Abstract
Background
Diverse and circumstantial evidence suggests that schizophrenia is a neurodevelopmental disorder. Genes contributing to neurodevelopment may be potential candidates for schizophrenia. The human SOX11 gene is a member of the developmentally essential SOX (Sry‐related HMG box) transcription factor gene family and mapped to chromosome 2p, a potential candidate region for schizophrenia.
Methods
Our previous genome‐wide association study (GWAS) implicated an involvement of SOX11 with schizophrenia in a Chinese Han population. To further investigate the association between SOX11 polymorphisms and schizophrenia, we performed an independent replication case‐control association study in a sample including 768 cases and 1348 controls.
Results
After Bonferroni correction, four SNPs in SOX11 distal 3′UTR significantly associated with schizophrenia in the allele frequencies: rs16864067 (allelic P = .0022), rs12478711 (allelic P = .0009), rs2564045 (allelic P = .0027), and rs2252087 (allelic P = .0025). The haplotype analysis of the selected SNPs showed different haplotype frequencies for two blocks (rs4371338‐rs7596062‐rs16864067‐rs12478711 and rs2564045‐rs2252087‐rs2564055‐rs1366733) between cases and controls. Further luciferase assay and electrophoretic mobility shift assay (EMSA) revealed the schizophrenia‐associated SOX11 SNPs may influence SOX11 gene expression, and the risk and non‐risk alleles may have different affinity to certain transcription factors and can recruit divergent factors.
Conclusions
Our results suggest SOX11 as a susceptibility gene for schizophrenia, and SOX11 polymorphisms and haplotypes in the distal 3′UTR of the gene might modulate transcriptional activity by serving as cis‐regulatory elements and recruiting transcriptional activators or repressors. Also, these SNPs may potentiate as diagnostic markers for the disease.
Keywords: association, neurodevelopment, schizophrenia, single‐nucleotide polymorphism (SNP), SOX11
1. INTRODUCTION
Schizophrenia is a chronic and devastating neuropsychiatric disorder and remains a long‐concerned unresolved public health problem with a steady prevalence of ~1% historically and worldwide. 1 With a peak onset at 18‐25 years old, schizophrenia inflicts physical and mental suffering on the affected individuals and their families with high medical and social costs. 2 Searching for specific biomarkers for schizophrenia plays an important role in identifying the disease state, contributing on underlying progression, and predicting the treatment response. 3 In the last centuries, studies for schizophrenia focused on the natural course of the disease, which various environmental and genetic risk factors are involved with the pathogenesis of schizophrenia. Family, twin, and adoption studies indicate an explicit hereditary contribution to the etiology of schizophrenia with heritability estimates of approximately 80%. 4 New experimental techniques, such as genome‐wide association study (GWAS), are providing insights into potential candidate genes involved. Since the first GWAS for schizophrenia was published in 2008, substantial polygenetic factors of small effect to the risk of schizophrenia have been identified, 5 resulting in the conclusion that schizophrenia is a polygenic and complex disorder.
Despite more than a century of research, the pathophysiological basis of schizophrenia remains undefined. By far, a vast majority of evidence revealed that schizophrenia is a neurodevelopmental disorder which indicates schizophrenia onset in late adolescence or early adulthood, risk factors operating mostly prenatally or during early childhood, general differences in intellect and behavior many years before onset, structural brain changes at or before onset, cognitive impairments, and functional alternatives of several neurodevelopmental genes (DISC1, NRG1, RELN, BDNF, and etc). 6 , 7 , 8 , 9 , 10 , 11 Neurodevelopment comprises multiple delicately tuned steps, including the proliferation, differentiation, migration, and integration of a variety of neural cell types. 12 The risks and insults to neurodevelopment occurring in the prenatal and postnatal stages have been related to the formation activation of aberrant neural circuits and emergence of schizophrenia symptoms. 13
In our previous GWAS performed in a Chinese Han population, 14 we identified, besides some prominent genes, several schizophrenia candidate genes with moderate significance, one of which was the human SOX11 gene. SOX11 is a member of the developmentally essential SOX (Sry‐related HMG box) transcription factor gene family and mapped to chromosome 2p, 15 , 16 a potential candidate region for schizophrenia. 17 , 18 Sox proteins have been indicated as major regulators for cell fate, survival, and differentiation across nearly all developing organ systems, and thus, dysfunction of Sox proteins can lead to all kinds of developmental diseases. 19 , 20 SOX11 plays a crucial role in neurodevelopment and organogenesis. Sox11 expression decreases along neurodevelopment progression and is absent in most normal adult tissues expect some niches locating stem cells with proliferation and differentiation potential. 21 A number of in vitro and in vivo experiments have substantiated the role of Sox11 as a regulator of various aspects of neuronal development. It has been revealed that Sox11 was co‐expressed with Doublecortin (DCX), a specific marker for neuronal precursor cells and immature neurons in neurogenic niches. 22 During cortical radial migration, suppression of dendrite morphogenesis by Sox11 is critical in cerebral cortex formation. In Xenopus, it has been confirmed that Sox11 was directly interacted with a MAP kinase NLK, linking Sox11 with Wnt signaling, in which many genes showed aberrant expression pattern and interrupted function in patients with schizophrenia. 23
In the present study, we aimed to examine the association of the human SOX11 gene with susceptibility for schizophrenia in an independent Chinese Han case‐control sample set and explore the potential for the polymorphisms of this gene as schizophrenia diagnosis biomarkers.
2. METHODS AND MATERIALS
2.1. Subjects
All subjects were unrelated Chinese Han nationality born and recruited in the Northern China. The sample set consisted of 768 patients affected by schizophrenia (360 males and 408 females; mean age: 33.5 ± 8.7 years) and 1348 healthy controls (658 males and 690 females; mean age: 31.1 ± 13.2 years). Consensus diagnoses were confirmed by at least two experienced psychiatrists according to the Diagnostic and Statistical Manual of Mental Disorders, fourth edition criteria (DSM‐IV, American Psychiatric Association, 2000). None of the patients had severe medical complications. The individuals with other severe psychiatric disorders, family history of other inherited diseases, and obvious somatic diseases were also excluded from the current study. The healthy controls included in the current study had no history of mental illness or any other neurological or medical condition that are suspected to be associated with schizophrenia; they were well matched to the patient group for gender, age, education, and ethnicity. All the healthy controls were recruited from communities by a simple none‐structured interview performed by psychiatrists.
2.2. SNP selection
The human SOX11 gene is a single‐exon gene with no introns, which means that most of the nucleotides in the gene region are nominated as the flanking sequences. The SNPs were selected by downloading the information of all the SNPs within and neighboring the human SOX11 gene from the International HapMap project database on dbSNP (http://www.ncbi.nlm.nih.gov/SNP/). The total coverage of the 15 selected SNPs was approximately 180kb.
2.3. Genotyping
Genomic DNA was extracted from venous blood using a commercially available QIAamp DNA Blood Mini Kit (QIAGEN). The SNPs were genotyped by either polymerase chain reaction (PCR) restriction fragment length polymorphism (RFLP) analysis or direct DNA sequencing. All primers were designed by software Oligo 6.0 (MBI Inc). PCR products were either completely digested with 4 U restriction enzyme overnight and then separated by agarose gel electrophoresis (2%‐3%) stained with ethidium bromide, or sequenced on an ABI PRISM 377‐96 DNA Sequencer (Applied Biosystems) after purifying them using a BigDye Terminator Cycle Sequencing Ready Reaction Kit. All the results were checked and confirmed by two experienced technicians independently.
2.4. Cell line culture and transfection
Human Neuroblastoma cell line SH‐SY5Y were maintained in DMEM (Invitrogen) supplemented with 10% FBS. Transient transfection of the cell lines was performed with LipofectamineTM 2000 (Invitrogen) according to the manufacturer's instructions.
2.5. Luciferase assay
Human SH‐SY5Y neuroblastoma cells with endogenous Sox11 expression were seeded in 24‐well plates and transiently transfected with equimolar amounts of various pGL3‐promoter vectors (Promega) containing different inserted SNP‐site(s) by using LipofectamineTM 2000 (Invitrogen). pRLCMV (Promega) was co‐transfected as an internal control of transfection efficiency. The transfected cells were harvested after 48 hours. Luminescence was measured by the dual‐luciferase reporter assay system (Promega) using a Centro LB960 96‐well luminometer (Berthold Technologies).
2.6. Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared as previous described. 24 Biotin‐labeled and corresponding unlabeled oligonucleotides containing the various genotypes were synthesized by BGI Genomics. Equal amounts of complementary oligonucleotides were heated at 95°C for 5 minutes and annealed by stepwise reducing the temperature to 25°C in 1 hours. EMSA was performed by using the commercial LightShift® Chemiluminescent EMSA kit (Pierce). Membrane was labeled with IRDye 800CW streptavidin and then detected by LI‐COR OdysseyH scanner and software (LI‐COR Biosciences).
2.7. Statistics
Deviation of the genotypes from the Hardy‐Weinberg equilibrium was examined by a chi‐squared (χ 2) goodness‐of‐fit test (Table 1). Distribution of gender and the difference of age between cases and controls were evaluated by Pearson's chi‐squared test and Student's t test. Statistical differences in genotypic and allelic distribution between patients and controls were evaluated by the Pearson's chi‐squared test at a significance level of 0.05. To analyze the association between SOX11 SNP genotypic distribution and schizophrenia, multiple logistic regression analyses were performed using the following: codominant 1 (major allele homozygotes vs. heterozygotes), codominant 2 (major allele homozygotes vs. minor allele homozygotes), dominant (major allele homozygotes vs. heterozygotes + minor allele homozygotes), recessive (major allele homozygotes + heterozygotes vs. minor allele homozygotes), and log‐additive (major allele homozygotes vs. heterozygotes vs. minor allele homozygotes). In codominant model, each genotype gives a different and non‐additive risk; therefore, we compared the major allele homozygotes (the most frequent alleles) to heterozygotes in codominant 1 and minor allele homozygotes to heterozygotes in codominant 2. In dominant model, a single copy of allele was enough to modify the risk with heterozygous and homozygous genotypes having the same risk. In recessive model, two copies of allele were necessary to change the risk. In overdominant model, heterozygotes were compared with both allele homozygotes. In the log‐additive model, each copy of allele modified the risk in an additive form. The haplotype frequencies were estimated by the expectation maximization algorithm. Pairwise linkage disequilibrium (LD) between any two alleles was evaluated by D′ and r 2 values. Odds ratio (OR) and their 95% confidence intervals (95% CI) were calculated to evaluate the effect of different alleles and haplotypes. The association analyses were performed by the SHEsis (http://analysis2.bio‐x.cn/myAnalysis.php) 25 , 26 and SPSS Statistics 17.0 program (SPSS Inc). The statistical power of the sample size was calculated by the genetic power calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/cc2.html). The sample had approximately 80% power to detect allele frequency differences assuming an OR of 1.5 with a minor allele frequency of 0.1. The Bonferroni correction for multiple testing was carried out to control inflation of the type I error rate. Results were considered significant at two tailed P < .05.
Table 1.
List of SNPs included in the present study
| rs code | Marker order | Position a | Distance from SNP1 (kb) | Allele Change | HCB b MAF | Sample set HWE P | Case HWE P | Control HWE P |
|---|---|---|---|---|---|---|---|---|
| rs1821797 | SNP1 | 5671640 | 0 | C>T | 0.227 | .160 | .457 | .233 |
| rs4485539 | SNP2 | 5724305 | 52.7 | T>C | 0.322 | .348 | .147 | .966 |
| rs7563508 | SNP3 | 5727884 | 56.2 | G>A | 0.389 | .268 | .259 | .665 |
| rs4547512 | SNP4 | 5739199 | 67.6 | T>G | 0.100 | .390 | .757 | .403 |
| rs4371338 | SNP5 | 5755481 | 82.8 | A>G | 0.478 | .159 | .383 | .255 |
| rs7596062 | SNP6 | 5772751 | 101.1 | T≥G | 0.500 | .517 | .426 | .793 |
| rs16864067 | SNP7 | 5777782 | 106.1 | G>A | 0.367 | .921 | .455 | .715 |
| rs12478711 | SNP8 | 5785238 | 113.6 | G>A | 0.389 | .982 | .371 | .524 |
| rs2564045 | SNP9 | 5787341 | 115.7 | A>G | 0.456 | .715 | .490 | .352 |
| rs2252087 | SNP10 | 5793753 | 122.1 | G>T | 0.244 | .443 | .285 | .803 |
| rs2564055 | SNP11 | 5803780 | 132.1 | A>C | 0.243 | .331 | .145 | .845 |
| rs1366733 | SNP12 | 5807622 | 136.0 | C>T | 0.267 | .626 | .209 | .771 |
| rs11892518 | SNP13 | 5818625 | 147.0 | C>T | 0.389 | .672 | .321 | .820 |
| rs10929818 | SNP14 | 5834699 | 163.1 | A>G | 0.400 | .571 | .566 | .807 |
| rs1429219 | SNP15 | 5854684 | 183.0 | C>T | 0.244 | .148 | .574 | .157 |
Abbreviations: HCB‐Han Chinese in Beijing; HWE‐Hardy‐Weinberg Equilibrium; MAF‐minor allele frequency.
From International HapMap database release#27.
Chinese Han population MAF from the International HapMap Project Database.
3. RESULTS
3.1. Single‐allele association of selected SOX11 SNPs with schizophrenia
None of the genotype distributions of the 15 selected SNPs in case and control groups deviated from Hardy‐Weinberg equilibrium. Detailed information and location of the selected SNPs are shown in Table 1 and Figure 1. Of the 15 SNPs, nine SNPs (rs4485539‐SNP2, rs7596062‐SNP6, rs16864067‐SNP7, rs12478711‐SNP8, rs2564045‐SNP9, rs2252087‐SNP10, rs2564055‐SNP11, rs1366733‐SNP12, and rs11892518‐SNP13) showed statistical differences in allele frequencies between cases and controls (Table 2). After rigorous Bonferroni correction, four SNPs located in the SOX11 3′UTR remained significantly associated with schizophrenia in the allele frequencies; SNP7 (allelic P = .0022), SNP8 (allelic P = .0009), SNP9 (allelic P = .0027), and SNP10 (allelic P = .0025) (Table 2).
Figure 1.
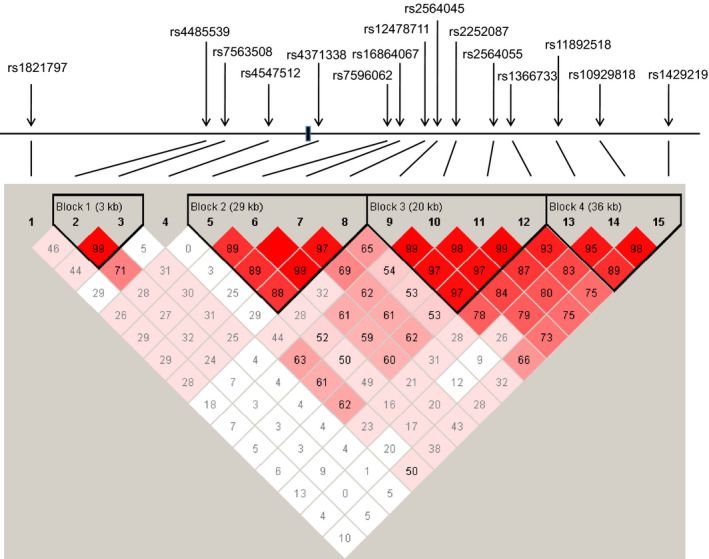
Genomic structure and linkage disequilibrium (LD) of human SOX11 gene. Human SOX11 is a single‐exonic gene. The positions of the 15 SNPs selected in the SOX11 gene are shown with arrows. LDs were computed for all possible combinations of the 15 SNPs using D′ values. Blocks were defined by a solid spine of LD
Table 2.
Genotype and allele frequencies of 15 SNPs in the human SOX11 gene between schizophrenia patients and controls
| No. | subjects | Allele and frequency a | χ 2 P(P b ) | OR (95% CI) | |
|---|---|---|---|---|---|
| C | T | ||||
| SNP1 | Case | 1209 (0.787) | 327 (0.213) | χ 2 = 0.9745 | 0.93 (0.79‐1.08) |
| Control | 2150 (0.800) | 538 (0.200) | p = .3236 | ||
| C | T | ||||
| SNP2 | Case | 520 (0.339) | 1016 (0.661) | χ2 = 4.7643 | 0.86 (0.76‐0.99) |
| Control | 1003 (0.372) | 1693 (0.628) | p = .0291 | ||
| A | G | ||||
| SNP3 | Case | 614 (0.400) | 922 (0.600) | χ2 = 3.3068 | 0.89 (0.78‐1.01) |
| Control | 1155 (0.428) | 1541 (0.572) | p = .0691 | ||
| G | T | ||||
| SNP4 | Case | 163 (0.106) | 1373 (0.894) | χ2 = 0.2670 | 0.95 (0.77‐1.16) |
| Control | 300 (0.111) | 2396 (0.889) | p = .6053 | ||
| A | G | ||||
| SNP5 | Case | 780 (0.508) | 756 (0.492) | χ2 = 1.8788 | 1.092 (0.96‐1.24) |
| Control | 1310 (0.486) | 1386 (0.514) | p = .1705 | ||
| G | T | ||||
| SNP6 | Case | 783 (0.510) | 753 (0.490) | χ2 = 6.9301 | 1.18 (1.04‐1.34) |
| Control | 1260 (0.468) | 1434 (0.532) | p = .0085 | ||
| A | G | ||||
| SNP7 | Case | 620 (0.404) | 914 (0.596) | χ2 = 9.3616 | 1.22 (1.07‐1.39) |
| Control | 962 (0.357) | 1734 (0.643) | p = .0022 (.033) | ||
| A | G | ||||
| SNP8 | Case | 640 (0.417) | 896 (0.583) | χ2 = 11.0119 | 1.24 (1.09‐1.41) |
| Control | 982 (0.365) | 1708 (0.635) | p = .0009 (.0135) | ||
| A | G | ||||
| SNP9 | Case | 857 (0.558) | 679 (0.442) | χ2 = 9.0128 | 1.21 (1.07‐1.38) |
| Control | 1374 (0.510) | 1320 (0.490) | p = .0027 (.0405) | ||
| G | T | ||||
| SNP10 | Case | 1117 (0.727) | 419 (0.273) | χ2 = 9.1470 | 1.23 (1.08‐1.42) |
| Control | 1841 (0.683) | 855 (0.317) | p = .0025 (.00375) | ||
| A | C | ||||
| SNP11 | Case | 1111 (0.723) | 425 (0.277) | χ2 = 7.5864 | 1.21 (1.06‐1.39) |
| Control | 1841 (0.683) | 855 (0.317) | p = .0059 | ||
| C | T | ||||
| SNP12 | Case | 1117 (0.727) | 419 (0.273) | χ2 = 6.1519 | 1.19 (1.04‐1.37) |
| Control | 1863 (0.691) | 833 (0.309) | p = .0132 | ||
| C | T | ||||
| SNP13 | Case | 911 (0.595) | 621 (0.405) | χ2 = 4.1815 | 0.87 (0.77‐0.99) |
| Control | 1689 (0.626) | 1007 (0.374) | p = .0409 | ||
| A | G | ||||
| SNP14 | Case | 869 (0.566) | 667 (0.434) | χ2 = 1.2700 | 1.08 (0.95‐1.22) |
| Control | 1477 (0.548) | 1219 (0.452) | p = .2598 | ||
| C | T | ||||
| SNP15 | Case | 1119 (0.729) | 417 (0.271) | χ2 = 0.4919 | 1.05 (0.91‐1.21) |
| Control | 1937 (0.718) | 759 (0.282) | p = .4831 | ||
Significant P values (<.05) are in boldface.
Frequencies are shown in parenthesis.
Significant P value after the strict Bonferroni correction.
3.2. Genotypic association of nine SNPs located in the SOX11 3′UTR
To further investigate the nine SNPs significantly associated with schizophrenia after single‐allele analysis, we examined the genotypic association of these SNPs under different genetic models. After analyzing the association of the SNPs by five genetic models (codominant, dominant, recessive, overdominant, and log‐additive models) with schizophrenia, the consequences demonstrated that all the SNP showed significant differences between cases and control under at least two different genetic models: (a) SNP2, codominant (P = .0087), dominant (P = .0080), overdominant (P = .0234), and log‐additive (P = .0279); (b) SNP6, codominant (P = .0426), recessive (P = .0126), and log‐additive (P = .0260); (c) SNP7, codominant (P = .0218), dominant (P = .0047), and overdominant (P = 0374); (d) SNP8, codominant (P = .0092), dominant (P = .0017), recessive (P = .0310), and log‐additive (P = .0279); (e) SNP 9, codominant (P = .0331), dominant (P = .0283), recessive (P = .0065), and log‐additive (P = .0097); (f) SNP10, dominant (P = .0116), recessive (P = .0133), and log‐additive (P = .0083); (g) SNP11, dominant (P = .0340), recessive (P = .0098), and log‐additive (P = .0131); (h) SNP12, codominant (P = .0289), recessive (P = .0096), and log‐additive (P = .0210); and (i) SNP13, codominant (P = .0260) and dominant (P = .0189) (Table 3).
Table 3.
Genotype Association of nine SNPs under different genetic models with schizophrenia
| No. | Genotype | Case a | Control a | Codominant | Dominant | Recessive | Overdominant | Log‐additive | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | P | ||||
| SNP2 |
TT CT CC |
345 (0.449) 326 (0.424) 97 (0.126) |
526 (0.390) 641 (0.476) 181 (0.134) |
1.290 (1.068‐1.558) 1.054 (0.793‐1.395) |
.0087 .7144 |
1.275 (1.064‐1.526) | .0080 | 1.073 (0.823‐1.397) | .6017 | 1.229 (1.029‐1.471) | .0234 | .0279 |
| SNP6 |
GG GT TT |
194 (0.253) 395 (0.514) 179 (0.233) |
294 (0.218) 672 (0.499) 381 (0.283) |
1.123 (0.899‐1.397) 0.799 (0.643‐0.9931) |
.3023 .0426 |
1.211 (0.985‐1.487) | .0714 | 0.771 (0.627‐0.945) | .0126 | 0.940 (0.788‐1.121) | .4947 | .0260 |
| SNP7 |
GG AG AA |
268 (0.349) 378 (0.493) 121 (0.158) |
555 (0.412) 624 (0.463) 169 (0.125) |
0.797 (0.658‐0.968) 1.182 (0.910‐1.540) |
.0218 .2180 |
0.767 (0.640‐ 0.922) | .0047 | 1.307 (1.014‐1.684) | .0374 | 0.887 (0.744 −1.058) | .1852 | .0085 |
| SNP8 |
GG AG AA |
256 (0.333) 384 (0.500) 128 (0.167) |
541 (0.402) 626 (0.465) 178 (0.132) |
0.771 (0.634‐0.938) 1.172 (0.905‐1.523) |
.0092 .2310 |
0.743 (0.617‐0.895) | .0017 | 1.311 (1.021‐1.672) | .0310 | 0.871 (0.730‐1.038) | .1259 | .0035 |
| SNP9 |
AA AG GG |
236 (0.307) 385 (0.501) 147 (0.191) |
354 (0.263) 666 (0.494) 327 (0.243) |
1.153 (0.937‐1.416) 0.778 (0.619‐0.982) |
.1770 .0331 |
1.244 (1.023‐1.513) | .0283 | 0.738 (0.594‐0.919) | .0065 | 0.973 (0.816‐1.160) | .7612 | .0097 |
| SNP10 |
GG GT TT |
401 (0.522) 315 (0.410) 52 (0.068) |
627 (0.465) 587 (0.435) 134 (0.099) |
1.192 (0.988‐1.433) 0.723 (0.5079‐1.021) |
.0638 .0673 |
1.256 (1.054‐1.499) | .0116 | 0.658 (0.474‐0.913) | .0133 | 1.109 (0.927‐1.328) | .2577 | .0083 |
| SNP11 |
AA AC CC |
394 (0.513) 323 (0.421) 51 (0.066) |
627 (0.465) 587 (0.435) 134 (0.099) |
1.142 (0.947‐1.372) 0.692 (0.484‐0.979) |
.1600 .0382 |
1.211 (1.016‐1.445) | .0340 | 0.644 (0.463‐0.897) | .0098 | 1.063 (0.889‐1.273) | .5060 | .0131 |
| SNP12 |
CC CT TT |
399 (0.520) 319 (0.415) 50 (0.065) |
647 (0.480) 569 (0.422) 132 (0.098) |
1.100 (0.912‐1.322) 0.676 (0.478‐0.963) |
.3135 .0289 |
1.172 (0.983‐1.398) | .0801 | 0.642 (0.458‐0.898) | .0096 | 1.028 (0.859‐1.230) | .7625 | .0210 |
| SNP13 |
CC CT TT |
264 (0.345) 383 (0.500) 119 (0.155) |
534 (0.396) 621 (0.461) 193 (0.143) |
0.802 (0.660‐0.974) 1.000 (0.768‐1.301) |
.0260 .9984 |
0.802 (0.667‐0.964) | .0189 | 1.101 (0.861‐1.406) | .4480 | 0.854 (0.716‐1.019) | .0818 | .0636 |
Significant P values (<.05) are in boldface.
Frequencies are shown in parenthesis.
3.3. Haplotype analysis of selected SOX11 SNPs with schizophrenia
To analyze the haplotype structure, the pairwise LD (linkage disequilibrium) of the 15 SNPs in our sample set was computed by the standardized measure D′ value. D′ value of two SNPs ranging between 0.8 and 1.0 indicated strong LD. Four strong LD haplotypes were constructed. The LD haplotype structure was shown in Figure 1. To investigate whether any haplotype would result in a higher risk for schizophrenia, all specific and global haplotypes of the 15 SNPs were tested. Specific P‐values for individual haplotype combinations, global P‐values for each haplotype and estimated haplotype frequencies in cases and controls are summarized in Table 4. The global association analyses revealed positive results for the second haplotype (χ 2 = 11.941, P = .0076) and third haplotype (χ 2 = 9.729, P = .0078). All the four haplotypes had specific haplotype combinations associated with schizophrenia. For the first haplotype, the C‐A haplotype combination was different in frequency between cases and controls (χ 2 = 4.683, P = .0305). For the second haplotype, the A‐G‐A‐A haplotype combination showed a distribution difference between cases and controls (χ 2 = 10.136, P = .0015). For the third haplotype, two haplotype combinations were associated with schizophrenia; A‐G‐A‐C (χ 2 = 8.502, P = .0036) and G‐T‐C‐T (χ 2 = 7.921, P = .0049). For the last haplotype, the T‐A‐C haplotype combination showed association with schizophrenia (χ 2 = 4.418, P = .0356).
Table 4.
Estimated haplotype frequencies and case‐control haplotype results of the human SOX11 gene
| Combinations | Haplotype | Haplotype frequency a | χ 2 | P | OR (95%CI) | global | ||
|---|---|---|---|---|---|---|---|---|
| Case | Control | χ 2 | P | |||||
| SNP2‐SNP3 |
C‐A T‐A T‐G |
518.94 (0.338) 95.06 (0.062) 920.94 (0.600) |
999.81 (0.371) 155.19 (0.058) 1537.81 (0.570) |
4.683 0.325 3.356 |
.0305 .5687 .0670 |
0.87 (0.76‐0.99) 1.08 (0.83‐1.41) 1.13 (0.99‐1.28) |
4.711 | .0950 |
|
SNP5‐SNP6‐ SNP7‐SNP8 |
A‐G‐A‐A A‐G‐G‐G A‐T‐G‐G G‐T‐G‐G |
576.50 (0.376) 126.03 (0.082) 47.62 (0.031) 703.19 (0.458) |
889.03 (0.331) 249.09 (0.093) 112.79 (0.042) 1315.78 (0.490) |
10.136 1.171 3.028 3.077 |
.0015 .2793 .0819 .0795 |
1.24 (1.09‐1.42) 0.88 (0.71‐1.10) 0.74 (0.52‐1.04) 0.89 (0.78‐1.01) |
11.941 | .0076 |
|
SNP9‐SNP10‐ SNP11‐SNP12 |
A‐G‐A‐C G‐T‐C‐T G‐G‐A‐C |
846.14 (0.551) 402.90 (0.262) 253.77 (0.165) |
1363.17 (0.506) 817.71 (0.304) 460.74 (0.171) |
8.502 7.921 0.207 |
.0036 .0049 .6492 |
1.21 (1.06‐1.37) 0.82 (0.71‐0.94) 0.96 (0.81‐1.14) |
9.729 | .0078 |
|
SNP13‐SNP14‐ SNP15 |
C‐A‐C C‐G‐C C‐G‐T T‐A‐C |
252.54 (0.165) 251.76 (0.164) 405.81 (0.265) 608.66 (0.397) |
488.28 (0.181) 464.55 (0.172) 734.91 (0.273) 982.62 (0.364) |
1.827 0.459 0.314 4.418 |
.1766 .4980 .5755 .0356 |
0.89 (0.75‐1.05) 0.94 (0.80‐1.12) 0.96 (0.83‐1.11) 1.15 (1.01‐1.31) |
4.852 | .1831 |
Significant P values (<.05) are in boldface.
Frequencies are shown in parenthesis.
3.4. Luciferase assay for the schizophrenia‐associated SNPs
Considering the distal 3′ location of positively associated SNPs, they may modulate SOX11 gene expression. Therefore, several luciferase reporter vectors were constructed by cloning DNA fragments of 200‐250 bps spanning each candidate SNPs into the 3′ side of the luciferase open reading frame (ORF) in the pGL3.0‐promoter reporter. The SNP‐site was in close proximity to the midpoint of the cloned sequence. The insert was sequenced, and a single site mutation was performed with the QuikChange II Site‐Directed Mutagenesis Kit (Agilent Technologies) to attain the allele of opposite risk. rs7596062‐SNP6, rs16864067‐SNP7, rs12478711‐SNP8, rs2564045‐SNP9, rs2252087‐SNP10, and rs2564055‐SNP11 were all positively associated with schizophrenia and scattered in the second or third haplotypes, which were the two significantly schizophrenia‐associated haplotypes globally. The reporter gene expression assays were performed using the Human neuroblastoma SH‐SY5Y cell line. The cloning site was to the 3′ side of luciferase gene, following SV40 polyA signaling sequence, which partially mimicked the SNPs genome environment. As shown in Figure 2, in SH‐SY5Y cells, 4 out of the 6 SNP pairs exhibited transcriptional regulatory effect on luciferase reporter gene compared with the empty control, which could be classified into 3 groups according to the regulatory tendency of non‐risk alleles: (a) upregulation, SNP6 (~1.27‐fold) and SNP10 (~2.15‐fold), while risk alleles exhibited no activation or a weak inhibitory effect; (b) no effect, SNP8, and SNP9; and (c) downregulation, SNP7 (~93%) and SNP11 (~78%), while the risk alleles exhibited stronger inhibitory effect, 52% and 45%, respectively. These results indicated that SOX11 3′ distal SNPs may modulate its expression.
Figure 2.
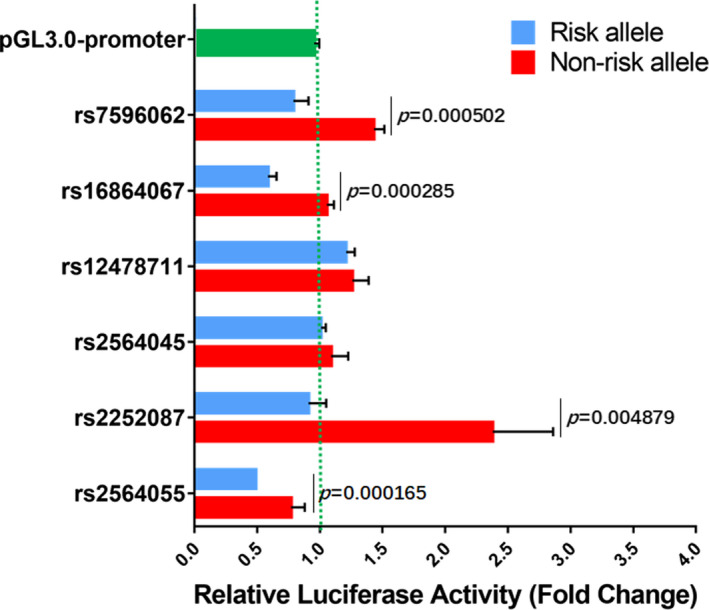
Luciferase analysis for the modulation of the 6 schizophrenia‐associated SNPs on gene expression. Of the 6, four SNP regions modulate Luciferase reporter gene expression with a significance difference between risk and non‐risk alleles. Non‐risk alleles have higher transcriptional activity than risk alleles
3.5. Electrophoretic Mobility Shift Assay (EMSA) for the Schizophrenia‐associated SNPs
According to these results, these SNP alleles may work as anchor points of regulatory DNA motifs, and the risk/non‐risk alleles have different affinity to certain transcription factors or can recruit divergent factors. Thus, EMSA with DNA sequences spanning ± 20 bps of each SNP alleles was performed, and the band shift differences between risk and non‐risk alleles (Figure 3), suggesting different potential of recruiting transcription factors (TFs). To further identify the corresponding TFs, the DNA sequences were subjected to MatInspector (www.genomatix.de/) for in‐silico analysis. We found that a single nucleotide substitution caused remarkable changes in TF prediction among all the 6 SNP pairs of DNA sequences and a total of 22 TFs were predicted binding to only one allele of these SNP pairs. Eight of the predicted TFs were ruled out as no mRNA expression detected in any brain regions or restricted to hindbrain (mRNA‐seq data from Allen brain institute, http://www.brain‐map.org/). The remaining 14 unambiguously expressed transcription factors could be grouped as followed: (a) constantly and ubiquitously expressed, including HBP1, MYT1L, ZNF239, DBP, POU3F3, POU6F1, CUX1, ESRRA, and NR1D1, (b) upregulated with development, NKX3‐1, and PPARG, (c) downregulated with development, PLAG1, and (d) regional specific, POU4F1, and DMRT3. Among all these transcription factors, we found that over‐expression of ZNF239 inhibited reporter gene expression, which is more effective on rs16864067‐SNP7 risk allele; while, on the contrary, ZNF239 knockdown released the inhibitory effect which was more effective on rs16864067‐SNP7 non‐risk allele (Figure 4A). EMSA performed with eukaryotic or prokaryotic over‐expressed ZNF239 protein showed higher affinity to risk allele probe (Figure 4B). These results indicated that SOX11 3′ distal SNPs may modulate its expression by serving as cis‐regulatory elements and recruiting transcriptional activators or repressors.
Figure 3.
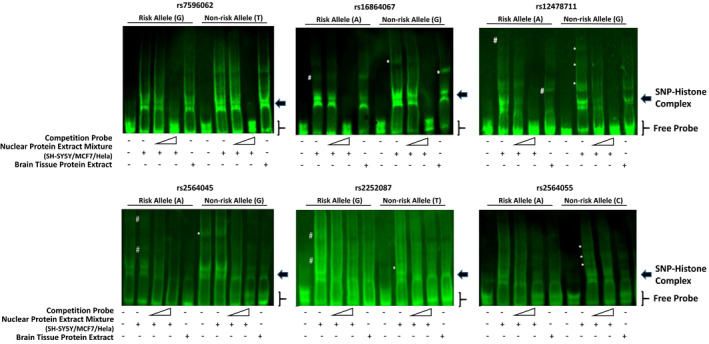
Detection of shift difference between risk and non‐risk alleles among the 6 schizophrenia‐associated SNPs by EMSA. Different allele of the schizophrenia‐associated SNPs showed different potential of recruiting transcription factors. Asterisk (*) indicated the risk allele specific shift, and pound sign (#) indicated the non‐risk allele specific shift
Figure 4.
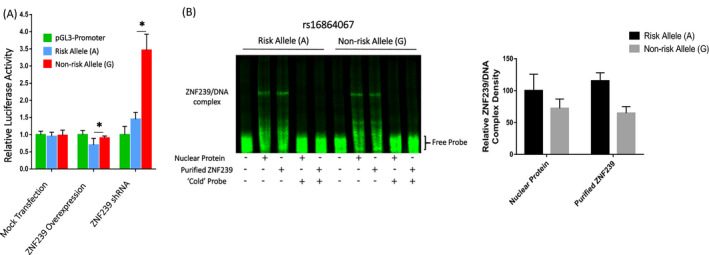
Influence of rs16864067 allele on affinity between transcription factor ZNF239 and DNA. A, Over‐expression of ZNP239 decrease transcription activity of rs16864067 region, which is more effective on risk allele. ZNF239 knockdown exhibits opposite effect. B, SNP rs16864067 recruits transcription repressor ZNF239, and light density quantification shows the risk allele have a higher affinity. *p < .05 and **p < .01
4. DISCUSSION
In the present research, we applied an association study for the human SOX11 gene to investigate the involvement of this gene with schizophrenia in an independent case‐control sample set including 768 schizophrenia patients and 1348 healthy controls. Allelic frequencies of 9 out of 15 selected SNPs covering the whole SOX11 gene region showed differences between cases and controls. After strict Bonferroni correction, 4 SNPs (rs16864067‐SNP7, rs12478711‐SNP8, rs2564045‐SNP9, and rs2252087‐SNP10) were still significantly associated with schizophrenia. The genotypic association of the 9 SNPs under different models by multiple logistic regression analyses showed that these SNPs were associated with schizophrenia significantly under at least two different genetic models. Two LD blocks containing the 4 Bonferroni‐significant SNPs also showed global differences in frequency between cases and controls. These data suggested SOX11 as a susceptibility gene for schizophrenia.
However, it is worth noting that, in the light of the construction of schizophrenia polygenic inheritance model, the most important and challenging task is to interpret the numerous potential schizophrenia genetic susceptibility loci. Therefore, in the present study, other than investigating the association between the SOX11 gene and schizophrenia, we also dissected the effect of the risk/non‐risk alleles on the expression of the SOX11 gene. These schizophrenia‐associated SNPs are in the distal 3′ UTR of SOX11 with potential transcriptional function. We examined whether the allelic variants of associated SNPs may modulate SOX11 gene expression by in vitro luciferase reporter assay and electrophoretic mobility shift assay (EMSA). The luciferase assays showed significant loss in promoter activity was related to risk alleles of the associated SNP, indicating that the risk alleles were less efficient in driving transcription than the non‐risk alleles and may inhibited the expression of SOX11 gene. Furthermore, we examined the different potential of the DNA sequences containing these SNPs to recruit TFs, and EMSA results showed a single nucleotide substitution caused remarkable changes in TF prediction among all examined SNP pairs of DNA sequences, indicating the different affinity of the SOX11 risk/non‐risk alleles in the 3′UTR region to TFs and solidating the functional prediction of schizophrenia‐associated SNPs in the regulation of SOX11 gene expression. Further examination discovered that a specific TF, ZNF239, had more effective reaction with risk allele of rs16864067‐SNP7 other than the non‐risk allele. The interaction of ZNF239 with 3′UTR region of SOX11 gene may inhibit its transcriptional level; therefore, high affinity of risk allele of rs16864067‐SNP7 with ZNF239 suggests the attenuated function of the SOX11 gene. These data showed that the associated SNPs may modulate SOX11 gene expression, indicating the schizophrenia‐associated SNPs located in SOX11 distal 3′UTR may contribute to normal neurodevelopment and fine response to environmental alterations by conditionally regulating SOX11 expression level.
The expression level of Sox11 is spontaneously upregulated in neurogenesis region with an indispensable role in neuronal regeneration and neuroplasticity. It has been reported that Sox11 expression increased immediately and significantly after electroconvulsive shock (ECS), an effective treatment for patients suffering from schizophrenia and major depression disorders. 27 Also, Sox11 and its putative binding partner Brn1 were induced in CA1 and/or DG of hippocampus following transient forebrain ischemia in rat. 28 BDNF, a pleiotropic growth factor influencing neuronal survival, differentiation, synaptic plasticity, and regeneration, brain‐derived neurotrophic factor, has been deeply investigated in schizophrenia and other disorders and modulated by Sox11 in a cell‐type and contextual specific manner. 29 It is notable that other than the role in early‐stage neurodevelopment, abnormal expression of Sox11 may lead to clinical symptoms due to adaptability decline of schizophrenia for adverse stimulus in external environment. However, more than one thousand schizophrenia candidate genes have been reported with poor repeatability and verification by thousands of candidate gene and genome‐wide association studies since 1965. 30 Thus, elaborate and convinced interpretation of SOX11 in schizophrenia emergence is required and critical for approval of its involvement in schizophrenia etiology by further functional studies of SOX11 in mice.
In conclusion, our independent case‐control association study confirmed a significant association between potential functional polymorphisms of the human SOX11 gene located in its distal 3′UTR and schizophrenia, indicating that SOX11 may contribute to schizophrenia risk, and SOX11 polymorphisms and haplotypes in the distal 3′UTR of the gene might modulate the SOX11 transcriptional activity by serving as cis‐regulatory elements and recruiting transcriptional activators or repressors. The functional polymorphisms may serve as the diagnostic targets for schizophrenia.
CONFLICT OF INTEREST
There are no conflict of interests to declare.
ETHICAL APPROVAL
All subjects in the current study provided written informed consent for the genetic study, which was approved by the Ethical Committee of the Institute of Mental Health, Peking University.
ACKNOWLEDGMENTS
We extend our gratitude to all the patients participating in this study. This work was supported by grants from the National Natural Science Foundation of China (81601174 and 31500764), and Natural Science Foundation of Liaoning Province, China (2019‐ZD‐0938).
Sun C‐P, Sun D, Luan Z‐L, et al. Association of SOX11 Polymorphisms in distal 3′UTR with Susceptibility for Schizophrenia. J Clin Lab Anal. 2020;34:e23306 10.1002/jcla.23306
Cheng‐Peng Sun and Dong Sun contributed equally to this work.
REFERENCES
- 1. Lewis DA, Lieberman JA. Catching up on schizophrenia: natural history and neurobiology. Neuron. 2000;28(2):325‐334. [DOI] [PubMed] [Google Scholar]
- 2. Wieronska JM, Zorn SH, Doller D, Pilc A. Metabotropic glutamate receptors as targets for new antipsychotic drugs: historical perspective and critical comparative assessment. Pharmacol Ther. 2016;157:10‐27. [DOI] [PubMed] [Google Scholar]
- 3. Goff DC, Romero K, Paul J, Mercedes Perez‐Rodriguez M, Crandall D, Potkin SG. Biomarkers for drug development in early psychosis: Current issues and promising directions. Eur Neuropsychopharmacol. 2016;26(6):923‐937. [DOI] [PubMed] [Google Scholar]
- 4. Cardno AG, Marshall EJ, Coid B, et al. Heritability estimates for psychotic disorders: the Maudsley twin psychosis series. Arch Gen Psychiatry. 1999;56(2):162‐168. [DOI] [PubMed] [Google Scholar]
- 5. Bergen SE, Petryshen TL. Genome‐wide association studies of schizophrenia: does bigger lead to better results? Curr Opin Psychiatry. 2012;25(2):76‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fatemi SH, Folsom TD. The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr Bull. 2009;35(3):528‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lewis DA, Levitt P. Schizophrenia as a disorder of neurodevelopment. Annu Rev Neurosci. 2002;25:409‐432. [DOI] [PubMed] [Google Scholar]
- 8. Perez JD, Rubinstein ND, Dulac C. New perspectives on genomic imprinting, an essential and multifaceted mode of epigenetic control in the developing and adult brain. Annu Rev Neurosci. 2016;39:347‐384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bradshaw NJ, Soares DC, Carlyle BC, et al. PKA phosphorylation of NDE1 is DISC1/PDE4 dependent and modulates its interaction with LIS1 and NDEL1. J Neurosci. 2011;31(24):9043‐9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perez‐Garcia CG. ErbB4 in laminated brain structures: a neurodevelopmental approach to schizophrenia. Front Cell Neurosci. 2015;9:472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lopez‐Bendito G, Cautinat A, Sanchez JA, et al. Tangential neuronal migration controls axon guidance: a role for neuregulin‐1 in thalamocortical axon navigation. Cell. 2006;125(1):127‐142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Budday S, Steinmann P, Kuhl E. Physical biology of human brain development. Front Cell Neurosci. 2015;9:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arnold SE. Neurodevelopmental abnormalities in schizophrenia: insights from neuropathology. Dev Psychopathol. 1999;11(3):439‐456. [DOI] [PubMed] [Google Scholar]
- 14. Yue WH, Wang HF, Sun LD, et al. Genome‐wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nat Genet. 2011;43(12):1228‐1231. [DOI] [PubMed] [Google Scholar]
- 15. Jay P, Goze C, Marsollier C, et al. The human SOX11 gene: cloning, chromosomal assignment and tissue expression. Genomics. 1995;29(2):541‐545. [DOI] [PubMed] [Google Scholar]
- 16. Azuma T, Ao S, Saito Y, et al. Human SOX11, an upregulated gene during the neural differentiation, has a long 3' untranslated region. DNA Res. 1999;6(5):357‐360. [DOI] [PubMed] [Google Scholar]
- 17. DeLisi LE, Mesen A, Rodriguez C, et al. Genome‐wide scan for linkage to schizophrenia in a Spanish‐origin cohort from Costa Rica. Am J Med Genet. 2002;114(5):497‐508. [DOI] [PubMed] [Google Scholar]
- 18. Straub RE, MacLean CJ, Ma Y, et al. Genome‐wide scans of three independent sets of 90 Irish multiplex schizophrenia families and follow‐up of selected regions in all families provides evidence for multiple susceptibility genes. Mol Psychiatry. 2002;7(6):542‐559. [DOI] [PubMed] [Google Scholar]
- 19. Prior HM, Walter MA. SOX genes: architects of development. Mol Med. 1996;2(4):405‐412. [PMC free article] [PubMed] [Google Scholar]
- 20. Pevny L, Placzek M. SOX genes and neural progenitor identity. Curr Opin Neurobiol. 2005;15(1):7‐13. [DOI] [PubMed] [Google Scholar]
- 21. Wang Y, Lin L, Lai H, Parada LF, Lei L. Transcription factor Sox11 is essential for both embryonic and adult neurogenesis. Dev Dyn. 2013;242(6):638‐653. [DOI] [PubMed] [Google Scholar]
- 22. Haslinger A, Schwarz TJ, Covic M, Lie DC. Expression of Sox11 in adult neurogenic niches suggests a stage‐specific role in adult neurogenesis. Eur J Neurosci. 2009;29(11):2103‐2114. [DOI] [PubMed] [Google Scholar]
- 23. Hyodo‐Miura J, Urushiyama S, Nagai S, Nishita M, Ueno N, Shibuya H. Involvement of NLK and Sox11 in neural induction in Xenopus development. Genes Cells. 2002;7(5):487‐496. [DOI] [PubMed] [Google Scholar]
- 24. Iuchi K, Yagura T. DNA binding activity of Ku during chemotherapeutic agent‐induced early apoptosis. Exp Cell Res. 2016;342(2):135‐144. [DOI] [PubMed] [Google Scholar]
- 25. Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97‐98. [DOI] [PubMed] [Google Scholar]
- 26. Li Z, Zhang Z, He Z, et al. A partition‐ligation‐combination‐subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio‐x.cn). Cell Res. 2009;19(4):519‐523. [DOI] [PubMed] [Google Scholar]
- 27. Sun W, Park KW, Choe J, et al. Identification of novel electroconvulsive shock‐induced and activity‐dependent genes in the rat brain. Biochem Biophys Res Commun. 2005;327(3):848‐856. [DOI] [PubMed] [Google Scholar]
- 28. Kim DK, Han SB, Hong ST, et al. Expression of Sox11 and Brn transcription factors during development and following transient forebrain ischemia in the rat. Neurosci Lett. 2008;433(3):259‐264. [DOI] [PubMed] [Google Scholar]
- 29. Salerno KM, Jing X, Diges CM, Cornuet PK, Glorioso JC, Albers KM. Sox11 modulates brain‐derived neurotrophic factor expression in an exon promoter‐specific manner. J Neurosci Res. 2012;90(5):1011‐1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Allen NC, Bagade S, McQueen MB, et al. Systematic meta‐analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40(7):827‐834. [DOI] [PubMed] [Google Scholar]


