The outer membrane (OM) of Gram-negative bacteria poses a barrier to antibiotic entry due to its high impermeability. Thus, there is an urgent need to study the function and biogenesis of the OM. In Enterobacterales, an order of bacteria with many pathogenic members, one of the components of the OM is enterobacterial common antigen (ECA). We have known of the presence of ECA on the cell surface of Enterobacterales for many years, but its properties have only more recently begun to be unraveled.
KEYWORDS: O-antigen, cross-reactivity, enterobacterial common antigen, outer membrane, surface antigens
ABSTRACT
The outer membrane (OM) of Gram-negative bacteria poses a barrier to antibiotic entry due to its high impermeability. Thus, there is an urgent need to study the function and biogenesis of the OM. In Enterobacterales, an order of bacteria with many pathogenic members, one of the components of the OM is enterobacterial common antigen (ECA). We have known of the presence of ECA on the cell surface of Enterobacterales for many years, but its properties have only more recently begun to be unraveled. ECA is a carbohydrate antigen built of repeating units of three amino sugars, the structure of which is conserved throughout Enterobacterales. There are three forms of ECA, two of which (ECAPG and ECALPS) are located on the cell surface, while one (ECACYC) is located in the periplasm. Awareness of the importance of ECA has increased due to studies of its function that show it plays a vital role in bacterial physiology and interaction with the environment. Here, we review the discovery of ECA, the pathways for the biosynthesis of ECA, and the interactions of its various forms. In addition, we consider the role of ECA in the host immune response, as well as its potential roles in host-pathogen interaction. Furthermore, we explore recent work that offers insights into the cellular function of ECA. This review provides a glimpse of the biological significance of this enigmatic molecule.
INTRODUCTION
Diverse environmental conditions on Earth (e.g., heat, pH, salinity, pressure, and osmotic activity) immensely affect the function of the cell, necessitating adaptation through structural modification. Gram-negative bacteria have an impermeable and strengthened outer membrane (OM) that allows them to withstand stress brought about by environmental factors, including other bacteria, antibiotics, and chemical stresses. The cell envelope structure of Gram-negative bacteria consists of the inner membrane, the periplasm containing the peptidoglycan cell wall, and the OM (1). The lipids of the OM form a barrier that is impermeable to large hydrophilic and hydrophobic molecules (2). Lipopolysaccharide (LPS) facilitates the formation of this barrier though (i) the high number of fatty acyl substituents per lipid molecule, which form a gel-like structure enhancing the rigidity of membrane (3, 4), (ii) strong lateral interaction between LPS molecules mediated by salt bridges with divalent cations (5), and (iii) modification of LPS structure in response to different environmental conditions (6). For example, in Salmonella, the PhoPQ two-component system causes antimicrobial peptide resistance after induction by divalent cation starvation by activating PagP (7). PagP facilitates the addition of palmitate chain to lipid A, altering the fluidity of the LPS molecules in the OM (7).
OM proteins (OMPs) are integral membrane proteins present in the membranes of Gram-negative bacteria, mitochondria, and chloroplasts. These proteins adopt a β-barrel architecture arranged in the membrane in anti-parallel patterns (8). Some of these proteins (porins) can form pores in the OM (2). These OMPs regulate the movement of small hydrophilic molecules across the outer membrane, such as nutrients, water, ions, and some small hydrophilic antibiotics (2). In fact, in Escherichia coli, β-lactam antibiotics, tetracyclines, chloramphenicol, and fluoroquinolones quickly diffuse through OmpF (9–12). Specific porins can also transport amphipathic substrates. For instance, transportation of long-chain fatty acid is facilitated by the lipid transporter FadL (13). Beyond its role controlling the entry of molecules into the cell, the OM plays a structural role, providing protection against mechanical and osmotic stresses (14, 15).
The Gram-negative OM is coated in highly variable molecules that can cause immune activation, known as antigens. Bacteria are divided into serotypes based on different antigen combinations (16). The three major types of antigens present on the cell surface are O (somatic), K (capsular), and H (flagellar) (17, 18). These antigens can play roles in motility (H-antigen), protection from a hostile environment (K-antigens and O-antigen), interaction with the environment (K-antigens and O-antigen), and increasing the ability of the OM to provide structural support to the cell (O-antigen) (15, 19–21). The outer leaflet of the OM is made mainly of lipopolysaccharide (LPS), which consists of lipid-A, the core polysaccharide, and O-antigen (1). O-antigen is a highly variable chain of carbohydrates and thus is serotype specific. K-antigens are the capsule, a coat on the surface of bacteria outside the cell envelope. They generally consist of high-molecular-weight polysaccharides with some exceptions (e.g., K-88 and K-99 of E. coli, which are protein antigens) (22–24). The H-antigen is a protein antigen based on flagellar structure (25). Enterobacterales is a bacterial order that is defined in part by the presence of an antigen known as enterobacterial common antigen (ECA) (26). ECA, a carbohydrate antigen, is located in the outer leaflet of the OM and in the periplasm (27–30). Although Enterobacterales express various antigens (e.g., K, O, and H) (31, 32), ECA is unique in that it is restricted to one order and in which it is invariant (Fig. 1A) allowing cross-reactivity among the members of Enterobacterales (33).
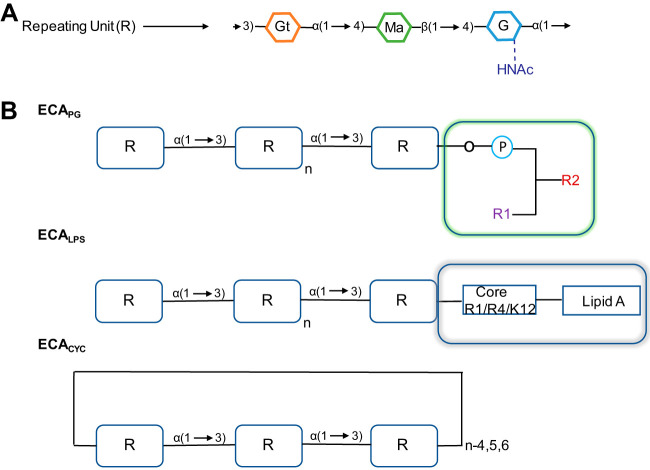
FIG 1.
The structure of ECA. (A) The structure of the repeating unit (R) of ECA is made up of amino sugars (G, N-acetylglucosamine; Ma; N-acetyl-d-mannosaminuronic acid; Gt, 4-acetamido-4,6-dideoxy-d-galactose). (B) Structural differences between the three ECA forms. ECAPG, phospholipid-linked ECA; ECALPS, lipopolysaccharide-linked ECA; ECACYC, cyclic form of ECA. In the ECAPG structure, R1 (-CH2OH group), and R2 (-CHOH group) indicate acyl chains. In the ECALPS structure, “core” represents the core polysaccharide of LPS, which is attached to lipid A, a hydrophobic lipid section that anchors LPS to the outer membrane. In the core region, the common tetrasaccharide structure is substituted for R1 (β-glucose) and R4 (β-galactose) compared to the K-12 core. n, a variable number of ECA repeating units. ECACYC generally consists of 4 to 6 repeating units (R) depending on the species. For example, in E. coli, 4 repeating units are present.
Calvin M. Kunin and colleagues first discovered ECA in 1962 (33). The discovery of ECA was a result of studying strains of E. coli causing urinary tract infections and observing the reaction between rabbit antisera generated against the strains and 102 homologous and heterologous E. coli strains. The authors used a standard procedure (passive hemagglutination) to detect O-antigen found in the LPS of the E. coli (33). While carrying out these experiments, they realized there was a cross-reacting specificity between the antisera and many strains of E. coli. Although various antisera demonstrated differing reactivities, anti-E. coli O14 sera reacted with a remarkable range of strains: anti-O14 serum had antibodies recognizing an antigen common to various E. coli strains. However, this antigen was not the LPS-attached O-antigen that Kunin and colleagues had been investigating (33). Furthermore, this cross-reacting antigen was also observed in most other enteric bacteria (33, 34). The antigen was, therefore, named enterobacterial common antigen (ECA) (35).
After the discovery of ECA, research was conducted to ascertain the dissemination of the new antigen among species, eventually aided by a monoclonal ECA antibody that enhanced ECA detection (36). ECA is present in wild-type strains of Enterobacterales and absent in both other Gram-negative bacteria and Gram-positive bacteria (Table 1). More studies need to be carried out on the unusual presence of enterobacterial common antigen in Aeromonas hydrophila 209A, as it is not present in the other strains belonging to the same species (37) and may be the result of horizontal gene transfer. Few exceptions to the ubiquitous expression of ECA in Enterobacterales exist. These species, which appear to have lost ECA expression, are the endosymbiotic members of Enterobacterales, which have a reduced genome size due to the loss of many genes rendered unnecessary by their obligate symbiotic life style (38, 39).
TABLE 1.
Distribution of ECA in Gram-negative bacteria
| Family | ECA positive | ECA negative |
|---|---|---|
| Enterobacterales (194) | ||
| Budviciaceae | Leminorella | |
| Pragia | ||
| Enterobacteriaceae | Atlantibacter | “Candidatus” |
| Buttiauxella | ||
| Cedecea | ||
| Citrobacter | ||
| Cronobacter | ||
| Enterobacter | ||
| Escherichia | ||
| Gibbsiella | ||
| Izhakiella | ||
| Klebsiella | ||
| Kluyvera | ||
| Kosakonia | ||
| Leclercia | ||
| Lelliottia | ||
| Limnobaculum | ||
| Metakosakonia | ||
| Pluralibacter | ||
| Raoultella | ||
| Salmonella | ||
| Shigella | ||
| Shimwellia | ||
| Erwiniaceae | Erwinia | Buchnera, Wigglesworthia |
| Mixta | ||
| Pantoea | ||
| Tatumella | ||
| Hafniaceae | Edwardsiella | |
| Hafnia | ||
| Obesumbacterium | ||
| Morganellaceae | Arsenophonus | “Candidatus Arsenophonus lipoptenae” |
| Morganella | ||
| Photorhabdus | ||
| Proteus | ||
| Providencia | ||
| Xenorhabdus | ||
| Pectobacteriaceae | Brenneria | |
| Dickeya | ||
| Lonsdalea | ||
| Pectobacterium | ||
| Sodalis | ||
| Yersiniaceae | “Candidatus Fukatsuia” | |
| Chania | ||
| Rahnella | ||
| Serratia | Serratia symbiotica | |
| Yersinia | ||
| Unclassified | Phytobacter | |
| Plesiomonas | ||
| Vibrionales | ||
| Vibrionaceae | Aeromonas hydrophila 209A | Vibrio |
| Other Gram-negative bacteria | ||
| Acidiferrobacter | ||
| Actinobacillus | ||
| Aeromonas | ||
| Alcaligenes | ||
| Bordetella | ||
| Campylobacter | ||
| Cardiobacterium | ||
| Chromatiaceae | ||
| Chromobacterium | ||
| Colwellia | ||
| Eikenella | ||
| Ferrimonas | ||
| Flavobacterium | ||
| Gardnerella | ||
| Haemophilus | ||
| Kingella | ||
| Moraxella | ||
| Moritella | ||
| Shewanella | ||
| Nitrobacteriaceae | ||
| Pasteurella | ||
| Pseudomonas | ||
| Rhodospirillaceae |
Antigens that are highly variable between strains of bacteria have served as the foundation for serological naming and grouping. For example, the Kauffmann-Perch scheme is used for Proteus, while the Kauffmann-White-Le Minor scheme is used for Salmonella (40–42). However, the importance of common antigens has often been overlooked. In recent times, the study of these antigens has increased given their potential significance in vaccine development, determination of phylogeny, and diagnosis. Furthermore, the invariance of common antigens suggests that they have important functions that do not allow for variability. ECA is a perfect example of an antigen that has undergone a recent resurgence of research despite its discovery many years ago. In this review, we explore the history of ECA, its interaction with the immune system, its isolation and biosynthesis, and finally its biological significance.
THE IMMUNOGENICITY OF ECA
Interactions of ECA with the immune response.
ECA has a complex interaction with the immune response. Initial studies elucidated that, while the antigen occurred across Enterobacterales, just a few sera had antibodies to ECA, for example, E. coli O14 (33, 43). Thus, all strains possessed antigenic ECA but very few possessed immunogenic ECA. The variance in immunogenicity of the strains studied could not be accounted for by differences in the amounts of ECA expressed (35, 44, 45). Therefore, something else must differentiate these types of ECAs. The elucidation of this difference came by separating ECA extracts with ethanol, in which LPS is not soluble, exposing a dissimilarity in the immunogenic types: an ethanol-insoluble immunogenic form and an ethanol-soluble nonimmunogenic form. The ethanol-insoluble form is not separable from LPS and signifies the immunogenic form of the enterobacterial common antigen (46). This form of ECA has ECA bound to the LPS core (ECALPS) (Fig. 1B). The ethanol-soluble form of ECA is not associated with LPS and, instead, consists of the ECA polysaccharide chain covalently linked to diacylglycerol through phosphodiester linkage (ECAPG) (47). There is a third form of ECA, cyclic ECA (ECACYC); however, this molecule is found in the periplasm and is not exposed to the environment (48, 49).
Still, why some strains made immunogenic ECALPS while others did not remained unclear, in part because of the classification of the traditional ECA immunogenic strain, E. coli O14, as an O-antigen-positive strain (50). In fact, the strain is an irregular type of the rough R4 strain disguised by the production of K7 capsular antigen (51). In combination with the production of immunogenic ECALPS by the R1, R4, and K-12 strains with rough LPS (43), this clarified that ECALPS is produced in significant amounts only by rough strains that do not make O-antigen. In nonimmunogenic strains, including O-antigen-producing smooth strains and rough strains with incomplete LPS cores or mutations in waaL, the O-antigen ligase, ethanol-soluble ECAPG is the predominant form of ECA on the cell surface (35). While some early studies suggested purified ECAPG could induce an antibody response (52), this was only true in strains that also produced significant amounts of ECALPS (53), suggesting that the antibody production may have resulted from contaminating ECALPS.
With our current knowledge of immunology, it can now be appreciated how the differences in structure between ECALPS and ECAPG would lead to differences in their immunogenicity. Antibody production is not efficiently stimulated without innate immune signaling (54). ECALPS possesses an intrinsic adjuvant to stimulate antibody production, as LPS is recognized by toll-like receptor 4 (TLR4), leading to the production of proinflammatory cytokines (55). Nevertheless, as a carbohydrate antigen, ECALPS mainly stimulates the production of IgM low-affinity, high-avidity antibodies (56). In contrast, the production of high-affinity IgG antibodies requires a protein antigen (57). As a proof of concept of the potential immunogenicity of ECAPG, purified ECAPG that contains proteins in addition to ECA can generate an immune response (52). Likewise, a conjugate of ECA and tetanus toxoid, a classic adjuvant, produces ECA antibodies mainly of the IgG isoform (56). Many of the initial studies on ECA immunogenicity were carried out with heat-killed bacteria (50). The difference in immunogenicity of ECALPS and ECAPG is less in live bacteria (50), likely because of the many pathogen-associated molecular patterns (PAMPs) linked with an active infection and an increased production of proinflammatory cytokines (58).
Prevalence of ECA antibodies.
Many early studies have reported a low titer of ECA antibodies present in human serum (33, 59, 60), with the caveat that these studies were conducted before the availability of an ECA knockout strain and so may report the combined titer of both ECA antibodies and antibodies to protein antigens shared among Enterobacterales (e.g., OmpA). These antibodies have been found in both healthy donors and, at higher levels, in patients with chronic urinary tract infections (61). The titers of ECA antibodies present in the blood have been reported to increase with age (62). A maternal vaccination study reported that the cord blood of a child has lower amounts of ECA antibodies than maternal serum, showing some low level of maternal transfer (61). In other mammals such as cats, dogs, horses, pigs, and mice, ECA antibodies have been reported in blood sera with the exception of rabbits, where no ECA antibody is found. The likely reason for this is due to the high colonization of rabbits by Gram-positive bacteria and low prevalence of E. coli (60). Among several strains of mice, C57B1/6HA mice have ample ECA antibody titers after responding to ECALPS immunogens (63–65).
Estimates of ECA antibody prevalence in various types of infections have indicated variable titers of ECA antibodies (35). In a few diseases such as enteritis, bacteremia, and acute urinary tract infections, low titers of ECA antibodies were detected. However, higher ECA antibody titers were found in shigellosis (61, 66), peritonitis (53, 67), and chronic urinary tract infections (61, 68, 69). Surprisingly consistent levels of ECA antibodies were observed in a rabbit pyelonephritis model immunized by ECALPS, irrespective of the route of infection and diagnosis (70–72).
The presence of ECA antibodies has been detected in human serum after infection from pathogenic bacteria such as E. coli, Yersinia enterocolitica O3 strains (26, 73–75), and in humans suffering from arthritis associated with Proteus mirabilis strains (76). The ECA immunogenicity is mainly due to ECALPS (26), with exception to the Rc mutant R4/O28 of P. mirabilis in which ECAPG provides immunogenicity. Therefore, the ECA can be used as a tool for serodetection in various infections caused by the members of the Enterobacterales order (77).
Role of ECA antibodies in host-pathogen interactions.
Several studies have attempted to determine whether ECA antibodies play a role, either protective or pathogenic, in various disease contexts. Under experimental conditions, an appreciable amount of ECA antibodies was found in mice immunized by E. coli O-antigen-negative strains and later challenged by pathogenic bacteria (63–65, 78), except in Swiss albino mice (65). However, protection from these immunizations was partial and temporary. The same partial protection was also observed in mice by passive immunization with serum from rabbits inoculated with E. coli O14 (63). Furthermore, a clinical study has demonstrated that passive immunization with a human monoclonal ECA antibody had no protective effect during sepsis caused by Enterobacterales (79).
Bridge et al. (80) reported that, in a mouse model of salmonellosis, infection with the ΔwecA strain of Salmonella enterica serovar Typhimurium SL1344 via the oral or intraperitoneal route provided cross-protection against this infection by the production of IgG antibodies. In addition, Huang et al. (81) reported cross-protection against heterologous Salmonella strains in mice by downregulating the expression of O-antigen (rmlB [rfbB]) and ECA (rmlBECA [rffG]) biosynthesis genes, allowing production of protein-recognizing antibodies. These studies suggest that, at least for Salmonella, ECA antibodies may not be protective and that ECA may “distract” the immune system from more efficacious targets. It should be mentioned that some studies have correlated the presence of antibodies to Enterobacterales, including to ECA, with rheumatoid arthritis (82–84); however, the causal relationships leading to the antibody production remains unclear.
FORMS AND BIOSYNTHESIS OF ECA
Over the years, several researchers have developed methods for isolating ECA (Table 2). However, the chemical composition of the antigen was initially difficult to identify, due in part to the existence of ECA in three forms (26) (Fig. 1B). These forms are ECA linked to diacylglycerol through phosphodiester linkage (ECAPG), LPS-linked ECA (ECALPS), and periplasmic cyclized ECA (ECACYC) (43, 47, 48). Observations made by Kunin, Neter, and Suzuki initially demonstrated that ECA occurred in two forms in the immunogenic E. coli O14. One of the forms was soluble in aqueous ethanol and was able to be separated from the LPS (ECAPG), while the other did not dissolve in ethanol and could not be separated from LPS (ECALPS) (35, 37). ECAPG is the predominant surface-exposed form of ECA, while ECALPS, the immunogenic form that allows antibody production, is predominantly found in rough mutants that do not make O-antigen (35). Later studies identified a third form of ECA, ECACYC (48), which is now thought to be present in all species that make ECA (48, 85–88). Initial studies done by Männel and Mayer in 1978 (89) reported that ECA consists of N-acetyl-d-glucosamine (GlcNAc) and N-acetyl-d-mannosaminuronic acid (ManNAcA). But in 1983, another component of ECA, 4-acetamido-4,6-dideoxy-d-galactose (Fuc4NAc), was identified in Shigella by Lugowski et al. (56), demonstrating that ECA consists of a trisaccharide repeating unit (Fig. 1A).
TABLE 2.
Methods used for study of ECA
| Methods useda | Representative species and strain(s) | Type of ECA | Representative reference(s) |
|---|---|---|---|
| Representative purification methods | |||
| ECALPS | |||
| Hot phenol-water extraction (water phase); dialysis; 90% ethanol precipitation (pellet); anion exchange chromatography | Escherichia coli O1, O14, O55; Shigella flexneri | ECALPS | 52 |
| LPS extraction and purification for analysis of ECALPS and other LPS forms | Yersinia enterocolitica O:3; Shigella sonnei phase II | ECALPS with LPS | 141, 195 |
| ECAPG | |||
| Lysis in boiling PBS (supernatant); 85% ethanol precipitation (supernatant) | Salmonella enterica serovar Typhimurium, Salmonella choleraesuis, Salmonella enteritidis, S. flexneri, E. coli O111, O55, O6, O75 | ECAPG | 44, 72 |
| Bacteria killed and dried with acetone; room temperature water extraction; picric acid precipitation (supernatant); acetone precipitation (pellet); Sephadex G200 column chromatography; preparative gel electrophoresis | Salmonella typhosa O901 | ECAPG | 196 |
| Hot phenol-water extraction (water phase); phenol-chloroform-petroleum ether extraction (phenol phase); ultracentrifugation (supernatant); anion exchange chromatography | S. Montevideo SH94, S. Typhimurium, S. sonnei phase I, Plesiomonas shigelloides | ECAPG | 81, 85, 89, 149, 197 |
| ECACYC | |||
| Sonication in EDTA and lysozyme; boiling water extraction; 85% ethanol precipitation (supernatant); acetone precipitation; column chromatography on silica gel and Sephadex LH-20 | S. sonnei | ECACYC | 48, 56, 140 |
| Cold trichloroacetic acid extraction; Sephadex G-50 chromatography; anion exchange chromatography | Yersinia pestis EV | ECACYC | 86 |
| Sonication in MgSO4; ultracentrifugation (supernatant); 75% ethanol precipitation (supernatant); drying and resuspension in ddH2O; desalting with ZipTipC18 | E. coli K-12 | ECACYC | 88, 111, 145 |
| Hot phenol-water extraction (water phase); DNase, RNase, and protease treatment; ultracentrifugation (supernatant); size exclusion chromatography; Biogel P-100 chromatography | E. coli O157:H− | ECACYC (no O-acetylation) | 198 |
| Common detection methods | |||
| Passive HA; detection of antigen (whole cell, cell lysates, purified) by coating erythrocytes and assaying agglutination caused by reacting antibodies | E. coli O6, O75, OS:K27, K-12; S. Typhimurium TV149 (Ra); S. Montevideo SH94 (S); S. sonnei; P. shigelloides; Plesiomonas rettgeri | ECALPS; ECAPG; ECACYC; O-acetylation required for strong reactivity of serum with ECA | 72, 85, 88, 89, 140, 197, 199 |
| HA inhibition; detection by supernatant antigen prevention of agglutination of antigen-coated erythrocytes in the presence of antigen-specific antibody | E. coli O14, O6, O75, K-12; S. Typhimurium TV149 (Ra); S. Montevideo SH94 (S); S. sonnei | ECALPS; ECAPG; ECACYC; O-acetylation required for strong reactivity of serum with ECA | 52, 72, 89, 111, 140, 199, 200 |
| Immunodiffusion precipitation; identifies antigens after gel electrophoresis through precipitation caused by reaction with antibodies | S. typhosa O901; S. Montevideo SH94 (S); S. sonnei | ECALPS; ECAPG | 89, 140, 196 |
| ELISA; quantification of antigens based on their reaction with antibodies | S. sonnei; S. Montevideo; P. shigelloides; E. coli OS:K27−, K-12; P. rettgeri | ECALPS; ECAPG | 85, 140, 197 |
| Immunoblot; including SDS-PAGE or dot blot followed by immunoblot analysis | S. Montevideo SH94; S. Typhimurium; E. coli R1, R4, OS:K27−, K-12; S. sonnei; P. shigelloides; P. rettgeri; Y. enterocolitica O:3 | ECALPS; ECAPG | 81, 141, 145, 197, 201 |
| LC; including liquid-gas chromatography, HPLC, reverse-phase HPLC | S. sonnei; Y. pestis; E. coli K-12 | ECALPS; ECAPG; ECACYC | 56, 86–88, 140 |
| NMR spectroscopy; including 1H, 13C, and 31P | S. sonnei; P. shigelloides; Y. pestis; S. Typhimurium LT2; E. coli K-12, O157:H | ECALPS; ECAPG; ECACYC | 56, 85–87, 149, 195, 198 |
| MS; including gas-LC-MS, gas chromatography-MS, matrix-assisted laser desorption–ionization time of flight | P. shigelloides; E. coli K-12, O157:H−; S. sonnei | ECALPS; ECAPG; ECACYC | 85, 88, 145, 195, 198 |
ddH2O, double-distilled water; ELISA, enzyme-linked immunosorbent assay; HA, hemagglutination; HPLC, high-pressure liquid chromatography; LC, liquid chromatography; MS, mass spectroscopy; PBS, phosphate-buffered saline.
Biosynthesis of the ECA polymer.
The synthesis of ECA is intricate, involving several phases (Fig. 2). The genes necessary for many steps in the synthesis of ECA are located within an operon known as the wec operon. The wec operon begins at position 85.4 centisomes on the E. coli K-12 chromosome (90), and the functions of each gene of the operon have been analyzed (90–98).
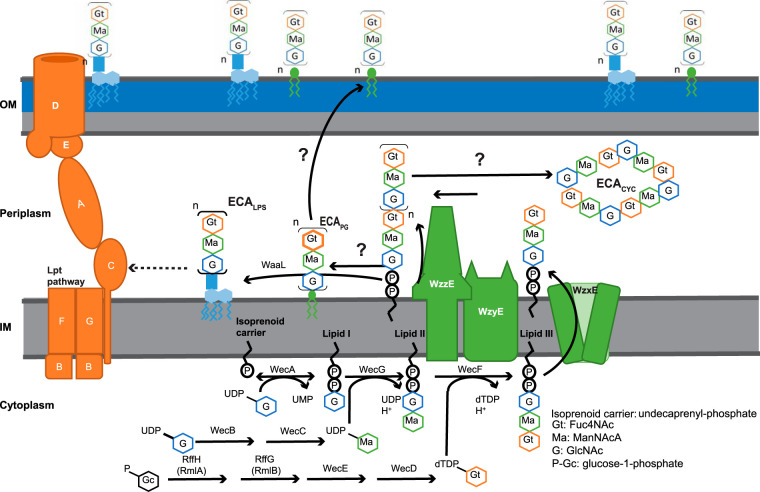
FIG 2.
Schematic representation of ECA biogenesis in E. coli. ECA biogenesis begins with synthesis of amino sugars and their attachment to an isoprenoid carrier (Und-P). After a complete subunit is made by series of enzymes namely, WecA, WecB, WecC, WecD, WecE, WecF, WecG, RmlAECA, and RmlBECA, the precursor is flipped across the inner membrane by WzxE, and the subunits are polymerized on the isoprenoid carrier by WzyE with the chain length controlled by WzzE. Three forms of ECA are made from the polymerized subunits: ECAPG, attached to diacylglycerol through phosphodiester linkage and surface exposed; ECACYC, which is periplasmic; and ECALPS, attached to LPS and surface exposed. This figure is adapted and modified from Mitchell et al. (145).
As for many other extracytoplasmic glycans (99), the assembly of the ECA trisaccharide repeat unit is carried out on an isoprenoid lipid carrier, undecaprenyl-phosphate (Und-P), a 55-carbon molecule made of isoprenoid units (93, 100–102). The assembly occurs on the inner side of the plasma membrane (103–107). The first step involves the formation of GlcNAc-pyrophosphoryl-undecaprenol, which is also known as lipid IECA (108). This step uses UDP-GlcNAc as a substrate to attach GlcNAc-1-phosphate to Und-P and is catalyzed by WecA (93, 101, 102). In silico predictions, cysteine accessibility experiments, and fusion-protein expression experiments have demonstrated that WecA has 11 transmembrane segments, with the N terminus in the periplasm and the C terminus in the cytoplasm (109, 110). Furthermore, mutational studies have shown that several conserved aspartate residues in cytoplasmic loops 2 and 3 are important for WecA catalytic activity and divalent cation binding (110). Fluorescence microscopy for green fluorescent protein (GFP)-tagged WecA revealed that it is localized to punctate regions on the cell surface (110). The punctate localization of WecA suggests that ECA and/or O-antigen biosynthesis is localized to discrete cellular regions. The GlcNAc residue of ECA is nonstoichiometrically O-acetylated by WecH, an inner membrane O-acetylase (111).
WecB and WecC are responsible for synthesizing UDP-ManNAcA from UDP-GlcNAc (103, 112, 113). Specifically, WecB (UDP-N-acetylglucosamine 2-epimerase) reversibly epimerizes at carbon position 2 between UDP-GlcNAc and UDP-N-acetylmannosamine (112, 113). Campbell et al. (114) solved the structure of WecB at a 2.4-Å resolution. This homodimeric enzyme is comprised of two similar sandwich domains with the active site located in the deep cleft at the domain interface. Several basic residues in the active site may have a role in proton transfer at the C-2 position or stabilization of the oxy-carbonium ions in the transition state (114). Residues in the active site have been found to be important for allosteric regulation of WecB as well as for binding and catalysis (115). WecC oxidizes UDP-N-acetylmannosamine in the presence of NAD+ to form UDP-ManNAcA (116). The UDP-ManNAcA is the substrate to attach ManNAcA to the lipid IECA, a reaction carried out by WecG (101, 117). This process results in ManNAcA-GlcNAc-pyrophosphoryl-undecaprenol, also known as lipid IIECA.
RmlAECA (RffH), RmlBECA (RffG), WecE, and WecD are responsible for synthesizing dTDP-Fuc4NAc from glucose-1-phosphate (103, 118, 119). The first reaction, carried out by RmlAECA (dTDP-glucose pyrophosphorylase 2), forms dTDP-glucose from glucose-1-phosphate, dTTP, and H+ (118, 120). Sivaraman et al. (120) solved the RmlAECA crystal structure in the presence of dTTP and Mg2+ ions at a 2.6-Å resolution. This enzyme is tetrameric with each monomer composed of an α/β fold with nucleotide-binding and sugar-binding domains. The active site was identified at the interface of two domains (120).
TDP-glucose acts as a substrate for second enzyme, RmlBECA (dTDP-glucose 4,6-dehydratase 2). RmlBECA converts dTDP-glucose to dTDP-4-keto-6-deoxy-d-glucose. This second reaction is a complex reaction that involves dehydrogenation, dehydration, and rereduction in the presence of cofactor NAD+ (121). Several active-site residues important for the function of RmlBECA have been identified based on similarity to UDP-galactose-4-epimerase and mutational analysis (122, 123).
The third reaction is catalyzed by WecE (dTDP-4-dehydro-6-deoxy-d-glucose transaminase), which converts dTDP-4-keto-6-deoxy-d-glucose to dTDP-4-amino-4,6-dideoxy-a-d-galactose (dTDP-Fuc4N) using glutamate as the amino donor (103, 124). A WecE crystal structure has been solved at a resolution of 2.24 Å (125). The structure indicates a homodimeric protein; however, a previous gel filtration experiment suggested a homotetrameric conformation (124). As is common for sugar aminotransferases, the WecE active site contains a conserved lysine that binds the catalytic cofactor, 5′-pyridoxal phosphate, an aspartate important for cofactor activation, and a conserved glutamine (125).
The last reaction is catalyzed by WecD (dTDP-fucosamine acetyltransferase) which uses acetyl coenzyme A (acetyl-CoA) as a cofactor to form dTDP-Fuc4NAc from dTDP-Fuc4N (103, 126). WecD has been crystalized at a resolution of 1.95 Å in its apo form and 1.66 Å in complex with acetyl-CoA (126). The structure shows a dimeric protein with each monomer adopting a GCN5-related N-acetyltransferase fold. WecF uses dTDP-Fuc4NAc to transfer Fuc4NAc to lipid IIECA, forming Fuc4NAc-ManNAcA-GlcNAc-pyrophosphoryl-undecaprenol (lipid IIIECA) (103).
The synthesis of lipid IIIECA occurs on the inner leaflet of the cytoplasmic membrane facing the cytosol (103); however, polymerization of the trisaccharide repeat unit to form polysaccharide chains occurs on the outer leaflet of the cytoplasmic membrane (127, 128). The flipping of lipid IIIECA to the periplasmic face of the membrane is mediated by a “flippase,” WzxE (127, 128). WzxE is a member of the polysaccharide-specific transporter family of proteins, which flip polysaccharides, including some O-antigens and capsular polysaccharides, across the inner membrane (IM) (128, 129). After translocation across the membrane, the ECA chain is polymerized by WzyE. The final chain length of the ECA polymer is determined by WzzE, the chain length regulator (91). These steps result in the formation of an ECA polymer attached to the isoprenoid carrier (Fig. 2).
Several studies have offered insights into the mechanism through which WzzE might control the chain length of ECA. Genetic evidence and cross-linking data support the notion that the flippase, polymerase, and chain length regulator work together as a complex (130, 131). Several structural studies have been performed with WzzE and other members of the class 1 polysaccharide copolymerase family (PCP-1) (132). A crystal structure of the periplasmic domain of WzzE, solved at 2.4 Å, revealed a bell-shaped homo-octameric structure (133); however, reports have suggested various oligomeric states for other PCP-1 family members depending on whether full-length protein was used and the method of analysis (134–137).
This structural inconsistent may be due to the lack of interactions with other complex members (i.e., WzyE). However, recent studies have again suggested octameric structure for both WzzE and WzzB (an E. coli O-antigen PCP-1) (136). The most recent structural data for PCP-1 proteins suggest a mechanism for PCP-1 chain length regulation where polymerization begins when the polymerase and PCP-1 form a complex with the growing polysaccharide chain wrapping either over the surface or through the cavity of the PCP-1 (137). The polymerization could then be terminated either when the PCP-1 and polymerase disassociate (a “stop-watch” mechanism) or when the polysaccharide-binding capacity of the PCP-1 is reached (a “molecular ruler” mechanism) (138–140).
Synthesis of the three forms of ECA.
The three forms of ECA are made from the ECA polymer. ECAPG is the dominant membrane-associated form of ECA and constitutes about 0.2% of the cellular dry weight of E. coli K-12 (26, 76, 140). This form is present in all Enterobacterales (26). The polysaccharide chain is transferred from the isoprenoid carrier to an unidentified lipid to form ECA linked to diacylglycerol through phosphodiester linkage (ECAPG) (47). In this molecule, ECA is the head group of the phospholipid (47). The newly synthesized ECAPG is then translocated to the outer membrane (27, 29). The genes and mechanisms involved in the synthesis and translocation of ECAPG remain unknown (87). ECALPS is synthesized by transferring the linear ECA polysaccharide chain to the core oligosaccharide of LPS (26, 46). This step is catalyzed by WaaL, which is also responsible for attaching O-antigen to the core of LPS (43, 51). However, data suggest that the method for attaching ECA to LPS in Yersinia enterocolitica may be different, allowing ECA and O-antigen to coexist (141). The last form of ECA, ECACYC, is a cyclic molecule consisting of polymerized ECA trisaccharide repeat units, and it is water soluble (26, 48). The ECACYC is localized in the periplasm (88). This polymer has a variable number of repeat units (4–6) depending on the species (91), and the chain length regulator, WzzE, is necessary for its synthesis (48, 87, 88). A cyclase has not been identified. In-depth structural analysis by crystallography, nuclear magnetic resonance (NMR), and molecular dynamics have suggested that ECACYC can exist in two three-dimensional conformations (142–144). In contrast to ECACYC, ECAPG and ECALPS consist of 1 to 14 repeat units, with a modal value of 5 to 7 units in E. coli K-12 (91). In addition, different modal chain lengths have been observed depending on the growth temperature (145).
Interactions with other biosynthetic pathways.
The use of isoprenoid carriers, such as Und-P, for the synthesis of extracytoplasmic glycans is highly conserved across the domains of life (99, 146, 147). Furthermore, these carriers are often utilized for the synthesis of multiple glycans in the same species. For instance, in E. coli, Und-P is used for the production of ECA, O-antigen, peptidoglycan, and the colanic acid capsule (43, 148–151). Thus, Und-P is a universal lipid carrier required for the synthesis of numerous glycan polymers (152), and this can lead to complex interactions between biosynthetic pathways.
Disruption of one Und-P pathway may lead to indirect consequences on other glycans by altering the amount of Und-P and other precursors available for their synthesis. For example, obstructing the O-antigen pathway in E. coli compromises peptidoglycan biosynthesis by sequestering Und-P (153). In relation to ECA, it was first observed that disruption of later steps in ECA biosynthesis that lead to the accumulation of lipid IIECA causes detergent sensitivity (154) and bile salt sensitivity (155). It was then found that these mutations also lead to the activation of extracytoplasmic stress responses, including Cpx, σE, and Rcs (156–158). Interestingly, in E. coli and Salmonella enterica, these stress responses are only activated with mutations that cause lipid IIECA accumulation (147, 156), but in Serratia marcescens, Rcs activation has been reported even in strains with mutations early in ECA biosynthesis (158). The link of detergent sensitivity and stress response activation to defects in peptidoglycan biosynthesis was conclusively established by the observation that mutations leading to the accumulation of lipid IIECA cause changes in cell shape due to sequestering of Und-P (159). While the accumulation of lipid IIECA is deleterious to the cell, in E. coli, the accumulation of lipid IIIECA has been shown to be lethal (88, 128). This has been observed with both loss of WzyE, the ECA polymerase, and loss of the capacity to flip lipid IIIECA across the inner membrane (see below) (88, 128). Avoiding lipid IIIECA accumulation by disrupting an earlier step in ECA biosynthesis prevents this lethality (88).
The ECA and O-antigen biosynthesis pathways are homologous in Enterobacterales. All members of Enterobacterales utilize the wec locus for the biosynthesis of ECA (102, 160). However, many Enterobacterales with GlcNAc as their first O-antigen residue (e.g., Salmonella enterica serovar Minnesota [161], Salmonella enterica serovar Montevideo [162], and E. coli [43]) also utilize wecA (rfe), the first gene in ECA biosynthesis, for the production of O-antigen chains (163). Therefore, disruptions of wecA result in loss of both O-antigen and ECA biosynthesis (43, 164). In addition, the functions of RmlAECA and RmlBECA, which function in the synthesis of dTDP-Fuc4NAc for ECA biosynthesis, are at least partially redundant with RmlA and RmlB, respectively, two enzymes involved in the synthesis of dTDP-l-rhamnose for O-antigen biosynthesis (118).
The absence of all Wzx family flippases (WzxO16, WzxC, and WzxE) in E. coli K-12 leads to a lethal accumulation of lipid IIIECA, which can be prevented by the expression of any of the flippases or by prevention of ECA synthesis at an earlier step (88, 128). However, with wild-type expression of wzxO16 and wzxC, deletion of wzxE is not lethal, and approximately wild-type levels of ECACYC are produced (88). These data can be explained by the complex specificity of Wzx family flippases, which has recently been thoroughly reviewed (165). Recent work from Liu et al. (166) suggests “structure-specific triggering,” where flipping is triggered by recognition of specific structural element(s) and other “incorrect” O-antigens may trigger flipping at low frequency when the “correct” substrate is absent (166).
Under normal conditions, the expression of Wzx flippases is low (167), and these flippases show specificity to their canonical substrates (166, 168–172). With various Wzx proteins, the specificity that triggers flipping has been found to be the first sugar residue attached to the Und-PP carrier (168, 169), the presence of terminal side branch residues (170, 171), or the identity or linkage of the terminal residue of the oligosaccharide (166, 171, 172). In the absence of their canonical substrate, the flippases can transport other Und-PP-linked oligosaccharides with various degrees of efficiency depending on their structural similarity, and this transport can be increased by overexpression (166, 170, 171). Thus, in E. coli K-12, which is O-antigen negative but would produce O-antigen with an initial GlcNAc and which does not produce colanic acid unless stressed, the O-antigen and colanic acid Wzx proteins can be utilized with enough efficiency by ECA to prevent a lethal accumulation of lipid IIIECA and to produce a measurable accumulation of ECA (88), although the process may be inefficient.
Finally, WaaL, the O-antigen ligase, is responsible for attaching both O-antigen and ECA to LPS (43). Thus, the level of ECALPS greatly depends on the presence or absence of O-antigen, with very little ECALPS produced in O-antigen plus strains in E. coli (35). However, Yersinia enterocolitica and Proteus mirabilis produce significant amounts of ECALPS even in the presence of O-antigen (76, 105). Thus, the interactions between ECA biosynthesis, O-antigen, and peptidoglycan biosynthesis pathways are highly complex. One consequence of these complex interactions is that it confounds the interpretation of high-throughput genetic screens identifying functions of genes in ECA biosynthesis (173–176).
BIOLOGICAL SIGNIFICANCE OF ECA
The order Enterobacterales contains many pathogens important to living organisms, including human beings. Early studies showed that when bacteria are subcultured for prolonged periods under laboratory conditions, the capability to synthesize O-chains diminishes, but there are no effects on the stability of ECA (37). Thus, ECA must have a vital role in Enterobacterales (Table 3). However, efforts to clarify the specific biological role of ECA for the enterobacterial cell have failed, partially because of the many interactions between the O-antigen, peptidoglycan (PG), and ECA biosynthesis pathways (43, 144, 156–159). Due to this, very few unambiguously assigned functions have been ascribed to ECA.
TABLE 3.
Biological significance of ECA in Enterobacterales
| Function | Type of ECA | Associated gene(s) | Reference(s) |
|---|---|---|---|
| Inhibition of P22 lysis in Salmonella enterica | Complete biosynthesis disruption, possible peptidoglycan stress | wecB, wecC, wecD, wecE, wecG, and wzxE | 176 |
| Virulence in S. Typhimurium | Loss of all forms of ECA | wecA, wecD | 147, 162 |
| Resistance to toxic molecules (e.g., bile salt, acetic acid, serum, and antibiotics) | Complete loss of ECA, loss O-antigen in some species | wecA | 150, 160, 178, 181, 182 |
| Resistance to gentamycin | Accumulation of lipid IIECA; peptidoglycan stress | wecE | 179 |
| Resistance to nalidixic and amikacin | Accumulation of lipid IIIECA; peptidoglycan stress | wzxE | 180 |
| Maintenance of OM permeability barrier and resistance to detergent and bile salt | ECACYC | 111, 145 | |
| Proposed regulation of Ca2+ ions in the cell | ECAPG | 52 | |
| Maintenance of cell membrane integrity in S. marcescens | Loss of all forms of ECA | 183 |
There has been no difference observed in both the autolysis and the viability in cells having ECA and their counterparts lacking the antigen (177). Barua et al. (178) have shown in the Shiga-toxin-producing E. coli strain O157:H7 that mutants in wecA and wecB resulting in the loss of ECA and O-antigen or ECA alone, respectively, are sensitive to acetic acid. In addition, there is an increased sensitivity to some antibiotics in mutants lacking ECA, especially aminoglycosides, for example, kanamycin and gentamicin (177). A similar observation was made in E. coli where a wecE mutant was found to be sensitive to gentamicin (179). In addition, Girgis et al. (180) observed that a wzxE mutant has no phenotype in neutral agar media but, in the presence of nalidixic acid and amikacin, the mutant is sensitive compared to the wild type. A large-scale chemical genetics screen suggested that a wecA mutant of E. coli K-12 lacking all ECAs was sensitive to azidothymidine, CHIR-90, minocycline, procaine, puromycin, triclosan, and trimethoprim and resistant to fusidic acid, isoniazid, amdinocillin, vancomycin, and polymyxin (181). A comparison between the ECAPG and the lipopolysaccharide of Salmonella Montevideo showed that the ECAPG has a higher Ca2+-to-Mg2+ ratio than lipopolysaccharide, hence the suggestion that enterobacterial common antigen is important for the supply of calcium ions in the cell (52). Random-transposon mutagenesis experiments performed in Salmonella enterica revealed that disruption of six of the ECA operon genes (wecB, wecC, wecD, wecE, wecG, and wzxE) led to increased speed of lysis by bacteriophage P22 (176). As no effect was observed for disruption of wecA, it is possible that this effect is due to the peptidoglycan stress caused by these mutations.
One of the critical roles of ECA is the pathogenicity of Enterobacterales, which has been found in some studies. For instance, Salmonella enterica lacking ECA (with mutations in either wecD or wecA) becomes less virulent and more sensitive to bile salts (147), although it does not use the wecA gene for O-antigen biosynthesis, as do many Enterobacterales. The phenotype of the wecD mutant was more severe than that for the wecA mutation, likely due to an accumulation of a Und-P-linked ECA precursor disrupting peptidoglycan synthesis. Gilbreath et al. (182) further validated this result. In vitro they found that a wecA null mutant of S. Typhimurium is deficient in ECA production but fully competent for O-antigen production and lacks stress response activation caused by peptidoglycan biosynthesis disruption. This mutant was highly attenuated in mice, causing a persistent low-level infection that did not kill the mice (182).
Interestingly, mutants defective in ECA biosynthesis trigger Rcs stress response activation in Serratia marcescens regardless of whether peptidoglycan biosynthesis is disrupted, suggesting that ECA may play an especially important role in envelope integrity for this species (150). A role in envelope integrity is also suggested by the overproduction of outer membrane vesicles (OMV) in the absence of ECA in Serratia marcescens, suggesting an instability in the OM (183). In contrast, a screen in E. coli K-12 revealed differences in OMV production in ECA biosynthesis mutants, but these phenotypes varied greatly depending on which gene was mutated, suggesting that the results may be indirect (184). Phan et al. (174) found that seven genes of the ECA operon are essential for serum resistance in E. coli; however, these effects may have been the result of the loss of O-antigen and/or isoprenoid carrier effects.
Recently, we determined that one of the forms of ECA, ECACYC, plays a significant role in the OM permeability barrier (145). We found that loss of ECACYC disrupts the OM permeability barrier, causing detergent and bile salt sensitivity. Furthermore, we determined that ECACYC genetically interacts with a protein of unknown function, YhdP, to carry out this activity. When yhdP is deleted, ECACYC takes on aberrant activity that damages the OM, despite greatly lowered ECACYC levels. Careful screening of different ECA mutants and screening for hallmarks of peptidoglycan stress allowed us to eliminate peptidoglycan stress as a cause of these phenotypes (145).
ECA has been considered as a vaccination against infections stemming from enterobacterial strains due to its prevalence within the order. In a mouse model, salmonellosis leads to the development of ECA antibodies. However, no insight into the role of ECA antibodies in protection from infection was provided (185). Results regarding the protective nature of ECA antibodies have been mixed (63–65, 78, 79, 81). Further investigation into the protective nature of ECA antibodies against Enterobacterales species is warranted, as ECA antibodies have the potential to protect against all enterobacterial pathogens. In an ECA vaccine candidate, it would likely be important that the O-acetylation of ECA be maintained. O-Acetylation of surface-exposed polysaccharide has been shown to be important for the generation of protective antibodies for many pathogens (186–191). Kajimura et al. (111) have reported that the absence of O-acetylation in ECACYC decreases the immunoreactivity of this ECA form toward a rabbit-generated antibody. A recent study has determined that a partially O-acetylated polysaccharide may be highly advantageous for vaccine use due to epitope exposure and hydrophobicity profiles (192). In this aspect, ECA may be ideal given its nonstoichiometric acetylation of GlcNAc (111).
CONCLUDING REMARKS
Since the discovery of ECA in 1962, the work of many investigators has elucidated considerable information about the structure and biogenesis of ECA, and yet our understanding of this fascinating molecule remains incomplete. Whereas many carbohydrate antigens on the bacterial cell surface are highly variable allowing for escape from immune surveillance, ECA remains invariant despite its presence in many pathogenic species. This suggest that the function of ECA must be incredibly important for the cell. Yet, this function is largely unknown. In the past, investigations of the function of ECA have been hampered by the complex genetic and biosynthetic interactions between ECA and other cytoplasmic glycans (i.e., peptidoglycan and O-antigen). Now that these interactions have been characterized, more in-depth studies of ECA functions will be possible.
It is quite likely that the functions of ECA will be found to vary between the types of ECA and to occur on the level of cellular function as well as interaction with the environment. For instance, it has become clear that ECACYC and ECAPG both play roles in maintaining the OM permeability barrier (145); however, given that antibiotic and detergent susceptibility differs between the loss of these two molecules and the different cellular location of the ECA forms, it is likely that the function of each is distinct. ECALPS, as of yet, has had no cellular function described. While it is clear that ECA is important for pathogenesis, at least in Salmonella, it has not yet been determined whether this is due to an alteration of Salmonella cellular function or an alteration of Salmonella’s interaction with the host.
Nevertheless, from the earliest studies of ECA it has been shown that ECA interacts with the host immune system, leading to the production of broadly cross-reactive antibodies. Yet, it is not known whether these ubiquitous antibodies play a role in the pathogenesis or protection from enterobacterial pathogens. Further evidence of the importance of ECA interactions with the environment can be gleaned from the regulation of ECA expression or chain length by temperature (141, 145, 193). Future investigations into the functions of ECA will lend important insights into the cellular function and host-pathogen interactions of an important group of bacteria.
ACKNOWLEDGMENTS
We thank members of the Mitchell Lab for productive discussions.
This work was supported by startup funds from Texas A&M University.
Footnotes
Citation Rai AK, Mitchell AM. 2020. Enterobacterial common antigen: synthesis and function of an enigmatic molecule. mBio 11:e01914-20. https://doi.org/10.1128/mBio.01914-20.
REFERENCES
- 1.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Labischinski H, Barnickel G, Bradaczek H, Naumann D, Rietschel ET, Giesbrecht P. 1985. High state of order of isolated bacterial lipopolysaccharide and its possible contribution to the permeation barrier property of the outer membrane. J Bacteriol 162:9–20. doi: 10.1128/JB.162.1.9-20.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plesiat P, Nikaido H. 1992. Outer membranes of Gram‐negative bacteria are permeable to steroid probes. Mol Microbiol 6:1323–1333. doi: 10.1111/j.1365-2958.1992.tb00853.x. [DOI] [PubMed] [Google Scholar]
- 5.Papahadjopoulos D, Portis A, Pangborn W. 1978. Calcium‐induced lipid phase transitions and membrane fusion. Ann N Y Acad Sci 308:50–66. doi: 10.1111/j.1749-6632.1978.tb22013.x. [DOI] [PubMed] [Google Scholar]
- 6.Simpson BW, Trent MS. 2019. Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol 17:403–416. doi: 10.1038/s41579-019-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189–198. doi: 10.1016/S0092-8674(00)81750-X. [DOI] [PubMed] [Google Scholar]
- 8.Fairman JW, Noinaj N, Buchanan SK. 2011. The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr Opin Struct Biol 21:523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harder KJ, Nikaido H, Matsuhashi M. 1981. Mutants of Escherichia coli that are resistant to certain beta-lactam compounds lack the ompF porin. Antimicrob Agents Chemother 20:549–552. doi: 10.1128/AAC.20.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hooper DC, Wolfson JS, Souza KS, Tung C, McHugh GL, Swartz MN. 1986. Genetic and biochemical characterization of norfloxacin resistance in Escherichia coli. Antimicrob Agents Chemother 29:639–644. doi: 10.1128/AAC.29.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pugsley AP, Schnaitman CA. 1978. Outer membrane proteins of Escherichia coli VII. Evidence that bacteriophage-directed protein 2 functions as a pore. J Bacteriol 133:1181–1189. doi: 10.1128/JB.133.3.1181-1189.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reeve EC, Doherty P. 1968. Linkage relationships of two genes causing partial resistance to chloramphenicol in Escherichia coli. J Bacteriol 96:1450–1451. doi: 10.1128/JB.96.4.1450-1451.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginsburgh CL, Black PN, Nunn WD. 1984. Transport of long chain fatty acids in Escherichia coli. Identification of a membrane protein associated with the fadL gene. J Biol Chem 259:8437–8443. [PubMed] [Google Scholar]
- 14.Berry J, Rajaure M, Pang T, Young R. 2012. The spanin complex is essential for lambda lysis. J Bacteriol 194:5667–5674. doi: 10.1128/JB.01245-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rojas ER, Billings G, Odermatt PD, Auer GK, Zhu L, Miguel A, Chang F, Weibel DB, Theriot JA, Huang KC. 2018. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559:617–621. doi: 10.1038/s41586-018-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Wuijckhuijse AL, van Baar BL. 2008. Recent advances in real-time mass spectrometry detection of bacteria, p 929–954. Principles of bacterial detection: biosensors, recognition receptors and microsystems. Springer, New York, NY. [Google Scholar]
- 17.Whitfield C, Roberts IS. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol 31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- 18.Robins‐Browne RM, Hartland EL. 2002. Escherichia coli as a cause of diarrhea. J Gastroenterol Hepatol 17:467–475. doi: 10.1046/j.1440-1746.2002.02769.x. [DOI] [PubMed] [Google Scholar]
- 19.Lerouge I, Vanderleyden J. 2002. O-antigen structural variation: mechanisms and possible roles in animal/plant–microbe interactions. FEMS Microbiol Rev 26:17–47. doi: 10.1111/j.1574-6976.2002.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Rothemund D, Curd H, Reeves PR. 2003. Species-wide variation in the Escherichia coli flagellin (H-antigen) gene. J Bacteriol 185:2936–2943. doi: 10.1128/JB.185.9.2936-2943.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams P, Lambert PA, Brown MR, Jones RJ. 1983. The role of the O and K antigens in determining the resistance of Klebsiella aerogenes to serum killing and phagocytosis. Microbiology 129:2181–2191. doi: 10.1099/00221287-129-7-2181. [DOI] [PubMed] [Google Scholar]
- 22.Ørskov I, Ørskov F, Sojka W, Leach J. 2009. Simultaneous occurrence of E. coli B and L antigens in strains from diseased swine: influence of cultivation temperature two new E. coli K antigens: K 87 and K 88. Acta Pathol Microbiol Scand 53:404–422. doi: 10.1111/j.1699-0463.1961.tb00424.x. [DOI] [PubMed] [Google Scholar]
- 23.Smith HW, Linggood MA. 1971. Observations on the pathogenic properties of the K88, Hly and Ent plasmids of Escherichia coli with particular reference to porcine diarrhoea. J Med Microbiol 4:467–485. doi: 10.1099/00222615-4-4-467. [DOI] [PubMed] [Google Scholar]
- 24.Ørskov I, Ørskov F, Smith HW, Sojka W. 2009. The establishment of K99, a thermolabile, transmissible Escherichia coli K antigen, previously called “Kco”, possessed by calf and lamb enteropathogenic strains. Acta Pathol Microbiol Scand B Microbiol 83B:31–36. doi: 10.1111/j.1699-0463.1975.tb00066.x. [DOI] [PubMed] [Google Scholar]
- 25.Ørskov F, Ørskov I. 1978. Chapter I. Serotyping of Enterobacteriaceae, with special emphasis on K antigen determination, p 1–77. Methods in microbiology, vol 11 Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 26.Kuhn H-M, Meier-Dieter U, Mayer H. 1988. ECA, the enterobacterial common antigen. FEMS Microbiol Rev 4:195–222. doi: 10.1111/j.1574-6968.1988.tb02743.x. [DOI] [PubMed] [Google Scholar]
- 27.Rinno J, Golecki J, Mayer H. 1980. Localization of enterobacterial common antigen: immunogenic and nonimmunogenic enterobacterial common antigen-containing Escherichia coli. J Bacteriol 141:814–821. doi: 10.1128/JB.141.2.814-821.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Acker G, Schmidt G, Mayer H. 1982. Accessibility of enterobacterial common antigen to antibodies in encapsulated and non-capsulated S and R forms of Escherichia coli. Microbiology 128:1577–1583. doi: 10.1099/00221287-128-7-1577. [DOI] [PubMed] [Google Scholar]
- 29.Acker G, Bitter-Suermann D, Meier-Dieter U, Peters H, Mayer H. 1986. Immunocytochemical localization of enterobacterial common antigen in Escherichia coli and Yersinia enterocolitica cells. J Bacteriol 168:348–356. doi: 10.1128/JB.168.1.348-356.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Acker G. 1988. Immunoelectron microscopy of surface antigens (polysaccharides) of Gram-negative bacteria using pre-and post-embedding techniques, p 147–174. Methods in microbiology, vol 20 Academic Press, San Diego, CA. [Google Scholar]
- 31.Jann K, Westphal O. 1975. Microbial polysaccharides, p 1–125, The antigens. Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 32.Weibull C, Högfeldt E, Sillén LG, Kinell P-O. 1950. Investigations on bacterial flagella. Acta Chem Scand 4:268–276. doi: 10.3891/acta.chem.scand.04-0268. [DOI] [Google Scholar]
- 33.Kunin CM, Beard MV, Halmagyi NE. 1962. Evidence for a common hapten associated with endotoxin fractions of E. coli and other Enterobacteriaceae. Proc Soc Exp Biol Med 111:160–166. doi: 10.3181/00379727-111-27734. [DOI] [Google Scholar]
- 34.Le Minor L, Chalon A, Veron M. 1972. Recherches sur la présence de l’antigène commun des” Enterobacteriaceae” (antigène Kunin) chez les” Yersinia” Levinea” Aeromonas” et” Vibrio. Ann Inst Pasteur (Paris) 123:761–774. [PubMed] [Google Scholar]
- 35.Mäkelä P, Mayer H. 1976. Enterobacterial common antigen. Bacteriol Rev 40:591–632. doi: 10.1128/MMBR.40.3.591-632.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Böttger E, Jürs M, Barrett T, Wachsmuth K, Metzger S, Bitter-Suermann D. 1987. Qualitative and quantitative determination of enterobacterial common antigen (ECA) with monoclonal antibodies: expression of ECA by two Actinobacillus species. J Clin Microbiol 25:377–382. doi: 10.1128/JCM.25.2.377-382.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer H, Schmidt G. 1979. Chemistry and biology of the enterobacterial common antigen (ECA), p 99–153, Current topics in microbiology and immunology. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 38.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, von Mering C. 2019. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCutcheon JP, Boyd BM, Dale C. 2019. The life of an insect endosymbiont from the cradle to the grave. Curr Biol 29:R485–R495. doi: 10.1016/j.cub.2019.03.032. [DOI] [PubMed] [Google Scholar]
- 40.Kauffmann F. 1966. The bacteriology of the Enterobacteriaceae, p 333–360. The Williams & Wilkins Co, Baltimore, MD. [Google Scholar]
- 41.Perch B. 2009. On the serology of the Proteus group. Acta Pathol Mic Sc 25:703–714. doi: 10.1111/j.1699-0463.1948.tb00712.x. [DOI] [Google Scholar]
- 42.Grimont PA, Weill FX. 2007. Antigenic formulae of the Salmonella serovars, 9th ed WHO Collaborating Centre for Reference and Research on; Salmonella, Institut Pasteur, Paris, France. [Google Scholar]
- 43.Schmidt G, Männel D, Mayer H, Whang H, Neter E. 1976. Role of a lipopolysaccharide gene for immunogenicity of the enterobacterial common antigen. J Bacteriol 126:579–586. doi: 10.1128/JB.126.2.579-586.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki T, Gorzynski E, Neter E. 1964. Separation by ethanol of common and somatic antigens of Enterobacteriaceae. J Bacteriol 88:1240–1243. doi: 10.1128/JB.88.5.1240-1243.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorzynski E, Whang H, Suzuki T, Neter E. 1963. Differences in antibody response of rabbit to enterobacterial antigen after intravenous and subcutaneous injection with adjuvant. Proc Soc Exp Biol Med 114:700–704. doi: 10.3181/00379727-114-28775. [DOI] [PubMed] [Google Scholar]
- 46.Kiss P, Rinno J, Schmidt G, Mayer H. 1978. Structural studies on the immunogenic form of the enterobacterial common antigen. Eur J Biochem 88:211–218. doi: 10.1111/j.1432-1033.1978.tb12440.x. [DOI] [PubMed] [Google Scholar]
- 47.Kuhn HM, Neter E, Mayer H. 1983. Modification of the lipid moiety of the enterobacterial common antigen by the “Pseudomonas factor”. Infect Immun 40:696–700. doi: 10.1128/IAI.40.2.696-700.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dell A, Oates J, Lugowski C, Romanowska E, Kenne L, Lindberg B. 1984. The enterobacterial common-antigen, a cyclic polysaccharide. Carbohydr Res 133:95–104. doi: 10.1016/0008-6215(84)85186-1. [DOI] [PubMed] [Google Scholar]
- 49.Dell A, Panico M. 1986. In Gaskel S. (ed), Mass spectrometry in biomedical research. Wiley, New York, NY. [Google Scholar]
- 50.Whang H, Mayer H, Schmidt G, Neter E. 1972. Immunogenicity of the common enterobacterial antigen produced by smooth and rough strains. Infect Immun 6:533–539. doi: 10.1128/IAI.6.4.533-539.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt G, Jann B, Jann K. 1974. Genetic and immunochemical studies on Escherichia coli O14:K7:H. Eur J Biochem 42:303–309. doi: 10.1111/j.1432-1033.1974.tb03340.x. [DOI] [PubMed] [Google Scholar]
- 52.Kuhn HM, Adamus G, Romanowska E, Mayer H. 1981. Effect of proteins on the immunogenicity of enterobacterial common antigen. Infect Immun 34:373–377. doi: 10.1128/IAI.34.2.373-377.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neter E, Whang HY, Mayer H. 1973. Immunogenicity and antigenicity of endotoxic lipopolysaccharides: reversible effects of temperature on immunogenicity. J Infect Dis 128:S56–S60. doi: 10.1093/infdis/128.Supplement_1.S56. [DOI] [PubMed] [Google Scholar]
- 54.Chaplin DD. 2010. Overview of the immune response. J Allergy Clin Immunol 125:S3–S23. doi: 10.1016/j.jaci.2009.12.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park BS, Lee JO. 2013. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med 45:e66. doi: 10.1038/emm.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lugowski C, Kułakowska M, Romanowska E. 1983. Enterobacterial common antigen-tetanus toxoid conjugate as immunogen. Infect Immun 42:1086–1091. doi: 10.1128/IAI.42.3.1086-1091.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schroeder HW Jr, Cavacini L. 2010. Structure and function of immunoglobulins. J Allergy Clin Immunol 125:S41–S52. doi: 10.1016/j.jaci.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mogensen TH. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev 22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kunin CM. 1963. Separation, characterization, and biological significance of a common antigen in Enterobacteriaceae. J Exp Med 118:565–586. doi: 10.1084/jem.118.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kunin CM, Beard MV. 1963. Serological studies of O antigens of Escherichia coli by means of the hemagglutination test. J Bacteriol 85:541–548. doi: 10.1128/JB.85.3.541-548.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whang H, Neter E. 1963. Study of heterogenetic (Kunin) antibodies in serum of healthy subjects and children with enteric and urinary tract infections. J Pediatr 63:412–419. doi: 10.1016/S0022-3476(63)80429-1. [DOI] [PubMed] [Google Scholar]
- 62.Moncevičiūtė-Eringienė E, Kazbarienė B, Milašienė V, Characiejus D, Kemeklienė R. 2006. Natural antibodies to endotoxin in experimental and clinical oncology. Exp Oncol 28:89–91. [PubMed] [Google Scholar]
- 63.Gorzynski EA, Ambrus JL, Neter E. 1971. Effect of common enterobacterial antiserum on experimental Salmonella Typhimurium infection of mice. Proc Soc Exp Biol Med 137:1209–1212. doi: 10.3181/00379727-137-35757. [DOI] [PubMed] [Google Scholar]
- 64.Gorzynski EA, Krasny SA. 1975. Immunologic mimicry between mouse tissue and enterobacterial common antigen. Immunol Comm 4:39–49. doi: 10.3109/08820137509055760. [DOI] [PubMed] [Google Scholar]
- 65.Gorzynski EA, Neter E, Ambrus JL. 1970. Differences in antibody responses of mouse strains to enterobacterial common antigen. Proc Soc Exp Biol Med 134:776–779. doi: 10.3181/00379727-134-34881. [DOI] [PubMed] [Google Scholar]
- 66.Dlaz F, Neter E. 1968. Antibody response to the common enterobacterial antigen of children with shigellosis, salmonellosis or urinary tract infection. Am J Med Sci 256:18–24. doi: 10.1097/00000441-196807000-00003. [DOI] [PubMed] [Google Scholar]
- 67.Neter E, Whang H, Nowotny A. 1972. The common antigen of Gram-negative bacteria Cellular antigens. Springer-Verlag, New York, NY. [Google Scholar]
- 68.Andersen HJ. 1966. Studies of urinary tract infections in infancy and childhood: VII. The relation of E. coli antibodies in pyelonephritis as measured by homologous and common (Kunin) antigens. J Pediatr 68:542–550. doi: 10.1016/S0022-3476(66)80391-8. [DOI] [Google Scholar]
- 69.Saito I. 1967. Serological study of chronic pyelonephritis, especially on the diagnostic value of the estimation of enterobacterial common antibody response. Fukushima J Med Sci 14:45–53. [PubMed] [Google Scholar]
- 70.Domingue G, Salhi A, Rountree C, Little W. 1970. Prevention of experimental hematogenous and retrograde pyelonephritis by antibodies against enterobacterial common antigen. Infect Immun 2:175–182. doi: 10.1128/IAI.2.2.175-182.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frentz G, Domingue G. 1973. Effects of immunization with ethanol-soluble enterobacterial common antigen on in vivo bacterial clearance and hematogenous pyelonephritis. Proc Soc Exp Biol Med 142:246–252. doi: 10.3181/00379727-142-36998. [DOI] [PubMed] [Google Scholar]
- 72.McLaughlin JC, Domingue G. 1974. The immunologic role of the ethanol-soluble enterobacterial common antigen versus experimental renal infection. Immunol Invest 3:51–75. doi: 10.3109/08820137409055745. [DOI] [PubMed] [Google Scholar]
- 73.Duda KT. 2007. Reactivity of polyclonal antisera against R mutants of Yersinia enterocolitica O:3. Dissertation. University of Silesia, Katowice, Poland. [Google Scholar]
- 74.Radziejewska-Lebrecht J, Skurnik M, Shashkov AS, Brade L, Rózalski A, Bartodziejska B, Mayer H. 1998. Immunochemical studies on R mutants of Yersinia enterocolitica O: 3. Acta Biochim Pol 45:1011–1019. doi: 10.18388/abp.1998_4358. [DOI] [PubMed] [Google Scholar]
- 75.Radziejewska-Lebrecht J, Kasperkiewicz K, Skurnik M, Brade L, Steinmetz I, Świerzko AS, Muszyński A. 2004. ECA-antibodies in antisera against R mutants of Yersinia enterocolitica O:3, p 215–218. The genus Yersinia. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 76.Duda KA, Duda KT, Beczała A, Kasperkiewicz K, Radziejewska-Lebrecht J, Skurnik M. 2009. ECA-immunogenicity of Proteus mirabilis strains. Arch Immunol Ther Exp (Warsz) 57:147–151. doi: 10.1007/s00005-009-0018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rastawicki W. 2007. Humoral response to selected antigens of Yersinia enterocolitica and Yersinia pseudotuberculosis in the course of yersiniosis in humans. I. Occurrence of antibodies to enterobacterial common antigen (ECA). Med Dosw Mikrobiol 59:93–102. [PubMed] [Google Scholar]
- 78.McCabe WR, Greely A. 1973. Common enterobacterial antigen II. Effect of immunization on challenge with heterologous Bacilli. Infect Immun 7:386–392. doi: 10.1128/IAI.7.3.386-392.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Albertson TE, Panacek EA, MacArthur RD, Johnson SB, Benjamin E, Matuschak GM, Zaloga G, Maki D, Silverstein J, Tobias JK. 2003. Multicenter evaluation of a human monoclonal antibody to Enterobacteriaceae common antigen in patients with Gram-negative sepsis. Crit Care Med 31:419–427. doi: 10.1097/01.CCM.0000045564.51812.3F. [DOI] [PubMed] [Google Scholar]
- 80.Bridge DR, Whitmire JM, Gilbreath JJ, Metcalf ES, Merrell DS. 2015. An enterobacterial common antigen mutant of Salmonella enterica serovar Typhimurium as a vaccine candidate. Int J Med Microbiol 305:511–522. doi: 10.1016/j.ijmm.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 81.Huang C, Liu Q, Luo Y, Li P, Liu Q, Kong Q. 2016. Regulated delayed synthesis of lipopolysaccharide and enterobacterial common antigen of Salmonella Typhimurium enhances immunogenicity and cross-protective efficacy against heterologous Salmonella challenge. Vaccine 34:4285–4292. doi: 10.1016/j.vaccine.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aoki S, Yoshikawa K, Yokoyama T, Nonogaki T, Iwasaki S, Mitsui T, Niwa S. 1996. Role of enteric bacteria in the pathogenesis of rheumatoid arthritis: evidence for antibodies to enterobacterial common antigens in rheumatoid sera and synovial fluids. Ann Rheum Dis 55:363–369. doi: 10.1136/ard.55.6.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aoki S, Mitsui T. 2005. Histological features of rheumatoid arthritis patients having antibodies to enterobacterial common antigens: correlation of antibody levels with semiquantitative histologic scores and laboratory markers. Clin Exp Rheumatol 23:8–13. [PubMed] [Google Scholar]
- 84.Rashid T, Jayakumar K, Binder A, Ellis S, Cunningham P, Ebringer A. 2007. Rheumatoid arthritis patients have elevated antibodies to cross-reactive and non-cross-reactive antigens from Proteus microbes. Clin Exp Rheumatol 25:259–267. [PubMed] [Google Scholar]
- 85.Basu S, Kuhn HM, Neszmelyi A, Himmelspach K, Mayer H. 1987. Chemical characterization of enterobacterial common antigen isolated from Plesiomonas shigelloides ATCC 14029. Eur J Biochem 162:75–81. doi: 10.1111/j.1432-1033.1987.tb10544.x. [DOI] [PubMed] [Google Scholar]
- 86.Vinogradov EV, Knirel YA, Thomas-Oates JE, Shashkov AS, L'vov VL. 1994. The structure of the cyclic enterobacterial common antigen (ECA) from Yersinia pestis. Carbohydr Res 258:223–232. doi: 10.1016/0008-6215(94)84088-1. [DOI] [PubMed] [Google Scholar]
- 87.Erbel PJ, Barr K, Gao N, Gerwig GJ, Rick PD, Gardner KH. 2003. Identification and biosynthesis of cyclic enterobacterial common antigen in Escherichia coli. J Bacteriol 185:1995–2004. doi: 10.1128/JB.185.6.1995-2004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kajimura J, Rahman A, Rick PD. 2005. Assembly of cyclic enterobacterial common antigen in Escherichia coli K-12. J Bacteriol 187:6917–6927. doi: 10.1128/JB.187.20.6917-6927.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Männel D, Mayer H. 1978. Isolation and chemical characterization of the enterobacterial common antigen. Eur J Biochem 86:361–370. doi: 10.1111/j.1432-1033.1978.tb12318.x. [DOI] [PubMed] [Google Scholar]
- 90.Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 91.Barr K, Klena J, Rick PD. 1999. The modality of enterobacterial common antigen polysaccharide chain lengths is regulated by o349 of the wec gene cluster of Escherichia coli K-12. J Bacteriol 181:6564–6568. doi: 10.1128/JB.181.20.6564-6568.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rahman A, Barr K, Rick PD. 2001. Identification of the structural gene for the TDP-Fuc4NAc:lipid II Fuc4NAc transferase involved in synthesis of enterobacterial common antigen in Escherichia coli K-12. J Bacteriol 183:6509–6516. doi: 10.1128/JB.183.22.6509-6516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rick P, Silver R. 1996. Enterobacterial common antigen and capsular polysaccharides, p 104–122. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC. [Google Scholar]
- 94.Daniels DL, Plunkett G, Burland V, Blattner FR. 1992. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science 257:771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- 95.Dodgson C, Amor P, Whitfield C. 1996. Distribution of the rol gene encoding the regulator of lipopolysaccharide O-chain length in Escherichia coli and its influence on the expression of group I capsular K antigens. J Bacteriol 178:1895–1902. doi: 10.1128/JB.178.7.1895-1902.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Franco AV, Liu D, Reeves PR. 1996. A Wzz (Cld) protein determines the chain length of K lipopolysaccharide in Escherichia coli O8 and O9 strains. J Bacteriol 178:1903–1907. doi: 10.1128/JB.178.7.1903-1907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Whitfield C. 1995. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol 3:178–185. doi: 10.1016/S0966-842X(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 98.Whitfield C, Amor PA, KöPlin R. 1997. Modulation of the surface architecture of Gram‐negative bacteria by the action of surface polymer: lipid A–core ligase and by determinants of polymer chain length. Mol Microbiol 23:629–638. doi: 10.1046/j.1365-2958.1997.2571614.x. [DOI] [PubMed] [Google Scholar]
- 99.Jones MB, Rosenberg JN, Betenbaugh MJ, Krag SS. 2009. Structure and synthesis of polyisoprenoids used in N-glycosylation across the three domains of life. Biochim Biophys Acta Gen Subj 1790:485–494. doi: 10.1016/j.bbagen.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barr K, Nunes-Edwards P, Rick PD. 1989. In vitro synthesis of a lipid-linked trisaccharide involved in synthesis of enterobacterial common antigen. J Bacteriol 171:1326–1332. doi: 10.1128/JB.171.3.1326-1332.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barr K, Rick P. 1987. Biosynthesis of enterobacterial common antigen in Escherichia coli. In vitro synthesis of lipid-linked intermediates. J Biol Chem 262:7142–7150. [PubMed] [Google Scholar]
- 102.Rick P, Mayer H, Neumeyer B, Wolski S, Bitter-Suermann D. 1985. Biosynthesis of enterobacterial common antigen. J Bacteriol 162:494–503. doi: 10.1128/JB.162.2.494-503.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meier-Dieter U, Starman R, Barr K, Mayer H, Rick P. 1990. Biosynthesis of enterobacterial common antigen in Escherichia coli. Biochemical characterization of Tn10 insertion mutants defective in enterobacterial common antigen synthesis. J Biol Chem 265:13490–13497. [PubMed] [Google Scholar]
- 104.Samuel G, Reeves P. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr Res 338:2503–2519. doi: 10.1016/j.carres.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 105.Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem 75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 106.Brown ED, Vivas EI, Walsh CT, Kolter R. 1995. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J Bacteriol 177:4194–4197. doi: 10.1128/JB.177.14.4194-4197.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Typas A, Banzhaf M, Gross CA, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rick P, Fung L, Ho C, Osborn M. 1977. Lipid A mutants of Salmonella Typhimurium. Purification and characterization of a lipid A precursor produced by a mutant in 3-deoxy-d-mannooctulosonate-8-phosphate synthetase. J Biol Chem 252:4904–4912. [PubMed] [Google Scholar]
- 109.Amer AO, Valvano MA. 2000. The N-terminal region of the Escherichia coli WecA (Rfe) protein, containing three predicted transmembrane helices, is required for function but not for membrane insertion. J Bacteriol 182:498–503. doi: 10.1128/JB.182.2.498-503.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lehrer J, Vigeant KA, Tatar LD, Valvano MA. 2007. Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide. J Bacteriol 189:2618–2628. doi: 10.1128/JB.01905-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kajimura J, Rahman A, Hsu J, Evans MR, Gardner KH, Rick PD. 2006. O acetylation of the enterobacterial common antigen polysaccharide is catalyzed by the product of the yiaH gene of Escherichia coli K-12. J Bacteriol 188:7542–7550. doi: 10.1128/JB.00783-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sala RF, Morgan PM, Tanner ME. 1996. Enzymatic formation and release of a stable glycal intermediate: the mechanism of the reaction catalyzed by UDP-N-acetylglucosamine 2-epimerase. J Am Chem Soc 118:3033–3034. doi: 10.1021/ja960266z. [DOI] [Google Scholar]
- 113.Morgan PM, Sala RF, Tanner ME. 1997. Eliminations in the reactions catalyzed by UDP-N-acetylglucosamine 2-epimerase. J Am Chem Soc 119:10269–10277. doi: 10.1021/ja971718q. [DOI] [Google Scholar]
- 114.Campbell RE, Mosimann SC, Tanner ME, Strynadka NC. 2000. The structure of UDP-N-acetylglucosamine 2-epimerase reveals homology to phosphoglycosyl transferases. Biochemistry 39:14993–15001. doi: 10.1021/bi001627x. [DOI] [PubMed] [Google Scholar]
- 115.Samuel J, Tanner ME. 2004. Active site mutants of the “non-hydrolyzing” UDP-N-acetylglucosamine 2-epimerase from Escherichia coli. Biochim Biophys Acta Proteins Proteom 1700:85–91. doi: 10.1016/j.bbapap.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 116.Kawamura T, Ishimoto N, Ito E. 1979. Enzymatic synthesis of uridine diphosphate N-acetyl-d-mannosaminuronic acid. J Biol Chem 254:8457–8465. doi: 10.1016/0014-5793(74)80013-X. [DOI] [PubMed] [Google Scholar]
- 117.Barr K, Ward S, Meier-Dieter U, Mayer H, Rick P. 1988. Characterization of an Escherichia coli rff mutant defective in transfer of N-acetylmannosaminuronic acid (ManNAcA) from UDP-ManNAcA to a lipid-linked intermediate involved in enterobacterial common antigen synthesis. J Bacteriol 170:228–233. doi: 10.1128/JB.170.1.228-233.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marolda CL, Valvano MA. 1995. Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7: K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J Bacteriol 177:5539–5546. doi: 10.1128/JB.177.19.5539-5546.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Meier-Dieter U, Barr K, Starman R, Hatch L, Rick P. 1992. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. Molecular cloning of the rfe-rff gene cluster. J Biol Chem 267:746–753. [PubMed] [Google Scholar]
- 120.Sivaraman J, Sauvé V, Matte A, Cygler M. 2002. Crystal structure of Escherichia coli glucose-1-phosphate thymidylyltransferase (RffH) complexed with dTTP and Mg2+. J Biol Chem 277:44214–44219. doi: 10.1074/jbc.M206932200. [DOI] [PubMed] [Google Scholar]
- 121.Gross JW, Hegeman AD, Gerratana B, Frey PA. 2001. Dehydration is catalyzed by glutamate-136 and aspartic acid-135 active site residues in Escherichia coli dTDP-glucose 4, 6-dehydratase. Biochemistry 40:12497–12504. doi: 10.1021/bi011138c. [DOI] [PubMed] [Google Scholar]
- 122.Gerratana B, Cleland WW, Frey PA. 2001. Mechanistic roles of Thr134, Tyr160, and Lys 164 in the reaction catalyzed by dTDP-glucose 4, 6-dehydratase. Biochemistry 40:9187–9195. doi: 10.1021/bi0108249. [DOI] [PubMed] [Google Scholar]
- 123.Hegeman AD, Gross JW, Frey PA. 2001. Probing catalysis by Escherichia coli dTDP-glucose-4, 6-dehydratase: identification and preliminary characterization of functional amino acid residues at the active site. Biochemistry 40:6598–6610. doi: 10.1021/bi010441a. [DOI] [PubMed] [Google Scholar]
- 124.Hwang BY, Lee HJ, Yang YH, Joo HS, Kim BG. 2004. Characterization and investigation of substrate specificity of the sugar aminotransferase WecE from E. coli K12. Chem Biol 11:915–925. doi: 10.1016/j.chembiol.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 125.Wang F, Singh S, Xu W, Helmich KE, Miller MD, Cao H, Craig AB, Jon ST, Phillips GN Jr.. 2015. Structural basis for the stereochemical control of amine installation in nucleotide sugar aminotransferases. ACS Chem Biol 10:2048–2056. doi: 10.1021/acschembio.5b00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hung MN, Rangarajan E, Munger C, Nadeau G, Sulea T, Matte A. 2006. Crystal structure of TDP-fucosamine acetyltransferase (WecD) from Escherichia coli, an enzyme required for enterobacterial common antigen synthesis. J Bacteriol 188:5606–5617. doi: 10.1128/JB.00306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Macpherson DF, Manning PA, Morona R. 1995. Genetic analysis of the rfbX gene of Shigella flexneri. Gene 155:9–17. doi: 10.1016/0378-1119(94)00918-I. [DOI] [PubMed] [Google Scholar]
- 128.Rick PD, Barr K, Sankaran K, Kajimura J, Rush JS, Waechter CJ. 2003. Evidence that the wzxE gene of Escherichia coli K-12 encodes a protein involved in the transbilayer movement of a trisaccharide-lipid intermediate in the assembly of enterobacterial common antigen. J Biol Chem 278:16534–16542. doi: 10.1074/jbc.M301750200. [DOI] [PubMed] [Google Scholar]
- 129.Paulsen IT, Beness AM, Saier MH. 1997. Computer-based analyses of the protein constituents of transport systems catalysing export of complex carbohydrates in bacteria. Microbiology 143:2685–2699. doi: 10.1099/00221287-143-8-2685. [DOI] [PubMed] [Google Scholar]
- 130.Marolda CL, Tatar LD, Alaimo C, Aebi M, Valvano MA. 2006. Interplay of the Wzx translocase and the corresponding polymerase and chain length regulator proteins in the translocation and periplasmic assembly of lipopolysaccharide O antigen. J Bacteriol 188:5124–5135. doi: 10.1128/JB.00461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nath P, Morona R. 2015. Detection of Wzy/Wzz interaction in Shigella flexneri. Microbiology 161:1797–1805. doi: 10.1099/mic.0.000132. [DOI] [PubMed] [Google Scholar]
- 132.Morona R, Van Den Bosch L, Daniels C. 2000. Evaluation of Wzz/MPA1/MPA2 proteins based on the presence of coiled-coil regions. Microbiology 146:1–4. doi: 10.1099/00221287-146-1-1. [DOI] [PubMed] [Google Scholar]
- 133.Tocilj A, Munger C, Proteau A, Morona R, Purins L, Ajamian E, Wagner J, Papadopoulos M, Van Den Bosch L, Rubinstein JL, Féthière J, Matte A, Cygler M. 2008. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat Struct Mol Biol 15:130–138. doi: 10.1038/nsmb.1374. [DOI] [PubMed] [Google Scholar]
- 134.Larue K, Kimber MS, Ford R, Whitfield C. 2009. Biochemical and structural analysis of bacterial O-antigen chain length regulator proteins reveals a conserved quaternary structure. J Biol Chem 284:7395–7403. doi: 10.1074/jbc.M809068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kalynych S, Yao D, Magee J, Cygler M. 2012. Structural characterization of closely related O-antigen lipopolysaccharide (LPS) chain length regulators. J Biol Chem 287:15696–15705. doi: 10.1074/jbc.M112.354837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kalynych S, Cherney M, Bostina M, Rouiller I, Cygler M. 2015. Quaternary structure of WzzB and WzzE polysaccharide copolymerases. Protein Sci 24:58–69. doi: 10.1002/pro.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Collins RF, Kargas V, Clarke BR, Siebert CA, Clare DK, Bond PJ, Whitfield C, Ford RC. 2017. Full-length, oligomeric structure of Wzz determined by cryo-electron microscopy reveals insights into membrane-bound states. Structure 25:806–815. doi: 10.1016/j.str.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 138.Bastin DA, Stevenson G, Brown PK, Haase A, Reeves PR. 1993. Repeat unit polysaccharides of bacteria: a model for polymerization resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol Microbiol 7:725–734. doi: 10.1111/j.1365-2958.1993.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 139.Morona R, van den Bosch L, Manning PA. 1995. Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J Bacteriol 177:1059–1068. doi: 10.1128/JB.177.4.1059-1068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lugowski C, Romanowska E. 1978. Enterobacterial common antigen: isolation from Shigella sonnei, purification and immunochemical characterization. Eur J Biochem 91:89–97. doi: 10.1111/j.1432-1033.1978.tb20941.x. [DOI] [PubMed] [Google Scholar]
- 141.Muszyński A, Rabsztyn K, Knapska K, Duda KA, Duda-Grychtoł K, Kasperkiewicz K, Radziejewska-Lebrecht J, Holst O, Skurnik M. 2013. Enterobacterial common antigen and O-specific polysaccharide coexist in the lipopolysaccharide of Yersinia enterocolitica serotype O:3. Microbiology 159:1782–1793. doi: 10.1099/mic.0.066662-0. [DOI] [PubMed] [Google Scholar]
- 142.Staaf M, Höög C, Stevensson B, Maliniak A, Widmalm G. 2001. Conformational investigation of a cyclic enterobacterial common antigen employing NMR spectroscopy and molecular dynamics simulations. Biochemistry 40:3623–3628. doi: 10.1021/bi002282l. [DOI] [PubMed] [Google Scholar]
- 143.Färnbäck M, Eriksson L, Senchenkova S, Zych K, Knirel YA, Sidorczyk Z, Widmalm G. 2003. Crystal structure of a cyclic enterobacterial common antigen. Angew Chem Int Ed Engl 42:2543–2546. doi: 10.1002/anie.200351250. [DOI] [PubMed] [Google Scholar]
- 144.Andersson A, Ahl Å, Eklund R, Widmalm G, Mäler L. 2005. Dynamics in the cyclic enterobacterial common antigen as studied by 13C NMR relaxation. J Biomol NMR 31:311–320. doi: 10.1007/s10858-005-1605-7. [DOI] [PubMed] [Google Scholar]
- 145.Mitchell AM, Srikumar T, Silhavy TJ. 2018. Cyclic enterobacterial common antigen maintains the outer membrane permeability barrier of Escherichia coli in a manner controlled by YhdP. mBio 9:e01321-18. doi: 10.1128/mBio.01321-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pazos M, Peters K. 2019. Peptidoglycan, p 127–168. Bacterial cell walls and membranes. Springer, New York, NY. [Google Scholar]
- 147.Kalynych S, Morona R, Cygler M. 2014. Progress in understanding the assembly process of bacterial O-antigen. FEMS Microbiol Rev 38:1048–1065. doi: 10.1111/1574-6976.12070. [DOI] [PubMed] [Google Scholar]
- 148.Wright A, Dankert M, Fennessey P, Robbins P. 1967. Characterization of a polyisoprenoid compound functional in O-antigen biosynthesis. Proc Natl Acad Sci U S A 57:1798–1803. doi: 10.1073/pnas.57.6.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rick PD, Hubbard GL, Kitaoka M, Nagaki H, Kinoshita T, Dowd S, Simplaceanu V, Ho C. 1998. Characterization of the lipid-carrier involved in the synthesis of enterobacterial common antigen (ECA) and identification of a novel phosphoglyceride in a mutant of Salmonella Typhimurium defective in ECA synthesis. Glycobiology 8:557–567. doi: 10.1093/glycob/8.6.557. [DOI] [PubMed] [Google Scholar]
- 150.Johnson JG, Wilson DB. 1977. Role of a sugar-lipid intermediate in colanic acid synthesis by Escherichia coli. J Bacteriol 129:225–236. doi: 10.1128/JB.129.1.225-236.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ikeda M, Wachi M, Jung H, Ishino F, Matsuhashi M. 1991. The Escherichia coli mraY gene encoding UDP-N-acetylmuramoyl-pentapeptide: undecaprenyl-phosphate phospho-N-acetylmuramoyl-pentapeptide transferase. J Bacteriol 173:1021–1026. doi: 10.1128/JB.173.3.1021-1026.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tytgat HL, Lebeer S. 2014. The sweet tooth of bacteria: common themes in bacterial glycoconjugates. Microbiol Mol Biol Rev 78:372–417. doi: 10.1128/MMBR.00007-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jorgenson MA, Young KD. 2016. Interrupting biosynthesis of O antigen or the lipopolysaccharide core produces morphological defects in Escherichia coli by sequestering undecaprenyl phosphate. J Bacteriol 198:3070–3079. doi: 10.1128/JB.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rick PD, Wolski S, Barr K, Ward S, Ramsay-Sharer L. 1988. Accumulation of a lipid-linked intermediate involved in enterobacterial common antigen synthesis in Salmonella Typhimurium mutants lacking dTDP-glucose pyrophosphorylase. J Bacteriol 170:4008–4014. doi: 10.1128/JB.170.9.4008-4014.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ramos-Morales F, Prieto AI, Beuzón CR, Holden DW, Casadesús J. 2003. Role for Salmonella enterica enterobacterial common antigen in bile resistance and virulence. J Bacteriol 185:5328–5332. doi: 10.1128/JB.185.17.5328-5332.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Danese PN, Oliver GR, Barr K, Bowman GD, Rick PD, Silhavy TJ. 1998. Accumulation of the enterobacterial common antigen lipid II biosynthetic intermediate stimulates degP transcription in Escherichia coli. J Bacteriol 180:5875–5884. doi: 10.1128/JB.180.22.5875-5884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Castelli ME, Fedrigo GV, Clementín AL, Ielmini MV, Feldman MF, Véscovi EG. 2008. Enterobacterial common antigen integrity is a checkpoint for flagellar biogenesis in Serratia marcescens. J Bacteriol 190:213–220. doi: 10.1128/JB.01348-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Castelli ME, Véscovi EG. 2011. The Rcs signal transduction pathway is triggered by enterobacterial common antigen structure alterations in Serratia marcescens. J Bacteriol 193:63–74. doi: 10.1128/JB.00839-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jorgenson MA, Kannan S, Laubacher ME, Young KD. 2016. Dead‐end intermediates in the enterobacterial common antigen pathway induce morphological defects in Escherichia coli by competing for undecaprenyl phosphate. Mol Microbiol 100:1–14. doi: 10.1111/mmi.13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jann K, Kanegasaki S, Goldemann G, Mäkelä PH. 1979. On the effect of rfe mutation on the biosynthesis of the O8 and O9 antigens of E. coli. Biochem Biophys Res Commun 86:1185–1191. doi: 10.1016/0006-291X(79)90242-0. [DOI] [PubMed] [Google Scholar]
- 161.Mäkelä PH, Mayer H, Whang H, Neter E. 1974. Participation of lipopolysaccharide genes in the determination of the enterobacterial common antigen: analysis of R mutants of Salmonella minnesota. J Bacteriol 119:760–764. doi: 10.1128/JB.119.3.760-764.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mäkelä PH, Jahkola M, Lüderitz O. 1970. A new gene cluster rfe concerned with the biosynthesis of Salmonella lipopolysaccharide. J Gen Microbiol 60:91–106. doi: 10.1099/00221287-60-1-91. [DOI] [PubMed] [Google Scholar]
- 163.Stocker BAD, Mäkelä P. 1978. Genetics of the (Gram-negative) bacterial surface. Proc R Soc Lond B Biol Sci 202:5–30. doi: 10.1098/rspb.1978.0055. [DOI] [PubMed] [Google Scholar]
- 164.Rick PD, Hubbard GL, Barr K. 1994. Role of the rfe gene in the synthesis of the O8 antigen in Escherichia coli K-12. J Bacteriol 176:2877–2884. doi: 10.1128/JB.176.10.2877-2884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hong Y, Liu MA, Reeves PR. 2017. Progress in our understanding of Wzx flippase for translocation of bacterial membrane lipid-linked oligosaccharide. J Bacteriol 200:e00154-17. doi: 10.1128/JB.00154-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu MA, Morris P, Reeves PR. 2019. Wzx flippases exhibiting complex O‐unit preferences require a new model for Wzx–substrate interactions. Microbiology Open 8:e00655. doi: 10.1002/mbo3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li GW, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Feldman MF, Marolda CL, Monteiro MA, Perry MB, Parodi AJ, Valvano MA. 1999. The activity of a putative polyisoprenol-linked sugar translocase (Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. J Biol Chem 274:35129–35138. doi: 10.1074/jbc.274.49.35129. [DOI] [PubMed] [Google Scholar]
- 169.Marolda CL, Vicarioli J, Valvano MA. 2004. Wzx proteins involved in biosynthesis of O antigen function in association with the first sugar of the O-specific lipopolysaccharide subunit. Microbiology 150:4095–4105. doi: 10.1099/mic.0.27456-0. [DOI] [PubMed] [Google Scholar]
- 170.Hong Y, Reeves PR. 2014. Diversity of O-antigen repeat unit structures can account for the substantial sequence variation of Wzx translocases. J Bacteriol 196:1713–1722. doi: 10.1128/JB.01323-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liu MA, Stent TL, Hong Y, Reeves PR. 2015. Inefficient translocation of a truncated O unit by a Salmonella Wzx affects both O-antigen production and cell growth. FEMS Microbiol Lett 362:fnv053. doi: 10.1093/femsle/fnv053. [DOI] [PubMed] [Google Scholar]
- 172.Hong Y, Cunneen MM, Reeves PR. 2012. The Wzx translocases for Salmonella enterica O‐antigen processing have unexpected serotype specificity. Mol Microbiol 84:620–630. doi: 10.1111/j.1365-2958.2012.08048.x. [DOI] [PubMed] [Google Scholar]
- 173.Klein KA, Fukuto HS, Pelletier M, Romanov G, Grabenstein JP, Palmer LE, Ernst R, Bliska JB. 2012. A transposon site hybridization screen identifies galU and wecBC as important for survival of Yersinia pestis in murine macrophages. J Bacteriol 194:653–662. doi: 10.1128/JB.06237-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Phan MD, Peters KM, Sarkar S, Lukowski SW, Allsopp LP, Moriel DG, Achard ME, Totsika M, Marshall VM, Upton M, Beatson SA, Schembri MA. 2013. The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet 9:e1003834. doi: 10.1371/journal.pgen.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Suezawa C, Yasuda M, Negayama K, Kameyama T, Hirauchi M, Nakai T, Okuda J. 2016. Identification of genes associated with the penetration activity of the human type of Edwardsiella tarda EdwGII through human colon epithelial cell monolayers. Microb Pathog 95:148–156. doi: 10.1016/j.micpath.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 176.Bohm K, Porwollik S, Chu W, Dover JA, Gilcrease EB, Casjens SR, McClelland M, Parent KN. 2018. Genes affecting progression of bacteriophage P22 infection in Salmonella identified by transposon and single gene deletion screens. Mol Microbiol 108:288–305. doi: 10.1111/mmi.13936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kuhn H, Meier U, Mayer H. 1984. ECA, dasgemeinsame antigender enterobacteriaceae stiefkindder mikrobiologie. Forum Mikrobiol 7:274–285. [Google Scholar]
- 178.Barua S, Yamashino T, Hasegawa T, Yokoyama K, Torii K, Ohta M. 2002. Involvement of surface polysaccharides in the organic acid resistance of Shiga Toxin‐producing Escherichia coli O157: H7. Mol Microbiol 43:629–640. doi: 10.1046/j.1365-2958.2002.02768.x. [DOI] [PubMed] [Google Scholar]
- 179.Tamae C, Liu A, Kim K, Sitz D, Hong J, Becket E, Bui A, Solaimani P, Tran KP, Yang H, Miller JH. 2008. Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J Bacteriol 190:5981–5988. doi: 10.1128/JB.01982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Girgis HS, Hottes AK, Tavazoie S. 2009. Genetic architecture of intrinsic antibiotic susceptibility. PLoS One 4:e5629. doi: 10.1371/journal.pone.0005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, Shales M, Lovett S, Winkler ME, Krogan NJ, Typas A, Gross CA. 2011. Phenotypic landscape of a bacterial cell. Cell 144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gilbreath JJ, Dodds JC, Rick PD, Soloski MJ, Merrell DS, Metcalf ES. 2012. Enterobacterial common antigen mutants of Salmonella enterica serovar Typhimurium establish a persistent infection and provide protection against subsequent lethal challenge. Infect Immun 80:441–450. doi: 10.1128/IAI.05559-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.McMahon KJ, Castelli ME, Vescovi EG, Feldman MF. 2012. Biogenesis of outer membrane vesicles in Serratia marcescens is thermoregulated and can be induced by activation of the Rcs phosphorelay system. J Bacteriol 194:3241–3249. doi: 10.1128/JB.00016-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kulp AJ, Sun B, Ai T, Manning AJ, Orench-Rivera N, Schmid AK, Kuehn MJ. 2015. Genome-wide assessment of outer membrane vesicle production in Escherichia coli. PLoS One 10:e0139200. doi: 10.1371/journal.pone.0139200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Saxén H, Hovi M. 1985. The effect of antibodies to the enterobacterial common antigen (ECA) on experimental mouse salmonellosis. FEMS Microbiol Lett 27:307–312. doi: 10.1111/j.1574-6968.1985.tb00687.x. [DOI] [Google Scholar]
- 186.Fattom AI, Sarwar J, Basham L, Ennifar S, Naso R. 1998. Antigenic determinants of Staphylococcus aureus type 5 and type 8 capsular polysaccharide vaccines. Infect Immun 66:4588–4592. doi: 10.1128/IAI.66.10.4588-4592.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Jennings HJ, Bhauacharjee AK, Bundle DR, Paul Kenny C, Martin A, Smith IC. 1977. Structures of the capsular polysaccharides of Neisseria meningitidis as determined by 13C-nuclear magnetic resonance spectroscopy. J Infect Dis 136:S78–S83. doi: 10.1093/infdis/136.Supplement.S78. [DOI] [PubMed] [Google Scholar]
- 188.Kooistra O, Lüneberg E, Lindner B, Knirel YA, Frosch M, Zähringer U. 2001. Complex O-acetylation in Legionella pneumophila serogroup 1 lipopolysaccharide. Evidence for two genes involved in 8-O-acetylation of legionaminic acid. Biochemistry 40:7630–7640. doi: 10.1021/bi002946r. [DOI] [PubMed] [Google Scholar]
- 189.Lemercinier X, Jones C. 1996. Full 1H NMR assignment and detailed. Carbohydr Res 296:83–96. doi: 10.1016/S0008-6215(96)00253-4. [DOI] [PubMed] [Google Scholar]
- 190.Lewis AL, Nizet V, Varki A. 2004. Discovery and characterization of sialic acid O-acetylation in group B Streptococcus. Proc Natl Acad Sci U S A 101:11123–11128. doi: 10.1073/pnas.0403010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.McNeely TB, Staub JM, Rusk CM, Blum MJ, Donnelly JJ. 1998. Antibody responses to capsular polysaccharide backbone and O-acetate side groups of Streptococcus pneumoniae type 9V in humans and rhesus macaques. Infect Immun 66:3705–3710. doi: 10.1128/IAI.66.8.3705-3710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Paavonen J, Valtonen V, Kasper D, Malkamäki M, Mäkelä PH. 1981. Serological evidence for the role of Bacteroides fragilis and Enterobacteriaceae in the pathogenesis of acute pelvic inflammatory disease. Lancet 317:293–295. doi: 10.1016/S0140-6736(81)91909-7. [DOI] [PubMed] [Google Scholar]
- 193.Rabsztyn K, Kasperkiewicz K, Duda K, Li C-M, Łukasik M, Radziejewska-Lebrecht J, Skurnik M. 2011. Characterization of anti-ECA antibodies in rabbit antiserum against rough Yersinia enterocolitica O:3. Biochemistry (Mosc) 76:832–839. doi: 10.1134/S0006297911070145. [DOI] [PubMed] [Google Scholar]
- 194.Adeolu M, Alnajar S, Naushad S, Gupta RS. 2016. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol 66:5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 195.Gozdziewicz TK, Lugowski C, Lukasiewicz J. 2014. First evidence for a covalent linkage between enterobacterial common antigen and lipopolysaccharide in Shigella sonnei phase II ECALPS. J Biol Chem 289:2745–2754. doi: 10.1074/jbc.M113.512749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Johns MA, Whiteside RE, Baker EE, McCabe WR. 1973. Common enterobacterial antigen: I. Isolation and purification from Salmonella typhosa 0:901. J Immunol 110:781–790. [PubMed] [Google Scholar]
- 197.Kuhn HM, Basu S, Mayer H. 1987. Comparison of enterobacterial common antigen from different species by serological techniques. Eur J Biochem 162:69–74. doi: 10.1111/j.1432-1033.1987.tb10543.x. [DOI] [PubMed] [Google Scholar]
- 198.Fregolino E, Ivanova R, Lanzetta R, Molinaro A, Parrilli M, Paunova-Krasteva T, Stoitsova SR, De Castro C. 2012. Occurrence and structure of cyclic enterobacterial common antigen in Escherichia coli O157:H−. Carbohydr Res 363:29–32. doi: 10.1016/j.carres.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 199.Marx A, Petcovici M. 1975. Immunochemical studies on purified common enterobacterial antigen (KUNIN). Zentralbl Bakteriol Orig A 233:486–494. [PubMed] [Google Scholar]
- 200.Hammarström S, Carlsson HE, Perlmann P, Svensson S. 1971. Immunochemistry of the common antigen of Enterobacteriaceae (Kunin) relation to lipopolysaccharide core structure. J Exp Med 134:565–576. doi: 10.1084/jem.134.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Peters H, Jürs M, Jann B, Jann K, Timmis KN, Bitter-Suermann D. 1985. Monoclonal antibodies to enterobacterial common antigen and to Escherichia coli lipopolysaccharide outer core: demonstration of an antigenic determinant shared by enterobacterial common antigen and E. coli K5 capsular polysaccharide. Infect Immun 50:459–466. doi: 10.1128/IAI.50.2.459-466.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]