A method is reported using ionic liquids for topical delivery of siRNA into skin for the treatment of psoriasis.
Abstract
Systemic antibodies targeting tumor necrosis factor–α (TNF-α) and interleukin-17A (IL-17A) are effective in plaque psoriasis. Despite their popularity, safety concerns pose a challenge for systemic biologics. While anti–TNF-α and anti–IL-17A antibodies effectively inhibit respective proteins, we hypothesize that an approach based on local silencing of an upstream target such as NFKBIZ can be advantageous for treating psoriasis. However, effective delivery of small interfering RNA (siRNA) into the skin is a substantial hurdle due to skin’s barrier function and poor stability of siRNA. Using ionic liquids as an enabling technology, we report on the effective delivery of NFKBIZ siRNA into the skin and its therapeutic efficacy in a psoriasis model. Treatment with IL-siRNA suppressed aberrant gene expression and resulted in down-regulation of psoriasis-related signals including TNF-α and IL-17A. These results provide a framework for a topical delivery platform for siRNA.
INTRODUCTION
Psoriasis is one of the most debilitating chronic skin diseases affecting more than 125 million people worldwide with an estimated economic burden of $135 billion/year in the United States (1). Its pathogenesis and the underlying mechanisms are still not fully understood. Nuclear factor κB (NF-κB), a ubiquitously expressed transcription factor, is considered as the master regulator of immune responses and is implicated in several autoimmune inflammatory diseases including psoriasis (2). Several therapeutics targeting NF-κB signaling pathways are available in the clinic; however, concerns regarding the lack of specificity and side effects pose a challenge (3). This is particularly challenging since systemic inhibition of pleotropic proteins like NF-κB might lead to serious side effects as they provide essential basal activity as survival factors. Network-centric approaches involving pathway-specific inhibitors have gained considerable therapeutic interests (4). In this regard, infliximab and adalimumab [both anti–tumor necrosis factor–α (TNF-α) monoclonal antibodies] as well as secukinumab [an anti–interleukin-17A (IL-17A) antibody] have been approved by the U.S. Food and Drug Administration and are claimed to mediate their therapeutic effects through the modulation of NF-κB activity (5).
NFKBIZ, a gene encoding atypical inhibitor of nuclear factor κB (IκB) protein IκBζ, has gained interests for therapeutic intervention due to its crucial role in the regulation of NF-κB complexes (6, 7). It is reported to be a direct transcription activator of TNF-α–, IL-17A–, and IL-36–inducible psoriasis-related gene products that are involved in inflammatory signaling, neutrophil chemotaxis, and leukocyte activation (8–11). In addition, strong expression of NFKBIZ in patients with psoriasis could be correlated to elevated IL-36– and IL-17A–type responses (12). Local silencing of NFKBIZ can be advantageous since it can potentially broaden the population of patients that can benefit from the treatment compared with that by a single antibody.
Silencing of NFKBIZ through topical applications of small interfering RNA (siRNA) offers a noninvasive and self-administered treatment option with minimal side effects (13). However, the greatest challenge of this route is that only a limited number of drugs with low molecular weights (up to few hundred daltons) and high octanol-water partition coefficients are usable for successful topical delivery (14). Transdermal and topical delivery of hydrophilic molecules, particularly macromolecules such as antibodies and nucleic acids, remains challenging, owing to their high molecular weights (15). Several reports have showcased topical siRNA delivery using techniques such as spherical nucleic acids (16) and self-assembling framework nucleic acids (17). Microneedles have also been explored for topical delivery of siRNA (18). Methods such as electroporation (19) and peptide carriers have also been explored (20–22). Strategies have also been developed to deliver siRNA to treat cutaneous wounds (23, 24). Recently, ionic liquids (ILs) have emerged as a class of versatile materials for topical and transdermal delivery (25). ILs provide a number of potential benefits including tunability, broad applicability, and excellent safety profile (26). ILs have shown potential for delivering siRNA, but their ability to induce a therapeutic effect by silencing a target in vivo has not been demonstrated.
Here, we report a modular IL-based siRNA delivery approach for silencing various genes of interest. Specifically, we have identified a combination of ILs that simultaneously stabilizes siRNA and enhances siRNA penetration into the skin following topical application. We demonstrate the efficacy of the formulation in silencing NFKBIZ in vivo in an imiquimod-induced psoriasis mouse model.
RESULTS
IL selection
A library of ILs was designed and synthesized to assess siRNA delivery into skin. Cholinium was used as the cation in all ILs due to its known biocompatibility and prior use in humans (27). Several different anions were used to synthesize ILs (fig. S1). Geranic acid was used as the reference anion in the IL library [that is, choline and geranic acid (CAGE) as a reference IL] due to its prior use for transdermal delivery of macromolecules (28–30). Other anions were chosen for several reasons. First, anions containing shorter linear carbon chains were chosen in contrast to geranic acid to assess the impact of the chain length on siRNA stability and delivery. Anions with aromatic groups were chosen since they might interact with the stacked RNA base pairs via electrostatic, hydrophobic, and polar interactions. All ILs were prepared at a stoichiometric ratio of 1:2 (cation:anion) and were assessed for stability and siRNA delivery. Of the ILs synthesized, CAGE, choline and dimethylacrylic acid (CADA), choline and isovaleric acid (CAVA), and choline and phenylpropanoic acid (CAPA) remained as a viscous liquid at room temperature (RT), whereas choline and 4-phenolsulfonic acid (CASA), choline and phenylphosphoric acid (CAPP), and choline and biphenyl-3-carboxylic acid (CABA) solidified or formed a gel (fig. S1). Representative 1H nuclear magnetic resonance (NMR) spectra can be found in fig. S1, confirming the successful synthesis and purity of the ILs. In addition, since both interleukin and ILs have been denoted as “IL,” for the purpose of clarity, all interleukins are referred by a numerical value throughout the manuscript.
Effect of ILs on siRNA stability
We first assessed the effect of ILs on siRNA stability. Circular dichroism (CD) spectroscopy of siRNA incubated with aqueous solutions of individual ILs at 50% (v/v) concentration revealed notable alteration in the α helix backbone (confirmed from the negative band at 210 nm) in the presence of CAGE, CADA, and CABA. On the other hand, CAVA and CAPA retained the secondary structure of siRNA (Fig. 1A). Bands obtained from the native gel electrophoresis complemented with the CD results (Fig. 1B). The improved stability of siRNA in the presence of CAPA suggested the possibility of synergistic effects between the ILs prepared from two structurally different anions. Consequently, we assessed the effect of IL mixtures on siRNA stability to determine whether the compatibility of CAPA with siRNA might offer additional protection against the adverse effects of CAGE and CABA on the siRNA structure. The combination of CAGE (25% v/v) and CAPA (25% v/v) led to a prominent band indicative of retention of siRNA structure (fig. S1).
Fig. 1. Screening of cholinium-based bioactive IL-RNA complex for enhanced epidermal accumulation.
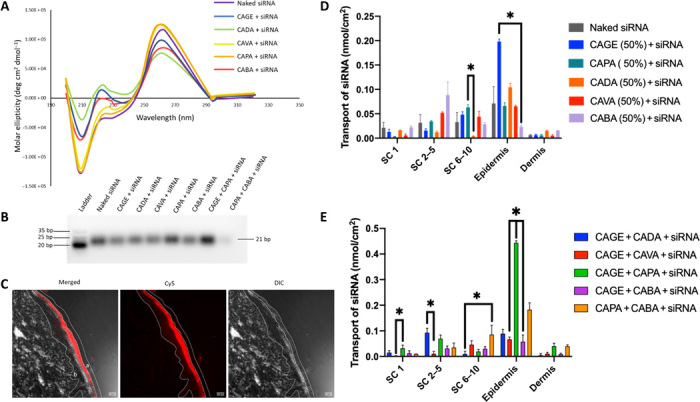
(A) CD spectra of siRNA in phosphate-buffered saline (PBS) following incubation with IL (50% v/v) for 30 min and dialysis for 72 hours. (B) Representative native gel image of siRNA following IL incubation. bp, base pair. (C) Representative confocal images of siRNA (red) in the different skin layers (a) stratum corneum (SC), (b) epidermis, and (c) dermis, in the presence of IL combination (CAGE + CAPA) mixed at a ratio 1:1 following 24 hours of incubation. Left to right: Merged, Cy5, differential interference contrast (DIC). Scale bars, 50 μm. (D and E) Transport of Cy5-labeled siRNA in the presence of individual ILs with concentration of 50% (v/v) (D) and combination of ILs at 50% (v/v) (E) into the different layers of skin determined by the tape-stripping method (n = 3). Data are averages ± SEM and were determined to be nonparametric by normality test and statistics by Kruskal-Wallis test for (D) and (E). *P < 0.05.
Screening of optimal IL combinations for siRNA delivery
The individual ILs and their combinations were then evaluated for epidermal permeation of Cy5-labeled siRNA into porcine skin in Franz diffusion cells (FDCs) (Fig. 1C). Some epidermal uptake for naked siRNA was seen in controls. CAGE exhibited the highest delivery among all tested ILs (Fig. 1D). About 0.20 nmol/cm2 of siRNA was delivered into the epidermis in the presence of CAGE (50% v/v) compared with 0.07 nmol/cm2 in case of naked siRNA. Since 50% CAGE had a potential effect on the siRNA structure, we also measured the ability of IL combinations to deliver siRNA into skin. A combination of CAPA and CAGE (25% v/v each) led to ~0.4 nmol/cm2 siRNA getting delivered into the skin (Fig. 1E). Because the CAGE + CAPA combination yielded the highest epidermal delivery as well as high stability, it was selected as the lead formulation for further studies (fig. S2).
IL-induced intercalation and solvating effects on RNA
Molecular dynamic (MD) simulations were performed to explore the mechanism by which the IL combination (CAGE + CAPA) stabilizes the RNA. It is evident from the snapshots of unit cells within 10 Å of RNA that geranic acid in CAGE is responsible for forming aggregated clumps, leading to separation of geranic acid from choline, water, and the RNA molecule (Fig. 2, A and B). Addition of phenylpropanoic acid to CAGE led to a more consistent distribution of the three molecular species/ions in the IL solution (Fig. 2, C and D). Furthermore, the proximity of phenylpropanoic acid molecules to the RNA molecules, possibly due to the presence of hydrophobic aromatic rings unlike its aliphatic counterpart (geranic acid), confirms its crucial role in intercalating between the stacked RNA base pairs contributing to the RNA solvation and stability.
Fig. 2. MD simulation identifies the extent of IL-siRNA interaction for enhanced solvation and stability.
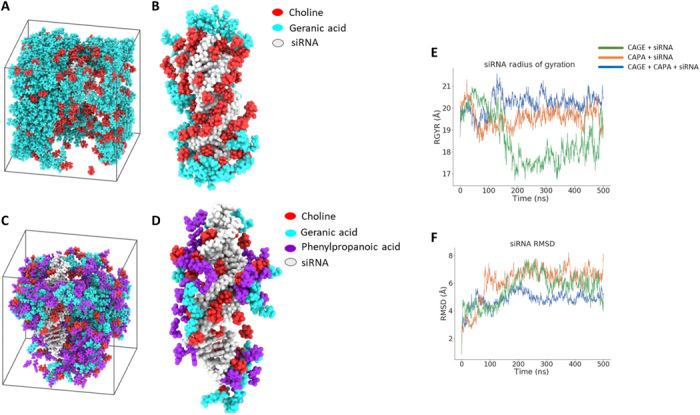
(A and B) Snapshot of simulation unit cell for CAGE and siRNA (A) and CAGE components found within 10 Å of siRNA (B) under periodic boundary conditions for 500 ns. (C and D) Snapshot of simulation unit cell for the optimized IL combination (CAGE and CAPA, 1:1) and siRNA (C) and IL species found within 10 Å of siRNA (B) under similar conditions. (E and F) Radius of gyration (RGYR) (E) and root mean square deviation (RMSD) (F) obtained over the course of 500 ns for CAPA and the IL combination (CAGE and CAPA) in contrast to CAGE (control).
Structural properties of RNA were assessed by performing simulations over the course of 500 ns and measuring the root mean square deviation (RMSD) and radius of gyration (RGYR). The RGYR obtained for the CAGE group was consistent up to 150 ns and started decreasing toward the end of the simulation, indicating the inconsistent compactness of the system (Fig. 2E). In contrast, the increased and consistent RGYR obtained for the IL combination (CAGE + CAPA) over 500 ns aligns well with the improved IL-RNA interaction results. Such improved interactions and compactness for the optimized IL system with the RNA could also be attributed to the increase in the relative molecular mobility or reduced local viscosity upon addition of phenylpropanoic acid to CAGE. In addition, lower viscosity of the IL system may weaken the intramolecular strain placed on the RNA by the IL and is a possible explanation for the reduced RMSD observed in the case of CAGE + CAPA (Fig. 2F).
IL-mediated lipid membrane dynamics modulation
To assess the insertion and translocation of the IL into the lipid bilayers, simulations of the lipid bilayer in the presence of IL were conducted (Fig. 3, A to C). In addition to improving the stability and solvation of the RNA, the compact packing of the ionic species leading to the formation of aggregates seems to augment the IL–lipid membrane interactions. The aggregates formed by the individual ionic moieties appear to enable a continuity between the IL system and the molecules, making up the lipid bilayer. It is possible that the collective mass of the ionic aggregates plays a crucial role in facilitating membrane permeation in addition to ILs, particularly geranic acid’s ability to extract or fluidize lipids as previously reported (26).
Fig. 3. MD simulations establish enhanced lipid bilayer interactions and translocation mechanisms of the IL combination.
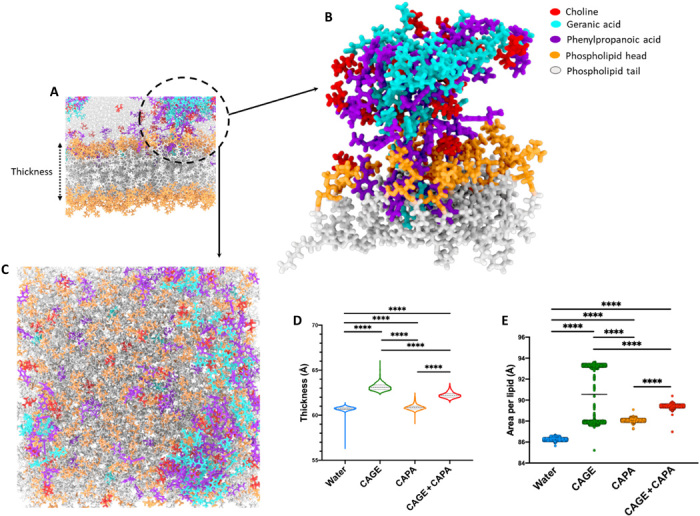
(A) Lipid bilayer simulation with the aggregates of choline, geranic acid, and phenylpropanoic acid highlighted with a circle. (B) Enlarged view of the ionic species from the circle depicting closed interaction of ionic species with the phospholipid heads and tails. The aggregate contains all three ionic species contributing to the interaction with the lipid membrane. (C) Representative snapshot viewing perpendicular to membrane in the plane of the lipid bilayer. (D and E) Average thickness of the lipid membrane (D) and average area per lipid (E) over the course of simulations in the presence of CAPA and the IL combination (CAGE and CAPA) in contrast to CAGE (control). All data are averages ± SEM and were determined to be nonparametric by normality test and statistics by Kruskal-Wallis test for (D) and (E). ****P < 0.0001.
The relative effect of the ILs including CAGE, CAPA, and CAGE + CAPA on membrane dynamics was assessed by measuring the average thickness of the lipid bilayer in the presence of ILs over a simulation time of 350 ns. The highest thickness was observed in the presence of CAGE (50% v/v), indicating greater IL intercalation within the lipid bilayer. Similar thickness was noted for the water and CAPA (50% v/v) groups, while CAGE (25% v/v) with CAPA (25% v/v) led to a higher thickness (Fig. 3D). The MD simulation snapshots highlight the dynamics of interactions of the individual ionic species in the IL with phospholipid membrane. Conclusive intercalation of the ionic species of the IL combination with the bilayer was detected (Fig. 3B). Furthermore, upon visualizing the trajectories of the individual ionic species within the CAGE + CAPA simulation, we were able to observe reduced mobility of geranic acid relative to phenylpropanoic acid (fig. S3). When focusing on an IL aggregate that consists of all three IL species (choline, geranic acid, and phenylpropanoic acid), we observed that each geranic acid molecule tends to remain in contact with the aggregate over the course of the simulation, while choline and phenylpropanoic acid are able to move between both the aggregate of heterogeneous species and the bulk solvent making up the rest of the system. This increase in mobility likely causes a change in the distribution of local viscosities across the system. When visualizing the head groups of lipids, which are in contact with the aggregate, we observe that the head groups occupy a larger area per lipid. This is demonstrated by a more “spread out” distribution of individual molecular trajectories within the area of the IL aggregate. This expansion of space between the lipids is caused by intercalation of the IL with the membrane and subsequent displacement of the lipid species. As the aggregation induces localization of the effects of IL on the bilayer membrane, it is likely that aggregation, with low constituent turnover with the bulk solvent, may lead to uneven membrane disruption as well as differences in the local viscosity. This heterogeneous distribution of membrane disruption may account for the wide distribution of area per lipid values seen over the course of the simulations in CAGE when compared with the other IL systems (Fig. 3E). Overall, these results signify the contribution of aggregate turnover for ILs in translocating RNA across lipid bilayers.
Biocompatibility of ILs in mice
The optimized CAGE + CAPA IL formulation was evaluated for toxicity in vivo in mice. IL–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) siRNA formulation (25 μl) was applied topically for four consecutive days to the dorsal skin of SKH-1 elite (SKH-1E) hairless mice (Fig. 4A). No signs of inflammation, redness, and/or irritation were observed for the IL-treated animals (fig. S4). Skin tissue was further harvested, and sections were cut and stained for histopathology and toxicology markers. Groups treated with the IL formulation exhibited no signs of epidermal thickening and keratinocyte hyperproliferation and were equivalent to the untreated and/or naked siRNA-treated animals (Fig. 4B and fig. S4). We also tested TNF-α gene expression levels in healthy mice. Animals treated with naked siRNA were statistically equivalent to the untreated animals. Mice treated with IL-GAPDH siRNA and IL–siCon (control siRNA used for subsequent experiments) demonstrated slightly lower TNF-α mRNA transcripts compared with the untreated group (fig. S4). Such inhibition of TNF-α expression might potentially arise from the inherent properties of the individual components of the ILs and need to be studied in future studies.
Fig. 4. IL-siRNA inhibits GAPDH expression following topical application without toxicity in mice.
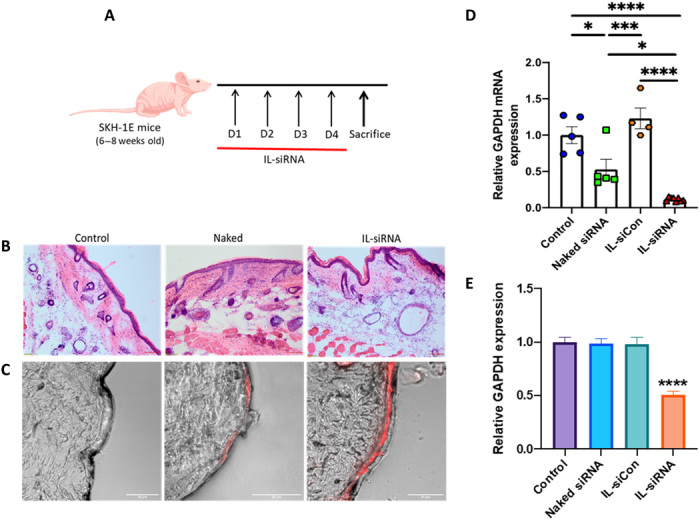
(A) Schematic illustration of the topical application schedule. (B) Representative histology [hematoxylin and eosin (H&E)] images of the skin tissue 5 days after topical application of IL-siRNA. Scale bars, 100 μm; magnification, ×10. (C) Confocal images of epidermal accumulation of Cy5-siRNA (red) with and without IL in a mouse skin tissue. Scale bars, 50 μm. (D) GAPDH mRNA expression was measured by qPCR. β-Actin mRNA expression was used for normalization. Data are averages ± SEM and were determined to be nonparametric by normality test and statistics by Kruskal-Wallis test. *P < 0.05, ***P < 0.001, and ****P < 0.0001. (E) GAPDH levels in the skin samples were determined using a GAPDH enzyme-linked immunosorbent assay. Data are averages ± SEM, statistics by one-way ANOVA with Tukey HSD posttest. ****P < 0.0001 (control, n = 5; naked siRNA, n = 5; IL-siCon, n = 4; IL-siRNA, n = 8). Photo credit: N.K., Harvard University.
IL-siRNA penetration and GAPDH silencing in healthy mice
Cy5 fluorescence within the epidermis was measured in healthy mice following transdermal application for four consecutive days. Confocal images revealed a marked increase in Cy5 fluorescence in the epidermis for the IL-treated group compared with the naked siRNA in mice (Fig. 4C). Upon determining the GAPDH gene silencing efficiency using quantitative polymerase chain reaction (qPCR), the expression levels of GAPDH were found to be reduced 4.5- and 8.6-fold for the IL-siRNA–treated group in contrast to the naked siRNA and untreated mice, respectively (Fig. 4D). A slight decrease in the GAPDH mRNA expression was also observed for the naked siRNA–treated group. Consecutively, it was necessary to ascertain if the change in GAPDH mRNA expression translated into protein reduction. Consistent with the gene knockdown results, the IL-siRNA–treated group demonstrated a statistically significant decay (⁓2-fold) in the GAPDH protein expression compared with all the other treatment groups (Fig. 4E). The reduced GAPDH mRNA expression for the naked siRNA–treated group did not down-regulate GAPDH protein expression.
Local NFKBIZ silencing in the skin inhibits imiquimod-induced psoriasis
The ability of NFKBIZ siRNA to treat psoriasis was tested using CAGE + CAPA as a topical formulation. Following induction of psoriasis and topical application of IL-NFKBIZ siRNA formulation (Fig. 5A), skin tissue was harvested and analyzed. Macroscopically, local knockdown of NFKBIZ in the dorsal skin markedly reduced imiquimod-induced inflammation, showing reduced erythema and scaling in the area where IL-NFKBIZ siRNA was applied compared with the untreated, IL-treated, and IL-siCon–treated groups (Fig. 5B and fig. S5). Hematoxylin and eosin (H&E) staining of skin sections from the mice revealed that the knockdown of NFKBIZ by IL-siRNA reduced epidermal thickening, acanthosis, hyperkeratosis, and club-shaped rete ridges (Fig. 5C and fig. S5). Likewise, immunohistochemistry (IHC) analysis revealed hyperproliferation of keratinocytes in the untreated, IL-treated, and IL-siCon–treated groups, whereas the group treated with IL-NFKBIZ siRNA exhibited lack of keratinocyte proliferation (Ki67 staining) (Fig. 5D and fig. S5). The common characteristic features of imiquimod-induced skin inflammation (erythema and scaling) were scored daily throughout the induction/application period. Individual scores for erythema and scaling demonstrated fair reduction starting from day 3 with topical IL-siRNA application (Fig. 5, E and F). Maximum cumulative scores were obtained for the untreated and IL-treated groups and were markedly lowered in the IL-siRNA–treated group (fig. S6). Double skin-fold thickness (DSFT) for measuring skin thickness did not yield major differences between the groups (fig. S6). In addition, the heat map and mRNA analyses indicated a substantial reduction in expression of NFKBIZ and other psoriasis-related gene products (fig. S7) in comparison with the untreated and IL-siCon–treated groups (Fig. 5, G to J, and fig. S8). Upon IL-siCon treatment, most genes were up-regulated, including NFKBIZ, TNF-α, cytokines (IL-17C, IL-19, IL-22, IL-36A, and IL-36G), chemokines (CCL20), and antimicrobial proteins (LCN2 and DEFB4) (Fig. 5G). Some down-regulation of TNF-α and IL-17A mRNA expression was observed in healthy mice upon treatment with IL alone (Fig. 5, I and J) and suggests additional inherent properties of ILs, which could be explored in future studies. Future studies should also focus on determining the dose-response curve of the therapeutic effect of siRNA-IL.
Fig. 5. Local inhibition of NFKBIZ by topical IL-siRNA suppresses imiquimod-induced psoriasis-like skin inflammation and other key psoriasis-associated genes.
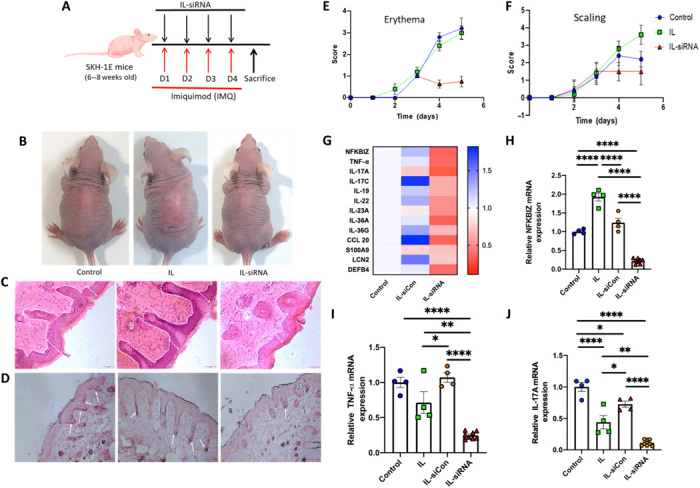
(A) Schematic illustration of the application schedule for disease induction and IL-siRNA topical administration. (B) Psoriasis-induced mice were treated topically with IL-NFKBIZ siRNA and were compared with untreated and IL-applied groups. (C) H&E staining of the psoriasis-induced skin sections from the mice with and without treatment. Scale bars, 50 μm; magnification, ×10. (D) Skin sections from the mice were analyzed for keratinocyte proliferation (proliferation marker, Ki67) by IHC. Scale bars, 100 μm. (E and F) Erythema and scaling scores obtained by blindly scoring using the human PASI scoring system daily on a scale from 0 (no alteration) to 4 (very distinct alteration). (G) Heat map for the expression levels of various psoriasis-associated genes following treatment with IL-NFKBIZ siRNA in comparison with the untreated (control) and IL-siCon–treated groups. (H to J) mRNA expression levels were measured by qPCR, and β-actin mRNA expression was used for normalization for NFKBIZ, TNF-α, and IL-17A, respectively. Data are averages ± SEM, statistics by one-way ANOVA with Tukey HSD posttest. *P < 0.05, **P < 0.01, and ****P < 0.0001 (control, n = 4; IL, n = 4; IL-siCon, n = 4; IL-siRNA, n = 8). Photo credit: (B) A.M., Harvard University; (C and D) N.K.
DISCUSSION
Limited understanding of key inflammatory signaling pathway regulators and the chronological order of the underlying mechanisms presents a challenge in the treatment of psoriasis. Signaling pathways including NF-κB, Janus kinase (JAK)/signal transducer and activator of transcription (STAT), and p38 mitogen-activated protein kinase have recently been found to play a major role in the pathogenesis of this complex disease (31). NFKBIZ, a gene encoding IκBζ, is a crucial transcriptional coactivator mediating downstream effects of an array of specific inflammatory cytokines and is particularly imperative in the light of recent findings by Johansen et al. (6) and Müller et al. (12), which indicated IκBζ to be a key modulator of IL-17A, IL-23, and IL-36 (32). Thus, targeting NFKBIZ/IκBζ to inhibit proinflammatory signaling pathways and production of psoriasis-related gene products is a viable strategy for psoriasis treatment. Clinically, antibodies targeting TNF-α and IL-17A have shown promise in meeting the primary endpoints and improving the disease condition (33). However, as biologics, these antibodies have challenges of potential systemic toxicity, generation of anti-antibodies, and high cost.
Here, we present an IL combination capable of improving epidermal accumulation and delivery of RNA through skin. We hypothesized that a combination of ILs would stabilize the siRNA and, at the same time, would improve its penetration. We validated this hypothesis in an imiquimod-induced psoriasis-like skin inflammation model that resembles plaque-type psoriasis in humans. Topical application of IL-siRNA for four consecutive days generated substantial reduction in the levels of inflammatory cytokines and an array of psoriasis-related gene products.
CAGE + CAPA IL formulation offers several advantages over other transdermal drug delivery systems. The components of the IL formulation, choline bicarbonate, geranic acid, and phenylpropanoic acid, have been proven safe or GRAS (generally recognized as safe) chemicals and provide a strong foundation for the safety of ILs. In addition, simple synthesis and scale-up processes, high solvating power, and tunability offer additional advantages over other volatile organic solvents. This system is particularly suitable for transdermal delivery of nucleic acids due to both its complex intercalation between the stacked RNA base pairs and aromatic rings of the IL, and enhanced interaction with the lipid bilayer.
Our results demonstrate that ILs can complex with nucleic acids without compromising the bioactivity, thus making them ideal for transdermal drug delivery. The salt metathesis or anion exchange reaction for IL synthesis is particularly advantageous because it does not require integration of harsh organic solvents for siRNA delivery. The individual IL components could be modulated to interact with nearly any nucleic acid based on the binding characteristics and molecular mechanism of interactions.
Tunable ion stoichiometry and physicochemical properties are other key features of IL-based systems. Previous work in our group has indicated the role of interionic interactions in solvation and partitioning of the active ingredient into the skin (34). In addition, Chandran et al. (35) have demonstrated the importance of electrostatic interactions and groove binding associations of ILs in DNA stability. Hitherto, the role of ILs in improving the stability and solvation of siRNA has not been comprehensively explored. We systematically varied the anionic component of the IL with structural similarity to geranic acid and/or containing an aromatic ring at a stoichiometry ratio of 1:2 and developed an in-house cholinium-based IL library. We observed that the anions of the ILs that contained aromatic rings generally solidified or formed a gel at RT except phenylpropanoic acid. Excellent siRNA stability was observed in the presence of CAVA, CAPA, and CAGE + CAPA in comparison to other ILs and combinations, possibly due to superior interactions with the siRNA. The IL combination CAGE + CAPA generated the highest epidermal accumulation of siRNA, notably higher than any individual ILs and/or combination.
The best performing IL combination that we identified in this study, CAGE + CAPA, demonstrated consistent distribution of the three ionic species through MD simulations, indicating improved molecular mobility and lower viscosity contributing to enhanced solvation effects. Furthermore, MD simulation snapshots revealed close association of phenylpropanoic acid with the RNA molecules, which could be possibly attributed to a combination of hydrophobic and polar interactions, π-π stacking, and/or intercalation between stacked RNA base pairs, leading to enhanced RNA stability. RGYR and RMSD measurements obtained from simulations over the course of 500 ns further confirmed improved IL-RNA interactions.
It is also important to understand the magnitude of IL-mediated lipid bilayer modulation. MD simulations revealed the crucial role of aggregation of ionic species in improving membrane permeation with the highest bilayer thickness obtained for CAGE (50% v/v) followed by CAGE + CAPA. Such observations from the simulations further establish geranic acid as the main driver in the translocation of the IL combination through the lipid bilayers, which is consistent with experimental results. While it seems that phenylpropanoic acid has a minor role in improving bilayer permeation by lowering the local viscosity of the overall IL system, we believe that it is also responsible for fluidizing the membrane with the formation of dynamic pores. It was earlier reported that deprotonated aromatic carboxylic acids, such as phenylpropanoic acid, permeate bilayers several orders of magnitude faster than that expected from the pH partition hypothesis, and their permeation is fully controlled by the anions at the physiological pH (36). On the basis of these results, we speculate that these ILs assist in crossing the cellular barriers to deliver siRNA into the cytosolic compartments.
To assess biocompatibility of CAGE + CAPA, we conducted a histological evaluation of skin on the fifth day, which coincided with the total duration of topical application. We did not observe any macroscopic changes in the skin structure, epidermal thickening, and keratinocyte proliferation in the IL-treated groups. Further investigation of inflammatory cytokine levels did not reveal any statistically significant increment in TNF-α mRNA compared with the untreated groups. Some of the IL-treated groups demonstrated a decrease in the TNF-α mRNA levels, which might be possibly due to the presence of IL and need to be further investigated. Marked inhibition of GAPDH mRNA and protein expression was observed in the IL-GAPDH siRNA–treated groups.
NFKBIZ has been previously demonstrated to play a crucial role in the gene transcription of several proinflammatory cytokines and antimicrobial peptides responsible for the pathogenesis of psoriasis (6, 12). Using an imiquimod-induced psoriasis model, we further demonstrated that local silencing of NFKBIZ following topical application of IL-NFKBIZ siRNA formulation impaired expression of psoriasis-related gene products under in vivo conditions. IL-siRNA–treated mice exhibited substantially reduced skin pathology including reduced erythema and scaling, less epidermal thickening, and keratinocyte proliferation. The local increase in mRNA levels of some of the inflammatory cytokines and related gene products for the IL-siCon– and IL-treated groups in comparison with the untreated group could be attributed to imiquimod. Local silencing of NFKBIZ resulted in a strong inhibition of crucial proinflammatory cytokine mRNA levels including IL-17A, IL-23, and IL-36. The downstream effects of local NFKBIZ silencing were also validated and are consistent with the previously reported effects of intradermal injection of IκBζ siRNA (6). Because mouse skin is generally much more permeable than human skin, detailed studies of quantification of skin penetration were not performed in vivo.
In summary, we have developed a transdermal IL platform capable of delivering RNA to the epidermis and have combined this framework with an array of gene screening to support NFKBIZ as a key signaling target gene in psoriasis treatment. The IL formulation retained the bioactivity of the siRNA and generated notable target gene abrogation upon topical application in an imiquimod-induced psoriasis-like skin inflammation model. The optimized IL formulation did not show toxicity and is acceptable for repeated applications. This platform is amenable to broad applications to nucleic acids and can be easily manufactured and scaled up. This platform could empower transdermal drug delivery for the treatment of dermatological conditions and help augmenting long-term therapeutic efficacy by targeting such common mediators. Upon further studies focused on assessment in large animals and comparison with clinical standard of care, the method described here opens a potential option for psoriasis treatment.
MATERIALS AND METHODS
Skin-penetrating IL-RNA complexes
The cholinium-based IL library was synthesized as described previously (26). Briefly, the cation, choline bicarbonate, and various anions were mixed at a 1:2 ratio to prepare ILs following salt metathesis reaction. The anions were dissolved in a minimum volume of ultrapure water or ethanol/methanol based on the solubility and were reacted with choline bicarbonate at 40°C for 24 hours. The resulting IL solution was dried using a rotary evaporator at 20 mbar at 60°C for 2 hours. The residual water was removed in a vacuum oven at 60°C for 48 hours. The ILs that were viscous at RT were characterized via NMR with dimethyl sulfoxide (DMSO)–d6 on an Agilent DD2 600-MHz spectrometer (Supplementary Materials and Methods). ILs were mixed with RNA (100 μM) at a volumetric ratio of 1:1 and incubated for 30 min at RT. The RNA-IL solutions (1 ml) were dialyzed against 10 mM sodium phosphate buffer for 72 hours using Dialysis Cassettes (10,000 molecular weight cutoff, Invitrogen). The concentration of RNA was confirmed and normalized using a NanoDrop instrument (Thermo Fisher Scientific). The stability of the RNA in the IL solution was determined using CD and gel electrophoresis (Supplementary Materials and Methods).
MD simulation studies
MD simulations were performed using OpenMM MD package and the AMBER force fields ff14SB and GAFF. Three-dimensional SD files for each of the IL species were downloaded from PubChem and parameterized with Antechamber before preparing simulation input topologies with LEaP. To generate starting coordinates for the lipid membrane simulations, PACKMOL was used to build a bilayer consisting of 100 phosphatidylcholine (POPC) molecules for each of the leaflets. The remaining contents of a 60-Å cube consisted of ~1:1 water (TIP3P) and IL, charge balanced with Na+ and Cl−. A 500-ns simulation was performed for each of the systems under periodic boundary conditions. For the simulations of siRNA, a helical starting structure for the nucleic acid was generated with Avogadro (37) before being placed in a simulation box consisting of ~1:1 water and IL for simulation under periodic boundary conditions for 350 ns. Analysis of MD trajectories was performed using the python library MDAnalysis for RGYR and RMSD of siRNA. Visual molecular dynamics (38) plugin MEMBPLUGIN (39) was used to perform analysis of membrane trajectories.
Skin penetration studies
Skin penetration studies were performed using porcine skin in FDCs, as described previously (40). A total volume of 20 μl of Cy5-labeled RNA (50 μM) in IL solutions was applied to the porcine skin surface and was incubated at 40°C for 24 hours under occlusive conditions with moderate stirring. The skin permeability of RNA was visualized and quantified using confocal microscopy and tape-stripping techniques, respectively (Supplementary Materials and Methods).
Animal studies
All animal studies were performed at the Harvard John A. Paulson School of Engineering and Applied Sciences, Harvard University. Procedures and studies conducted were approved by the Institutional Animal Care and Use Committee of the Faculty of Arts and Sciences, Harvard University, and were consistent with all applicable regulations. ILs carrying GAPDH (custom siRNA, sense seq: 5′-GUGUGAACCACGAGAAAUAUU-3′’, antisense seq: 5′- AAUAUUUCUCGUGGUUCACAC-3′; Dharmacon), siCon (catalog no. D-001810-02-50; Dharmacon), and NFKBIZ siRNAs (catalog no. J-040680-06-0050; Dharmacon) were applied topically to healthy and imiquimod-treated SKH-1E hairless mice (Charles River), respectively. A blind scoring system similar to the human Psoriasis Area and Severity Index (PASI) score was used to measure the degree of severity, erythema, and scaling on the back of mice. In addition, skin thickness was monitored by the DSFT of dorsal skin of the mice with caliper measurements throughout the disease induction and treatment period (Supplementary Materials and Methods).
Quantification of mRNA transcripts
Flash-frozen skin tissues were pulverized to form a powder and homogenized in QIAzol Lysis Reagent to prepare the tissue lysates for qPCR. The mRNA levels were quantified and normalized following the manufacturer’s protocol. The relative abundance of mRNA transcripts and silencing in treated groups was normalized to the housekeeper gene (β-actin). The mean normalized siRNA treatment values were then plotted with their SEM (Supplementary Materials and Methods).
Statistical analysis
One-way analysis of variance (ANOVA) and statistical analyses were performed using GraphPad Prism software (GraphPad Software Inc.). Results are depicted as average ± SEM. Two-tailed Student’s t test was used for comparison between two groups. Parametric data were analyzed by one-way ANOVA followed by Tukey’s honestly significant difference (HSD) post hoc tests. Kruskal-Wallis tests were performed for nonparametric data. Statistical tests are indicated in the figures. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We acknowledge the assistance from the Department of Stem Cell and Regenerative Biology, Bauer Core Facility, and Wyss Institute for Biologically Inspired Engineering for the core facilities. Special thanks to R. Bronson for histological analyses, Y. Gao for NMR analyses, and J. Kim, Z. Zhao, and V. Krishnan for scientific discussions. Schematic representations were generated with the support of Biorender (©BioRender - biorender.com) and Servier Medical Art (smart.servier.com). Funding: This work was financially supported by Leo Foundation and Wyss Institute for Biologically Inspired Engineering at Harvard University. Author contributions: A.M. and S.M. designed the experiments and wrote the manuscript. A.M., N.K.,and V.D. conducted the experiments. A.M. and N.K. contributed to animal studies and subsequent analyses. A.M., A.U., and S.M. analyzed the experiments. C.R. performed MD simulation studies and subsequent analyses with support from D.E.I. Competing interests: S.M. and A.M. are inventors on a patent application filed by Harvard University that covers the ILs described herein. S.M. is a shareholder and board member/consultant to Liquideon, CAGE Bio, and i2O Therapeutics. A.M. is an employee and shareholder of CAGE Bio. There is a provisional patent application related to this work (U.S. patent application no. 62/939,088) on which S.M. and A.M. are inventors (owned by Harvard University), and a U.S. patent 10,449,254 on which S.M. is an inventor (owned by the University of California, Santa Barbara, CA). In addition to the competing interests mentioned (S.M. and A.M.), the authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/30/eabb6049/DC1
REFERENCES AND NOTES
- 1.Brezinski E. A., Dhillon J. S., Armstrong A. W., Economic burden of psoriasis in the United States: A systematic review. JAMA Dermatol. 151, 651–658 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Smale S. T., Hierarchies of NF-κB target-gene regulation. Nat. Immunol. 12, 689–694 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan S., Xu Z., Lou F., Zhang L., Ke F., Bai J., Liu Z., Liu J., Wang H., Zhu H., Sun Y., Cai W., Gao Y., Su B., Li Q., Yang X., Yu J., Lai Y., Yu X.-Z., Zheng Y., Shen N., Chin Y. E., Wang H., NF-κB-induced microRNA-31 promotes epidermal hyperplasia by repressing protein phosphatase 6 in psoriasis. Nat. Commun. 6, 7652 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pabon N. A., Zhang Q., Cruz J. A., Schipper D. L., Camacho C. J., Lee R. E. C., A network-centric approach to drugging TNF-induced NF-κB signaling. Nat. Commun. 10, 860 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veilleux M. S., Shear N. H., Biologics in patients with skin diseases. J. Allergy Clin. Immunol. 139, 1423–1430 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Johansen C., Mose M., Ommen P., Bertelsen T., Vinter H., Hailfinger S., Lorscheid S., Schulze-Osthoff K., Iversen L., IκBζ is a key driver in the development of psoriasis. Proc. Natl. Acad. Sci. U.S.A. 112, E5825–E5833 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sano S., Chan K. S., Carbajal S., Clifford J., Peavey M., Kiguchi K., Itami S., Nickoloff B. J., DiGiovanni J., Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nat. Med. 11, 43–49 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Tsoi L. C., Spain S. L., Ellinghaus E., Stuart P. E., Capon F., Knight J., Tejasvi T., Kang H. M., Allen M. H., Lambert S., Stoll S. W., Weidinger S., Gudjonsson J. E., Koks S., Kingo K., Esko T., Das S., Metspalu A., Weichenthal M., Enerback C., Krueger G. G., Voorhees J. J., Chandran V., Rosen C. F., Rahman P., Gladman D. D., Reis A., Nair R. P., Franke A., Barker J. N. W. N., Abecasis G. R., Trembath R. C., Elder J. T., Enhanced meta-analysis and replication studies identify five new psoriasis susceptibility loci. Nat. Commun. 6, 7001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaffen S. L., Jain R., Garg A. V., Cua D. J., The IL-23–IL-17 immune axis: From mechanisms to therapeutic testing. Nat. Rev. Immunol. 14, 585–600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson N. J., Boniface K., Chan J. R., McKenzie B. S., Blumenschein W. M., Mattson J. D., Basham B., Smith K., Chen T., Morel F., Lecron J.-C., Kastelein R. A., Cua D. J., McClanahan T. K., Bowman E. P., de Waal Malefyt R., Development, cytokine profile and function of human interleukin 17–producing helper T cells. Nat. Immunol. 8, 950–957 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Zheng Y., Danilenko D. M., Valdez P., Kasman I., Eastham-Anderson J., Wu J., Ouyang W., Interleukin-22, a TH17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature 445, 648–651 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Müller A., Hennig A., Lorscheid S., Grondona P., Schulze-Osthoff K., Hailfinger S., Kramer D., IκBζ is a key transcriptional regulator of IL-36–driven psoriasis-related gene expression in keratinocytes. Proc. Natl. Acad. Sci. U.S.A. 115, 10088–10093 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prausnitz M. R., Langer R., Transdermal drug delivery. Nat. Biotechnol. 26, 1261–1268 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy S. D., Flynn G. L., Transdermal delivery of narcotic analgesics: comparative permeabilities of narcotic analgesics through human cadaver skin. Pharm. Res. 6, 825–832 (1989). [DOI] [PubMed] [Google Scholar]
- 15.Prausnitz M. R., Mitragotri S., Langer R., Current status and future potential of transdermal drug delivery. Nat. Rev. Drug Discov. 3, 115–124 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Zheng D., Giljohann D. A., Chen D. L., Massich M. D., Wang X.-Q., Iordanov H., Mirkin C. A., Paller A. S., Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proc. Natl. Acad. Sci. U.S.A. 109, 11975–11980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiraja C., Zhu Y., Lio D. C. S., Yeo D. C., Xie M., Fang W., Li Q., Zheng M., Van Steensel M., Wang L., Fan C., Xu C., Framework nucleic acids as programmable carrier for transdermal drug delivery. Nat. Commun. 10, 1147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng Y., Chen J., Zhao Y., Yan X., Zhang L., Choy K., Hu J., Sant H. J., Gale B. K., Tang T., Transdermal delivery of siRNA through microneedle array. Sci. Rep. 6, 21422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang G., Zhang Y., Gu Z., Punching and electroporation for enhanced transdermal drug delivery. Theranostics 8, 3688–3690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan R., Chen M., Sun S., Wei P., Zou L., Liu J., Gao D., Wen L., Ding W., Topical and targeted delivery of siRNAs to melanoma cells using a fusion peptide carrier. Sci. Rep. 6, 29159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen M., Zakrewsky M., Gupta V., Anselmo A. C., Slee D. H., Muraski J. A., Mitragotri S., Topical delivery of siRNA into skin using SPACE-peptide carriers. J. Control. Release 179, 33–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin C.-M., Huang K., Zeng Y., Chen X.-C., Wang S., Li Y., A simple, noninvasive and efficient method for transdermal delivery of siRNA. Arch. Dermatol. Res. 304, 139–144 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Thanik V. D., Greives M. R., Lerman O. Z., Seiser N., Dec W., Chang C. C., Warren S. M., Levine J. P., Saadeh P. B., Topical matrix-based siRNA silences local gene expression in a murine wound model. Gene Ther. 14, 1305–1308 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Kasiewicz L. N., Whitehead K. A., Lipid nanoparticles silence tumor necrosis factor α to improve wound healing in diabetic mice. Bioeng. Transl. Med. 4, 75–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agatemor C., Ibsen K. N., Tanner E. E. L., Mitragotri S., Ionic liquids for addressing unmet needs in healthcare. Bioeng. Transl. Med. 3, 7–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zakrewsky M., Lovejoy K. S., Kern T. L., Miller T. E., le V., Nagy A., Goumas A. M., Iyer R. S., Del Sesto R. E., Koppisch A. T., Fox D. T., Mitragotri S., Ionic liquids as a class of materials for transdermal delivery and pathogen neutralization. Proc. Natl. Acad. Sci. U.S.A. 111, 13313–13318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Institute of Medicine (U.S.) Commitee on Military Nutrition Research, in Food Components to Enhance Performance: An Evaluation of Potential Performance-Enhancing Food Components for Operational Rations, B. M. Marriott, Ed. (National Academies Press, Washington (DC, 1994). [PubMed] [Google Scholar]
- 28.Qi Q. M., Mitragotri S., Mechanistic study of transdermal delivery of macromolecules assisted by ionic liquids. J. Control. Release 311–312, 162–169 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Tanner E. E. L., Ibsen K. N., Mitragotri S., Transdermal insulin delivery using choline-based ionic liquids (CAGE). J. Control. Release 286, 137–144 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Banerjee A., Ibsen K., Brown T., Chen R., Agatemor C., Mitragotri S., Ionic liquids for oral insulin delivery. Proc. Natl. Acad. Sci. U.S.A. 115, 7296–7301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowes M. A., Suárez-Fariñas M., Krueger J. G., Immunology of psoriasis. Annu. Rev. Immunol. 32, 227–255 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahil S. K., Catapano M., di Meglio P., Dand N., Ahlfors H., Carr I. M., Smith C. H., Trembath R. C., Peakman M., Wright J., Ciccarelli F. D., Barker J. N., Capon F., An analysis of IL-36 signature genes and individuals with IL1RL2 knockout mutations validates IL-36 as a psoriasis therapeutic target. Sci. Transl. Med. 9, eaan2514 (2017). [DOI] [PubMed] [Google Scholar]
- 33. Trial watch: Targeting IL-17A shows broad promise in autoimmune diseases. Nat. Rev. Drug Discov. 9, 908 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Tanner E. E. L., Curreri A. M., Balkaran J. P. R., Selig-Wober N. C., Yang A. B., Kendig C., Fluhr M. P., Kim N., Mitragotri S., Design principles of ionic liquids for transdermal drug delivery. Adv. Mater. 31, e1901103 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Chandran A., Ghoshdastidar D., Senapati S., Groove binding mechanism of ionic liquids: A key factor in long-term stability of DNA in hydrated ionic liquids? J. Am. Chem. Soc. 134, 20330–20339 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Thomae A. V., Wunderli-Allenspach H., Krämer S. D., Permeation of aromatic carboxylic acids across lipid bilayers: The pH-partition hypothesis revisited. Biophys. J. 89, 1802–1811 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanwell M. D., Curtis D. E., Lonie D. C., Vandermeersch T., Zurek E., Hutchison G. R., Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J Cheminform. 4, 17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Guixà-González R., Rodriguez-Espigares I., Ramírez-Anguita J. M., Carrió-Gaspar P., Martinez-Seara H., Giorgino T., Selent J., MEMBPLUGIN: Studying membrane complexity in VMD. Bioinformatics 30, 1478–1480 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Hsu T., Mitragotri S., Delivery of siRNA and other macromolecules into skin and cells using a peptide enhancer. Proc. Natl. Acad. Sci. U.S.A. 108, 15816–15821 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/30/eabb6049/DC1


