Abstract
Studies over the past several decades have identified numerous epigenetic mechanisms associated with pathological states in psychiatric and neurological disease. Until recently, studies investigating chromatin-regulatory proteins, using overexpression or knockdown approaches, did not establish causal roles for epigenetic modifications at specific genes because these techniques typically affect hundreds or thousands of genomic loci. In this Review, we describe recent efforts in using locus-specific neuroepigenome editing in vivo to, for the first time, define causal relationships between a single chromatin modification at a specific gene in a defined cell population and downstream measures at the molecular, cellular, circuit and behavioural levels. We briefly introduce three epigenome-editing platforms: zinc-finger proteins, transcriptional activator-like effectors and clustered regularly interspaced short palindromic repeats (CRISPR). We then explore the development of in vivo neuroepigenome-editing tools and their applications to resolve epigenetic contributions to the pathophysiology of brain diseases. We also discuss technical considerations for in vivo neuroepigenome-editing experiments and ongoing innovations in the field, including new tools to investigate chromatin marks, manipulate chromatin topology and induce epigenetic modifications at multiple genes in the same cell. Lastly, we explore the potential clinical applications of in vivo neuroepigenome editing for treating brain pathology.
Studies of neuroepigenetics seek to define the mechanisms by which various environmental stimuli and other factors, such as ageing, induce lasting changes in neuronal or glial function through many types of chromatin modifications, which alter the expression of specific genes without affecting the base pair sequence of DNA. In the nucleus, DNA wraps around an octamer of histone proteins to form a nucleosome — the fundamental unit of chromatin. Chromatin exists along a spectrum from a densely packaged (heterochromatic) and transcriptionally silent state to a more open (euchromatic) state available for transcriptional machinery binding and the dynamic activation or suppression of gene expression in response to external stimuli. The diverse mechanisms that regulate chromatin state are reviewed extensively elsewhere, and include post-translational modifications to histones, DNA methylation and non-coding RNAs1–3.
The ability to precisely delineate the mechanisms by which these neuroepigenetic factors regulate cellular function in the brain in vivo is limited by the molecular techniques available to manipulate chromatin modifications. Historically, a histone- or DNA-modifying enzyme would be overexpressed or knocked down, but these manipulations — even if done inducibly in the adult brain in a cell type-specific manner — would affect that histone or DNA modification at hundreds or thousands of genomic loci, making it difficult to understand the effects of the targeted modification at an individual locus of interest and its downstream functional effects at the transcriptional, cellular, circuit and behavioural levels.
Genome engineering is a pioneering molecular technique to study a gene’s function by inducing precise modifications to the base pair sequence of DNA. Well-established gene-editing platforms include zinc-finger nucleases (ZFNs), transcriptional activator-like effector (TALE) nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR). ZFNs and TALENs recognize specific sequences of DNA via direct protein–DNA interactions to induce site-specific DNA cleavage. In CRISPR systems, Cas9 forms a complex with a CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA) — which can be fused together to produce a single guide RNA (gRNA) — that undergoes complementary base pairing with the targeted genomic locus to guide DNA cleavage. These tools are used extensively in basic research to study the genetic basis of cell function and disease, including in the brain4. CRISPR-based therapies are also entering the clinic for a range of disorders5.
More recently, numerous studies have demonstrated that each of these gene-editing approaches can be adapted to control transcription at a given gene, rather than altering the base pair sequence of DNA6–8. These tools were modified to remove functional nuclease domains and are instead fused to strong transcriptional activators or repressors, such as VP64 (a viral transcriptional activator) or a Krüppel-associated box (KRAB) domain (present in numerous repressive zinc-finger proteins (ZFPs))9. Guiding the transcriptional effector domain to the regulatory region of a targeted gene (such as the promoter or enhancer) provides a method to selectively modify gene expression. These tools continue to be improved to enable greater specificity, tunable gene expression and simultaneous targeting of more than one gene.
However, these gene-editing and transcription-regulation approaches provide limited insight into endogenous epigenetic mechanisms. For instance, VP64 and KRAB represent artificial means of activating or repressing a given gene of interest, and often produce changes in gene expression that are far greater in magnitude than those seen under physiological or pathological conditions in vivo. Similarly, chromatin immunoprecipitation followed by sequencing and related approaches that correlate the enrichment of a given chromatin modification with gene expression do not provide causal information. This is particularly important because dozens of proteins and epigenetic modifications work in concert to drive expression of a given gene, making it impossible to study the role of a single epigenetic mark in isolation. Therefore, these approaches fall short in defining causal relationships between a specific chromatin modification at a single gene and downstream experimental measures.
In this Review, we focus on the very recent development and applications of in vivo locus-specific neuroepigenome-editing tools to study endogenous epigenetic mechanisms in the brain. These tools make it possible to induce a single type of chromatin-regulatory event (such as histone acetylation or methylation, DNA methylation or binding of a transcription factor (TF)) at a single gene of interest in a specific cell population in a single brain region of awake, behaving animals.
These new approaches offer four main advances for the field. First, they dramatically increase the quality of proof that causally links an epigenetic mechanism to a functional end point, overcoming the limitations of traditional overexpression or knockout approaches stated above. Second, locus-specific neuroepigenome editing mimics endogenous mechanisms of epigenetic regulation at a given locus, enabling the study of causal roles of such naturally occurring mechanisms. Third, conventional overexpression, knockout and CRISPR-based gene regulation strategies in the brain often induce high-magnitude changes in gene expression that exceed physiologically relevant degrees of gene regulation10–13. By contrast, because locus-specific neuroepigenome-editing tools capture normal mechanisms of regulation, they have the potential to produce smaller changes in a gene’s expression levels in the brain, better reflecting what happens under physiological or pathological conditions10,14,15. To date, few studies offer direct comparisons between conventional approaches and neuroepigenome editing. Fourth, these tools enable the manipulation of suites of genes in a single cell type and thereby advance beyond the still predominant approach of studying the actions of individual genes, one at a time.
Zinc-finger proteins
Synthetic ZFPs were first introduced more than 30 years ago as a novel gene-editing tool16. ZFPs are composed of a series of zinc ion-regulated Cys2-His2 domains and bind to specific 18-bp sequences of DNA8,16. However, engineering the ZFP domains to target specific DNA sequences is time-consuming and technically challenging. This process includes bioinformatic analysis to predict binding interactions between ZFPs and the target DNA, followed by expensive and labour-intensive empirical validation.
Among the most commonly used ZFPs are ZFNs. For example, a ZFP is fused to the DNA-cleavage domain of FokI endonucleases and, on binding to its target DNA sequence, disrupts (that is, knocks out) the encoding gene17. In addition, ZFPs can be fused with a histone modifier, such as a histone methyltransferase (HMT) or histone acetyltransferase (HAT), or with a TF, which then induces epigenetic changes at the targeted gene18–20 (FIG. 1).
Fig. 1 |. In vivo neuroepigenome-editing tools.
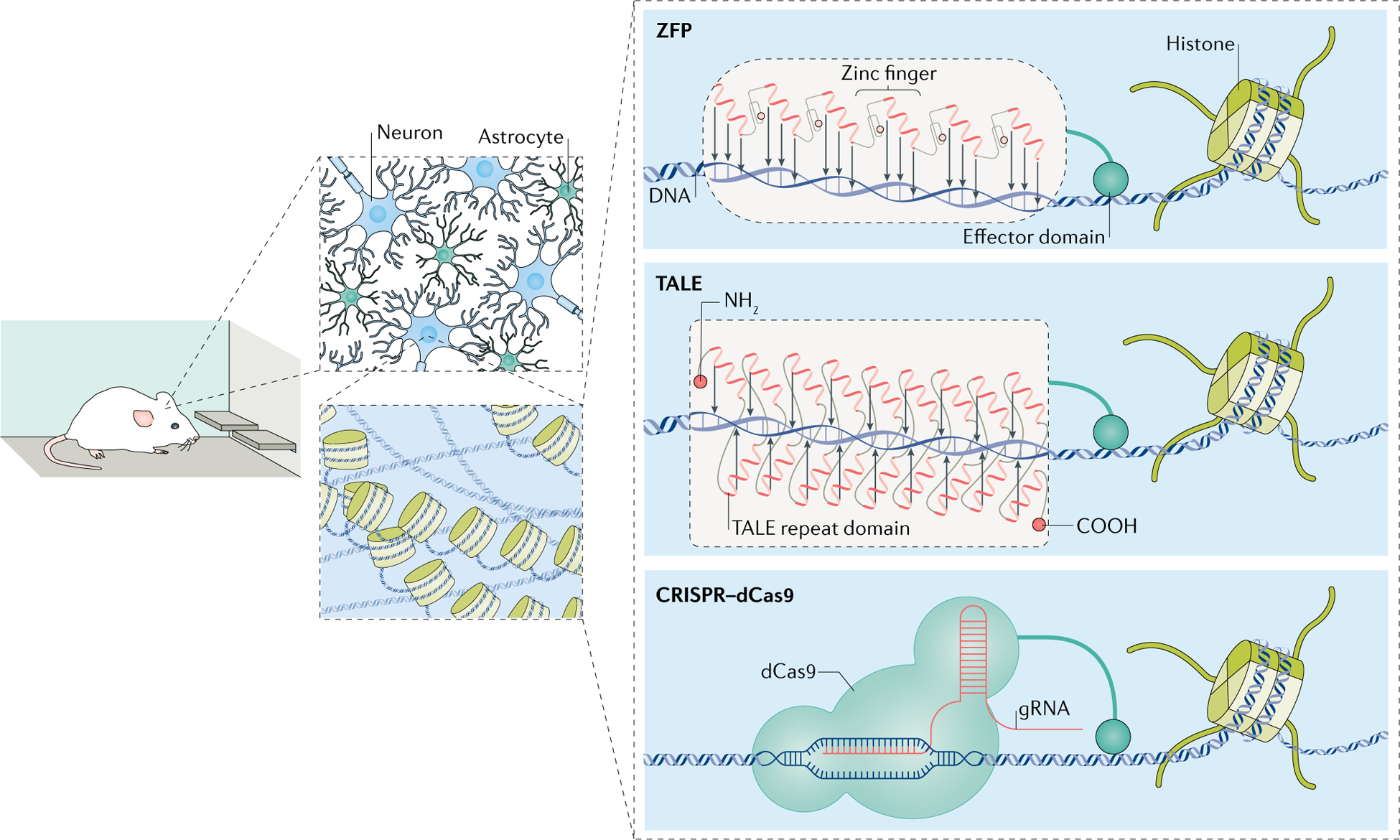
Zinc-finger proteins (ZFPs) and transcriptional activator-like effectors (TALEs) are DNA-binding proteins that can be engineered to target specific genomic loci. Fusing the DNA-binding domain to transcription effector proteins offers a tool to selectively regulate gene transcription. In the clustered regularly interspaced short palindromic repeats (CRISPR) system, a nuclease-dead CRISPR-associated protein 9 (dCas9) molecule fused to an effector domain forms a complex with a guide RNA (gRNA) that undergoes base pairing with the homologous DNA sequence. Pairing these various tools with transgene delivery methods offers a method to regulate the expression of specific genes in a defined cell population in the brains of awake, behaving rodents. These tools can be applied to a wide range of behavioural paradigms, electrophysiology experiments and biochemical analyses to understand the functional roles of a given gene in the brain.
In combination with viral vectors, ZFP–HAT complexes, ZFP–HMT complexes, and ZFP–TF complexes can be delivered into a discrete brain region to study neuroepigenetic mechanisms in vivo. In one study, a ZFP was fused to p65, an activation domain in the TF nuclear factor-κB (NF-κB) to assess the neuroprotective effects of the activation of the gene encoding glial cell line-derived neurotrophic factor (GDNF) in the 6-hydroxydopamine rat model of Parkinson disease21. Virus-mediated delivery of ZFP–p65 into the striatum of adult rats produced an approximately 60% increase in striatal GDNF levels and attenuated neurodegeneration of dopaminergic nerve terminals in the injection region. In another study, ZFPs were fused either to VP64 or to an HMT called ‘SUVDEL76’ and were directed to the promoter of the Dlg4 gene (which encodes postsynaptic density protein 95 (PSD95)) in the rat hippocampus with use of a herpes simplex virus (HSV) vector. The researchers demonstrated that bidirectional regulation of Dlg4 expression with these tools altered the maturation of hippocampal synapses and spines in vivo22. Moreover, increasing Dlg4 expression in APP/PS1 mice (a model of Alzheimer disease in which the genes encoding amyloid precursor protein and presenilin 1 are mutated), using adeno-associated virus (AAV) encoding ZFP–VP64, rescued memory deficits in these mice22.
This technique was applied to elucidate how drug- or stress-induced activation of Fosb (which encodes a FOS-family TF) in the nucleus accumbens (NAc), a portion of the ventral striatum that has a key role in reward and motivation, is controlled by histone post-translational modifications23. Previously, induction of Fosb in the NAc was shown to increase the reinforcing effects of drugs of abuse and, depending on the cell type involved, to promote either susceptibility or resilience to chronic social stress24–26. Regulation of Fosb in the NAc is associated with changes in histone acetylation and histone H3 lysine 9 (H3K9) methylation27,28 at this gene, but previous studies of the role of these histone modifications in regulating Fosb expression were limited to conventional overexpression or knockout approaches26,29. To obtain more direct evidence of the role played by these chromatin mechanisms in controlling Fosb expression and its downstream functional consequences, ZFPs targeting Fosb were fused either to the p65 domain of NF-κB, which promotes acetylation at nearby histones by recruiting a HAT, or to G9a, a repressive HMT that catalyses histone H3K9 dimethylation. Selective expression of ZFP–p65 in NAc neurons using HSVs induced histone acetylation, but not other histone modifications, at the Fosb locus, and this epigenetic modification increases Fosb expression23. Conversely, ZFP–G9a induced H3K9 dimethylation selectively at the Fosb locus and repressed Fosb expression in the NAc. These manipulations bidirectionally controlled cocaine- or stress-induced behavioural outcomes, thus linking an individual histone modification at a single locus to downstream transcriptional and behavioural outcomes23,30. A later study showed that suppressing Fosb expression in the NAc using ZFP–G9a also attenuates aggression30.
An important insight from these experiments is that they provide direct evidence that histone acetylation or methylation is sufficient to control the expression of a gene and is not simply a downstream consequence of transcriptional regulation mediated by TFs. ZFP–G9a-mediated deposition of dimethylated H3K9 at Fosb suppressed cocaine-evoked induction of Fosb in the NAc by preventing the phosphorylation of cAMP response element-binding protein (CREB), which was already bound to the Fosb promoter23.
A follow-up study demonstrated the ability to induce such histone changes in a cell type-specific manner in the NAc using Cre-dependent viral vectors in transgenic mice expressing Cre recombinase under the control of the genes encoding dopamine D1 receptor (D1R; Drd1) or D2R (Drd2). Here, inducing Fosb expression in D1R-expressing medium spiny neurons (MSNs) using ZFP–p65 promoted stress resilience, whereas Fosb suppression in D1R-expressing MSNs using ZFP–G9a increased stress susceptibility. In D2R-expressing MSNs, the opposite phenotypes were observed31. This study establishes the principle of targeting neuroepigenome editing in vivo to a given neuronal cell type in a mouse model of neuropsychiatric disease.
The general applicability of this approach was demonstrated by the use of different ZFP–p65 and ZFP–G9a constructs to bidirectionally control the expression of another gene, Cdk5 (which encodes cyclin-dependent kinase 5), in the NAc and its downstream control of drug- and stress-induced behavioural outcomes32. CDK5 is a member of the serine/threonine cyclin-dependent kinase (CDK) family and is involved in cocaine- and stress-related behaviour as well as fear-memory formation, regulating both the expression and the magnitude of fear-related memory and depressive-like phenotypes through its actions in the forebrain33, hippocampus34 and striatum35. HSV-mediated delivery of ZFP–p65 targeting Cdk5 in the NAc increased cocaine-induced locomotor behaviour and resilience to social stress in male mice32. In a later study, ZFP–p65 was applied to target Cdk5 in the female mouse hippocampus and was shown to attenuate fear-memory retrieval36.
These ZFP studies highlight two main challenges. First, synthesizing a ZFP that targets a single gene of interest in vivo is extremely labour-intensive and time-consuming. This is due to the lack of a convenient bioinformatics method to design functional ZFPs, the technical expertise required to perform protein engineering and the observation that the effectiveness of a ZFP in cultured Neuro2A cells (a line of mouse neuro-blastoma cells) is not predictive of its in vivo activity in the brain14,31. Primary neuronal cultures may improve the predictive validity of ZFP screens in vitro, but this has not yet been tested. The ZFP approach therefore requires extensive in vivo screening of numerous constructs. For these reasons, ZFPs are no longer the preferred neuroepigenome-editing tool. Another challenge, pertinent to ZFPs and all other neuroepigenome-editing tools, relates to the technical difficulty in confirming their locus-specificity in vivo (BOX 1).
Box 1 |. Experimental validation of in vivo locus-specific neuroepigenome editing.
The effective use of locus-specific neuroepigenome editing requires empirical validation that the targeted locus is affected with high selectivity across the entire genome. this is essential because zinc-finger proteins (ZFPs), transcriptional activator-like effectors (TALEs) and nuclease-dead clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (dCas9) can bind to off-target sites in the genome, and the binding of these proteins, even without a fused functional domain, can be sufficient to affect transcription; for example, via steric hindrance of RNA polymerase II (REF.102). Moreover, studies using native Cas9 for DNA base pair editing show that the guide RNA (gRNA) sequence tolerates one or two mismatched base pairs and exhibits widespread off-target effects96. Despite this considerable potential for nonspecific effects, there are no well-established guidelines for validating the single-locus specificity of neuroepigenome-editing tools, especially in vivo. Here we discuss the advantages and potential pitfalls of current validation techniques.
A commonly used approach for validating the targeting specificity of epigenome-editing tools is RNA sequencing9. this technique allows researchers to identify differentially expressed genes on the basis of experimenter-set thresholds for fold change and P value. the key advantage of RNA sequencing is that it provides an unbiased quantification of all RNA transcripts. Off-target activity is detected on the basis of differential expression of genes besides the targeted locus — particularly those with a few mismatches compared with the gRNA used. However, the differential expression of off-target genes may be due not to promiscuous binding of a ZFP, TALE or dCas9, but rather may be to the physiological consequences of altered expression levels of the targeted gene. this is particularly the case when the targeted gene is a transcription factor or another transcription-regulatory protein.
Chromatin immunoprecipitation (ChiP) followed by sequencing is a method to quantify genome-wide binding of a ZFP, TALE or dCas9 and better establish the single-locus specificity of epigenome-editing approaches. However, validating epigenome-editing approaches using ChiP–sequencing is extremely challenging: if epigenome-editing tools are perfectly selective, there would be only two binding sites per cell, compared with the many thousands of sites normally characterized by ChiP–sequencing for chromatin modifications or transcription factor binding. this would be even more challenging when applied to the brain because of the limited amount of tissue and the requirement for especially selective ChiP-grade antibodies. Modified ChiP–sequencing procedures, such as CUT&RUN (cleavage under targets and release using nuclease), provide an alternative method that may reduce the amount of input tissue required105. Nevertheless, to date, it has not been possible to validate any epigenome-editing approach by use of ChiP–sequencing on brain tissue. instead, researchers rely on demonstrating induction or depletion of the targeted histone or DNA modification at the target locus and not at other loci most homologous in DNA sequence8,17,38.
Studies of native Cas9 identify several methods that may increase the single-locus specificity of neuroepigenome editing. One study showed that shorter gRNAs (18–19 base pairs) exhibit fewer off-target effects, while preserving efficient binding at the target gene106. Cas9 specificity can also be increased by altering the secondary structure of the gRNA by adding a hairpin loop to its 5′ end107. the hairpin structure may destabilize Cas9 binding at off-target loci, where energetic favourability of binding is lower owing to mismatches between the gRNA and the DNA strand. in addition, analysis of the crystal structure of CRISPR–Cas9 reveals a positively charged groove that probably stabilizes interactions with DNA97. By inducing point mutations to neutralize the positively charged amino acids in the groove, only highly complementary RNA–DNA interactions would be able to stabilize Cas9 binding. Last, different Cas9 orthologues have distinct protospacer-adjacent motif (PAM) sequences, which may be less tolerant of mismatched base pairs98. Overall, despite efforts to enhance Cas9 specificity, it remains essential to consider off-target interactions in the experimental design.
Transcriptional activator-like effectors
TALEs are DNA-binding proteins derived from pathogenic bacteria (Xanthomonas) that regulate the transcription of specific target genes. The central TALE domains comprise a series of highly conserved tandem repeats that are approximately 34 amino acids long37. These repeat sequences differ from each other mainly at amino acid positions 12 and 13, referred to as the ‘repeat variable diresidues’, which dictate the sequence specificity of the TALE. These repeat variable diresidues can be engineered to target specific genomic loci, providing an additional method to achieve locus-specific regulation of gene expression (FIG. 1).
Compared with ZFPs, TALEs are more easily engineered and highly selective and can target multiple genes in parallel38. Still, TALEs have a much more limited track record in DNA-targeting applications, and few in vivo studies have used TALEs as neuroepigenome-editing tools6,39. In one, light-inducible transcriptional effectors (LITEs) were developed to optogenetically regulate the expression of metabotropic glutamate receptor 2 (mGluR2) in the brain. In the LITE system, light-sensitive cryptochrome 2 (CRY2) from Arabidopsis thaliana is tethered to the carboxy terminus of the TALE, and calcium and integrin-binding protein 1 (CIB1), the interacting partner of CRY2, is tethered to VP64. Stimulation with blue light (with a wavelength of approximately 466 nm) induces a conformational change in CRY2 and facilitates heterodimerization of CRY2 and CIB1. This study demonstrated that Virus-mediated delivery of LITEs into the infralimbic cortex achieves light-inducible mGluR2 activation.
Despite the numerous advantages of TALEs over ZFPs, TALEs are scarcely used for gene editing or for regulating gene expression owing to recent advances in CRISPR-based techniques.
CRISPR
CRISPR gene editing.
CRISPR is an important component of the adaptive immune system in bacteria and archaea40. CRISPR loci comprise a set of CRISPR-associated (Cas) genes and the CRISPR array, which contains a series of unique spacers separated by repeat sequences. There are three main CRISPR systems (types I–III), which differ in their composition of Cas genes and mechanisms of adaptive immunity41. In the type II CRISPR system, Cas endonucleases recognize short fragments of bacteriophage DNA (protospacers) downstream of the protospacer-adjacent motif (PAM) and integrate the foreign sequence into the CRISPR array as spacers. A tracrRNA initiates RNA processing of the CRISPR array to release the spacer sequences as DNA-targeting crRNAs. The tracrRNA and crRNA form a complex with the Cas9 endonuclease, producing an RNA-guided endonuclease complex that cleaves the foreign nucleic acid sequence.
Early studies showing that CRISPR is a highly specific, RNA-guided, DNA-cleaving complex led to the hypothesis that it could be repurposed as a genome-editing tool. In 2013, two studies published in parallel provided the first evidence of genome engineering in mammalian cells using the type II CRISPR system42,43. Soon afterwards, gene-editing technology was paired with plasmid-delivery methods to study the functional consequences of altering the base pair sequence of DNA in vivo, including in the brain44.
Now, the applications of CRISPR go far beyond inducing double-strand breaks in DNA. The CRISPR toolbox rapidly expanded to include single-strand nicking, single base pair modification, fluorescent tagging of genomic loci, regulation of gene expression and light-inducible genetic manipulations. Here we focus on the recent development of CRISPR-based tools to regulate gene expression and their applications in neuroscience research (FIG. 1).
Regulating gene expression.
Following the emergence of CRISPR-based gene editing, work from numerous laboratories demonstrated the ability to either induce or repress the expression of a given gene by fusing nuclease-dead Cas9 (dCas9) with VP64 or with a KRAB protein9. In the literature, these techniques are referred to as CRiSPR activation (CRISPRa) and CRiSPR interference (CRISPRi), respectively.
The versatility of the CRISPRa/CRISPRi platform has expanded dramatically over the last several years (FIG. 2). For example, by combining multiple gRNAs into a single vector (multiplexing), several studies demonstrated that CRISPRa/CRISPRi can be used to regulate the expression of multiple genes simultaneously9,45. This can be achieved by expressing dCas9–VP64 or dCas9–KRAB with multiple different gRNAs. By contrast, multiplexing with ZFPs or TALEs is far more challenging, and would require extensive optimization of single-locus specificity for each gene as well as the expression of numerous ZFPs and TALEs simultaneously.
Fig. 2 |. Overview of CRISPRa/CRISPRi techniques.
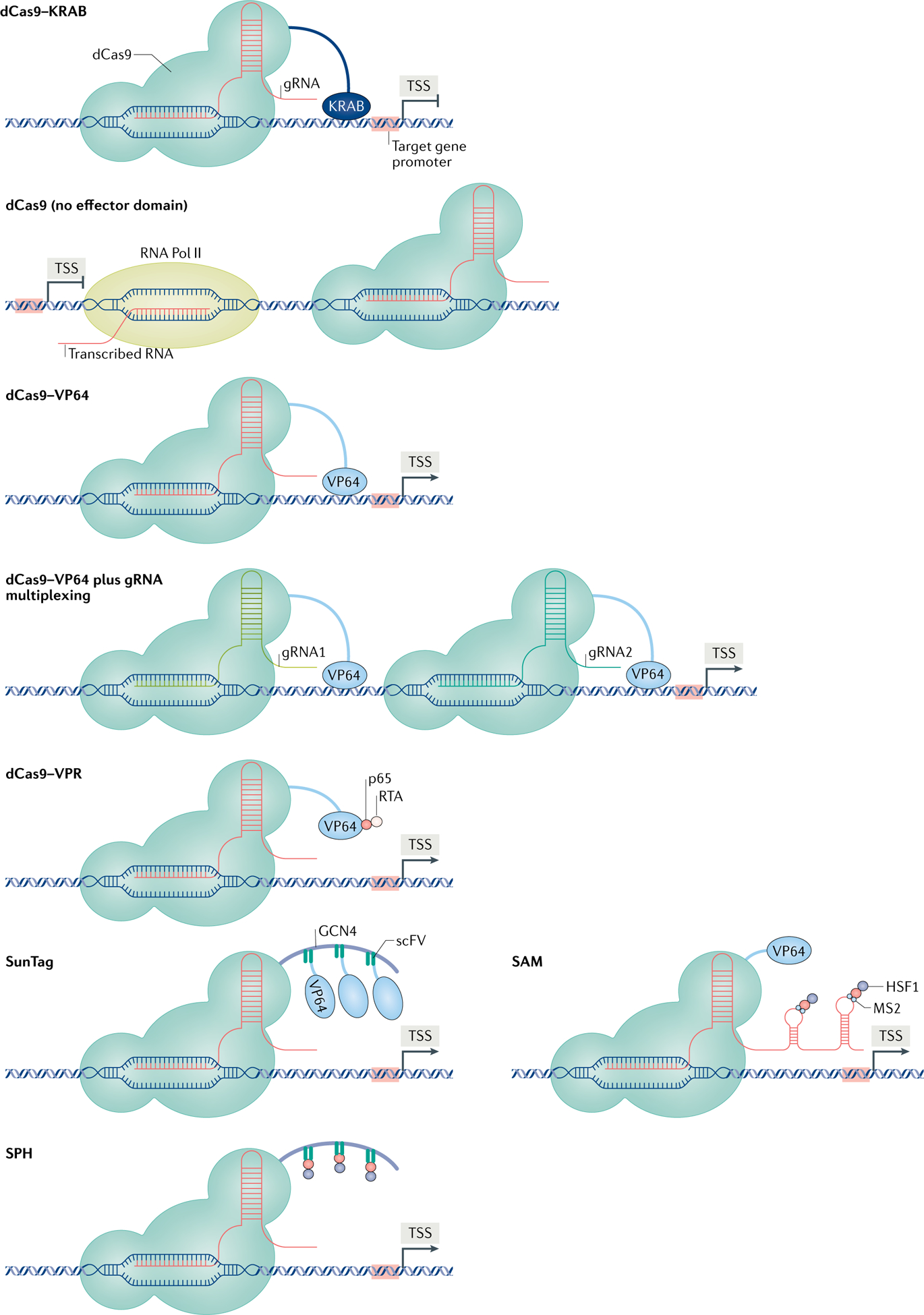
Clustered regularly interspaced short palindromic repeats (CRISPR) activation (CRISPRa)/CRISPR interference (CRISPRi) provides a powerful set of tools to study gene function through precise and modulatory regulation of gene expression. CRISPRi techniques include empty nuclease-dead CRISPR-associated protein 9 (dCas9) (that is, with no effector domain), which is thought to attenuate gene expression via steric hindrance of RNA polymerase II (Pol II) binding to DNA102, and dCas9 tethered to the Krüppel-associated box (KRAB) domain, a potent transcriptional repressor. The CRISPRa techniques were developed using a fusion between dCas9 and VP64, a potent viral transcriptional activator. This toolbox continues to expand to include new methods to increase effector recruitment to a single gene and increase the magnitude of gene induction. Combining dCas9–VP64 with multiplexed guide RNAs (gRNAs) directed towards several locations along the same promoter synergistically activates transcription45. In addition, dCas9 fused to multiple transcriptional effectors, including VP64, p65 and RTA (dCas9–VPR), substantially increases the magnitude of gene induction12,13. Synergistic activation mediator (SAM) combines dCas9–VP64 with a gRNA scaffold that selectively recruits an RNA-binding protein tethered to the p65 and HSF1 activation domains. Last, SunTag and SunTag–p65–HSF1 (SPH) use a dCas9-bound protein scaffold that acts as a binding site for single-chain variable fragment (scFv)-fused transcriptional activators. The effector domains for SunTag and SPH are VP64 and p65–HSF1, respectively. The CRISPRa/CRISPRi methods are illustrated on a spectrum showing their estimated repression and activation of gene expression. TSS, transcription start site.
The CRISPRa/CRISPRi system can also be adapted to fine-tune the level of gene induction or repression by recruiting more than one transcription effector to a single promoter. In one study, researchers increased effector recruitment to the target gene in vitro by directing multiple gRNAs towards different locations along the same promoter45, leading to synergistic activation of gene transcription. In another study, researchers amplified effector recruitment in vitro using a novel CRISPR system called ‘synergistic activation mediator’ (SAM)46. In this method, dCas9–VP64 is combined with a modified gRNA that recruits the RNA-binding protein MS2 tethered to p65 and heat shock protein 1 (HSP1), another activation domain. Together, these potent activation domains induce gene expression far beyond that achieved with the traditional dCas9–VP64 fusion. This study showed further that SAM is compatible with gRNA multiplexing and that targeting multiple genes induces a similar magnitude of gene induction as does targeting a single gene.
In addition, SunTag augments effector recruitment by using single-chain variable fragment (scFv)-fused effectors that selectively bind to a protein scaffold tethered to dCas9 (REF.47). The SunTag method may recruit up to 24 effectors to a single gene and achieves far greater gene induction than does dCas9–VP64. Elements from the SAM and SunTag techniques have been combined to produce the most potent transcriptional activator to date, termed ‘SunTag–p65–HSF1’ (SPH), in which a p65–HSF1 fusion is recruited to the protein scaffold12. This approach may also reactivate highly heterochromatic regions of DNA, thereby offering a tool to study the consequences of activating genes that are normally quiescent. Together, these techniques allow researchers to set the dial for the magnitude of gene induction. In future studies, the various techniques to amplify effector recruitment could be paired with transcription repressors to achieve more robust knockdowns.
CRISPRa/CRISPRi techniques have been applied to study gene function in the brains of awake, behaving rodents. In one study, researchers used CRISPRa/CRISPRi to bidirectionally regulate expression of the Nr4a1 gene (which encodes the TF nuclear receptor 77) and study the downstream effects on behavioural responses to cocaine48. CRISPR-mediated activation of Nr4a1 in the NAc reduced an animal’s sensitivity to the rewarding properties of cocaine as well as drug-seeking behaviour. Small molecule-mediated activation of Nr4a1 recapitulated this effect on cocaine reward, suggesting that Nr4a1 may be a promising therapeutic target for cocaine-use disorder. Another study packaged dCas9–KRAB into a lentivirus and demonstrated a robust and highly specific knockdown of synaptotagmin 1 (SYT1), a protein involved in evoked neurotransmitter release, in glutamatergic and GABAergic neurons of the hippocampal dentate gyrus49. Knockdown of SYT1 in this brain region altered the physiological properties of the neurons and performance in learning- and memory-associated tasks. The study demonstrated further that pairing dCas9–KRAB with multiplexed gRNAs enabled simultaneous downregulation of several genes in vivo. Similarly, the simultaneous induction of more than one gene in vivo was achieved with SPH and dCas9 fused to the activation domains VP64, p65 and RTA, a replication and transcription activator protein encoded by Orf50 (dCas9–VPR)12,13. In summary, CRISPRa/CRISPRi is a highly specific and tunable method to study the effects of gene induction or repression within specific cell populations in the brain.
CRISPR-based epigenome editing.
CRISPR emerged as a leading platform for epigenome editing with the advent of novel fusion proteins composed of dCas9 tethered to various chromatin-modifying proteins (FIG. 3). With these tools, it is possible to mimic endogenous epigenetic mechanisms by recruiting an epigenetic effector to a single genomic locus. The ease of swapping epigenetic effector proteins tethered to dCas9 led to the rapid development of a growing library of CRISPR-based epigenome-editing tools. To date, the epigenetic effectors tethered to dCas9 include HATs, histone deacetylases (HDACs), HMTs, histone demethylases, DNA methyltransferases (DNMTs), ten–eleven translocation methylcytosine dioxygenase 1 (TET1), DNA glycosylase protein ROS1 (REFS50,51) and TFs10,52–60.
Fig. 3 |. CRISPR-based neuroepigenome-editing tools.
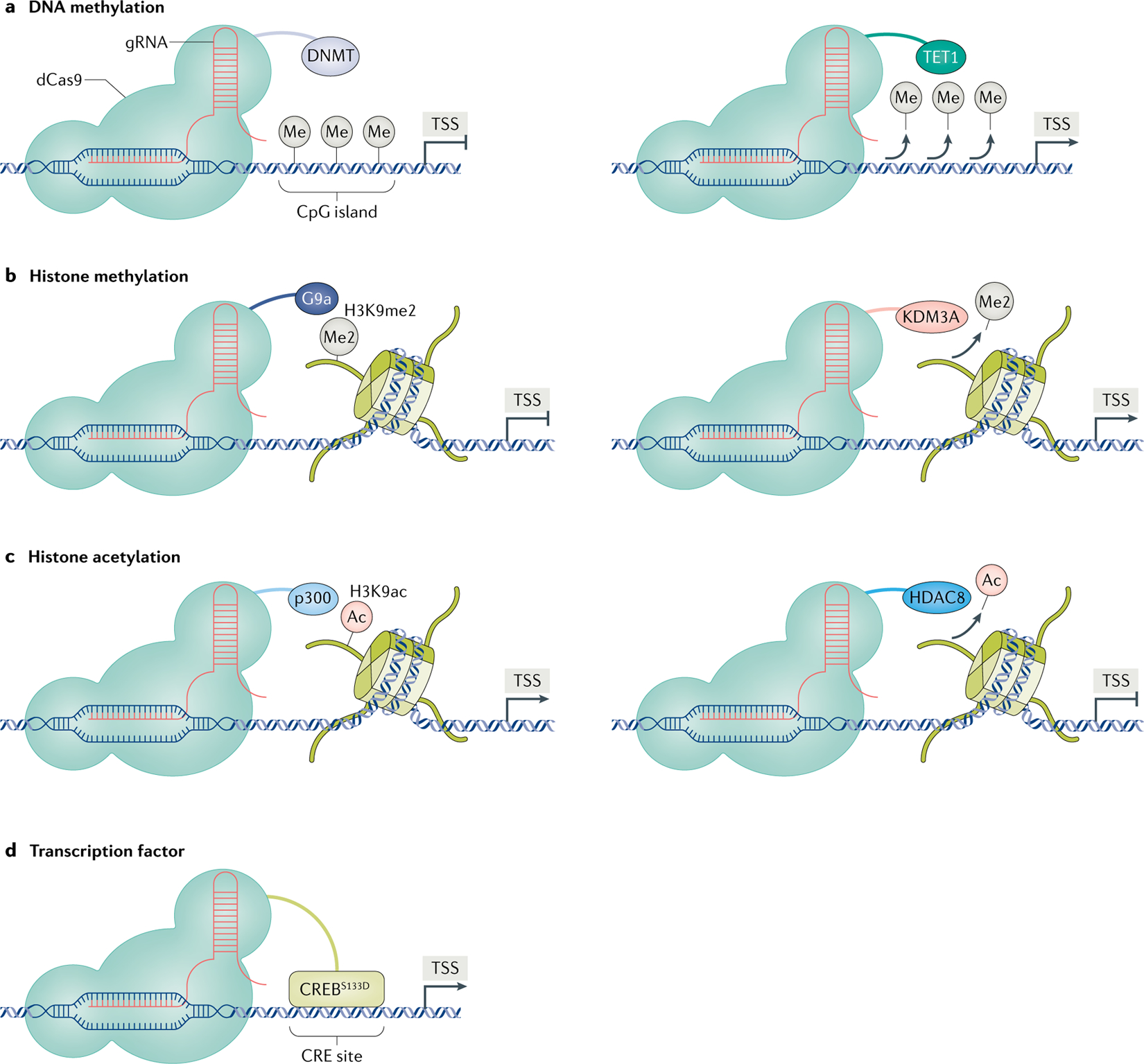
The neuroepigeneome-editing toolbox continues to expand to incorporate a wider range of chromatin-modifying proteins. a | Clustered regularly interspaced short palindromic repeats (CRISPR)–nuclease-dead CRISPR-associated protein 9 (dCas9) fused to a DNA methyltransferase (DNMT) or ten–eleven translocation methylcytosine dioxygenase 1 (TET1) provides a method to bidirectionally regulate DNA methylation at a single genomic locus53. CRISPR-based neuroepigenome editing also includes methods to add or remove various post-translational histone modifications, such as histone methylation10 (part b) and histone acetylation61 (part c). d | More recently, work from our laboratory used CRISPR tools to selectively guide the phosphomimetic (active) form of the transcription factor cAMP response element (CRE)-binding protein (CREB) (CREBS133D) to a CRE site at a single target gene10. Ac, acetyl group; gRNA, guide RNA; HDAC8, histone deacetylase 8; H3K9ac, acetylated histone H3 lysine 9; H3K9me2, dimethylated histone H3 lysine 9; Me, methyl group; Me2, dimethyl group; TSS, transcription start site.
As mentioned above, epigenome-editing tools offer several key advantages over CRISPRa/CRISPRi and conventional gene-regulation approaches. Epigenome editing recapitulates endogenous mechanisms regulating gene expression and causally links a specific epigenetic manipulation with a functional output. Further, overexpression and knockdown techniques, as well as the highly potent synthetic effector domains discussed already, induce high-magnitude changes in gene expression in the brain, which fail to recapitulate the subtler changes in gene expression that occur under most normal and pathophysiological conditions10–13. The current literature on in vivo neuroepigenome editing consistently shows that this approach induces more physiologically relevant degrees of gene regulation10,14,15,61. For example, HSV-mediated overexpression of ZFP189 (encoded by Zfp189) in the mouse prefrontal cortex (PFC) induces Zfp189 mRNA expression 20-fold10. By contrast, inducing or suppressing endogenous Zfp189 expression using neuroepigenome editing more closely recapitulates the smaller (less than 50%) changes in gene expression that are observed in the PFC of depressed humans or chronically stressed mice.
There remains, however, a lack of literature on direct side-by-side comparisons among conventional knockout and overexpression, synthetic transcriptional effectors and endogenous epigenome editing. Such comparisons will be required to understand the relative changes in gene expression achieved with each of these methods and are important because the degree of a gene’s regulation can affect functional outcome measures. For example, knocking down CDK5 expression using a genetic knockout approach or using epigenomic suppression in the same brain region has opposite effects on cocaine-induced behavioural plasticity32,62.
CRISPR-based in vivo locus-specific neuroepigenome editing.
A growing number of studies are testing a wide range of CRISPR-based epigenome-editing tools in rodent models (TABLE 1). One study used a CRISPR-based method for in vivo neuroepigenome editing by developing a method to precisely manipulate the methylation status of DNA in the brain using dCas9 fused to either DNMT3A or TET1 (REF.53) (FIG. 3a). Recruitment of dCas9–TET1 to the Snrpn promoter (which drives expression of small nuclear ribonucleoprotein-associated polypeptide N) in the mouse forebrain reduced DNA methylation at this locus and increased gene expression, whereas recruitment of dCas9–DNMT3A had the opposite effects. Whereas previous work studying the effect of DNA methylation on gene expression was limited to correlational relationships, these studies causally link DNA methylation in the brain with the bidirectional control of gene expression. These tools will be essential for delineating the role of aberrant DNA methylation patterns observed across many neurological and psychiatric diseases63–65.
Table 1 |.
Studies using in vivo locus-specific neuroepigenome editing in the brain
| DNA-binding region | Packaging | Brain region | Summary | Ref. |
|---|---|---|---|---|
| ZFP–p65 | AAV | Striatum | Gdnf activation reduces neurodegeneration in a rat model of PD | 21 |
| TALE–CIB1 plus CRY2–VP64 | AAV | Infralimbic cortex | Light-induced transcriptional activation using TALEs | 6 |
| ZFP–p65and ZFP–G9a | HSV | Nucleus accumbens | Fosb regulates addictive-like and anxiety-like behaviours | 23 |
| ZFP–p65and ZFP–G9a | HSV | Nucleus accumbens | Cdk5 regulates behavioural responses to cocaine and CSDS | 32 |
| ZFP–VP64 and ZFP–SUVDEL76 | HSV | Hippocampus | Dlg4 activation rescues memory deficits in a mouse model of AD | 22 |
| ZFP–C9a | HSV | Nucleus accumbens | Suppression of Fosb reduces aggressive behaviour | 30 |
| ZFP–p65and ZFP–C9a | HSV | Nucleus accumbens | Fosb oppositely regulates anxiety-like behaviour in DIR-expressing and D2R-expressing MSNs | 31 |
| ZFP–p65 | HSV | CA1 of the hippocampus | Cdk5 has sex-specific effects on fear memory | 36 |
| dCas9–DNMT3A and dCas9–TETl | LV | Forebrain | Demonstrated epigenetic editing of DNA methylation in the brain | 53 |
| dCas9–KRAB | LV | Dentate gyrus | CRISPRi multiplexing in the brain | 49 |
| dCas9–SPH | AAV | Cerebral cortex | Highly potent CRISPRa method for multiplex activation in the brain | 12 |
| dCas9–CREBsl33D and dCas9–G9a | HSV | Prefrontal cortex | CREB-mediated Zfpl89 induction promotes stress resilience | 10 |
| dCas9–VPR | LV | CA1 of the hippocampus, nucleus accumbens and prefrontal cortex | LV-mediated CRISPRa approach for targeting neurons in vivo | 13 |
| dCas9–p300and dCas9–HDAC8 | LV | Hippocampus | Histone acetylation regulates neuronal physiology via Fos induction | 61 |
| dCas9–CCN4and scFv–C11orf46 | IUE | Somatosensory cortex | Sema6a suppression rescues neuronal arborization deficits after Cl lor/46 knockdown | 14 |
| dCas9–DNMT3A | AAV | Hippocampus | Mecp2 methylation induces ASD-like phenotypes | 15 |
| dCas9–VP64and dCas9–KRAB | Transfection reagent | Nucleus accumbens | Activating transcription factor gene Nr4al suppresses addictive-like behaviour | 48 |
AAV, adeno-associated virus; AD, Alzheimer disease; ASD, autism spectrum disorder; CIB1, calcium and integrin-binding protein 1; CREB, cAMP response element-binding protein; CRISPRa, clustered regularly interspaced short palindromic repeats (CRISPR) activation; CRISPRi, CRISPR interference; CRY2, cryptochrome 2; CSDS, chronic social defeat stress; dCas9, nuclease-dead CRISPR-associated protein 9; D1R, dopamine D1 receptor; D2R, dopamine D2 receptor; DNMT3A, DNA methyltransferase 3A; HSV, herpes simplex virus; IUE, in utero electroporation; KRAB, Krüppel-associated box; LV, lentivirus; MSNs, medium spiny neurons; PD, Parkinson disease; scFv, single-chain variable fragment; SPH, SunTag–p65–HSF1; TALE, transcriptional activator-like effector; TET1, ten–eleven translocation methylcytosine dioxygenase 1; ZFP, zinc-finger protein.
Similar in vivo neuroepigenome-editing tools were used to study the consequences of increased DNA methylation at the methylated CpG-binding protein 2 gene (Mecp2) promoter, which is observed in the brains of some individuals with autism spectrum disorder (ASD)15. Virus-mediated delivery of dCas9–DNMT3A into the hippocampus of mice suppressed MeCP2 expression and induced ASD-like behavioural phenotypes. These data suggest that DNA methylation at the Mecp2 promoter causally contributes to ASD pathology, with reversal of DNA methylation at this gene possibly improving treatment outcomes in individuals with ASD. In addition, this work demonstrates that neuroepigenome-editing tools are well suited to study epigenetic mechanisms that drive pathology in neuro-developmental disorders and identify opportunities for therapeutic intervention.
Work from our laboratory applied another in vivo neuroepigenome-editing approach to study genes in the PFC that causally contribute to stress resilience10. Transcriptomic network analysis of RNA-sequencing data collected from the PFC after chronic social defeat stress identified Zfp189 as a key driver of genes associated with a resilient phenotype, downstream of CREB signalling. To study the role of Zfp189 in stress resilience, we generated novel fusion constructs composed of dCas9 tethered to the phosphomimetic (activated) form of CREB (dCas9–CREBS133D) or to G9a (FIG. 3b,d) Previous studies investigating CREB function using overexpression or knockdown techniques were limited, because these approaches induce transcriptional changes at hundreds of CREB target genes66. By injecting an HSV expressing dCas9–CREBS133D into the PFC, we selectively recruited CREB to the Zfp189 gene and causally linked this manipulation to behavioural outputs. CREB-mediated activation of Zfp189 in PFC neurons induced a more resilient behavioural phenotype after chronic social defeat stress, whereas knockdown of Zfp189 with dCas9–G9a had the opposite effect. Delivery of dCas9 fused to a mutant form of CREB that prevents its activation, CREBS133A, had no effect on Zfp189 expression or stress responses. Importantly, dCas9–CREBS133D also selectively induced genes in the Zfp189 module as compared with those in all other gene modules. The behavioural and transcriptional effects of CRISPR-mediated Zfp189 induction were lower in magnitude compared with those induced by conventional, Virus-mediated overexpression of Zfp189.
We further demonstrated that delivery of dCas9–CREBS133D, but not dCas9–CREBS133A, into the NAc similarly induced expression of Fosb, another CREB target gene67. This work provides evidence that CRISPR-based neuroepigenome editing provides a powerful set of tools to study endogenous TFs in the brain and will be crucial for elucidating the role of individual target genes controlled by TF function.
Another recent study used CRISPR tools to establish a causal role for histone acetylation in regulating the physiological properties of hippocampal neurons via immediate early gene induction61. Neuronal depolarization induces histone H3 lysine 27 acetylation at several enhancers in close proximity to Fos, an immediate early gene commonly used as a proxy for measuring neuronal activity. Recruitment of dCas9 tethered to p300 (a HAT) to a Fos enhancer increased Fos expression, whereas recruitment of dCas9 tethered to HDAC8 had the opposite effect (FIG. 3c). LentiVirus-mediated delivery of dCas9–HDAC8 into the dentate gyrus markedly reduced Fos expression, thereby establishing the use of in vivo neuroepigenome editing of histone acetylation with CRISPR tools.
Lastly, another study investigated the function of the chromosome 11 open reading frame 46 gene (C11orf46), which encodes a chromatin-regulatory protein, in neuronal arborization in the somatosensory cortex using CRISPR-based neuroepigenome editing14. Mutations in C11orf46 are associated with intellectual disability and reduced corpus callosum volume68. Knockdown of C11orf46 substantially increases the expression of the semaphorin 6A gene (Sema6a), a putative C11orf46 target gene associated with axonal development. To establish a causal role for the interaction between C11orf46 and Sema6a, the researchers used the dCas9–SunTag platform to recruit multiple copies of C11orf46 to the Sema6a promoter. C11orf46 recruitment reduced Sema6a expression and rescued the marked reduction in neuronal arborization in the somatosensory cortex seen after C11orf46 knockdown.
As with all epigenome-editing approaches, the field needs to pay more attention to ensuring the functionality and selective targeting of epigenome-editing tools. This includes validation of selective dCas9 binding, quantitative measurements of epigenetic modifications at a target gene and genome-wide measures of gene expression. This poses a major technical challenge for the field (BOX 1).
Innovations in neuroepigenome editing
To date, neuroepigenome-editing tools applied to the brain include histone- and DNA-modifying enzymes and TFs. This list represents a small fraction of known epigenetic mechanisms that govern gene expression in the nervous system. The field thus needs to dramatically expand the suite of neuroepigenome-editing tools to capture the diversity of endogenous epigenetic-regulatory events. These tools will be crucial to reveal causal relationships between epigenetic modifications at individual genomic loci and their downstream effects at the molecular, cellular, circuit and behavioural levels.
Ongoing innovations in neuroepigenome editing go beyond increasing the types of epigenetic manipulations that can be applied in the brain. For example, gRNA multiplexing can be used to model more complex transcriptional programmes by inducing epigenetic modifications at several loci simultaneously (FIG. 4a). Multiplexing gRNAs has already been accomplished using CRISPRa/CRISPRi approaches but have not yet been implemented using epigenome-editing tools that target endogenous regulatory mechanisms12,13,49. Such multiplexing approaches are crucial for studying multiple gene targets of a given TF. For example, it would be required to determine the functional effect achieved when CREB is targeted to both Zfp189 and Fosb (which are both induced by cocaine in the same population of NAc MSNs) as opposed to when CREB activates either gene alone. Multiplexing is also instrumental for characterizing the function of gene networks — for example, targeting multiple driver genes in a single module, or key drivers in different modules — in neuropsychiatric disease models.
Fig. 4 |. Innovative neuroepigenome-editing tools.
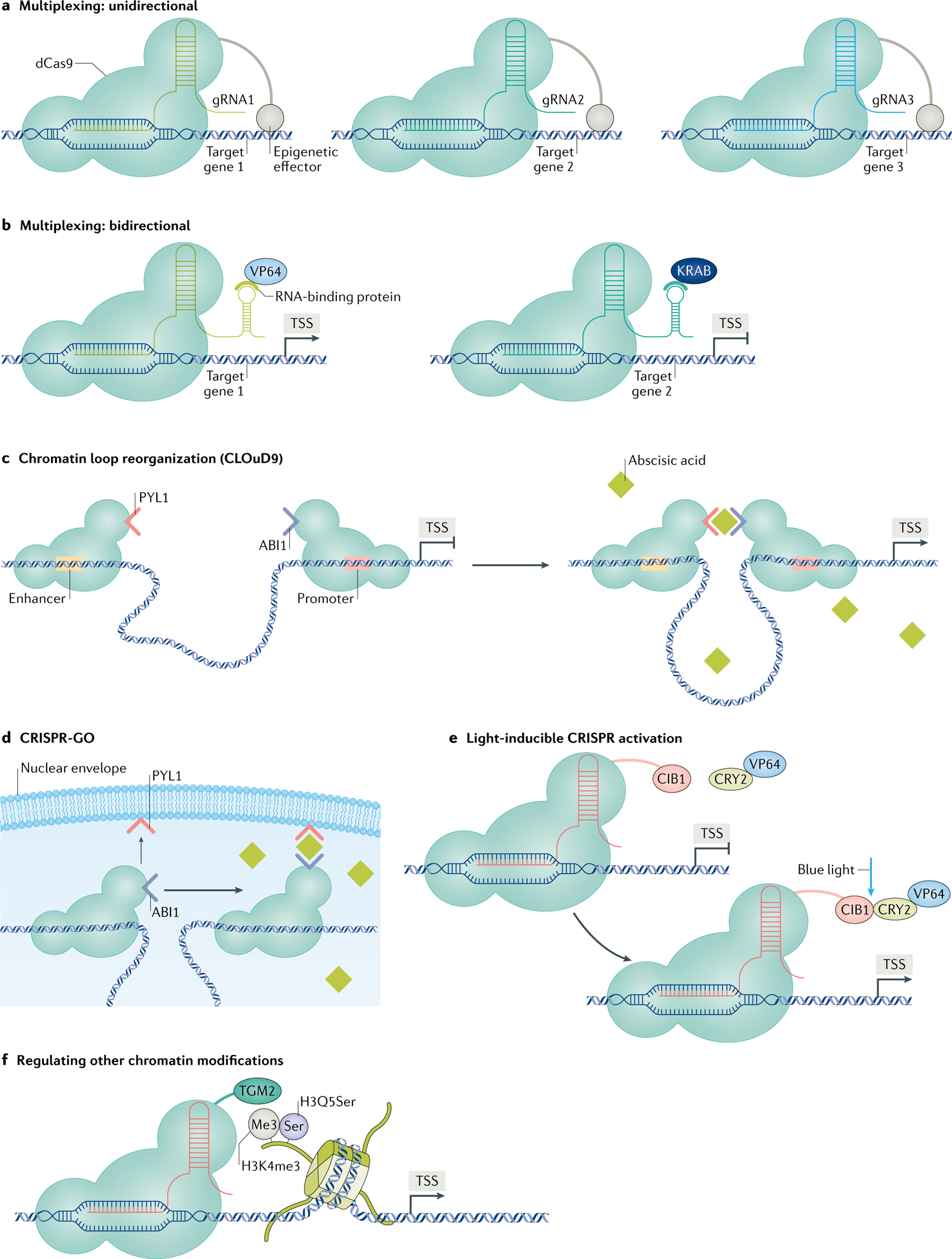
Ongoing innovations in the neuroepigenome-editing field include novel clustered regularly interspaced short palindromic repeats (CRISPR)–nuclease-dead CRISPR-associated protein 9 (dCas9) tools that are poised for applications in neuroscience research. a | Neuroepigenome editing can be paired with guide RNA (gRNA) multiplexing to regulate the chromatin state at multiple genes simultaneously. b | Novel CRISPR tools have been applied in vitro to activate the expression of one gene and suppress another in the same cell. This system uses two distinct gRNAs that are directed towards separate genes and contain a unique RNA scaffold69. These RNA scaffolds are recognized by distinct RNA-binding protein–effector fusions. Pairing this method with use of chromatin-modifying proteins offers a means of regulating multiple genes in the same cell, but with different chromatin modifications. c | Chromatin loop reorganization using CRISPR–dCas9 (CLOuD9) is a method to force chromatin interactions between two genomic loci of interest77. The presence of abscisic acid induces dimerization between the domains tethered to dCas9. d | CRISPR genome organization (CRISPR-GO) uses a similar principle of inducible ligation, but instead forces proximity between a genomic locus and a nuclear subcompartment of interest, such as Cajal bodies or the nuclear envelope78. e | Light-inducible CRISPR activation uses blue light to induce heterodimerization between dCas9-bound calcium and integrin-binding protein 1 (CIB1) and cryptochrome 2 (CRY2) tethered to an effector protein, such as VP64103. Similar light-inducible systems have been achieved with VP64 in the brain in vivo but have not yet been achieved with endogenous chromatin-modifying enzymes. f | Lastly, ongoing work seeks to expand the library of epigenome-editing tools to capture the diversity of endogenous epigenetic mechanisms. This includes a wider range of chromatin modifications, such as the newly discovered serotonylation of histones, in which serotonin (Ser) is added by transglutaminase 2 (TGM2) to the glutamine at position 5 (Q5) of histone H3 trimethylated at lysine 4 (H3K4me3)104. KRAB, Krüppel-associated box; TSS, transcription start site.
Another innovation uses a novel CRISPR–dCas9 platform that recruits RNA-binding proteins — tethered to transcription effectors — to distinct scaffolds on gRNAs that are directed towards separate genes69. This method makes it possible to simultaneously control multiple genes but with different chromatin modifications (FIG. 4b) — for example, inducing histone acetylation at one gene and removing histone acetylation at another in the same cell population. This approach has been validated in cell culture experiments and is poised to be applied to the brain.
Besides targeting multiple genes, epigenome-editing tools can be expressed exclusively in a specific cell type in a single brain region. This is a major frontier in the field given recent single-cell sequencing experiments that increasingly highlight the numerous neuron and glia subtypes involved in nervous system function and pathophysiology70. As noted earlier, specific cell populations can be targeted by coupling Cre-dependent viral constructs with Cre driver transgenic animals, such that the CRISPR–dCas9 components are expressed exclusively in Cre-expressing cells. Work from our laboratory used Cre-dependent vectors to target D1R versus D2R subpopulations of NAc MSNs in mice31. Similarly, Cre-inducible CRISPRa and Cre-inducible gRNA mouse lines have been developed for various other cell types, including dopaminergic neurons and astrocytes, allowing cell type-specific expression of CRISPR tools71,72. Alternatively, cell-type specificity could potentially be achieved by use of promoters that target transgene expression to a specific cell population of interest, such as the human synapsin 1 gene (SYN1) promoter to target neurons12,73,74 or the glial fibrillary acidic protein (Gfap) promoter to target astrocytes75. Despite these examples, the track record of using promoters to target specific subtypes of neurons has been problematic to date, perhaps because promoters are often active across numerous cell types in the nervous system. Recent work suggests that using enhancers might offer superior cell-type specificity, and their small size is compatible with virus-mediated transgene delivery, although only a few cell type-specific enhancers have been identified and empirically validated to date76. Expanding the capability of cell type-specific transgene expression will be crucial to interrogate the role of individual cell types in normal brain function and disease.
Another active area of research in the epigenome-editing field seeks to regulate gene expression by precisely manipulating the 3D chromatin conformation in the nucleus. The arrangement of chromatin alters gene expression through various mechanisms, including the proximity of distant enhancers to the regulatory regions of a gene, localization in specific nuclear subcompartments and the density of chromatin packaging. One group developed the chromatin loop reorganization using CRISPR–dCas9 (CLOuD9) method to force chromatin looping between two specified genomic loci in cultured cells77 (FIG. 4c). In this method, gRNAs are directed towards two genes of interest, and application of abscisic acid induces heterodimerization of the domains tethered to dCas9. A similar study, using the same principles of chemically induced ligation, introduced a method called ‘CRISPR genome organization’ (CRISPR-GO) to localize a single gene to a nuclear subcompartment of interest, such as the nuclear envelope or Cajal bodies, again in cultured cells78 (FIG. 4d). CLOuD9 and CRISPR-GO are powerful tools to interrogate the causal relationships between 3D chromatin topology and gene expression. These systems both depend on the presence of abscisic acid, a plant-derived chemical that facilitates dimerization, making it more difficult to apply in the brain, however. Presumably, abscisic acid could be introduced to the brain via cannulation, but the technical difficulty and potential for toxicity and off-target effects pose considerable challenges.
Last, recent work introduced methods to overcome the temporal constraints of in vivo neuroepigenome-editing experiments. Light- or chemical-inducible epigenome-editing systems offer improved spatiotemporal control of genetic manipulations (FIG. 4e). These techniques are technically challenging to conduct in the brain but may be useful for experiments that require high temporal resolution (for a detailed discussion, see REF.79). In addition, viral delivery approaches dictate the window of transgene expression but are limited in their packaging capacity80. HSVs are ideal for packaging dCas9, owing to their large carrying capacity, but HSVs can be neurotoxic unless carefully purified and their in vivo expression window is less than 7 days, making it difficult to study the long-term consequences of epigenetic perturbations81. Lentiviruses offer sufficient packaging capacity and long-term expression in vivo, but their poor safety profile and relatively low infection rates hinder experimental insight and the potential for clinical translation82. AAVs exhibit stable in vivo expression and low toxicity, making them a suitable delivery platform with translational potential. Nevertheless, the small packaging capacity of AAVs makes it impossible to package the traditional CRISPR–dCas9 components into a single vector82. Recent studies have demonstrated that smaller dCas9 orthologues, such as dCas9 from Staphylococcus aureus (dSaCas9), tethered to a transcriptional effector can be packaged into AAVs83,84. The size of the effector moiety and other transgene components may still be a limiting factor for AAV packaging, however.
Clinical potential
Studies of the molecular mechanisms of neurological and psychiatric disease, using post-mortem tissue and animal models, show widespread changes in the transcriptome across many brain regions. Equipped with new tools to casually link endogenous epigenetic mechanisms to a specific pathological state, neuroscientists will inevitably identify new gene targets with therapeutic potential.
Ongoing clinical trials are evaluating the efficacy of gene-editing approaches, including ZFPs and CRISPR, for treating various diseases in peripheral organs. In the past decade, these tools have been preclinically and clinically tested for several ocular, skin, neuromuscular, cardiovascular, hepatic, respiratory, gastric and haematologic disorders85. For example, ZFPs are used to engineer T cells to control human immunodeficiency virus (HIV) infection. HIV-infected patient T cells are treated ex vivo with ZFNs to knock out CCR5 to remove a co-receptor needed for HIV infection86. Moreover, CRISPR–Cas9 is used to generate chimeric antigen receptor T cells, whereby T cells from a patient or from a healthy donor are modified ex vivo to more effectively target cancer cells and are then administered to the patient87. Studies are also under way to use CRISPR tools to treat Leber congenital amaurosis type 10 (LCA10), a form of congenital blindness88. LCA10 is caused by mutation of the CEP290 gene, which encodes a centrosomal protein. Cas nucleases targeting CEP290 are being administered directly into the eye to excise the mutated region of the gene and restore normal levels of CEP290 function. These clinical trials establish the therapeutic potential of genome-engineering tools. Moreover, the recent discovery of protein inhibitors of CRISPR–Cas systems, termed ‘anti-C RISPR–Cas proteins’, adds potential to more precisely regulate CRISPR–Cas9-based gene editing89–91. We are much further from applying such approaches to patients with neurological or psychiatric disorders, however, owing to challenges in delivering the constructs to the brain.
Developing a safe and minimally invasive approach to deliver neuroepigenome-editing tools to the brains of human patients would mark a critical turning point for treating pathology in the CNS. An in-d epth discussion of the obstacles that must be overcome to progress towards clinical applications of neuroepigenome-e diting tools is beyond the scope of this Review, but we refer the reader to literature addressing methods to deliver transgenes to the brain, such as use of AAVs and nanoparticles92–94, to circumvent the adaptive immune response to Cas proteins95 and to mitigate off-t arget effects96–98 (BOX 1).
Conclusions
Psychiatric and neurological conditions are associated with profound changes in gene expression in the CNS that directly contribute to disease pathology. The complex epigenetic mechanisms that govern gene expression in pathological states remain incompletely understood. In this Review, we introduced in vivo locus-specific neuroepigenome editing as the gold standard for establishing a causal link between a given epigenetic modification at a single gene and functional output measures.
Ongoing innovations hold promise for further elucidating the epigenetic basis of nervous system development, cellular differentiation and psychiatric and neurological disease by expanding the library of epigenome-editing tools, targeting specific populations of cells in the CNS and modelling more complex transcriptional programmes involving the upregulation and downregulation of numerous genes99–101. For example, CRISPR is capable not only of identifying genes that contribute to neuronal differentiation but also of establishing a causal relationship between epigenetic modifications at these genes and cell fate. Moreover, in combination with single-c ell RNA sequencing and use of non-mammalian organisms that permit high-throughput screening (such as Drosophila and zebrafish)101, in vivo locus-specific neuroepigenome editing can identify cell type-specific gene signatures across large panels of genes implicated in psychiatric and neurological diseases100. Last, the recent success of CRISPR-based genome-editing therapies in clinical trials for peripheral and ocular disturbances demonstrates that epigenome editing is poised for clinical applications, with the ultimate frontier being the treatment of CNS diseases.
Acknowledgements
The authors are very grateful to J. K. Gregory for her help with the figures in this Review.
Glossary
- Transcription activator-like effector (TALE)
DNA-binding protein derived from bacteria (Xanthomonas) that regulates gene expression
- Clustered regularly interspaced short palindromic repeats (CRISPR)
A component of the adaptive immune system in bacteria and archaea that cleaves foreign nucleic acid sequences. it is used routinely in the laboratory to enable targeted genetic and epigenetic manipulations
- Guide RNA (gRNA)
A synthetic RNA that guides clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9) to a specific DNA sequence in the genome
- VP64
A complex of four copies of VP16 (a viral protein sequence of 16 amino acids) that activates gene transcription
- Zinc-finger proteins (ZFPs)
Proteins consisting of zinc ion-regulated Cys2-His2 domains that recognize specific 18-bp sequences of DNA. These proteins can be fused to various effector proteins, including nucleases and chromatin-modifying proteins
- Transcription factor (TF)
Protein that binds to specific sequences of DNA and regulates gene expression through the recruitment of chromatin-modifying enzymes and other proteins
- Fosb
An immediate early gene that encodes full-length FOSB and a truncated splice variant ΔFOSB, and that has served as a useful target for the development of novel neuroepigenome-editing tools
- cAMP response element-binding protein (CREB)
A ubiquitously expressed transcription factor implicated in diverse functions in the central nervous system and periphery
- Protospacer adjacent motif (PAM)
A short DNA sequence upstream of the target gene that is recognized by clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9)
- CRISPR activation (CRISPR)
A clustered regularly interspaced short palindromic repeats (CRISPR) system that uses potent activation domains, such as the viral transcription factor VP64, to increase gene expression
- CRISPR interference (CRISPR i)
A clustered regularly interspaced short palindromic repeats (CRISPR) system that uses repressive domains, such as the Krüppel-associated box (KRAB) domain, to suppress gene expression
- SunTag
A clustered regularly interspaced short palindromic repeats (CRISPR)-based method that uses a repeating peptide array to recruit multiple copies of single-chain variable fragment (scFv)-fused effector proteins to a target gene
- ZFP189
A putative transcription factor whose gene is a target of cAMP response element-binding protein (CREB). Recent studies suggest that this protein is involved in regulating synaptic plasticity and behavioural responses to stress
- DNMT3A
Enzyme that catalyses the addition of methyl groups to DNA
- Chromatin loop reorganization using CRISPR–dCas9 (CLOuD9)
A clustered regularly interspaced short palindromic repeats (CRISPR) system that uses chemically induced ligation to selectively and reversibly establish chromatin loops
- CRISPR genome organization (CRISPR-GO)
A clustered regularly interspaced short palindromic repeats (CRISPR) system that uses chemically induced ligation to bring loci in close proximity to nuclear subcompartments
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Jenuwein T & Allis CD Translating the histone code. Science 293, 1074–1080 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Suzuki MM & Bird A DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet 9, 465–476 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Mercer TR & Mattick JS Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol 20, 300–307 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Liu XS & Jaenisch R Editing the epigenome to tackle brain disorders. Trends Neurosci. 42, 861–870 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Fellmann C, Gowen BG, Lin PC, Doudna JA & Corn JE Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat. Rev. Drug. Discov 16, 89–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konermann S et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maeder ML et al. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10, 977–979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dreier B, Beerli RR, Segal DJ, Flippin JD & Barbas CF 3rd Development of zinc finger domains for recognition of the 5’-ANN-3’ family of DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem 276, 29466–29478 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Gilbert LA et al. CRISPR-mediated Modular RNA-guided regulation of transcription in Eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorsch ZS et al. Stress resilience is promoted by a Zfp189-driven transcriptional network in prefrontal cortex. Nat. Neurosci 22, 1413–1423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; Neuroepigenome editing with CRISPR is used to direct CREB to the Zfp189 promoter (a CREB target) in PFC neurons. The resulting induction of endogenous Zfp189 promotes resilience to behavioural stress.
- 11.Cates HM et al. Transcription factor E2F3a in nucleus accumbens affects cocaine action via transcription and alternative splicing. Biol. Psychiatry 84, 167–179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou HB et al. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat. Neurosci 21, 440–446 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Savell KE et al. A neuron-optimized CRISPR/dCas9 activation system for robust and specific gene regulation. eNeuro 10.1523/ENEURO.0495-18.2019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peter CJ et al. In vivo epigenetic editing of Sema6a promoter reverses transcallosal dysconnectivity caused by C11orf46/Arl14ep risk gene. Nat. Commun 10, 4112 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Z et al. Locus-specific DNA methylation of Mecp2 promoter leads to autism-like phenotypes in mice. Cell Death Dis. 11, 85 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; DNA methylation of the Mecp2 promoter in the hippocampus is achieved by targeting dCas9 fused to a DNMT catalytic domain to the locus. This action suppresses Mecp2 expression and is sufficient to induce autism-l ike behavioural phenotypes.
- 16.Klug A & Rhodes D Zinc fingers: a novel protein fold for nucleic-acid recognition. Cold Spring Harb Symp. Quant. Biol 52, 473–482 (1987). [DOI] [PubMed] [Google Scholar]
- 17.Kim YG, Cha J & Chandrasegaran S Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl Acad. Sci. USA 93, 1156–1160 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camenisch TD, Brilliant MH & Segal DJ Critical parameters for genome editing using zinc finger nucleases. Mini Rev. Med. Chem 8, 669–676 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Gordley RM, Smith JD, Graslund T & Barbas CF III. Evolution of programmable zinc finger-recombinases with activity in human cells. J. Mol. Biol 367, 802–813 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Kolb AF et al. Site-directed genome modification: nucleic acid and protein modules for targeted integration and gene correction. Trends Biotechnol. 23, 399–406 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Laganiere J et al. An engineered zinc finger protein activator of the endogenous glial cell line-derived neurotrophic factor gene provides functional neuroprotection in a rat model of Parkinson’s disease. J. Neurosci 30, 16469–16474 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bustos FJ et al. Epigenetic editing of the Dlg4/PSD95 gene improves cognition in aged and Alzheimer’s disease mice. Brain 140, 3252–3268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heller EA et al. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat. Neurosci 17, 1720–1727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; ZFPs fused to p65 or to G9a are targeted to the Fosb locus in NAc neurons; they bidirectionally control Fosb expression as well as downstream behavioural responses to social stress or cocaine.
- 24.Lobo MK et al. ΔFosB induction in striatal medium spiny neuron subtypes in response to chronic pharmacological, emotional, and optogenetic stimuli. J. Neurosci 33, 18381–18395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robison AJ & Nestler EJ Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci 12, 623–637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maze I et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327, 213–216 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renthal W et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron 62, 335–348 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar A et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48, 303–314 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Covington HE III et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron 71, 656–670 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aleyasin H et al. Cell-type-specific role of ΔFosB in nucleus accumbens in modulating intermale aggression. J. Neurosci 38, 5913–5924 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamilton PJ et al. Cell-type-specific epigenetic editing at the Fosb gene controls susceptibility to social defeat stress. Neuropsychopharmacology 43, 272–284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; ZFPs developed by Heller et al. (2014) are expressed in the D1R-e xpressing or D2R-expressing MSNs in the NAc, where they are shown to produce opposite effects on depression-l ike behaviours.
- 32.Heller EA et al. Targeted epigenetic remodeling of the Cdk5 gene in nucleus accumbens regulates cocaine- and stress-evoked behavior. J. Neurosci 36, 4690–4697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su SC, Rudenko A, Cho S & Tsai LH Forebrain-specific deletion of Cdk5 in pyramidal neurons results in mania-like behavior and cognitive impairment. Neurobiol. Learn. Mem 105, 54–62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hawasli AH et al. Cyclin-dependent kinase 5 governs learning and synaptic plasticity via control of NMDAR degradation. Nat. Neurosci 10, 880–886 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong P et al. Cyclin-dependent kinase 5 in the ventral tegmental area regulates depression-related behaviors. J. Neurosci 34, 6352–6366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sase AS et al. Sex-specific regulation of fear memory by targeted epigenetic editing of Cdk5. Biol. Psychiatry 85, 623–634 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Moscou MJ & Bogdanove AJ A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501–1501 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Sanjana NE et al. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc 7, 171–192 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toegel M et al. A multiplexable TALE-based binary expression system for in vivo cellular interaction studies. Nat. Commun 8, 1663 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu PD, Lander ES & Zhang F Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makarova KS et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol 9, 467–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mali P et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heidenreich M & Zhang F Applications of CRISPR-Cas systems in neuroscience. Nat. Rev. Neurosci 17, 36–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng AW et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23, 1163–1171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konermann S et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS & Vale RD A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carpenter MD et al. Nr4a1 suppresses cocaine-induced behavior via epigenetic regulation of homeostatic target genes. Nat. Commun 11, 504 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng Y et al. CRISPR interference-based specific and efficient gene inactivation in the brain. Nat. Neurosci 21, 447–454 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Devesa-Guerra I et al. DNA methylation editing by CRISPR-guided excision of 5-methylcytosine. J. Mol. Biol 432, 2204–2216 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Engmann O et al. Cocaine-induced chromatin modifications associate with increased expression and three-dimensional looping of Auts2. Biol. Psychiatry 82, 794–805 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hilton IB et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol 33, 510–517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu XS et al. Editing DNA methylation in the mammalian genome. Cell 167, 233–247 e217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stepper P et al. Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic Acids Res 45, 1703–1713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu XS et al. Rescue of fragile X syndrome neurons by DNA methylation editing of the FMR1 gene. Cell 172, 979–992 e976 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lei Y et al. Targeted DNA methylation in vivo using an engineered dCas9-MQ1 fusion protein. Nat. Commun 8, 16026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Geen H et al. Ezh2-dCas9 and KRAB-dCas9 enable engineering of epigenetic memory in a context-dependent manner. Epigenetics Chromatin 12, 26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwon DY, Zhao YT, Lamonica JM & Zhou Z Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC. Nat. Commun 8, 15315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kearns NA et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat. Methods 12, 401–403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vojta A et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 44, 5615–5628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen LF et al. Enhancer histone acetylation modulates transcriptional bursting dynamics of neuronal activity-inducible genes. Cell Rep. 26, 1174–1188 e1175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benavides DR et al. Cdk5 modulates cocaine reward, motivation, and striatal neuron excitability. J. Neurosci 27, 12967–12976 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Massart R et al. Role of DNA methylation in the nucleus accumbens in incubation of cocaine craving. J. Neurosci 35, 8042–8058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klengel T, Pape J, Binder EB & Mehta D The role of DNA methylation in stress-related psychiatric disorders. Neuropharmacology 80, 115–132 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Al-Mahdawi S, Virmouni SA & Pook MA The emerging role of 5-hydroxymethylcytosine in neurodegenerative diseases. Front. Neurosci 8, 397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McClung CA & Nestler EJ Regulation of gene expression and cocaine reward by CREB and ΔFosB. Nat. Neurosci 6, 1208–1215 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Lardner CK et al. in Neuroscience 2019 Vol. Program Number 415.17 (Society for Neuroscience, 2019). [Google Scholar]
- 68.Najmabadi H et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478, 57–63 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Zalatan JG et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160, 339–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; A novel CRISPRa/CRISPRi approach is used to recruit distinct transcriptional effector domains to separate genes within the same cell.
- 70.Ofengeim D, Giagtzoglou N, Huh D, Zou C & Yuan J Single-cell RNA sequencing: unraveling the brain one cell at a time. Trends Mol. Med 23, 563–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Back S et al. Neuron-specific genome modification in the adult rat brain using CRISPR-Cas9 transgenic rats. Neuron 102, 105–119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Daigle TL et al. A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality. Cell 174, 465–480 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kugler S, Kilic E & Bahr M Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area. Gene Ther. 10, 337–347 (2003). [DOI] [PubMed] [Google Scholar]
- 74.Shevtsova Z, Malik JM, Michel U, Bahr M & Kugler S Promoters and serotypes: targeting of adeno-associated virus vectors for gene transfer in the rat central nervous system in vitro and in vivo. Exp. Physiol 90, 53–59 (2005). [DOI] [PubMed] [Google Scholar]
- 75.Merienne N, Le Douce J, Faivre E, Deglon N & Bonvento G Efficient gene delivery and selective transduction of astrocytes in the mammalian brain using viral vectors. Front. Cell Neurosci 7, 106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blankvoort S, Descamps LAL & Kentros C Enhancer-driven gene expression (EDGE) enables the generation of cell type specific tools for the analysis of neural circuits. Neurosci. Res 152, 78–86 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Morgan SL et al. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat. Commun 8, 15993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; CRISPR tools are used to chemically induce, in a reversible manner, the chromatin looping between two genes.
- 78.Wang H et al. CRISPR-mediated programmable 3D genome positioning and nuclear organization. Cell 175, 1405–1417 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Day JJ Genetic and epigenetic editing in nervous system. Dialogues Clin. Neurosci 21, 359–368 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hamilton PJ, Lim CJ, Nestler EJ & Heller EA Neuroepigenetic editing. Methods Mol. Biol 1767, 113–136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carlezon WA Jr., Nestler EJ & Neve RL Herpes simplex virus-mediated gene transfer as a tool for neuropsychiatric research. Crit. Rev. Neurobiol 14, 47–67 (2000). [DOI] [PubMed] [Google Scholar]
- 82.Nelson CE & Gersbach CA Engineering delivery vehicles for genome editing. Annu. Rev. Chem. Biomol. Eng 7, 637–662 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Thakore PI et al. RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nat. Commun 9, 1674 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vora S et al. Rational design of a compact CRISPR-Cas9 activator for AAV-mediated delivery. bioRxiv 10.1101/298620 (2018). [DOI] [Google Scholar]
- 85.Maeder ML & Gersbach CA Genome-editing technologies for gene and cell therapy. Mol. Ther 24, 430–446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tebas P et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med 370, 901–910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ren J & Zhao Y Advancing chimeric antigen receptor T cell therapy with CRISPR/Cas9. Protein Cell 8, 634–643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wood EH et al. Stem cell therapies, gene-based therapies, optogenetics, and retinal prosthetics: current state and implications for the future. Retina 39, 820–835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Borges AL, Davidson AR & Bondy-Denomy J The discovery, mechanisms, and evolutionary impact of anti-CRISPRs. Annu. Rev. Virol 4, 37–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marino ND, Pinilla-Redondo R, Csorgo B & Bondy-Denomy J Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat. Methods 17, 471–479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pawluk A, Davidson AR & Maxwell KL Anti-CRISPR: discovery, mechanism and function. Nat. Rev. Microbiol 16, 12–17 (2018). [DOI] [PubMed] [Google Scholar]
- 92.Chan KY et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stavarache MA et al. Safe and stable noninvasive focal gene delivery to the mammalian brain following focused ultrasound. J. Neurosurg 130, 989–998 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Lee B et al. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng 2, 497–507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moreno AM et al. Immune-orthogonal orthologues of AAV capsids and of Cas9 circumvent the immune response to the administration of gene therapy. Nat. Biomed. Eng 3, 806–816 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu X et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol 32, 670–676 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Slaymaker IM et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hu JH et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu Y et al. CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Cell Stem Cell 23, 758–771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jin X et al. In vivo Perturb-Seq reveals neuronal and glial abnormalities associated with autism risk genes. bioRxiv 10.1101/791525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thyme SB et al. Phenotypic landscape of schizophrenia-associated genes defines candidates and their shared functions. Cell 177, 478–491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qi LS et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Polstein LR & Gersbach CA A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol 11, 198–200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; A light-inducible CRISPRa/CRISPRi approach using heterodimerization domains tethered to dCas9 and the transcriptional effector domain is developed.
- 104.Farrelly LA et al. Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 567, 535–539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; Serotonylation at histone H3 glutamine 5, in close proximity to the well-characterized histone H3 lysine 4 trimethylation mark, was shown to regulate gene expression in the brain.
- 105.Meers MP, Bryson TD, Henikoff JG & Henikoff S Improved CUT&RUN chromatin profiling tools. eLife 10.7554/eLife.46314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fu Y, Sander JD, Reyon D, Cascio VM & Joung JK Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol 32, 279–284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kocak DD et al. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol 37, 657–666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


