitaxel has been approved for prostate cancer, the major clinical concerns associated with cabazitaxel are its poor solubility and relatively high toxicity (18). Therefore, numerous efforts have been made to increase its solubility and reduce systemic toxicity. For example, encapsulation of cabazitaxel in polymeric micelles or conjugation to albumin can significantly reduce the systemic toxicity (18).
In our study, we developed a hybrid nanocomplex to encapsulate the IKBKE siRNA and cabazitaxel simultaneously. As shown in fig. S3B, encapsulation of cabazitaxel in the nanocomplex reduced the half-maximal inhibitory concentration (IC50) from 99.0 to 38.2 nM, which is consistent with a report about cabazitaxel-loaded micelle (18). Meanwhile, the blank nanocomplex did not show any cytotoxicity (fig. S3A), suggesting good safety of the system. This result is consistent with the enhanced cellular uptake (Fig. 3) and in vivo antitumor activity of cabazitaxel encapsulated in the nanocomplex (Fig. 6).
While two siRNA therapeutics have been approved by the FDA, both of them are targeted to the liver. There remains a great challenge to deliver siRNAs to solid tumors. In this study, we modified the nanocomplex with HA to target CD44, which is highly expressed in numerous cancer cells including TNBC (34, 35). HA is a natural acidic polysaccharide and has been widely used as a CD44-specific ligand in various drug delivery systems (24, 34). The size of HA is critical to its binding affinity to CD44 and following biological functions (36). While ligands with small size are preferred for targeted drug delivery, the binding affinity of HA to CD44 decreases rapidly when the size is smaller than 30 kDa (37). In this study, a low–molecular weight HA (50 kDa) was selected as the CD44-specific ligand. It is conjugated to cholesterol to enhance its coating efficacy on the surface of the hybrid siRNA nanocomplex. Coating of the hybrid nanocomplex with HA increased cellular uptake (Fig. 3), tumor penetration (Fig. 5, A to F), and in vivo tumor accumulation (Fig. 6, A to C) of the IKBKE siRNA and cabazitaxel. The HA-coated nanocomplex exhibited superior antitumor activity in an orthotopic TNBC mouse model (Fig. 6, D to I).
CONCLUSIONS
We found that silencing the IKBKE gene using siRNA markedly inhibited the proliferation, migration, and invasion of TNBC cells. Peritumoral injection of the IKBKE siRNA suppressed tumor growth in an MDA-MB-231 subcutaneous xenograft tumor model. We developed an HA-modified peptide-based hybrid nanocomplex to coencapsulate the IKBKE siRNA and cabazitaxel to achieve optimal antitumor efficiency. The nanocomplex showed higher cellular uptake and better tumor penetration of the encapsulated siRNA and cabazitaxel. Surface modification of the nanocomplex with HA enhanced the accumulation of the nanocomplex in TNBC cells in vivo. The hybrid nanocomplex exhibited a potent antitumor effect in an orthotopic TNBC model after systemic administration. Our results clearly demonstrated that targeting IKBKE with siRNA is a potential therapeutic strategy for TNBC. Encapsulation of cabazitaxel in the hybrid nanocomplex also enhances the activity of the IKBKE siRNA. Furthermore, combination therapy has become a mainstay in cancer therapy, but it is difficult to combine siRNAs and chemotherapy agents in the same formulation due to their very different physical and chemical properties. Therefore, the peptide-based hybrid nanocomplex provides a novel and versatile platform for combination therapies using siRNAs and chemotherapeutic agents.
MATERIALS AND METHODS
Material
Lipofectamine RNAiMAX was ordered from Invitrogen (Carlsbad, CA). Cabazitaxel was purchased from MedKoo Biosciences (Morrisville, NC). Mouse and human sera were obtained from BD Biosciences (San Jose, CA). Human IKBKE ELISA kit was ordered from LifeSpan BioSciences Inc. (Seattle, WA). CellTiter-Glo Luminescent Cell Viability Assay kit was purchased from Promega (Madison, WI). BD Matrigel, annexin V–FITC (fluorescein isothiocyanate) apoptosis kit, and other chemical reagents were obtained from Thermo Fisher Scientific (Pittsburgh, PA).
Synthesis of cholesterol-peptide
A cholesterol-modified peptide containing the cathepsin B–liable dipeptide (Val-Cit) was synthesized using the solid-phase peptide synthesis method (fig. S1A) (19, 20). Briefly, 10 mmol of protected Val-Cit-HHHKKHHHKK peptide conjugated to Wang resin was obtained from United Peptide (Herndon, VA), and the Fmoc (9-fluorenyl methoxycarbonyl) protecting group on Val was deprotected with 20% piperidine at room temperature for 30 min. After washing with dichloromethane (DCM) for three times, the deprotected peptide was added into 10 ml of DCM containing cholesteryl chloroformate (12.5 mmol) and N,N′-diisopropylethylamine (DIPEA) (37.5 mmol), followed by adding 2-(6-chloro-1H-benzotriazol-1-yl)-1,1,3,3-tetramethylaminium-hexafluorophosphat (HCTU) (12.5 mmol), and stirring continuously for 72 hours at room temperature under N2 protection. After the reaction was complete, the cholesterol-conjugated peptide was cleaved from the resin with TFA (trifluoroacetic acid)/water/TIPS (triisopropylsilane) (95:2:3, v/v/v) at room temperature for 2 hours. The Boc protecting groups on lysine and the Trt protecting groups on histidine were removed at the same time. The cholesterol-peptide was purified by high-performance liquid chromatography (HPLC) (Shimadzu LC-20; Kyoto, Japan), and the molecular weight (MW, 2022.5) was confirmed by mass spectrometry (fig. S1C).
Synthesis of cholesterol-HA
Cholesterol-HA was synthesized using a modified version of a reported method as illustrated in fig. S1B (38). First, a cholesteryl linker was synthesized by mixing 2,2′-(ethylenedioxy)-bis-ethylamine (30 mmol) with 7.5 mmol of triethylamine (TEA) in DCM, followed by adding 7.5 mmol of cholesteryl chloroformate dropwise under N2 protection. After stirring overnight at room temperature, the product was purified on a silica gel column, and the MW (560.5) was confirmed by mass spectrometry.
Four hundred milligrams of HA (MW, 50 kDa; Creative PEGworks, NC) was dissolved in formamide and incubated at 50°C for 1 hour with stirring. TEA (0.3 mmol) and 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxide hexafluorophosphate (0.3 mmol) were added into the solution. The cholesteryl linker (0.128 mmol) was dissolved in DMF, added to the HA solution at a 50/50 (v/v) ratio, and the mixture was stirred at 50°C for 24 hours and then at room temperature for another 24 hours. The final product was purified by dialysis [molecular weight cutoff (MWCO), 3.4 kDa] against water and lyophilized. Cholesterol-HA was confirmed by recording the 1H nuclear magnetic resonance (NMR) spectrum in d-DMSO (fig. S1D).
Fabrication and characterization of the HA-modified siRNA nanocomplex
The IKBKE siRNA (sense strand sequence: 5′-GGUCUUCAACACUACCAGCtt-3′) was purchased from GE Dharmacon (Lafayette, CO). For the HA-modified siRNA nanocomplex, the siRNA was first condensed with the cholesterol-peptide (containing the Val-Cit dipeptide) at different N/P ratios (1:1, 2:1, 3:1, 4:1, 5:1, and 10:1) at room temperature for 30 min to form the cholesterol-peptide/siRNA nanocomplex. The formation of the nanocomplex was confirmed using a gel retardation assay on a 2% agarose gel. Particle sizes and zeta potentials of the nanocomplexes were determined using a Malvern Zetasizer Nano ZS (Westborough, MA) after dilution in deionized (DI) water. Cholesterol-HA was added to coat the surface of the cholesterol-peptide/siRNA nanocomplex (N/P ratio of 10:1) at different molar ratios of cholesterol-HA to cholesterol-peptide at room temperature for 1 hour. Agarose gel was used to confirm the condensation after coating with cholesterol-HA. Zeta potential was measured to evaluate the changes in the surface charge of the nanocomplexes coated with different concentrations of cholesterol-HA. A CM12 TEM (Philips, Germany) was used to study the morphological structures of these nanocomplexes.
The CMC of the HA-modified siRNA nanocomplex was determined using iodine as a probe (33). Briefly, a series of sample solutions ranging in concentration from 0.001 to 1 mg/ml were prepared in DI water, and the iodine solution was prepared by mixing iodine and potassium iodide (w/w; 1:2) in DI water. A 200-μl sample was added to a 1.5-ml centrifuge tube, mixed with 10 μl of the iodine solution, and incubated at room temperature for 15 hours in dark. Then, the samples were transferred to a 96-well plate with black wall, and the absorbance of iodine in the core of the micelles was determined at 366 nm using a SpectraMax 190 microplate spectrophotometer (San Jose, CA).
Preparation of cabazitaxel-loaded siRNA nanocomplex
Cabazitaxel was encapsulated into the cholesterol-peptide micelle as reported (39). Twenty milligrams of the cholesterol-peptide was dissolved in 4 ml of DI water, followed by adding 16 ml of cabazitaxel solution (30 mg/ml in ethanol) dropwise and stirring for 30 min. The mixture was sonicated for 30 min in an ice bath and centrifuged at 3000 rpm for 10 min. The supernatant was dialyzed against DI water for 24 hours using a dialysis bag (MWCO, 3.4 kDa), followed by filtration through a 0.45-μm pore–sized membrane and lyophilized. The drug loading and entrapment efficiency of cabazitaxel in the micelle were measured using HPLC after dissolving cabazitaxel and micelles in ethanol (39).
Cell culture
MDA-MB-231 cell line was purchased from the American Type Culture Collection. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS, penicillin (100 U/ml), and streptomycin (100 μg/ml). The cells were incubated in a humidified atmosphere with 5% CO2 at 37°C.
Silencing activity study
Five thousand MDA-MB-231 cells were seeded in a 24-well plate and incubated at 37°C for 12 hours. The IKBKE siRNA was condensed with Lipofectamine RNAiMAX or cholesterol-peptide nanocomplex as described above and transfected into the cells at a final siRNA concentration of 50 nM for 24 hours. A scrambled siRNA was used as a negative control. Total RNA was isolated using a Direct-zol RNA isolation kit (Zymo Research, CA), and real-time reverse transcription PCR was performed as previously reported (3). For Western blot assays, the cells were first transfected with the IKBKE siRNA in Opti-MEM for 24 hours, replaced with fresh medium, and incubated for another 24 hours. Total proteins were isolated, and the silencing activity was quantified using Western blot as previously reported (3, 40).
Migration and invasion studies
MDA-MB-231 cells were transfected with the IKBKE siRNA using Lipofectamine RNAiMAX. After 48 hours of incubation, treated cells were detached and resuspended in serum-free DMEM at a concentration of 1 × 106 cells/ml for the migration assay. Then, 100 μl of the resuspended cells was seeded into a Transwell chamber with serum-free DMEM. For the invasion study, the Transwell chamber was precoated with 50 μg of Matrigel, and 2 × 105 cells were seeded in the precoated Transwell with serum-free DMEM. After 6 hours for the migration assay and 24 hours for the invasion assay, the migrated cells were fixed with 10% paraformaldehyde, stained with 0.05% crystal violet, and counted as previously described (29, 40).
Cell proliferation study
Two thousand five hundred MDA-MB-231 cells were seeded in a 96-well plate, incubated at 37°C for 12 hours, and treated with the IKBKE siRNA/Lipofectamine RNAiMAX for 24 hours. After replacement of the medium was DMEM supplemented with 10% FBS, the cells were incubated at 37°C for 24 or 72 hours. Then, 100 μl of CellTiter-Glo buffer (Promega, WI) was added to the 96-well plate, and the luminescence intensity was measured using a SpectraMax M5e spectrophotometer (San Jose, CA).
In vitro apoptosis study
Approximately 2 × 105 MDA-MB-231 cells were seeded in a six-well plate, incubated at 37°C for 12 hours, and treated with the IKBKE siRNA/Lipofectamine RNAiMAX. After 48 hours of incubation, the cells were treated according to the protocol of the Alexa Fluor 488 Annexin V/Dead Cell Apoptosis Kit (Thermo Fisher Scientific, NY) and examined using a FACS (fluorescence-activated cell sorting) II flow cytometer. The same study was performed for free cabazitaxel, HA-modified siRNA nanocomplex, and cabazitaxel-loaded HA-modified siRNA nanocomplex for 48 hours of incubation.
Serum stability of the siRNA nanocomplex
Serum stability study was performed with human and mouse sera as previously reported (40). The siRNA nanocomplex was incubated with 50% human or mouse serum for different time intervals at 37°C. Encapsulated siRNAs were then released from the nanocomplex by incubation with heparin (40 μM) for 10 min and then analyzed by electrophoresis in a 20% native polyacrylamide gel electrophoresis gel. To study the premature release of encapsulated siRNA in the serum, mixtures of the siRNA nanocomplex and serum were directly analyzed by electrophoresis in a 2% agarose gel. siRNA was visualized by staining with GelRed.
Cellular uptake study
Approximately 1× 105 MDA-MB-231 cells were seeded in a 24-well plate and incubated at 37°C for 12 hours. The Cy5-labeled IKBKE siRNA (50 nM) was used to form the siRNA nanocomplex. Two, 4, and 6 hours after transfection, the cells were treated with 40 μM heparin, and cellular uptake was measured using a FACS II flow cytometer (BD Instrument, NJ) (40). Confocal microscopy was performed 2 and 6 hours after transfection. To determine the HA targeting efficiency, the cells were pretreated with free HA (5 mg/ml) for 2 hours.
3D spheroid penetration study
3D tumor spheroids were prepared with Cultrex spheroid formation ECM (Trivigen, Gaithersburg, MD) as previously reported (29, 33). Three thousand MDA-MB-231 cells were suspended in 50 μl of spheroid formation ECM and added into a Corning 96-well ultralow attachment microplate, followed by centrifugation at 200g for 3 min at 4°C. The plate was incubated at 37°C for 120 hours to induce the formation of spheroids. The tumor spheroids were incubated with Coumarin 6–encapsulated siRNA nanocomplexes and Cy5-labeled siRNA nanocomplexes to evaluate the tumor penetration capability of Coumarin 6 and siRNA, respectively.
3D tumor spheroid invasion study
3D MDA-MB-231 spheroid invasion assay was performed as previously reported (29). The spheroids were embedded in invasion matrix from the Cultrex Spheroid Cell Invasion Assay kit (Trivigen, Gaithersburg, MD) at 37°C for 60 min and incubated with the HA-modified siRNA nanocomplexes diluted in Opti-MEM medium for 24 hours. The medium was then replaced by DMEM containing 10% FBS, and the spheroids were incubated at 37°C for up to 3 days. Invasion of the spheroids in the matrix was photographed every 24 hours using a microscope (Leica DMI3000B, Germany) and analyzed using ImageJ. The spheroid areas of each day were normalized to the areas measured at day 1, and the spheroid invasion index was presented as the percentage of spheroid areas on day 1.
In vitro cytotoxicity assay
Ten thousand MDA-MB-231 cells were seeded in a 96-well plate and incubated at 37°C for 12 hours and then treated with free cabazitaxel or cabazitaxel-loaded siRNA nanocomplexes in DMEM supplemented with 10% FBS for 48 hours. Cytotoxicity was evaluated using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay as previously described (29).
In vivo biodistribution, pharmacokinetic, and antitumor activity study
The animal protocol was approved by the Institutional Animal Care and Use Committee of the University of Missouri-Kansas City. For the subcutaneous xenograft tumor model, 1 × 106 MDA-MB-231 cells were implanted into the back of female nude mice. The IKBKE siRNA or scrambled siRNA were condensed with the cholesterol-peptide lacking the Val-Cit dipeptide and injected into the mice via peritumoral injection at a dose of 0.3 mg siRNA/kg. The siRNAs were administered every 4 days for a total number of four injections.
For the biodistribution and antitumor activity studies of the HA-modified siRNA nanocomplex, 5 × 105 MDA-MB-231 cells were implanted into the right mammary gland of female mice to establish an orthotopic TNBC model as previously reported (29). For biodistribution study, nine mice were randomly divided into three groups and intravenously injected free siRNA or siRNA nanocomplex at a dose of 1.5 mg siRNA/kg. Fluorescence intensity of the Cy5-labeled siRNA was monitored at different time intervals using a Bruker MS FX PRO imaging system (Billerica, CA). After 48 hours, the mice were euthanized, and major organs (tumor, liver, spleen, lungs, kidneys, heart, and muscle) were harvested for imaging analysis.
For the antitumor activity study, 35 mice were randomly divided into five groups and intravenously injected with saline, HA-modified IKBKE siRNA nanocomplex, cabazitaxel, HA-modified IKBKE siRNA nanocomplex plus free cabazitaxel, and cabazitaxel-loaded HA-modified IKBKE siRNA nanocomplex. The treatments were performed every 4 days for five injections with a dose of 1.5 mg siRNA/kg and 5 mg cabazitaxel/kg. Tumor volumes were calculated using the formula 0.5 × (longest diameter) × (shortest diameter)2. The expressions of IKBKE in tumors were measured using a human IKBKE ELISA kit (LifeSpan Bioscience, WA) and normalized to the total amount of protein in each sample. A TUNEL assay kit (Invitrogen, CA) was used to determine apoptotic cells in tumor specimen as previously described (33). Images from each treatment group were evaluated using an inverted fluorescence microscope (Leica DMI3000B, Germany) and quantified using ImageJ.
For in vivo pharmacokinetic study, eight Balb/c mice were randomly divided into two groups and intravenously injected with siRNA or HA-modified siRNA nanocomplex at a siRNA dose of 1.5 mg/kg (contains 10% Cy5-labeled siRNA). Blood was collected from the tail vein at various time points after injection and dissolved into lysis buffer. Fluorescence intensity was measured and the AUC was calculated as described before (29).
Statistical analysis
Data were presented as the means ± SD. Statistical analysis was performed using a one-way analysis of variance (ANOVA) with Tukey’s post hoc test. A P value less than 0.05 was considered statically significant.
Supplementary Material
Acknowledgments
Funding: This work is supported by the American Cancer Society–Lee National Denim Day Research Scholar Grant (RSG-15-132-01-CDD) and a School of Graduate Studies Research Grant Award from the University of Missouri-Kansas City. The work is also partially supported by the awards (1R01GM121798, 5R01AA021510, and 1R01CA23109901) from the NIH. Author contributions: Z.Z., Y.L., and K.C. designed all the experiments in this study and interpreted the results. Z.Z. and K.C. wrote the paper. Z.Z. and Y.L. performed most of the experiments and analyzed the results. H.L. and A.J. assisted in the tumor activity study. P.V.P. performed mass spectrometry and NMR studies. All authors read and approved the final manuscript. Competing interest: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested form the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/29/eabb0616/DC1
REFERENCES AND NOTES
- 1.Wang C., Zheng X., Shen C., Shi Y., MicroRNA-203 suppresses cell proliferation and migration by targeting BIRC5 and LASP1 in human triple-negative breast cancer cells. J. Exp. Clin. Cancer Res. 31, 58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang W., Nag S. A., Zhang R., Targeting the NFκB signaling pathways for breast cancer prevention and therapy. Curr. Med. Chem. 22, 264–289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin B., Cheng K., Silencing of the IKKε gene by siRNA inhibits invasiveness and growth of breast cancer cells. Breast Cancer Res. 12, R74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boehm J. S., Zhao J. J., Yao J., Kim S. Y., Firestein R., Dunn I. F., Sjostrom S. K., Garraway L. A., Weremowicz S., Richardson A. L., Greulich H., Stewart C. J., Mulvey L. A., Shen R. R., Ambrogio L., Hirozane-Kishikawa T., Hill D. E., Vidal M., Meyerson M., Grenier J. K., Hinkle G., Root D. E., Roberts T. M., Lander E. S., Polyak K., Hahn W. C., Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 129, 1065–1079 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Liu T., Gao X., Xin Y., Identification of an IKBKE inhibitor with antitumor activity in cancer cells overexpressing IKBKE. Cytokine 116, 78–87 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Tian Y., Hao S., Ye M., Zhang A., Nan Y., Wang G., Jia Z., Yu K., Guo L., Pu P., Huang Q., Zhong Y., MicroRNAs let-7b/i suppress human glioma cell invasion and migration by targeting IKBKE directly. Biochem. Biophys. Res. Commun. 458, 307–312 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Barbie T. U., Alexe G., Aref A. R., Li S., Zhu Z., Zhang X., Imamura Y., Thai T. C., Huang Y., Bowden M., Herndon J., Cohoon T. J., Fleming T., Tamayo P., Mesirov J. P., Ogino S., Wong K.-K., Ellis M. J., Hahn W. C., Barbie D. A., Gillanders W. E., Targeting an IKBKE cytokine network impairs triple-negative breast cancer growth. J. Clin. Invest. 124, 5411–5423 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montagut C., Tusquets I., Ferrer B., Corominas J. M., Bellosillo B., Campas C., Suarez M., Fabregat X., Campo E., Gascon P., Serrano S., Fernandez P. L., Rovira A., Albanell J., Activation of nuclear factor-κ B is linked to resistance to neoadjuvant chemotherapy in breast cancer patients. Endocr. Relat. Cancer 13, 607–616 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Kim S.-H., Park H.-J., Moon D.-O., Sulforaphane sensitizes human breast cancer cells to paclitaxel-induced apoptosis by downregulating the NF-κB signaling pathway. Oncol. Lett. 13, 4427–4432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark K., Peggie M., Plater L., Sorcek R. J., Young E. R., Madwed J. B., Hough J., McIver E. G., Cohen P., Novel cross-talk within the IKK family controls innate immunity. Biochem. J. 434, 93–104 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Petherick K. J., Conway O. J., Mpamhanga C., Osborne S. A., Kamal A., Saxty B., Ganley I. G., Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J. Biol. Chem. 290, 11376–11383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martín M., Rodríguez-Lescure A., Ruiz A., Alba E., Calvo L., Ruiz-Borrego M., Santaballa A., Rodríguez C. A., Crespo C., Abad M., Domínguez S., Florián J., Llorca C., Méndez M., Godes M., Cubedo R., Murias A., Batista N., García M. J., Caballero R., de Alava E., Molecular predictors of efficacy of adjuvant weekly paclitaxel in early breast cancer. Breast Cancer Res. Treat. 123, 149–157 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Sparano J. A., Wang M., Martino S., Jones V., Perez E. A., Saphner T., Wolff A. C., Sledge G. W. Jr., Wood W. C., Davidson N. E., Weekly paclitaxel in the adjuvant treatment of breast cancer. N. Engl. J. Med. 358, 1663–1671 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vrignaud P., Semiond D., Benning V., Beys E., Bouchard H., Gupta S., Preclinical profile of cabazitaxel. Drug Des. Devel. Ther. 8, 1851–1867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vrignaud P., Sémiond D., Lejeune P., Bouchard H., Calvet L., Combeau C., Riou J.-F., Commercon A., Lavelle F., Bissery M.-C., Preclinical antitumor activity of cabazitaxel, a semisynthetic taxane active in taxane-resistant tumors. Clin. Cancer Res. 19, 2973–2983 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Gottesman M. M., Fojo T., Bates S. E., Multidrug resistance in cancer: Role of ATP–dependent transporters. Nat. Rev. Cancer 2, 48–58 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Zhong T., He B., Cao H.-Q., Tan T., Hu H.-Y., Li Y.-P., Zhang Z.-W., Treating breast cancer metastasis with cabazitaxel-loaded polymeric micelles. Acta Pharmacol. Sin. 38, 924–930 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H., Lu Z., Wang L., Guo T., Wu J., Wan J., Zhou L., Li H., Li Z., Jiang D., Song P., Xie H., Zhou L., Xu X., Zheng S., New generation nanomedicines constructed from self-assembling small-molecule prodrugs alleviate cancer drug toxicity. Cancer Res. 77, 6963–6974 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Qin B., Chen Z., Jin W., Cheng K., Development of cholesteryl peptide micelles for siRNA delivery. J. Control. Release 172, 159–168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang L., Lin S.-W., Dai W., Lu J.-K., Yang T.-Y., Xiang Y., Zhang Y., Li R.-T., Zhang Q., Novel cathepsin B-sensitive paclitaxel conjugate: Higher water solubility, better efficacy and lower toxicity. J. Control. Release 160, 618–629 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Yang K.-M., Bae E., Ahn S. G., Pang K., Park Y., Park J., Lee J., Ooshima A., Park B., Kim J., Jung Y., Takahashi S., Jeong J., Park S. H., Kim S.-J., Co-chaperone BAG2 determines the pro-oncogenic role of cathepsin B in triple-negative breast cancer cells. Cell Rep. 21, 2952–2964 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Chu D. S., Johnson R. N., Pun S. H., Cathepsin B-sensitive polymers for compartment-specific degradation and nucleic acid release. J. Control. Release 157, 445–454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattheolabakis G., Milane L., Singh A., Amiji M. M., Hyaluronic acid targeting of CD44 for cancer therapy: From receptor biology to nanomedicine. J. Drug Target. 23, 605–618 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Lv Y., Xu C., Zhao X., Lin C., Yang X., Xin X., Zhang L., Qin C., Han X., Yang L., He W., Yin L., Nanoplatform assembled from a CD44-targeted prodrug and smart liposomes for dual targeting of tumor microenvironment and cancer cells. ACS Nano 12, 1519–1536 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Perez A. P., Cosaka M. L., Romero E. L., Morilla M. J., Uptake and intracellular traffic of siRNA dendriplexes in glioblastoma cells and macrophages. Int. J. Nanomedicine 6, 2715–2728 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nyga A., Cheema U., Loizidou M., 3D tumour models: Novel in vitro approaches to cancer studies. J. Cell Commun. Signal. 5, 239–248 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H., Zhao Z., Zhang L., Li Y., Jain A., Barve A., Jin W., Liu Y., Fetse J., Cheng K., Discovery of low-molecular weight anti-PD-L1 peptides for cancer immunotherapy. J. Immunother. Cancer 7, 270 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waite C. L., Roth C. M., PAMAM-RGD conjugates enhance siRNA delivery through a multicellular spheroid model of malignant glioma. Bioconjug. Chem. 20, 1908–1916 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Z., Li Y., Shukla R., Liu H., Jain A., Barve A., Cheng K., Development of a biocompatible copolymer nanocomplex to deliver VEGF siRNA for triple negative breast cancer. Theranostics 9, 4508–4524 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dent R., Hanna W. M., Trudeau M., Rawlinson E., Sun P., Narod S. A., Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res. Treat. 115, 423–428 (2009). [DOI] [PubMed] [Google Scholar]
- 31.T. A. Martin, L. Ye, A. J. Sanders, J. Lane, W. G. Jiang, Cancer invasion and metastasis: Molecular and cellular perspective, in Metastatic Cancer: Clinical and Biological Perspectives, J. Rahul, Ed. (Landes Bioscience, Austin, TX, 2013), pp. 135–168. [Google Scholar]
- 32.Fusella F., Secli L., Busso E., Krepelova A., Moiso E., Rocca S., Conti L., Annaratone L., Rubinetto C., Mello-Grand M., Singh V., Chiorino G., Silengo L., Altruda F., Turco E., Morotti A., Oliviero S., Castellano I., Cavallo F., Provero P., Tarone G., Brancaccio M., The IKK/NF-κB signaling pathway requires Morgana to drive breast cancer metastasis. Nat. Commun. 8, 1636 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y., Zhao Z., Liu H., Fetse J. P., Jain A., Lin C.-Y., Cheng K., Development of a tumor-responsive nanopolyplex targeting pancreatic cancer cells and stroma. ACS Appl. Mater. Interfaces 11, 45390–45403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dosio F., Arpicco S., Stella B., Fattal E., Hyaluronic acid for anticancer drug and nucleic acid delivery. Adv. Drug Deliv. Rev. 97, 204–236 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Jin J., Krishnamachary B., Mironchik Y., Kobayashi H., Bhujwalla Z. M., Phototheranostics of CD44-positive cell populations in triple negative breast cancer. Sci. Rep. 6, 27871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cyphert J. M., Trempus C. S., Garantziotis S., Size matters: Molecular weight specificity of hyaluronan effects in cell biology. Int. J. Cell Biol. 2015, 563818 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolny P. M., Banerji S., Gounou C., Brisson A. R., Day A. J., Jackson D. G., Richter R. P., Analysis of CD44-hyaluronan interactions in an artificial membrane system: Insights into the distinct binding properties of high and low molecular weight hyaluronan. J. Biol. Chem. 285, 30170–30180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei X., Senanayake T. H., Bohling A., Vinogradov S. V., Targeted nanogel conjugate for improved stability and cellular permeability of curcumin: Synthesis, pharmacokinetics, and tumor growth inhibition. Mol. Pharm. 11, 3112–3122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou L., Yao J., Zhou J., Zhang Q., Pharmacokinetics of a paclitaxel-loaded low molecular weight heparin-all-trans-retinoid acid conjugate ternary nanoparticulate drug delivery system. Biomaterials 33, 5431–5440 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Zhao Z., Li Y., Jain A., Chen Z., Liu H., Jin W., Cheng K., Development of a peptidemodified siRNA nanocomplex for hepatic stellate cells. Nanomedicine 14, 51–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/29/eabb0616/DC1