Abstract
Identifying and visualizing transcriptionally similar cells is instrumental for accurate exploration of cellular diversity revealed by single-cell transcriptomics. However, widely used clustering and visualization algorithms produce a fixed number of cell clusters. A fixed clustering “resolution” hampers our ability to identify and visualize echelons of cell states. We developed TooManyCells, a suite of graph-based algorithms for efficient and unbiased identification and visualization of cell clades. TooManyCells introduces a novel visualization model built on a concept intentionally orthogonal to dimensionality reduction methods. TooManyCells is also equipped with an efficient matrix-free divisive hierarchical spectral clustering wholly different from prevalent single-resolution clustering methods. Together, TooManyCells enables multi-resolution and multifaceted exploration of single-cell clades. An advantage of this paradigm is the immediate detection of rare and common populations that outperforms popular clustering and visualization algorithms as demonstrated using existing single-cell transcriptomic data sets and new data modeling drug resistance acquisition in leukemic T cells.
Introduction
Transcription is an important contributor to phenotypic and functional cell states. Emergent technologies such as single-cell RNA sequencing (scRNA-seq) have markedly improved identification and characterization of cell state heterogeneity. To this end, algorithms for unsupervised delineation and visualization of cells with similar expression patterns have improved the understanding of cell lineage complexity, tumor heterogeneity, and diversity of response to oncology drugs1–5. Nevertheless, it remains challenging to simultaneously stratify rare and common cell populations and explore their relationships.
Clustering algorithms have been proposed to partition scRNA-seq data to identify groups of cells with related transcriptional programs1,6–10. In most scRNA-seq analyses, the identified cell clusters are visualized using dimensionality reduction algorithms such as t-SNE or UMAP11–13. These workflows produce and visualize single-resolution cell clustering using methods that mostly lack quantitative presentation of relationships among the clusters.
Resolution of cell state stratification unduly influences findings in scRNA-seq experiments. For instance, a resolution separating lymphocytes from monocytes may not readily subdivide various lymphocyte lineages. Given that varying cell states are inherently nested, we postulated that algorithms delineating hierarchies of groups and visualizing their relationships can be used to effectively interrogate echelons of cell states. To this end, we developed TooManyCells for scRNA-seq data visualization and exploration. TooManyCells implements a suite of novel graph-based algorithms and tools for efficient, global, and unbiased identification and visualization of cell clades. TooManyCells maintains and presents cluster relationships within and across varying clustering resolutions, and enables delineation of context-dependent rare and abundant cell populations.
We demonstrated the effectiveness of TooManyCells in reliably identifying and clearly visualizing abundant and rare subpopulations using several analyses. Three publicly available scRNA-seq data sets, synthetic data, and controlled subsetting and mixing experiments of single-cell populations were used for comparative benchmarking. TooManyCells outperforms other popular methods to detect and visualize rare populations down to the smallest tested benchmark of 0.5% prevalence in several controlled cell admixtures and simulated data. Additionally, TooManyCells assisted in a fine-grain B cell lineage stratification within mouse splenocytes and was able to identify rare plasmablasts14 that were overlooked by popular Louvain-based clustering and projection-based visualization algorithms.
We further used TooManyCells to explore the effect of dosage on acquiring resistance to a gamma-secretase inhibitor (GSI), a targeted Notch signaling antagonist. While other popular methods failed, TooManyCells revealed a rare resistant-like subpopulation of parental cells. TooManyCells and its individual components are available through https://github.com/faryabib/too-many-cells.
Results
TooManyCells for visualization of cell clade relationships.
Clear visualization is critical for scRNA-seq data exploration and is dominated by projection-based algorithms such as t-SNE and UMAP. For large and complex cell admixtures, projection methods suffer from rendering many overlapping cells that overwhelms the single-cell resolution visualization. More importantly, these algorithms generally do not report quantitative inter-cluster relationships and lack interpretable visualizations across clustering resolutions. To address these limitations, we developed TooManyCells for fully customizable visualization of inter-cluster relationships in a tree data abstraction (Figure 1).
Figure 1:
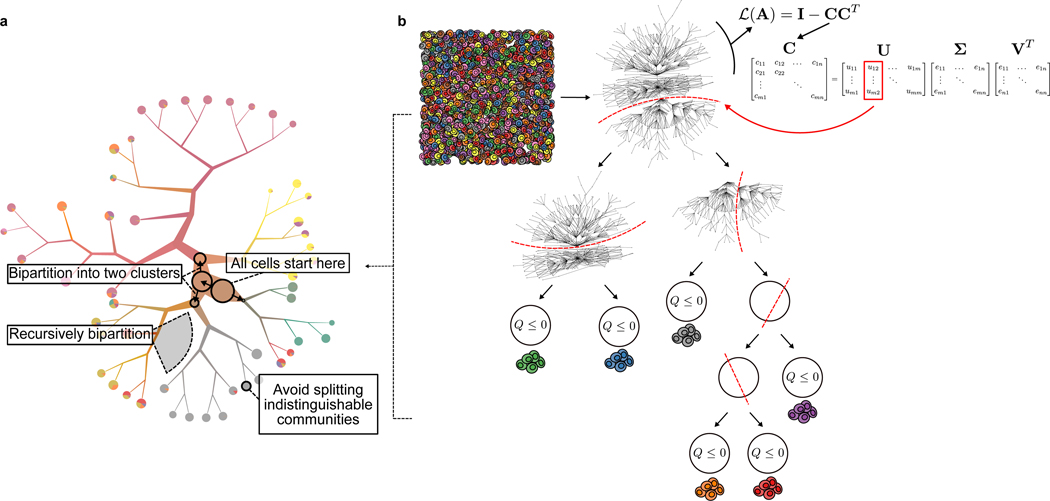
The TooManyCells visualization and clustering algorithms. (a) TooManyCells visualizes inter-cluster relationships while providing many capabilities and options including, but not limited to, weighted average blending of colors, scaling branches, modularity overlays, smart tree pruning, and several leaf node visualizations. Cells from 11 mouse organs are color coded based on their organ-of-origin. (b) TooManyCells matrix-free divisive hierarchical spectral clustering. TooManyCells is conceptually similar to recursive separation of cells based on their color (state/type) similarities, first separating green and blue from red, purple, orange, and gray cells, followed by separation of green from blue, gray from red, purple, and orange, etc. The network of cells (nodes) connected by their cosine similarities (edges) is recursively bipartitioned (red dashed lines) using truncated singular value decomposition (SVD) of the transformed matrix C that is directly calculated from the gene expression matrix. Here, truncated SVD only calculates the first two left singular vectors corresponding to the two largest singular values instead of full matrix factorization. This “matrix-free” process eliminates the need for the explicit calculation of cell-cell similarity (A) and the normalized Laplacian (L(A)) matrices followed by full eigenvalue decomposition (calculation of all the matrices on the right hand side of the equation instead of only the red-marked column) at each bipartitioning. Recursive bipartitioning is terminated when a candidate split results in non-positive Newman-Girvan modularity (Q).
Multiple algorithms use traditional dendrogram plots to infer cell clades from scRNA-seq profiles15–18. Yet, robust cell clade inference remains challenging. Alternatively, outputs of flat clustering algorithms at different resolutions can be related in a tree structure18; however, this method relies on arbitrary numbers of resolutions and tuning parameters. We reasoned that divisive hierarchical spectral clustering can overcome these limitations by using all information embedded in the cell-cell similarity graph. To enable efficient generation of the hierarchy, TooManyCells implements a transformation of the gene expression matrix that eliminates the explicit calculation of cell-cell similarity and Laplacian matrices followed by full matrix factorization, which were otherwise required for finding the most informative bipartition of cells at each branching point (Figure 1b). This novel “matrix-free” approach substantially improves the memory and time requirements of divisive clustering and recursively identifies candidate bipartitions to create a hierarchy of cell clades. By using Newman-Girvan modularity19 as a stopping criteria instead of an optimization parameter, TooManyCells bypasses limitations associated with heuristic global optimization-based clustering such as Louvain-based algorithms20,21, avoids creating arbitrarily small clusters, and allows simultaneous detection of large and small clusters (see Online Methods and Supplementary Note 1).
For clear and interpretable displays of cell clades, TooManyCells is designed with many features that facilitate data exploration and assist with finding relevant populations, including branch scaling, weighted average color blending, and statistically-driven tree pruning (Figure 2 and Supplementary Note 1). To enhance data visualization versatility and complement existing single-resolution methods, TooManyCells can display any tree data structure and outputs of other clustering algorithms (Figures S1 – S4). To this end, TooManyCells produces visually informative hierarchies of nested cell clusters. Inner nodes are clusters at a given resolution and leaf nodes are finer-grain clusters, where additional bipartitioning would be as informative as random bipartitioning. To enable an end-to-end built-in scRNA-seq analysis solution, we also equipped TooManyCells with a suite of tools and functionalities including, but not limited to, data normalizations, data filtrations, similarity measure calculation, subtree generation, differential expression, data import/export, and novel algorithms for scRNA-seq diversity quantification and rarefaction analysis (Figure S5, Online Methods, and Supplementary Note 2).
Figure 2:
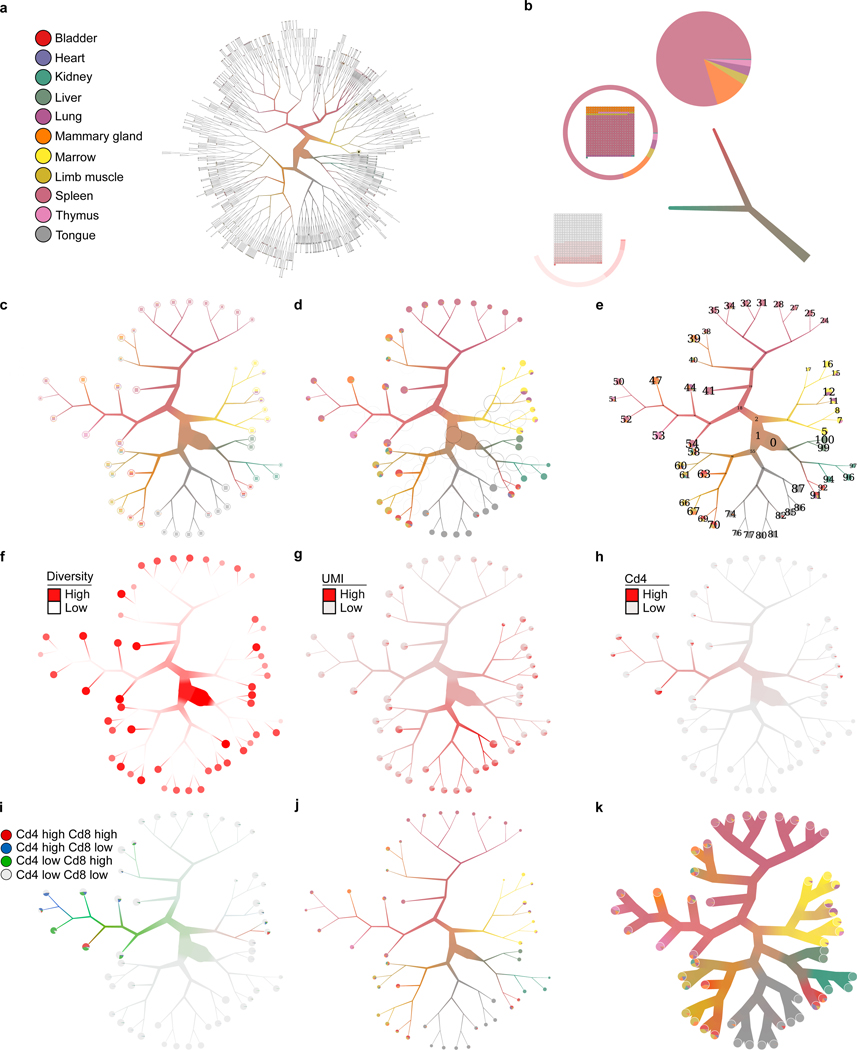
Example of TooManyCells visualization capabilities using 11 mouse organs. (a) The complete tree with default settings. (b) Different leaf rendering options (clockwise from bottom: gene expression, “pie ring”, pie chart) and example of scaling and average weighted color blending for branches. (c) Tree from (a) pruned with median(node size)+3*MAD(node size), which is used in panels d to k. (d) Tree with modularity of bipartitioning at each internal node displayed as black circles, where higher modularity is represented by darker circumference intensity. (e) Tree with numbered nodes. (f) Color-coded tree with a continuous variable (e.g. cell diversity of organs, increasing color intensity represents increasing diversity). For clarity, inner and leaf nodes use different intensity scales. (g) Color-coded tree with a discrete variable presenting UMI counts. (h) Color-coded tree with expression level of a specific gene (Cd4 expression level). (i) Color-coded tree with expression level of multiple genes (Cd4 and Cd8 expression levels). (j) Tree with non-default scaling width. (k) Tree with disabled branch scaling.
TooManyCells efficiently identifies pure cell clusters.
To assess TooManyCells’ performance, we first used the Tabula Muris data sets22 to examine the extent of cell homogeneity in cluster identification. As part of the Tabula Muris, 11 organs of three month old mice were profiled by scRNA-seq, and their cell type composition was determined using organ-specific optimized analyses22. TooManyCells clusters were compared with the clusters generated by widely used Cell Ranger23, Monocle8, Phenograph6, Seurat7, RaceID24, CIDR16, and BackSPIN15 algorithms, the latter two being agglomerative and divisive hierarchical algorithms, respectively (Figures 3a–d and Supplementary Note 3).
Figure 3:
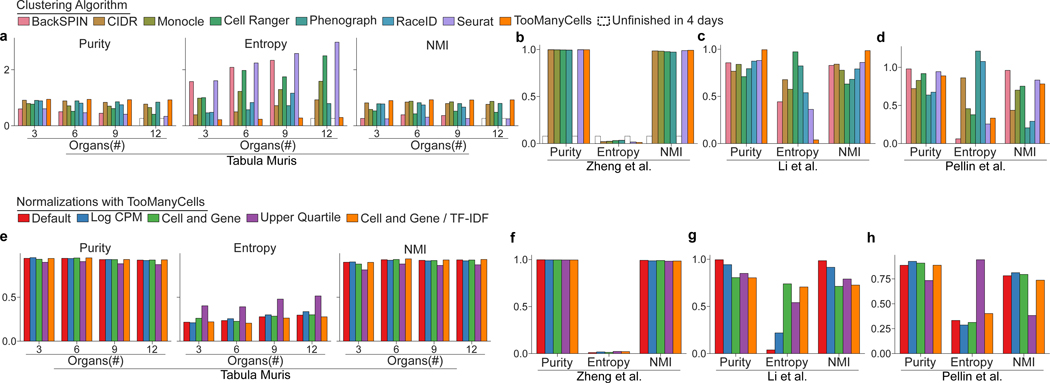
Comparative analysis of clustering performance and scalability. (a) From Tabula Muris22 (n = 41,689 cells), the population analyzed is represented by the number of organs on the x-axis taken from the left of the ordering {Thymus, Spleen, Bone marrow, Limb muscle, Tongue, Heart, Lung, Mammary gland, Bladder, Kidney, Liver}. Purity (left), entropy (middle), or normalized mutual information (NMI, right) performance measures are calculated. Higher purity and NMI represent higher accuracy, while lower entropy indicates better performance. Default or suggested filterings and parameters were used for all algorithms (see Online methods). A dashed border indicates the algorithm did not complete within four days. (b-d) Same measures as (a) are used to compare clustering accuracy based on data from Zheng et al.23 (n = 23,910 cells) (b), Li et al.17 (n = 561 cells) (c), and Pellin et al.25 (n = 2,815 cells) (d). (e-h) Equivalent data sets as (a-d) for different TooManyCells normalization procedures: the TooManyCells default normalization (Default) term frequency-inverse document frequency (tf-idf), Log Counts-Per-Million (Log CPM), total and gene median count normalization (Cell and Gene), upper quartile normalization (Upper Quartile), total and gene median count normalization followed by tf-idf normalization (Cell and Gene / TF-IDF).
For each algorithm, default or suggested filters and parameters were considered (see Online Methods). The first comparative analysis was performed based on an increased level of cell mixture complexity, where the first 3, 6, 9, and finally all 11 data sets from thymus, spleen, bone marrow, limb muscle, tongue, heart, lung, mammary gland, bladder, kidney, and liver were considered (Figures 3a and S6). Further comparisons were carried out using three additional data sets of cell lines or fluorescence activated cell sorting (FACS)-purified cells: CD14+ monocytes, CD19+ B, and CD4+ T cells23 (Figure 3b), seven cancer lines17 (Figure 3c), and B lymphocytes/natural killer, megakaryocyte-erythroid, and granulocyte–monocyte progenitors25 (Figure 3d).
Rare-cell-clustering RaceID and hierarchical CIDR and BackSPIN methods failed to finish analyses of the high complexity data sets of ~30,000 to 40,000 cells within four days (Figures 3a and 3b). Across all complexities and evaluation metrics in the Tabula Muris data sets, TooManyCells was the most successful in separating cell type labels (Figure 3a). All the scalable algorithms that clustered the immune cells generally performed well. However, TooManyCells again marginally outperformed all others (Figure 3b). Similarly, TooManyCells performed the best in separating seven distinct cancer cell lines (Figure 3c). However, TooManyCells was close with Seurat and Cell Ranger in separating lineage negative hematopoietic progenitor cells (Figure 3d). We note that these cells are highly heterogeneous and their population structures, defined by a few cell surface markers, remain enigmatic25,26. Comparison of different normalization procedures showed that TooManyCells’ performance was only marginally influenced by normalization choice (Figures 3e–h and Supplementary Note 4).
While not scalable to large data sets (Figure 3a), BackSPIN, another divisive clustering algorithm, exhibited the best performance in separating highly diverse hematopoietic progenitor cells (Figure 3d). Importantly, all the scalable algorithms only report single-resolution cluster outputs at a time, while TooManyCells’ multilayer output identifies context-dependent clades from the entire presented cluster hierarchy. The TooManyCells-rendered cluster tree further guides the choice of clustering granularity by contextualizing cluster features such as relative size, modularity (Figure 2d), and distance from the root. This unique TooManyCells feature sets it apart from existing visualization algorithms that lack interpretable rendering of relationships across varying clustering resolutions. Furthermore, the run time of TooManyCells’ multi-resolution clustering was comparable to run times of single-resolution clustering algorithms for small data sets (Figure S7 and Supplementary Note 5), and markedly outperformed them for large data sets (Figures 3a and 3b). Together, these data show that in contrast to rare-cell-detection (RaceID) and hierarchical clustering (BackSPIN, CIDR), TooManyCells provides accurate and scalable clustering.
TooManyCells accurately delineates both rare and common subpopulations of controlled admixtures.
Simultaneous detection of rare and common cell populations is a major challenge in scRNA-seq analysis. While many clustering algorithms claim to identify rare populations, few have explicitly benchmarked this ability. To rigorously assess each algorithm’s affinity to delineate rare populations, we simulated different levels of rare and common populations based on cells from different mouse organs. An accurate clustering is expected to not only detect the rare populations from the common but also distinguish the rare populations from each other. To this end, two equal-size “rare” populations were mixed with a “common” cell population. TooManyCells recapitulated known relationships between cell types within mouse organs (Figures S8 – S18) and showed that T cells were dissimilar from both macrophages and dendritic cells, as expected (Figure S19). Based on these data, ten different cell admixtures with different ratios of “common” T and “rare” macrophage and dendritic cell populations were generated (see Online Methods).
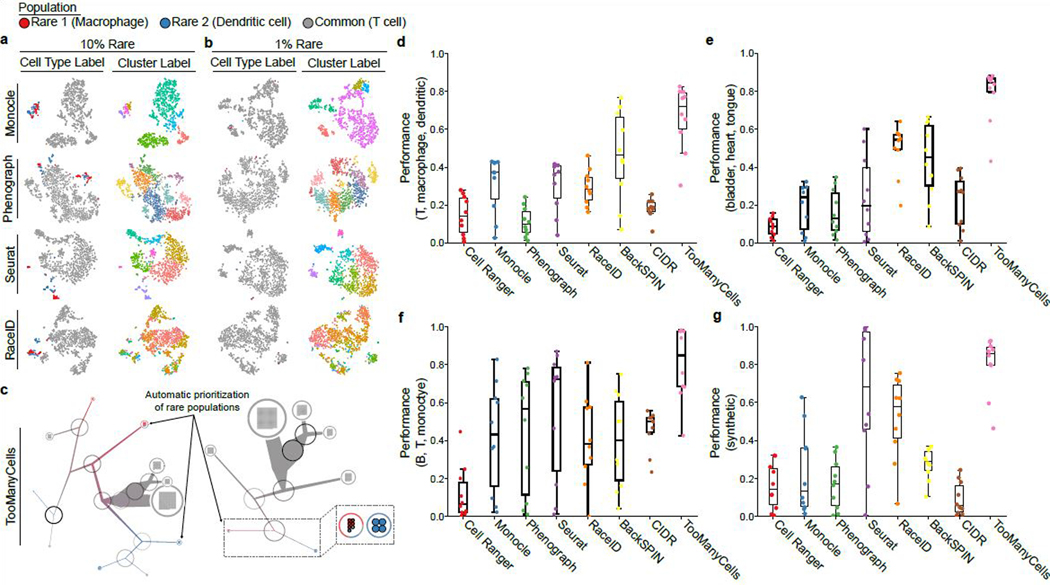
Visual inspection of t-SNE projections showed discrepancies between the actual cell types and their cluster labels (Figures 4a, 4b, and S20). Regardless of the clustering algorithm, t-SNE plots were limited in clearly distinguishing the two rare populations in an admixture. t-SNE plots’ visual inspections identified numerous small islands (Figures 4a, 4b left columns, and S20). However, it was impossible to visually localize the true rare populations in the absence of cell type labels. This issue is inherent to t-SNE, where distance and density are converted to local density. UMAP projections had similarly poor performance (Figure S21). By contrast, TooManyCells is specifically designed to plot cluster relationships and thus readily presented the rare populations (Figures 4c, S22, and S23). In the 10% rare populations admixture, TooManyCells separated the rare and common populations followed by splitting the two rare groups, keeping the common cells in large clusters (Figure 4c left panel). Interestingly, rare populations would have been easily identifiable even in the absence of cell type labels as the branch thickness and modularity values (shown by black circles) pointed out the rare subpopulations (Figure 4c left panel). In 1% rare population mixing experiment, TooManyCells again delineated the rare populations and readily presented them with the help of a drastically smaller subtree (Figure 4c right panel). Similar observations were made for eight other mixing experiments with different admixture ratios (Figures S22 and S23).
Figure 4:
Detection of cells from two “rare” populations mixed with a “common” population was benchmarked for widely used clustering algorithms. (a, b) Columns from left to right: cells labeled by actual cell types and assigned clusters of a clustering algorithm. Rows from top to bottom: Monocle, Phenograph, Seurat, and RaceID t-SNE projections. Each projection used the corresponding package’s implementation of t-SNE with the same seed (see Online Methods). Analysis for 900 common (T cells) and 100 rare cells (50 macrophages and 50 dendritic cells) (a) and 990 common and 10 rare cells (5 macrophages and 5 dendritic cells) (b) are presented. (c) TooManyCells with priority given to rare cells (pruning median(modularity)+5*MAD(modularity)). Left: 900 common and 100 rare cells. Right: 990 common and 10 rare cells. Magnified rare-population-containing subtree showed in insert. Black to white colored circles represent high to low modularity. (d-g) Box-Whisker plots (center line, median; box limits, upper (75th) and lower (25th) percentiles; whiskers, 1.5x interquartile range; points, outliers) quantifying accuracy of rare population detection in admixtures from various data sets (m = 10 admixtures, n = 1,000 cells): (d) T cells (Common), macrophages (Rare1), and dendritic cells (Rare2); (e) mouse bladder (Common), heart (Rare1), and tongue cells (Rare2); (f) human PBMC FACS-purified CD19+ B (Common), CD14+ monocytes (Rare1) and CD4+ T (Rare2) cells; and (g) three subpopulations of synthetic data. Each point represents average performance of ten experiments from an admixture (ten admixtures overall, from 90% common to 99% common). Performance indicates true rare pairs (i.e. Rare1 with Rare1 in the same cluster) / total rare pairs (true rare pairs, and Rare1 with Rare2). TooManyCells was evaluated with default normalization. To accommodate the Splatter model in (g), TooManyCells was run with PCA and relaxed modularity cutoff to account for the transformation.
We next quantitatively compared the performance of TooManyCells in the detection of rare populations (Figures S20 – S23) with other commonly used clustering algorithms (see Online methods). These analyses showed that regardless of the purity benchmark (Figure 3a), TooManyCells frequently outperformed other algorithms (Figure 4d).
Given that the organ-of-origin would provide an unbiased cell labeling, we further quantified how TooManyCells and other algorithms simultaneously segregated common and rare subpopulations in controlled admixtures consisting of cells from distinct mouse organs. In both the “common” bladder cells with “rare” cells from heart and tongue (Figure 4e) and “common” tongue cells with more dissimilar (Figure S19) “rare” bone marrow and mammary gland cells (Figure S24), TooManyCells more accurately separated common and rare cells from different mouse organs.
Furthermore, controlled admixtures of FACS-purified CD14+ monocytes, CD19+ B, and CD4+ T cells from healthy human peripheral blood mononuclear cells (PBMCs)23 confirmed that TooManyCells produces the best segregation of “common” B cells, and “rare” monocytes and T cells (Figure 4f). More importantly, while t-SNE and UMAP embeddings lacked clear guidance toward the location of rare cells (Figures S25 and S26), structural features of the TooManyCells tree highlighted the rare subpopulations (Figures S27 and S28).
Lastly, we sought to characterize performance using synthetic data. Not only did TooManyCells accurately identify the number of populations in a controlled synthetic admixture (Figure S29), the algorithm also outperformed all other tested methods (Figures 4f, S30 and Supplementary Note 6). Together, these data suggest that TooManyCells robustly outperformed the other algorithms in stratifying both common and rare subpopulations, and further revealed that the performance of BackSPIN, RaceID, and Seurat markedly varied across benchmarking experiments.
TooManyCells identifies rare plasmablasts in mouse spleen.
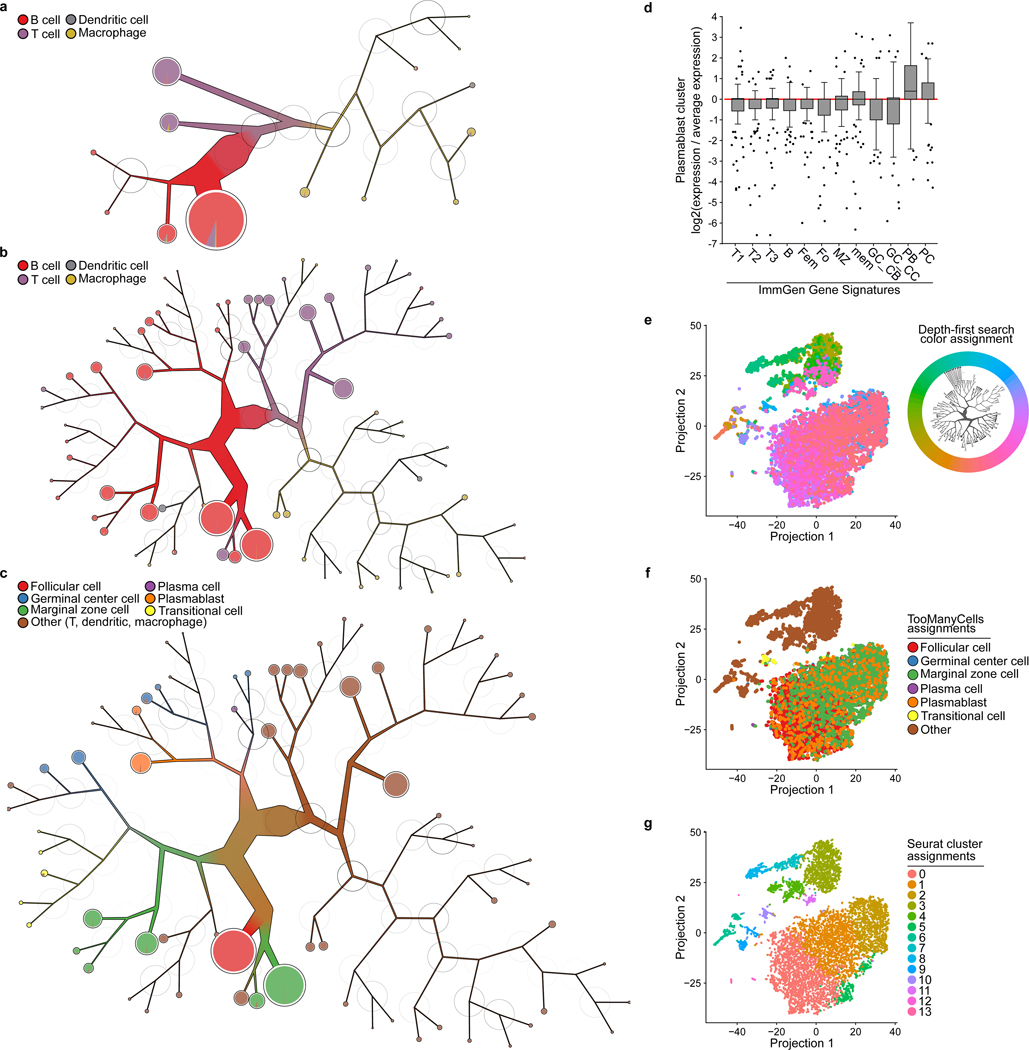
To further demonstrate TooManyCells’ ability to simultaneously stratify rare and common cell populations de novo, we analyzed the immune cell composition of the C57BL/6 mouse spleen. TooManyCells with a restricted modularity pruning threshold (Figure S31) readily separated B cells, T cells, macrophages, and dendritic cells (Figure 5a). As expected, B and T cells comprised the majority of profiled splenocytes, and were mostly separated at the first bifurcation. The macrophages were less abundant and were separated from the T cells and further subgrouped. High modularity throughout the macrophage subtree suggested heterogeneity of splenic resident macrophages, confirming flow cytometry analysis27,28. Similarly, heterogenous and relatively rare dendritic cells were also partitioned in high modularity locations (Figure 5a), as expected29.
Figure 5:
TooManyCells stratifies rare plasmablasts in mouse spleen. (a, b) TooManyCells clustering tree of the mouse splenocytes labeled with major immune cell lineages based on predefined lineage markers22 with (a) more (0.1 modularity) and (b) less (0.025 modularity) restricted modularity pruning thresholds, respectively. (c) Tree from (b) colored with newly identified B cell subtypes (see Online Methods). (d) ImmGen MyGeneSet30 gene expressions for the top n = 100 differentially expressed genes of the plasmablast node from (c) compared to all other B cell subtypes (box-whisker plots: center line, median; box limits, upper (75th) and lower (25th) percentiles; whiskers, 1.5x interquartile range; points, outliers). (e) Cells from Figure S16 projected using Seurat’s processing and t-SNE, colored by TooManyCells clustering tree leaves, where each leaf is assigned a different color (top-right insert). Similar colors represent nearby locations within the tree, for example pink and purple are closer in the tree than pink and green. (f) Coordinates from t-SNE projection in (e) colored by subset populations from (c). Orange color-coded plasmablasts are indistinguishable from other B lymphocytes. (g) Coordinates from the t-SNE projection in (e) colored by Seurat-generated cluster labels fails to separate plasmablasts. Circles colored from black to white base on high to low modularity. Definitions of x-axis ticks from (d), T1: Splenic T1 (transitional), T2: Splenic T2, T3: Splenic T3, B: Splenic B cells, Fem: Female Splenic B cells, Fo: Splenic Follicular, MZ: Splenic Marginal Zone, mem: Splenic Memory, GC_CB: Splenic Germinal Center Centroblasts, GC_CC: Splenic Germinal Center Centrocytes, PB: Splenic Plasmablasts, PC: Splenic Plasma Cells. n = 9,552 cells in all the panels.
Given the diversity of lymphocytes, we repeated the TooManyCells analysis with less restricted modularity pruning threshold (Figures 5b and S31). Traversing further along the TooManyCells clustering hierarchy, T and B cells separated into more refined clusters (Figure 5b). Interestingly, TooManyCells successfully separated CD4+ and CD8+ T cells (Figures 5b and S32), and stratified more common marginal zone, germinal center and follicular B lymphocytes (Figure 5c). Importantly, labeling of the splenic TooManyCells tree by B cell subtype signatures30 identified two branches enriched for rare splenic14 Igj-expressing plasma and plasmablasts B cells (Figures 5b–d and Online Methods). Together, these analyses showed TooManyCells’ ability to stratify both rare and common cell types in mouse spleen and showcased TooManyCells-enabled multilayer exploration of single-cell clades de novo.
To further assess the ability of popular methods to identify rare plasmablasts in mouse spleen, we used Seurat to generate t-SNE plots and cluster splenocytes. Overlaying cells in the t-SNE projection with their respective leaves from the TooManyCells tree (Figure S16) showed that for the most part cells nearby in the tree were nearby in the t-SNE projection (Figure 5e). However, there were some discrepancies where cells farther apart in the tree were proximal on the t-SNE plot (e.g. mixing of green and pink labeled cells on the top-right of the t-SNE plot). Overlaying the B cell subtypes as defined by TooManyCells and validated by B cell subtype signatures (Figures 5c and 5d) onto the t-SNE coordinates failed to visually separate plasmablasts from other B cell subtypes (Figure 5f). Furthermore, default Seurat clustering was unable to identify the distinct cluster of rare splenic plasmablasts (Figure 5g). Together, these results further support the advantage of TooManyCells’ visualization and clustering over widely used algorithms in guiding simultaneous detection of rare and common splenocyte subpopulations.
Different GSI treatment regimens lead to distinct drug-resistant T-ALL populations.
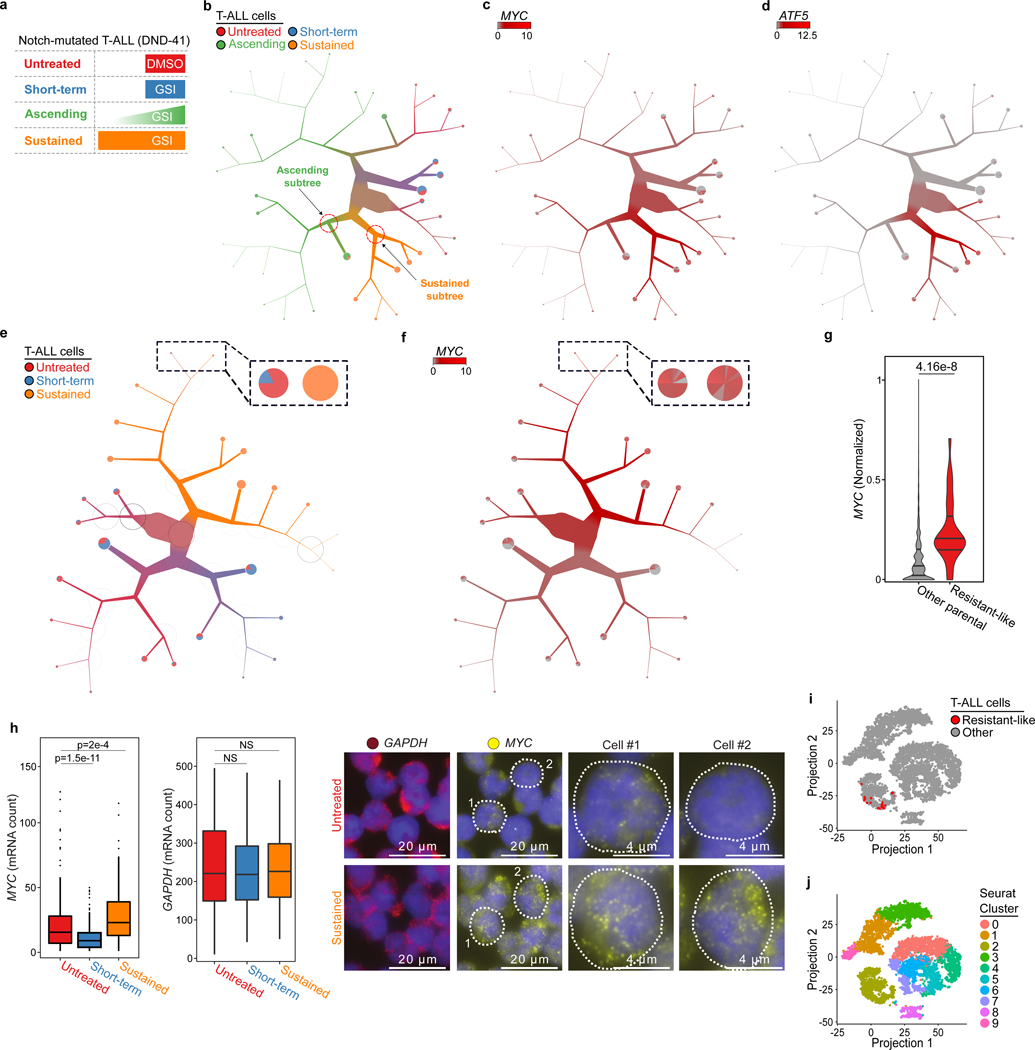
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive malignancy in children and adults31,32. Identification of Notch as the most frequently mutated gene in T-ALL led to clinical testing of gamma-secretase inhibitor (GSI), a targeted Notch signaling antagonist33. However, GSI-resistance development may limit its clinical efficacy34. We generated new scRNA-seq data and used TooManyCells to investigate the effect of GSI on individual resistant DND-41 T-ALL cells that were selected under two distinct treatment regimens. Ascending-dose GSI-resistant cells (referred to as ascending resistant) were selected by gradually doubling the GSI dose from ∼200% to 1,600% of the DND-41 IC50, while sustained high-dose GSI-resistant cells (referred to as sustained resistant) were selected by a prolonged treatment with ~1,600% of the IC50 (Figures 6a and 33a). Transcriptomes of ~10,000 DND-41 cells from ascending resistant, sustained resistant, untreated parental, and short-term (24 hours) GSI-treated parental populations were profiled.
Figure 6:
TooManyCells identifies GSI-resistant cell heterogeneity and detects resistant-like T-ALL cells. (a) Treatment strategies for untreated (n = 2,338 cells), short-term (n = 2,616 cells), ascending (n = 2,727 cells), and sustained (n = 2,417 cells) DND-41 populations. (b) TooManyCells tree showing distinct GSI-resistant populations (n = 10,098 cells). (c, d) Upper quartile normalized (UQ) MYC (c) and ATF5 (d) expressions overlaid onto (b). Gray to red: low to high expression. (e) TooManyCells tree of parental and sustained populations (n = 7,371 cells). Magnified resistant-like subtree in insert. (f) UQ MYC expression overlaid onto (e). Magnified resistant-like subtree in insert. Black to white circles represent high to low modularity. (g) Violin plots (center line, median; upper and lower lines, 75th and 25th percentiles; lower and upper bounds, minimum and maximum) normalized MYC expression of resistant-like (n = 28 cells) and other parental (n = 4,926 cells) cells (two-tailed Mann-Whitney U test, p = 4.16 × 10−8). (h) Box-Whisker plots (center line, median; box limits, upper (75th) and lower (25th) percentiles; whiskers, 1.5x interquartile range; points, outliers) showing single-cell MYC (left) and GAPDH (center) RNA FISH signal distributions for untreated (n = 250 cells), short-term (n = 261 cells; two-tailed t test, MYC: p = 1.5 × 10−11), and sustained (n = 222 cells; two-tailed t test, MYC: p = 2 × 10−4) populations. Cell images (right) of RNA FISH signals for GAPDH (pseudo-color red) and MYC (pseudo-color yellow) in untreated (top) and sustained (bottom) cells. Top 3rd and 4th columns showing two untreated cells with high MYC and low MYC expression, respectively. Bottom 3rd and 4th columns showing two sustained cells with high MYC expression. Cell nuclei in purple. NS: p > 0.005 (i) Cells from (e) projected using Seurat (n = 4,954 cells), colored by resistant-like population (red) from (e). (j) Coordinates from (i) colored by Seurat-generated clusters.
The TooManyCells tree of these four populations showed mixing of untreated and short-term treated parental cells and the heterogeneity of response to GSI in genetically homogeneous DND-41 parental cells (Figure 6b), which was not due to technical biases (Figure S33b and independent bulk RNA-seq (data not shown)). While the sustained resistant population occupied a distinct part of the tree (Figure 6b), the ascending resistant cells showed markedly diverse gene expression profiles (Figure 6b) and were significantly more heterogeneous (Figure S33c, p = 0.0140). Visualizing and quantifying relationships among the populations further showed that ascending resistant cells partially resembled both sustained resistant and parental cells (Figures 6b and 33d). TooManyCells revealed that ~40% of the ascending resistant cells were transcriptionally similar to the parental cells (Figure 6b) and the remaining ascending resistant cells were more closely related to the sustained resistant population. Nevertheless, the expressions of several genes in this group of ascending resistant cells, including proto-oncogene MYC and anti-apoptotic gene ATF535–39, were significantly different from the sustained resistant population (Figures 6c, 6d, S33e, S33f, and Table S1). Together, these single-cell resolution analyses identified a subpopulation of ascending resistant cells that, despite similarities with their sustained resistant counterparts, evolved differently to acquire GSI resistance and exhibited significantly lower expression of pro-survival genes — potentially enabling gradual adaptation to elevated GSI.
TooManyCells identifies a rare GSI-resistant-like subpopulation.
To investigate the underpinning GSI-resistance mechanisms, we next focused on the sustained resistant cells (Figure 6e), which were more distinct from the parental cells (Figures 6b and S33d). GSI treatment equally blunted expression of Notch and its known targets in drug-responsive and sustained resistant cells (Figures S33f–j and Tables S2 and S3). By contrast, while short-term GSI treatment significantly reduced expression of MYC and its known targets in most of the parental cells (Figures S33f and S33i), it had no significant effect on their expression in the sustained resistant cells (Figures S33f and S33j). Together, these data imply that Notch-independent elevated MYC expression contributes to high GSI dosage tolerance.
To further test this hypothesis, we compared individual resistant and parental cells. Interestingly, this single-cell resolution analysis revealed a rare (< 1%) parental subpopulation that was transcriptionally similar to sustained resistant cells and localized at their encompassing subtree (Figure 6e).This rare resistant-like subpopulation showed markedly elevated MYC levels compared to the other parental cells (Figures 6f and 6g, 2.85 fold change, p = 4.01 × 10−8). Furthermore, Gene Set Enrichment Analysis (GSEA)40 showed that known MYC targets41 were the most differentially expressed pathways in the rare resistant-like cells compared to both other parental (Figure S33k, Table S4) and sustained resistant cells (Figure S33l, Table S5). Single-molecule RNA fluorescence in situ hybridization (FISH) analysis independently showed the prevalence and rarity of high MYC levels in sustained resistant and parental DND-41 cells, respectively (Figure 6h, Table S6).
Having verified the existence of high MYC-expressing resistant-like cells, we sought to find this rare parental subpopulation using other algorithms to compare against TooManyCells. These analyses showed that both t-SNE projection (Figure 6i) and Seurat clustering (Figure 6j) were unable to visually and algorithmically stratify this rare resistant-like subpopulation from the rest of the parental cells (Figure S33m). Together, these analyses demonstrate the unique ability of TooManyCells to guide discovery of a rare DND-41 subpopulation that could potentially tolerate high GSI doses, and hint at underpinning resistance mechanisms.
Discussion
Popular single-cell clustering and visualization methods have been firmly set in variations of single-resolution clustering and projection-based visualization algorithms. While these methods are inherently useful for single-cell analysis, they may be unsuitable for certain applications as demonstrated in this study. Here, we developed TooManyCells which provides complementary algorithms for clustering and visualization. TooManyCells uses a recursive technique to repeatedly identify subpopulations whose relationships are maintained in a tree. Compared to projection-based algorithms, the TooManyCells visualization model is fundamentally different and, in conjunction with an array of visualization features, enables a flexible platform for cell state stratification, exploration, and rare population detection. In addition to clustering and visualization, TooManyCells also provides other capabilities including, but not limited, to heterogeneity assessment, clumpiness measurement, and diversity and rarefaction statistics. In addition to synthetic data, the superior performance of TooManyCells to simultaneously identify rare and common cell populations was demonstrated in three independent contexts. In controlled settings, TooManyCells not only separated the two rare cell populations from an admixture of common and rare cells, but successfully sequestered the two rare populations from each other. Applying TooManyCells to cell lineage identification showed its ability to isolate rare plasmablasts from total mouse splenocytes, while a popular single-cell tool and visualization failed to do so. Lastly, TooManyCells was able to detect a resistant-like subclone in DND-41 cells with exceptionally high MYC levels that was separately verified by single-molecule RNA FISH and could potentially tolerate high doses of Notch inhibitor GSI, leading to the development of drug resistance in Notch-mutated T-ALL.
In addition to performance, scalability, and usability, we considered flexibility and versatility in the TooManyCells design. TooManyCells is a generic framework consist of several algorithms that may be interchanged with other existing algorithms. The TooManyCells clustering and visualization modules, ClusterTree and BirchBeer respectively, can be potentially used for analysis of other single-cell genomic or observation-feature data. Together, our studies suggest that further improvement of clustering and visualization techniques are warranted to fully explore outputs of various single-cell measurement technologies. TooManyCells is a step in that direction.
Online Methods
Clustering
TooManyCells implements a generalized adaptation of a matrix-free hierarchical spectral clustering process originally proposed for text mining42. Spectral clustering using normalized cuts is a technique to partition data into groups, or clusters, where the items within a cluster are more similar to each other than they are to items in other clusters43. This analysis is based on the pairwise similarity between items, leading to a computational complexity of O(m2) with m items42. Let A be a similarity matrix where A(i, j) represents the similarity between items i and j and D = diag(A1) be the diagonal matrix where 1 is a column vector of 1’s. Then
defines the normalized Laplacian of A. A partition into two clusters denoted by 0 and 1 labels can be defined as
where V is the eigenvector corresponding to the second smallest eigenvalue of ℒ(A)43. Alternatively, the eigenvector corresponding to the second largest eigenvalue of the shifted Laplacian,
can be used instead of the second smallest eigenvalue of the Laplacian matrix. While this process bipartitions the data into two clusters, its inefficiency in both time and space makes the algorithm impractical for recurrent clustering of a large number of single cells. To improve the speed of spectral clustering while retaining the original accuracy, TooManyCells implements a generalized version of an algorithm that was originally proposed for text mining42 and can be used with sparse scRNA-seq matrices or any other observation / feature matrix. This implementation explicitly circumvents calculating A and the complete singular vector decomposition (SVD) of ℒ(A).
To this end, let B1 be an m × n matrix with m rows of cells and n columns of read counts. TooManyCells takes as input a transpose of this matrix to conform to the current single-cell matrix file format standards where the cells are columns. By default, TooManyCells offers the option to remove columns (genes) with no reads and rows (cells) with < 250 read counts. Then, for all 1 ≤ i ≤ m, 1 ≤ j ≤ n,
where and δ(x) is 1 if x ≥ 1 and 0 if x = 0, for all This normalization transforms B1 into a term frequency-inverse document frequency (tf-idf) matrix B244,45, where the importance of common genes is de-emphasized for clustering. Intuitively, a ubiquitously expressed gene is unlikely to be as important for cell clustering compared to a gene only expressed in a given subpopulation. Other data normalizations can be performed prior to this transformation or replace the tf-idf process entirely. For instance, one may normalize each cell based on its total read count followed by the normalization of each gene by that gene’s median positive read count. In order to relate cells in a matrix-free manner, cosine similarity was used46. It has been shown42 that the similarity matrix A can be derived from B2 with
However, in order to lower the computational complexity, TooManyCells does not calculate this matrix. Instead, a new matrix B is defined as
where is the Euclidean norm of B2 row i.
To prepare the matrix as a form of a normalized Laplacian, let
and
Then the eigenvector of ℒ(A) corresponding to the second smallest eigenvalue is the second left singular vector corresponding to the second largest singular value of C, which can be found using truncated SVD42. It has been shown that the computation complexity of this process is O(Jm), the number of non-zero entries of C, where J is the average number of expressed genes within a cell. This bipartition can be recursively applied to each delineated cluster until a stopping criteria is reached, which results in a divisive hierarchical cluster structure.
In accordance with the original implementation42, TooManyCells uses Newman-Girvan modularity (Q)19 as a stopping criteria. Modularity is a measure from community detection which has also been used in single-cell clustering through optimization using the Louvain method6,7,21. Let G = (V, E) be a weighted graph of m nodes (cells) with e edges. Then, as A represents the connectivity strength among nodes, Newman-Girvan modularity measures the strength of the partition of nodes. For a bipartition,
where is the total degree of nodes in cluster Ck, if is the degree of node i then is the total degree of nodes in Ck, and is the degree of all nodes in the network. Q measures the distance of edges within clusters to the random distribution of clusters, such that Q > 0 denotes non-random communities and Q ≤ 0 demonstrates communities randomly found19,42.
TooManyCells uses Q to assess a candidate bipartition of cells to determine whether to continue the recursion or stop as a leaf in the divisive hierarchical clustering. That is, at each bipartion, if Q > 0 then continue the recursion, otherwise stop. Thus, the end result of this top-down clustering is a tree structure of clusters, where each inner node is a cluster and the leaves are the most fine-grain clusters where any additional splitting would lead to random partitioning of cells. This process has O(Jm log m) computational complexity42. The code for the TooManyCells implementation of this algorithm is available at https://github.com/faryabib/too-many-cells.
Visualization
The TooManyCells clustering algorithm results in a tree structure, where each inner node is a coarse cluster and each leaf is the most refined cluster per modularity measure. The BirchBeer rendering method was developed for displaying single-cell cluster hierarchies. To this end, BirchBeer utilizes graphviz for node coordinate placement and the Haskell diagrams library as rendering engine.
BirchBeer provides a multitude of graphical features to assist in the detection and interpretation of cell clusters. The tree leaves can be displayed in various ways. Single-cell resolution exploration is facilitated by drawing color-coded individual cells at the tree leaves. Alternatively, a pie chart can be shown to visualize a summary of the cell composition of the clusters at the tree leaves. Both single-cell resolution and statistical summarization can be shown using a “pie ring”. Each tree branch can be scaled to the relative number of cells within each subtree, allowing for quick inspection of cell population sizes of various clustering levels and visualizing clusters of rare and common populations. Furthermore, colors can be applied to each branch such that the weighted average blend of the colors of each label in the subtree is used, allowing for immediate detection of subtrees with large differences or similarities. Cluster numbers can be displayed on each node, tracing the data back into a human readable interpretation of differences between the clusters at various hierarchy levels. Furthermore, the modularity of each candidate split can be displayed at each node as a black circle with varying darkness to demonstrate the dissimilarity of cell populations encompassing that assay. Large trees may result in busy figures, much like large t-SNE plots, so options to prune the tree are available. Cutting the tree at certain levels, node sizes, or modularity are some options, but additionally there is a statistically driven option called --smart-cutoff which cuts the tree depending on the median absolute deviation (MAD). For instance, a stopping criteria of four MADs from the median node size to keep the structure of the tree but prune smaller branches. BirchBeer accepts JSON trees as a standard input. The code for BirchBeer is available at https://github.com/faryabib/birch-beer.
Differential expression
Given multiple cluster identification numbers, TooManyCells can perform differential expression analysis to identify the difference between the gene expression of cells in these clusters. TooManyCells interfaces with edgeR for differential expression analysis47. Cells were processed using the recommended edgeR settings for single-cell analysis: genes with at least 1 count per million (cpm) in at least two cells were kept, normalized with calcNormFactors, and analyzed with estimateDisp, glmFit, and glmLRT respectively. To visually facilitate this analysis, BirchBeer can label clusters with their identification numbers. All the presented differential expression analyses and statistics use this feature of TooManyCells.
Diversity analysis
While Shannon entropy is frequently used as a measure of “diversity”, the effective number of species is a more meaningful measure of diversity in biological settings. For example, a population with 16 equally abundant species should be twice as diverse as a population with 8 equally abundant species. Assuming each cell is an “organism” belonging to a “species” group defined by the clustering algorithm, then a diversity index can be applied to find the effective number of cell states in a population.
The diversity satisfying such a property can be defined as48
| (1) |
where pi is the frequency of species i, R is the total number of species in the population, and q is the “order” of diversity. q > 1 gives additional weight towards common species, while more weight is given to rare species when q < 1. q = 1 gives equal weight to all the species regardless of their commonality and is defined as
Several diversity measures can be derived from equation (1) For instance, 0D defines richness, or the number of species, in the population. 1D relates to exp (Shannon entropy) and 2D is the inverse of the Simpson index. Various diversity measures have been used previously in domains such as lymphocyte receptor repertoires and cell clones49–51. Here, we use the diversity in TooManyCells to quantitate the effective number of cell states within a population.
TooManyCells implements the concept of rarefaction curve from ecology52 to estimate the number of detectable species in a given number of profiled single cells. Briefly, the estimated number of species in a population can be calculated from a given number of samples taken from a population through random subsampling. The estimated number of species in a subsample of size n representing Xn species can be calculated as
| (2) |
where N is the total number of cells, R is the total number of cell states in all samples, and Ni is the number of cells belonging to state i. For the interval [0, R], equation (2) generates a rarefaction curve that shows the estimated number of species for a given number of profiled cells. The steepness of the rarefaction curve may represent the heterogeneity of a population. For a given number of subsamples, the estimated number of species across multiple populations can be compared based on their respective rarefaction curves. This property is useful for comparing populations with different sample sizes. A plateau in the curves indicates no substantial increase in the number of new cell states, implying a sufficient sampling to observe all the cell states in a sample. TooManyCells implements this procedure to rarefy populations.
Cluster purity
To compare the accuracy of clustering algorithms, we used measures that quantify the extent of clustering output “purity”. We considered cluster output “purity” measures since they mitigate lack of information about markers accurately defining “true” cell identity. Moreover, these measures are robust to cluster size variability. For instance, FACS-purified CD4+ cells lack the resolution to accurately define “ground truth” cell types, as these cells comprised of several functionally well-characterized subtypes (e.g. various Th1, Th2, Th17, Treg, and many more CD4+ T cell types). To assess cluster “purity”, three measures were used: purity, entropy, and normalized mutual information (NMI). All three measures are commonly used in scRNA-seq comparative analysis53–55.
Purity is based on the frequency of the most abundant class (e.g. cell type) in a cluster. Let Ω = {ω1, ω2, . . . , ωK} be the set of clusters and be the set of classes. Then purity is defined as
where N is the total number of cells, ωk is the set of cells in cluster k, and cj is the set of cells in class j45. This measure ranges from 0, poor clustering, to 1, perfect clustering.
Entropy as a measure of cluster accuracy uses Shannon entropy to measure the expected amount of information from the clusters. The entropy of each cluster k is defined by
where ωkj is the set of cells from ωk ∩ cj. Then the entropy for the entire clustering is56
Here, lower entropy of a clustering indicates higher accuracy.
Normalized mutual information (NMI) measures the normalized dependency of the class labels on the cluster labels, or the amount of information about the class labels gained when the cluster labels are given. Mutual information is defined by
To compare mutual information across clusterings, I(Ω; ) is normalized to the interval [0, 1]. As I(Ω; ) is bounded by min[H(Ω), H()], where
is the entropy of Ω along with the analogous H(), total normalization NMI can be defined by
where higher values indicate more accurate clustering based on 57.
For Tabula Muris, four data sets were generated based on organ admixture complexity: either the first 3, first 6, first 9, or all 11 organs were considered from thymus, spleen, bone marrow, limb muscle, tongue, heart, lung, mammary gland, bladder, kidney, and liver. Other data sets were not subsampled as the complexity was lower or controlled.
Each algorithm was run on each data set with default or suggested settings. Suggested settings: for Monocle, densityPeak method was used. For Seurat, Louvain clustering after K-nearest neighbor graph construction was used with 10 dimensions from PCA (as in the PBMC3k vignette, which was followed as the recommended Seurat processes). More lenient filtering thresholds from the Tabula Muris Organ Annotation Vignette were used for data sets with fewer cells. For BackSPIN, the number of levels was set to 4, as shown in the documentation.
Rare population benchmark
Rare population detection was determined by the ability of algorithms to separate two known rare populations from each other. Three cell types were considered for one common and two rare cell populations. As T cells cells were dissimilar from both macrophage and dendritic cells (Figure S19), T cells were chosen as the common population with macrophages and dendritic cells as the rare populations, all from mouse spleen. To benchmark clustering accuracy in separating rare cells, we also performed two additional experiments based on mixings cells from different mouse organs: 1) tongue (common), mammary (rare), and bone marrow (rare); 2) bladder (common), heart (rare), and tongue (rare). Likewise for the immune population data set: CD4+ T (common), CD14+ monocytes (rare), CD19+ B (rare) cells were used23. 100 data sets of 1,000 cells were generated by randomly subsampling from each cell type or organ. These 1,000 cells per data set ranged from 900 to 990 common cells and 100 to 10 rare cells (e.g. half macrophages and half dendritic cells), with ten runs each. For instance, the smallest common data set was comprised of 900 common cells (90%) and 100 rare cells (10%, 5% for each rare population). The largest common data set was comprised of 990 common cells (99%) and 10 rare cells (1%, 0.5% for each rare population). All algorithms were run on these data sets with default or suggested settings in the same fashion as in the cluster purity benchmark. These results were visualized using t-SNE for each package (Monocle: reduceDimension with t-SNE method, Phenograph: TSNE from scikit-learn which is not included in Phenograph, BackSPIN: TSNE from scikit-learn which is not included in BackSPIN, Seurat: RunTSNE with dim.use of 10 dimensions, CIDR: Rtsne from Rtsne which is not included in CIDR, RaceID: comptsne, and Cell Ranger: output t-SNE projections). UMAP visualization was calculated with the umap-learn python package. TooManyCells output was visualized using BirchBeer trees and given rare population priority with --smart-cutoff 5 --min-distance-search 1.
To quantify these benchmarks, a contingency table of the fraction of pairwise labels was used. For all rare cell pairs, a true pair was called if the two cells were of the same cell type (e.g. a macrophage with another macrophage or a dendritic cell with another dendritic cell), while a false pair was called if the two cells were of different cell types (e.g. a macrophage with a dendritic cell). Then, the measure for accuracy in this benchmark was the fraction of true pairs in all pairs.
For the simulated rare population benchmark, Splatter58 with default settings was used to generate data sets of 1,000 cells in three groups, identical in composition to the previous subsampled rare population benchmark. Here, TooManyCells was run with --pca 50 (in concordance with Seurat) to account for the synthetic nature of the Splatter model, and --min-modularity −0.05 to accommodate the PCA transformation. BackSPIN, RaceID, and Phenograph did not use dimensionality reduction by default, as with TooManyCells, so additional benchmarks were run with dimensionality reduction through the TooManyCells PCA matrix for BackSPIN and Phenograph (which do not have any function for reduction in their libraries), and CCcorrect for RaceID.
Timing benchmark
1,000 cells were used to benchmark clustering algorithm times in order to accommodate RaceID, CIDR, and BackSPIN, which did not finish on larger data sets from the purity benchmark after 4 days. Each algorithm was run 10 times to determine an average runtime.
Distribution-based pruning and stopping criteria
TooManyCells can prune the tree by including a stopping criteria in a variety of ways, including specific nodes, the minimum size of a node (i.e. number of cells) , and the proportion of cells in each child node. To simultaneously identify both rare and common cell populations, TooManyCells uses modularity to guide the tree pruning. TooManyCells quantifies the distribution of modularity for all non-leaf nodes and chooses a value of modularity based on the specified number of median absolute deviations from the median (or a chosen value). The algorithm preserves all paths to all nodes of this value or greater, and cuts all levels below. This results in large nodes with low modularity in their descendents and small nodes with high modularity.
Clumpiness
The hierarchical structure generated from any hierarchical clustering, both divisive and agglomerative, holds cells in the leaf nodes. Each cell can be assigned a label, such as an organ of origin, cell type, or expression level of high or low. In order to quantify the level of aggregation within the tree, a measure of “clumpiness” is needed59. For instance, the degree of how “clumped”, or co-localized, are CD4 T cells and CD8 T cells within the tree. Here, one would expect those T cells to be grouped together more closely than CD4 T cells with B cells. A clumpiness measure enables the quantification of this similarity.
The clumpiness measure used here was specifically designed for hierarchical structures and was previously described in more detail59. Briefly, consider a rooted k-ary tree. The clumpiness of the set of leaves M when partitioned according to L = {L1, L2, . . . , Ln} is defined as
| (3) |
This measure takes the geometric mean of x weighted by yi. x represents the weighted number (weighted by distance to the descendant leaves) of “viable” non-root inner nodes, and yi is the frequency of leaves in Li in all leaves not connected to the root node. Viable nodes are comprised of inner nodes that have at least one vertex of each label in their descendant leaves. The clumpiness of a label Li with itself is simply considering an Lʹ containing two sets — leaves in Li and all other leaves. Then the clumpiness of Li with itself is 1 − C(Lʹ)59.
Splenic cell markers
Branches of the TooManyCells tree were defined in two ways. First, differential expression analysis was carried out for each node, and the following lineage markers were used to designate enriched cell type in each leaf node. Second, listed populations were classified using ImmGen: the top 100 differential genes in those nodes were used as input to ImmGen MyGeneSet in order to find enrichment for markers from the designated cell type30.
| Cell type | Genes |
|---|---|
| Plasma cell | Igj |
| Germinal center cell | Classified using ImmGen |
| Follicular cell | Fcer2a, Klf2 |
| Marginal zone cell | Tcf4, Crebl2 |
| Transitional cell | Gfi1, Myb, Uhrf1 |
| Plasmablast | Classified using ImmGen |
Lineage-specific transcription factors in addition to cell surface markers were used, since scRNA-seq can not differentiate between cytoplasmic and surface expression of markers.
GSI-resistant T-ALL cell culture
DND-41 cells (DSMZ, cat# ACC525) were purchased from the Leibniz-Institute DSMZ-German Collection of Microorganisms and Cell Lines. Cells were cultured in RPMI 1,640 (Corning, cat# 10–040-CM) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, cat# SH30070.03), 2 mM L-glutamine (Corning, cat# 25–005-CI), 100 U/mL and 100 μg mL−1 penicillin/streptomycin (Corning, cat# 30–002-CI), 100 mM nonessential amino acids (Gibco, cat# 11140–050), 1 mM sodium pyruvate (Gibco, cat# 11360–070) and 0.1 mM of 2-mercaptoethanol (Sigma, cat# M6250). All cells were grown at 37 °C and 5% CO2 with media refreshed every 3–4 days. Cells were regularly tested for mycoplasma contamination.
IC50 values for gamma-secretase inhibitor (GSI) compound E (Calbiochem, cat# 565790) were calculated from dose-response curves using CellTiter Glo Luminescent Cell Viability Assay (Promega, cat# G7571). Briefly, 1,000 treatment-naïve DND-41 cells in 5 replicates/condition were plated in 96-well plates with vehicle or increasing concentrations of GSI (0.016, 0.031, 0.062, 0.125, 0.25, 0.5, 1, 2 μM). Luminescence was measured on day 7 with CellTiter Glo Luminescent Cell Viability Assay according to the manufacturer’s instructions. DND-41 IC50 of GSI was determined to be 5 nM.
To generate ascending GSI-resistant cells, DND-41 treatment-naïve cells were cultured in the presence of 10, 20, 40, 80, 125 nM GSI with concentration increasing every week for six weeks and maintained in 125 nM GSI. To generate sustained high-dose GSI-resistant cells, DND-41 treatment-naïve cells were cultured in the presence of 125 nM GSI for at least six weeks. The establishment of GSI-resistance was determined with IC50 assay as described above. Both ascending and sustained high-dose GSI-resistant DND-41 cells can tolerate 10 μM GSI with less than 20% cell death. Short-term DMSO/GSI treatment was performed on treatment-naïve DND-41 cells with 125 nM DMSO/GSI for 24 hours.
GSI-resistant T-ALL single-cell RNA sequencing
Prior to single–cell transcriptomic profiling, cells were washed with 1 x PBS (Corning, cat# 21031CV) and stained with DAPI (Sigma-Aldrich, cat# D9542) and live cells were sorted on BD FACS Aria II using 100 μm nozzle. Cells were washed twice with RPMI, counted and single-cell RNA-seq was performed using 10X Genomics Single Cell 3’ Library and Gel Bead Kit v2 (10 x Genomics, cat# 1000092) following the manufacturer’s instruction. Briefly, cells were loaded onto independent channels of a Chromium Controller (10 x Genomics) for targeted recovery of 3,000 cells/condition. Complementary DNA was synthesized and amplified with PCR for 13 cycles. Amplified cDNA was assessed for QC and quantified on Agilent TapeStation using High sensitivity D5000 chip and subsequently used for library construction. Libraries were quantified using KAPA Library Quantification Kits for Illumina® platform (KAPA Biosystems, Roche, cat# KK4824) and pair-end sequenced on NextSeq 550 using 150 cycles High Output kit.
FASTQ file generation and alignment to GRCh38 were performed using Cell Ranger v2.1.1 with default arguments. In total, 10,109 cells passed the Cell Ranger QC and showed the typical “knee” plots indicating high quality from untreated (2,340), short-term (2,618), ascending (2,734), and sustained high-dose (2,417). These cell were aggregated using Cell Ranger. The fraction of reads in cells was 94.1%. The total number of post-normalization reads was 786,185,264, with mean reads per cell at 66,768 and median genes per cell of 3,333. Multiplets were identified with Scrublet60 and removed from the Cell Ranger filtered matrix, which was then used as input to TooManyCells or Seurat with default settings.
RNA FISH
Parental DND-41 cells treated with 125 nM DMSO or GSI and sustained GSI-resistant cells were harvested and resuspended in PBS at a concentration of 4.5 × 106 cells mL−1. 80 μL of the cells in each condition were added to the same polysine microscope slide (Thermo Scientific, cat# P4981) using silicone isolators (Electron Microscopy Sciences, cat# 7033905) and adhered to the slide for 30 min at room temperature in a humidified chamber. Cells were then fixed in 4% formaldehyde (Fisher Scientific, cat# PI28908) in 1xPBS for 10 min, and then dipped in 1xPBS. Cells were permeabilized in 0.5% Triton (Sigma-Aldrich Roche, cat# 10789704001) in 1xPBS for 15 min and dehydrated with an ethanol row of 70%, 80%, and 100% ethanol for 2 min each. Cells were washed in wash buffer containing 2X SSC, 10% formamide (Thermo Fisher, cat# 3442061L), in Nuclease-free water (Ambion, cat# AM9937) to remove remaining ethanol. 50 μL of hybridization mix (10% dextran sulfate, 10% formamide, 2X SSC) and 1 μL of RNA FISH probes against MYC (Alexa594) and GAPDH (Alexa 647) (gift from Dr. Arjun Raj) were added to a 24×50 coverslip, attached to the slide and sealed with no-wrinkle rubber cement (Elmer/s). Hybridization was performed overnight in a 37 °C humidified chamber. Rubber cement was removed and cells were washed for 30 min in wash buffer. Cells were then stained with 0.1 μg mL−1 DAPI in 2XSSC for 15 min in a coplin jar with shaking. Slide was allowed to completely dry before mounting on coverslip with Slowfade Gold Antifade Reagent (Invitrogen, cat# S36936) and sealing with transparent nail polish.
Imaging was carried out on a Nikon widefield fluorescent microscope (Nikon Ti-E with a 60xPlan-Apo objective) and z stack size of 10 μM with a z step size of 330 nM (Nikon Elements software). DAPI signal was used for manual nuclei segmentation and the number of MYC or GAPDH mRNA in each cell were determined as described in61 (https://bitbucket.org/arjunrajlaboratory/rajlabimagetools/wiki/Home). 250, 261, and 222 DMSO-, GSI-treated parental and sustained resistant cells were analyzed, respectively. The number of MYC or GAPDH RNA FISH count were compared by t.test in R. Example images of DMSO-treated parental and sustained GSI-resistant cells were selected on the brightest z plane and adjusted in ImageJ such that the brightness of each channel is comparable across the two conditions.
Reporting Summary
Further information on research design is available in the Nature Life Sciences Reporting Summary linked to this article.
Data availability
The accession number for the new datasets reported in this paper is GEO: GSE138892. Microfluidics single-cell RNA-seq count data from 11 organs in 3 female and 4 male, C57BL/6 NIA, three-month-old mice were obtained from https://figshare.com/articles/_/5715025, removing P8 libraries due to outlier cell counts22. FACS-purified CD14+ monocytes, CD19+ B, and CD4+ T cells were obtained from https://support.10xgenomics.com/single-cell-gene-expression/datasets23. Data for seven cancer cell lines were obtained from GSE8186117. FACS-purified B lymphocytes/natural killer, megakaryocyte-erythroid, and granulocyte–monocyte progenitors were obtained from GSE11749825.
Code availability
TooManyCells is available at https://github.com/faryabib/too-many-cells or as a Docker image https://cloud.docker.com/repository/docker/gregoryschwartz/too-many-cells/. An R wrapper for TooManyCells is available at https://cran.r-project.org/web/packages/TooManyCellsR.BirchBeer is available at https://github.com/faryabib/birch-beer or as a Docker image https://cloud.docker.com/repository/docker/gregoryschwartz/birch-beer. Codes necessary to reproduce the presented analyses are available at https://github.com/faryabib/NatMethods_TooManyCells_analysis.
Supplementary Material
Acknowledgments
This work was supported by T32-CA-009140 (to G.W.S.), LLS-5456–17 (to J.P.), R01-CA-215518 (to W.S.P.), R01-HL-145754, the Penn Epigenetics pilot award, and the Sloan Foundation Grant (to G.V.), Therapeutics Translational Medicine and Therapeutics program for Transdisciplinary Awards Program in Translational Medicine and Therapeutics, Concern Foundation’s The Conquer Cancer Now Award, Susan G. Komen CCR185472448, and R01-CA-230800 (to R.B.F.).
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Lafzi A, Moutinho C, Picelli S. & Heyn H. Tutorial: Guidelines for the Experimental Design of Single-Cell RNA Sequencing Studies. Nat. Protoc 13, 2742 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Trapnell C. Defining Cell Types and States with Single-Cell Genomics. Genome Res. 25, 1491–1498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Packer J. & Trapnell C. Single-Cell Multi-Omics: An Engine for New Quantitative Models of Gene Regulation. Trends in Genet. 34, 653–665 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu S. & Trapnell C. Single-Cell Transcriptome Sequencing: Recent Advances and Remaining Challenges. F1000Res 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svensson V, Vento-Tormo R. & Teichmann SA. Exponential Scaling of Single-Cell RNA-Seq in the Past Decade. Nat. Protoc 13, 599–604 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Levine JH et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells That Correlate with Prognosis. Cell 162, 184–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler A, Hoffman P, Smibert P, Papalexi E. & Satija R. Integrating Single-Cell Transcriptomic Data across Different Conditions, Technologies, and Species. Nat. Biotechnol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu X. et al. Reversed Graph Embedding Resolves Complex Single-Cell Trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azizi E, Prabhakaran S, Carr A. & Pe’er D. Bayesian Inference for Single-Cell Clustering and Imputing. Genomics Comput. Biol 3, 46 (2017). [Google Scholar]
- 10.Ho Y-J et al. Single-Cell RNA-Seq Analysis Identifies Markers of Resistance to Targeted BRAF Inhibitors in Melanoma Cell Populations. Genome Res. 28, 1353–1363 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van der Maaten L. & Hinton G. Visualizing Data Using T-SNE. J. Mach. Learn. Res 9, 2579–2605 (November 2008). [Google Scholar]
- 12.McInnes L, Healy J. & Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction (2018). [Google Scholar]
- 13.Becht E. et al. Dimensionality Reduction for Visualizing Single-Cell Data Using UMAP. Nat. Biotechnol 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Nutt SL, Hodgkin PD, Tarlinton DM & Corcoran LM The Generation of Antibody-Secreting Plasma Cells. Nat. Rev. Immunol 15, 160–171 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Zeisel A. et al. Cell Types in the Mouse Cortex and Hippocampus Revealed by Single-Cell RNA-Seq. Science 347, 1138–1142 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Lin P, Troup M. & Ho JWK CIDR: Ultrafast and Accurate Clustering through Imputation for Single-Cell RNA-Seq Data. Genome Biology 18, 59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H. et al. Reference Component Analysis of Single-Cell Transcriptomes Elucidates Cellular Heterogeneity in Human Colorectal Tumors. Nat. Genet 49, 708–718 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Zappia L. & Oshlack A. Clustering Trees: A Visualization for Evaluating Clusterings at Multiple Resolutions. Gigascience 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman MEJ & Girvan M. Finding and Evaluating Community Structure in Networks. Phys. Rev. E 69 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Lancichinetti A. & Fortunato S. Limits of Modularity Maximization in Community Detection. Phys. Rev. E 84, 066122 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Blondel VD, Guillaume J-L, Lambiotte R. & Lefebvre E. Fast Unfolding of Communities in Large Networks. J. Stat. Mech 2008, P10008 (2008). [Google Scholar]
- 22.The Tabula Muris Consortium et al. Single-Cell Transcriptomics of 20 Mouse Organs Creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng GXY et al. Massively Parallel Digital Transcriptional Profiling of Single Cells. Nat. Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman JS, Sagar & Grün D. FateID Infers Cell Fate Bias in Multipotent Progenitors from Single-Cell RNA-Seq Data. Nat. Methods 15, 379–386 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Pellin D. et al. A Comprehensive Single Cell Transcriptional Landscape of Human Hematopoietic Progenitors. Nat Commun 10, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahlin JS et al. A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in Kit mutant mice. Blood 131, e1 e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borges da Silva H. et al. Splenic Macrophage Subsets and Their Function during Blood-Borne Infections. Front. Immunol 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Den Haan JMM & Kraal G. Innate Immune Functions of Macrophage Subpopulations in the Spleen. JIN 4, 437–445 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hey YY & O’Neill HC Murine Spleen Contains a Diversity of Myeloid and Dendritic Cells Distinct in Antigen Presenting Function. J. Cell. Mol. Med 16, 2611–2619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jojic V. et al. Identification of Transcriptional Regulators in the Mouse Immune System. Nat. Immunol 14, 633–643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winter SS et al. Improved Survival for Children and Young Adults With T-Lineage Acute Lymphoblastic Leukemia: Results From the Children’s Oncology Group AALL0434 Methotrexate Randomization. J Clin Oncol 36, 2926–2934 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marks DI et al. T-Cell Acute Lymphoblastic Leukemia in Adults: Clinical Features, Immunophenotype, Cytogenetics, and Outcome from the Large Randomized Prospective Trial (UKALL XII/ECOG 2993). Blood 114, 5136–5145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aster JC, Pear WS & Blacklow SC The Varied Roles of Notch in Cancer. Annu. Rev. Pathol. Mech. Dis 12, 245–275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knoechel B. et al. An Epigenetic Mechanism of Resistance to Targeted Therapy in T Cell Acute Lymphoblastic Leukemia. Nat. Genet 46, 364–370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dluzen D, Li G, Tacelosky D, Moreau M. & Liu DX BCL-2 Is a Downstream Target of ATF5 That Mediates the Prosurvival Function of ATF5 in a Cell Type-Dependent Manner. J. Biol. Chem 286, 7705–7713 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamazaki T. et al. Regulation of the Human CHOP Gene Promoter by the Stress Response Transcription Factor ATF5 via the AARE1 Site in Human Hepatoma HepG2 Cells. Life Sci. 87, 294–301 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Liu DX, Qian D, Wang B, Yang J-M & Lu Z. P300-Dependent ATF5 Acetylation Is Essential for Egr-1 Gene Activation and Cell Proliferation and Survival. Mol. Cell. Biol 31, 3906–3916 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angelastro JM Targeting ATF5 in Cancer. Trends Cancer 3, 471–474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karpel-Massler G. et al. A Synthetic Cell-Penetrating Dominant-Negative ATF5 Peptide Exerts Anticancer Activity against a Broad Spectrum of Treatment-Resistant Cancers. Clin. Cancer. Res 22, 4698–4711 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian A. et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liberzon A. et al. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Systems 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shu L, Chen A, Xiong M. & Meng W. Efficient SPectrAl Neighborhood Blocking for Entity Resolution in (IEEE, 2011), 1067–1078. [Google Scholar]
- 43.Shi J. & Malik J. Normalized Cuts and Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell 22, 18 (2000). [Google Scholar]
- 44.Sparck Jones K. A Statistical Interpretation of Term Specificity and Its Application in Retrieval. Journal of Documentation 28, 11–21 (1972). [Google Scholar]
- 45.Manning CD, Raghavan P. & Schütze H. Introduction to Information Retrieval OCLC: ocn190786122. 482 pp. (Cambridge University Press, New York, 2008). [Google Scholar]
- 46.Salton G, Wong A. & Yang CS. A Vector Space Model for Automatic Indexing. Commun. ACM 18, 613–620 (1975). [Google Scholar]
- 47.Robinson MD, McCarthy DJ & Smyth GK edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 26, 139– 140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hill MO Diversity and Evenness: A Unifying Notation and Its Consequences. Ecology 54, 427 (1973). [Google Scholar]
- 49.Schwartz GW & Hershberg U. Conserved Variation: Identifying Patterns of Stability and Variability in BCR and TCR V Genes with Different Diversity and Richness Metrics. Phys. Biol 10, 035005 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Schwartz GW & Hershberg U. Germline Amino Acid Diversity in B Cell Receptors Is a Good Predictor of Somatic Selection Pressures. Front. Immunol 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng W. et al. An Atlas of B-Cell Clonal Distribution in the Human Body. Nat. Biotechnol (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heck KL, van Belle G. & Simberloff D. Explicit Calculation of the Rarefaction Diversity Measurement and the Determination of Sufficient Sample Size. Ecology 56, 1459 (1975). [Google Scholar]
- 53.Tian L. et al. Benchmarking Single Cell RNA-Sequencing Analysis Pipelines Using Mixture Control Experiments. Nat Methods 16, 479–487 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Ronen J. & Akalin A. netSmooth: Network-Smoothing Based Imputation for Single Cell RNA-Seq. F1000Res 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dai H, Li L, Zeng T. & Chen L. Cell-Specific Network Constructed by Single-Cell RNA Sequencing Data. Nucleic Acids Res 47, e62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan P-N, Steinbach M, Karpatne A. & Kumar V. Introduction to Data Mining Second edition. 839 pp. (Pearson, NY NY, 2019). [Google Scholar]
- 57.Kvålseth TO On Normalized Mutual Information: Measure Derivations and Properties, 14 (2017). [Google Scholar]
- 58.Zappia L, Phipson B. & Oshlack A. Splatter: Simulation of Single-Cell RNA Sequencing Data. Genome Biology 18, 174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwartz GW, Shokoufandeh A, Ontañón S. & Hershberg U. Using a Novel Clumpiness Measure to Unite Data with Metadata: Finding Common Sequence Patterns in Immune Receptor Germline V Genes. Pattern Recognit. Lett 74, 24–29 (2016). [Google Scholar]
- 60.Wolock SL, Lopez R. & Klein AM Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Systems 8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A. & Tyagi S. Imaging Individual mRNA Molecules Using Multiple Singly Labeled Probes. Nat. Methods 5, 877– 879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the new datasets reported in this paper is GEO: GSE138892. Microfluidics single-cell RNA-seq count data from 11 organs in 3 female and 4 male, C57BL/6 NIA, three-month-old mice were obtained from https://figshare.com/articles/_/5715025, removing P8 libraries due to outlier cell counts22. FACS-purified CD14+ monocytes, CD19+ B, and CD4+ T cells were obtained from https://support.10xgenomics.com/single-cell-gene-expression/datasets23. Data for seven cancer cell lines were obtained from GSE8186117. FACS-purified B lymphocytes/natural killer, megakaryocyte-erythroid, and granulocyte–monocyte progenitors were obtained from GSE11749825.