The ongoing outbreak of coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a major threat to global health [1]. The mechanism of cellular entry by SARS-CoV-2 is through binding to angiotensin-converting enzyme 2 (ACE-2) [2, 3], a metalloproteinase ectoenzyme that primarily functions in the regulation of angiotensin II, but also has non-catalytic roles such as intestinal neutral amino acid transport. The level of ACE-2 protein and its subcellular localisation in the respiratory tract may be a key determinant of susceptibility to infection, symptoms and outcomes in COVID-19. In humans, ACE-2 protein is broadly expressed in the lung, kidney and small intestine [4]. Pathological analysis of COVID-19 post mortem samples shows substantial damage in the lung [5], suggesting that the airway is the principal entry and target of SARS-CoV-2. However, analysis of multiple single cell RNA-seq datasets reveal overall low ACE-2 RNA transcription in nasal airway epithelium, with further reduced expression in lower airway club cells and rare expression in alveolar epithelial cells [6]. This pattern of ACE-2 expression provides evidence that the upper, rather than the lower, airway is the initial site of SARS-CoV-2 infection.
Short abstract
ACE2 protein is expressed at high levels in the human olfactory epithelium relative to upper airway epithelial cells. This may explain COVID-19-associated olfactory dysfunction, and suggests a SARS-CoV-2 reservoir site and potential intranasal therapy. https://bit.ly/3hxT0qm
To the Editor:
The ongoing outbreak of coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a major threat to global health [1]. The mechanism of cellular entry by SARS-CoV-2 is through binding to angiotensin-converting enzyme 2 (ACE-2) [2, 3], a metalloproteinase ectoenzyme that primarily functions in the regulation of angiotensin II, but also has non-catalytic roles such as intestinal neutral amino acid transport. The level of ACE-2 protein and its subcellular localisation in the respiratory tract may be a key determinant of susceptibility to infection, symptoms and outcomes in COVID-19. In humans, ACE-2 protein is broadly expressed in the lung, kidney and small intestine [4]. Pathological analysis of COVID-19 post mortem samples shows substantial damage in the lung [5], suggesting that the airway is the principal entry and target of SARS-CoV-2. However, analysis of multiple single cell RNA-seq datasets reveal overall low ACE-2 RNA transcription in nasal airway epithelium, with further reduced expression in lower airway club cells and rare expression in alveolar epithelial cells [6]. This pattern of ACE-2 expression provides evidence that the upper, rather than the lower, airway is the initial site of SARS-CoV-2 infection.
There is growing interest in a presentation of SARS-CoV-2 infection characterised by olfactory loss without concomitant nasal inflammatory symptoms. Disturbances in the sense of smell have been widely reported in COVID-19 patients internationally, with a reported prevalence as high as 85% in a large, multicentre European survey [7]. These reports show, importantly, that some COVID-19 patients manifest olfactory loss as their initial or only symptom. As this presentation is largely not recognised or thought to mandate isolation, this patient group may be a source of continued viral spread and a target population for early intervention and mitigation. The loss of the sense of smell suggests the possibility of direct targeting by SARS-CoV-2 of the olfactory system. However, the cellular location of ACE-2 protein in the olfactory epithelium has not been previously demonstrated.
In this study, we performed an immunohistological analysis to determine the location of ACE-2 protein in human nasal and tracheal specimens. Nasal tissue included olfactory epithelial or respiratory epithelial samples collected from chronic rhinosinusitis (CRS) patients and control subjects undergoing endonasal surgical approaches for non-CRS disease processes [8]. Four control (two females and two males; age range 45–63 years old) and 19 CRS biopsies (10 females and nine males; age range 37–74 years old) were used for detailed immunohistochemistry analysis. Seven tracheal specimens were collected from tracheal stenosis patients who undergoing bronchoscopy. The immunostaining process was carried out after an antigen retrieval step using the following primary antibodies: Goat anti-ACE-2 (1:40, AF933; R&D), Rabbit anti-ACE-2 (1:100, MA5-32307; Thermo), Goat anti-GFP (1:100, ab6673; Abcam, IgG negative control), Mouse anti-Keratin 18 (1:500; Pierce MA1-39483), Rabbit anti-DCX (1:500; GeneTex, GTX134052), Rabbit anti-PGP9.5 (1:500; Ultraclone RA95101), and Mouse anti-Mucin 5AC (1:200; Abcam ab3649). The research protocol involving human specimens was approved by the Johns Hopkins institutional review board, and all subjects provided signed informed consent.
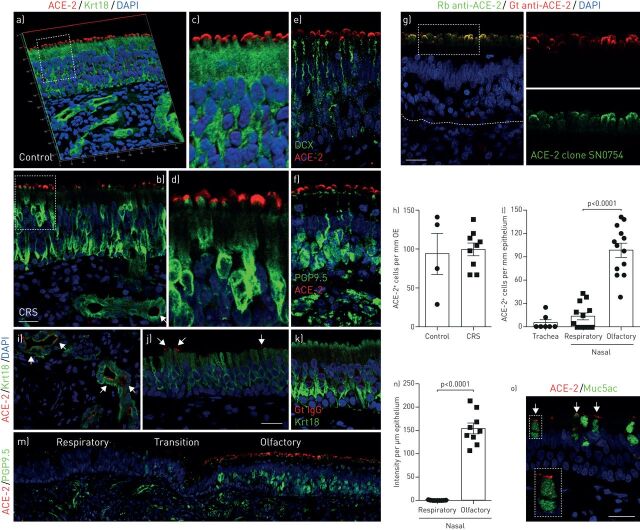
Within the accessible portion of the olfactory cleft in the nasal cavity, islands of olfactory mucosa are often found surrounded by respiratory epithelium. The specialised olfactory neuroepithelium has an apical surface consisting mainly of sustentacular cells, which support neuronal dendritic projections containing the odour-sensing cilia. Confocal images demonstrated that the majority of ACE-2-staining is localised to the apical surface of Krt18+ sustentacular cells in the olfactory neuroepithelium (figure 1a, b and g). This distribution of ACE-2 protein is similar to that reported in bat nasal epithelium [9]. We further quantified olfactory ACE-2 expression and found the number of ACE-2-positive cells to be comparable between healthy controls and CRS (figure 1a, b and h), a common inflammatory disease of the nasal mucosa that affects the olfactory mucosa [8]. ACE-2 is not present in olfactory neurons, demonstrated by co-staining with the immature and mature olfactory neuron marker DCX (figure 1e) and PGP9.5 (figure 1f), respectively. We further confirmed the apical location of ACE-2 in olfactory epithelium using another ACE-2 antibody recognising a different epitope (figure 1g). The specificity of ACE-2 staining was verified by using a Goat IgG isotype control (figure 1k).
FIGURE 1.
Cellular location of angiotensin-converting enzyme 2 (ACE-2) in human nasal and tracheal biopsies. a–d) Confocal image of ACE-2 (red) and Krt18 (green) immunostaining in the olfactory neuroepithelium from healthy control (a and c) and chronic rhinosinusitis (CRS) patient (b and d). The three-dimensional image (a) shows that the ACE-2 is localised to the apical surface of Krt18-positive sustentacular cells in the olfactory epithelium. Confocal images were obtained under Z stack mode which covered 8 µm in depth. The boxed area in panel (a and b) was highlighted in (c and d), respectively. e and f) The location of ACE-2- and DCX-positive immature (e) or PGP9.5-positive mature (f) olfactory sensory neurons in control. g) Confocal image verified the apical expression pattern of ACE-2 by co-staining of Goat anti-ACE-2 and another Rabbit anti-ACE-2 antibody (clone SN0754). The boxed area in (g) was highlighted in right panels. h) Quantification of ACE-2-positive cells per mm olfactory epithelium (OE). At least three images were collected from each specimen (four controls and nine CRS biopsies) using 40×objectives under the Z stack scan mode at same depth. i and j) Expression of ACE-2 in glands (i) and nasal respiratory epithelium (j). k) No detectable signal in Goat IgG isotype control. l) Quantification of ACE-2-positive cells per mm epithelium. The positive cells in seven tracheal biopsies and 13 nasal specimen that contained both respiratory and olfactory epithelium were counted. m) Representative image of respiratory-olfactory mucosa adjacent area. PGP9.5 and ACE-2 co-staining image was obtained using confocal microscope under the tile scan mode. n) Quantification of the ACE-2 fluorescence intensity per µm epithelium. Nasal specimen that contained both respiratory and olfactory epithelium were quantified using Image J. o) Co-staining of ACE-2 and secretory cell marker Muc5ac in tracheal airway epithelium. The inset represents magnification of the selected area. Dots in graph represent independent specimens (h, l and n). Data are represented as mean±sem. p-value was calculated by unpaired two-tailed Student's t-test. Differences were considered significant when p<0.05. Scale bars: 20 µm.
High-intensity ACE-2 staining was detected in all 13 olfactory mucosal biopsies. In addition, ACE-2 is frequently observed in Bowman's glands and duct cells (figure 1i). In the adjacent nasal respiratory epithelium, ACE-2 is also located on the apical surface (figure 1j), with a significantly lower level of expression than the olfactory epithelium (figure 1l). As shown in figure 1m, in adjacent areas, there is intense ACE-2 expression in the PGP9.5+ olfactory region, but ACE-2 can barely be detected in the PGP9.5− respiratory epithelium. Only 47.4% of nasal respiratory epithelial biopsies (nine in 19) contained ACE-2-positive epithelial cells. We quantified the intensity of ACE-2 fluorescence and observed striking enrichment (200–700 fold) in the olfactory epithelium (figure 1n). This cellular tropism of SARS-CoV-2 may be associated with olfactory dysfunction and underlie its high transmissibility. Given the supporting function of Krt18+ sustentacular cells for olfactory sensory neurons and sensory cilia, the absence of ACE-2 expression in neurons indicates an indirect effect of SARS-CoV-2 infection on COVID-19-associated anosmia.
We further examined ACE-2 protein in tracheal epithelium. In two of seven specimens, we detected low expression of ACE-2 in Muc5ac+ secretory cells (figure 1o). The expression of ACE-2 by secretory cells is reminiscent of the recent findings that increased ACE-2 expression in small airway epithelium in COPD patients [10], a disease characterised by secretory cell hyperplasia [11]. Together, the comparatively enhanced human airway expression of ACE-2 localised to the olfactory neuroepithelium (figure 1m and n) suggests a mechanism of olfactory loss and a potential entry point of SARS-CoV-2 into the central nervous system and causes neurological symptoms in COVID-19 patients [12].
Recent studies have suggested a correlation between ACE-2 expression level and COVID-19-associated clinical traits. For example, the increased severity of COVID-19 in obese young patients may be linked to increased ACE-2 expression in lung epithelial cells [13, 14], and the lower ACE-2 gene expression in the nasal epithelium of children relative to adults may help explain the lower prevalence of COVID-19 in children [15]. The cohort presented in the current study are all over 30 years old, limiting the ability to assess the role of age as a factor in the level of ACE-2 expression. However, the relatively increased ACE-2 expression in olfactory versus respiratory epithelium is similar across all individuals. Notably, like in animals [16], the olfactory area in adults may be significantly larger compared to children, with a different geometry and airflow pattern.
Understanding of the pattern of viral load in tissues of COVID-19 patients is critical for diagnosis, management of transmission, and potential treatment strategies. Detection of SARS-CoV-2 in clinical specimens shows that the highest viral copy number is found in nasal swabs (∼200 fold), compared to bronchoalveolar lavage or pharyngeal swabs [17, 18]. In the early stages of SARS-CoV-2 infection, viral RNA can readily be detected in upper respiratory specimens but not in blood, urine or stool [19]. These findings, taken together with ACE-2 protein cellular localisation presented here, suggests that active virus infection and replication occurs in the apical layer of nasal and olfactory mucosa. The differential expression of ACE-2 in the olfactory neuroepithelium and respiratory epithelium may help to account for the spectrum of nasal-related symptoms, while also raising the intriguing possibility that COVID-19 may be amenable to novel therapeutic approaches. Whether nasal saline irrigation, a common treatment for sinonasal conditions, is beneficial or counter-productive in SARS-CoV-2 infection remains to be determined; however, consideration should be given to the delivery of topical anti-viral additives, such as detergent or povidone-iodine, directed at the nasal and nasopharyngeal viral reservoirs.
Shareable PDF
Footnotes
Conflict of interest: M. Chen has nothing to disclose.
Conflict of interest: W. Shen has nothing to disclose.
Conflict of interest: N.R. Rowan has nothing to disclose.
Conflict of interest: H. Kulaga has nothing to disclose.
Conflict of interest: A. Hillel has nothing to disclose.
Conflict of interest: M. Ramanathan Jr has nothing to disclose.
Conflict of interest: A.P. Lane has nothing to disclose.
Support statement: This work was funded by NIH grants R01 AI132590, R01 DC016106 (A.P. Lane). Funding information for this article has been deposited with the Crossref Funder Registry
References
- 1.Guan WJ, Ni ZY, Hu Y, et al. . Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382: 1708–1720. doi: 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, Yang XL, Wang XG, et al. . A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579: 270–273. doi: 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M, Kleine-Weber H, Schroeder S, et al. . SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181: 271–280. doi: 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamming I, Timens W, Bulthuis ML, et al. . Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 2004; 203: 631–637. doi: 10.1002/path.1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Z, Shi L, Wang Y, et al. . Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020; 8: 420–422. doi: 10.1016/S2213-2600(20)30076-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sungnak W, Huang N, Becavin C, et al. . SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 2020; 26: 681–687. doi: 10.1038/s41591-020-0868-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lechien JR, Chiesa-Estomba CM, De Siati DR, et al. . Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur Arch Otorhinolaryngol 2020; 277: 2251–2261. doi: 10.1007/s00405-020-05965-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen M, Reed RR, Lane AP. Chronic inflammation directs an olfactory stem cell functional switch from neuroregeneration to immune defense. Cell Stem Cell 2019; 25: 501–513 e5. doi: 10.1016/j.stem.2019.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Doremalen N, Schafer A, Menachery VD, et al. . SARS-like coronavirus WIV1-CoV does not replicate in Egyptian fruit bats (Rousettus aegyptiacus). Viruses 2018; 10: 727. doi: 10.3390/v10120727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leung JM, Yang CX, Tam A, et al. . ACE-2 expression in the small airway epithelia of smokers and COPD patients: implications for COVID-19. Eur Respir J 2020; 55: 2000688. doi: 10.1183/13993003.00688-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Randell SH. Airway epithelial stem cells and the pathophysiology of chronic obstructive pulmonary disease. Proc Am Thorac Soc 2006; 3: 718–725. doi: 10.1513/pats.200605-117SF [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao L, Jin H, Wang M, et al. . Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol 2020; 77: 1–9. doi: 10.1001/jamaneurol.2019.3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kass DA, Duggal P, Cingolani O. Obesity could shift severe COVID-19 disease to younger ages. Lancet 2020; 395: 1544–1545. doi: 10.1016/S0140-6736(20)31024-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al Heialy S, Hachim MY, Senok A, et al. . Regulation of angiotensin converting enzyme 2 (ACE2) in obesity: implications for COVID-19. bioRxiv 2020; preprint [ 10.1101/2020.04.17.046938]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bunyavanich S, Do A, Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA 2020; 323: 2427–2429. doi: 10.1001/jama.2020.8707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bilinska K, Jakubowska P, Von Bartheld CS, et al. . Expression of the SARS-CoV-2 entry proteins, ACE2 and TMPRSS2, in cells of the olfactory epithelium: identification of cell types and trends with age. ACS Chem Neurosci 2020; 11: 1555–1562. doi: 10.1021/acschemneuro.0c00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Xu Y, Gao R, et al. . Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020; 323: 1843–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou L, Ruan F, Huang M, et al. . SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med 2020; 382: 1177–1179. doi: 10.1056/NEJMc2001737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfel R, Corman VM, Guggemos W, et al. . Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581: 465–469. doi: 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This one-page PDF can be shared freely online.
Shareable PDF ERJ-01948-2020.Shareable (149.1KB, pdf)