Abstract
Background/Aim: Mechanical loading of differentiated myoblasts in vitro may mimic loading patterns of skeletal muscle in vivo. However, it is still uncharacterized the loading conditions that can produce the most effective muscle cells’ biological responses, in vitro. This study investigated the effects of different loading protocols on the expression of myogenic regulatory factors, anabolic, atrophy and pro-apoptotic factors in skeletal myotubes. Materials and Methods: C2C12 myoblasts were differentiated and underwent various stretching protocols by altering their elongation, frequency and duration, utilizing an in vitro cell tension system. The loading-induced expression changes of MyoD, Myogenin, MRF4, IGF-1 isoforms, Murf1, Atrogin, Myostatin, Foxo and Fuca were measured by Real Time-PCR. Results: Stretching by 2% elongation at 0.25 Hz for 12 h was overall the most effective in inducing beneficial responses. Conclusion: A low strain, low frequency intermediate duration stretching can most effectively up-regulate myogenic/anabolic factors and down-regulate pro-apoptotic and atrophy genes in myotubes.
Keywords: Mechanical loading, mechanotransduction, myogenesis, myotubes
Skeletal muscle tissue has the ability to adapt to mechanical stimuli by changing its mass and overall contractile phenotype via the activation of mechanotransduction and intracellular signaling mechanisms (1,2). Mechanosensitive complexes (mechanosensors) situated in the muscle cell membrane are able to sense alterations and appear to have an important role in promoting muscle cell growth, metabolism and survival (3). Indeed, cellular mechanotransduction can induce and modulate a wide range of biological responses, such as gene expression, protein synthesis and secretion, cell proliferation, survival and apoptosis (3,4). Moreover, mechanical signals that regulate muscle growth and regeneration, in both physiological and pathological conditions, can also affect the differentiation of myoblasts to mature myotubes during myogenesis (5).
Myogenic differentiation is driven by multiple signal transduction pathways which coordinate the balance between muscle growth and atrophy, or protein synthesis and protein degradation (6,7). Specifically, the myogenic program is regulated by four, structurally similar transcriptional activators, the Myogenic Regulatory Factors (MRFs) Myf5, MyoD, Myogenin and MRF4 (8,9). In addition, Insulin-like growth factor-1 (IGF-1) signaling has been implicated in the process of myogenesis (10,11) as it is a key factor in the regulation of skeletal muscle development and growth (12-14). Its up-regulation leads to muscle hypertrophy (15) while, interestingly, potentially differential actions of IGF-1 isoforms in skeletal muscle growth and regeneration have been proposed (16-19).
On the other hand, muscle-specific atrophy factors along with pro-apoptotic factors are negative regulators of muscle growth and development (20). In particular, muscle protein degradation occurs by the activation of muscle-specific ubiquitin ligases, Atrogin-1 (MaFbx) and muscle ringer finger-1 (MuRF1), along with the down-regulation of pro-growth genes (21-23). Moreover, Atrogin-1 promotes MyoD degradation and deregulates transcription factors and mitochondrial enzymes (24,25). Similarly, MuRF1 has been reported to interact with and control the half-life of many important muscle structural proteins (26). In addition, increased myostatin activity prevents muscle growth, inhibits myoblast differentiation and protein synthesis, reduces myotube size and inhibits IGF-1−induced myotube hypertrophy (27-29). Furthermore, it has been reported that when skeletal muscle cells undergo atrophy, a subset of myoblasts exhibits apoptosis (30) and important pro-apoptotic factors, such as FoxO and p53, have been negatively implicated in muscle cell growth and differentiation. In particular, overexpression of FoxO has been found to dramatically inhibit C2C12 myoblasts differentiation (31). Also, p53 and its downstream effectors are required for an inflammatory cytokine-mediated inhibition of myogenic differentiation in vitro (32).
The expression of muscle growth/atrophy-associated factors such as the afore-mentioned has been implicated in the unloading-induced atrophy mechanisms and found to be influenced by the lack of external load of skeletal muscle in vivo (33). Thus, understanding the contribution of the particular inputs of such factors on muscle cell adaptation to mechanical loading has been challenging. However, defining the molecular responses of muscle cells to mechanical stimuli in vivo is technically very difficult (34,35), because skeletal muscle contains several cell types that influence its responses to loading. Given the complexity of the in vivo myogenic adaptation in response to exercise-induced external loading, in vitro models of mechanical loading applied on muscle cells are crucial for understanding the cellular and molecular mechanisms that mediate loading-induced adaptations. The myogenic differentiation of myoblast cell lines into myotubes represents a well-established model of skeletal muscle differentiation and in the last decade in vitro models have been adopted to particularly mimic in vivo muscle loading conditions (1,36).
The aim of this study was to characterize the efficacy and efficiency of various in vitro mechanical loading protocols on gene expression responses of differentiated myoblasts, associated with anabolic, atrophy and pro-apoptotic factors involved in their myogenic lineage. We set the hypothesis that expression responses elicited would differ depending on the loading characteristics of the protocols used, thus indicating loading-specific, detrimental or beneficial effects on myotubes in vitro.
Materials and Methods
C2C12 cell culture. The C2C12 cell line of mouse myoblasts was obtained from American Type Culture Collection (Manassas, VA, USA) and cultured as previously described (37). Briefly, cells were grown in Dubecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), plus 1% penicillin/streptomycin at 37˚C in a humidified atmosphere of 5% CO2 in air, while medium was changed every other day. The C2C12 myoblasts were seeded onto 6-well flexible-bottomed culture plates coated with Collagen I (Flex I Culture Plates Collagen I; Flexcell International, Hillborough, NC, USA) and maintained in growth media until 70% to 80%confluent, then switched to differentiation media (2% horse serum, 1% of penicillin/streptomycin in DMEM). Myoblasts were allowed to differentiate into multinucleated myotubes for a 10-day period during which media was changed every other day before stretching as described below.
Myotube mechanical loading. Differentiated myotubes were stretched using the Flexcell FX-4000 strain unit (Flexcell International) that produces isotropic two-dimensional (biaxial) strain of cells cultured on the flexible surface (silicone membrane) of the culture plates, again at 37˚C in a humidified atmosphere of 5% CO2. Briefly, myotubes were subjected to five different stretching protocols: a) 15% elongation (strain) at a frequency of 1Hz for 15 min, b) 10% strain at 1 Hz for 1 h, c) 10% elongation at 0.25 Hz for 1 h, d) 2% strain at 0.25 Hz for 12 h, or e) 2% strain at 0.25 Hz for 24 h.
Cell lysis and RNA extraction. Cell extracts were obtained by cell lysis using NucleoZOL (Mecherey-Nagel, Duren, Germany) 12 h after the completion of the stretching protocol, while control (non-stretched) myotubes were also harvested 12 h after the end of each stretching protocol. Total RNA was isolated from the lysates according to the manufacturer’s recommendations. The extracted RNA was dissolved in RNAases free water (Invitrogen, Carlsbad, CA, USA) and the concentration and purity were determined spectrophotometrically (Thermo Nanodrop 2000, Thermo Scientific™, Waltham, MA, USA) by absorption at 260 and 280 nm. The integrity of total RNA was confirmed by visual inspection of the electrophoretic pattern of 18S and 28S ribosomal RNA in ethidium bromide-stained 1% agarose gels under ultraviolet (UV) light. The total RNA samples were stored at –80˚C until further analyses for the determination of the mRNA levels of the genes of interest by reverse transcription and semi-quantitative real-time PCR procedures.
Reverse transcription and real-time PCR. Total RNA from each sample was used to produce single-stranded cDNA by reverse transcription using reverse transcriptase ProtoScript II (NEB, Ipswich, MA, USA) and the resultant cDNAs were utilized in real-time PCR. More specifically, for the reverse transcription 1 μg of total RNA from each sample was mixed with random primers mix (300 ng/reaction), oligod(T)23VN (300 ng/reaction) and nuclease-free water in a total volume of 8 μl, heated at 65˚C for 5 min and then placed on ice. Next, the samples were mixed with 10 μl ProtoScript II Reaction Mix and 2 μl Protoscript II Enzyme mix and incubated consecutively at 25˚C for 5 min and at 45˚C for 1 h according to manufacturer’s recommendations. At the final step of the reverse transcription, the samples were heated at 80˚C for 5 min, to inactivate the enzyme, and stored at –20˚C.
Real-time PCR analyses were performed using the Bio-Rad 96-well iCycler thermal cycler (Bio-Rad iQ5 Real-Time PCR Detection System, Hercules, CA, USA) and Bio-Rad reagents (iQ™ SYBR Green Supermix). The primer set sequences used for the specific detection of IGF-1 isoforms (IGF-1Ea, IGF-1Eb), MRFs (MyoD, Myogenin, Mrf4), atrophy (Myostatin, MuRF1, Atrogin-1) and pro-apoptotic factors (FoxO, FUCA, p53), are shown in Table I. To prevent detection of genomic DNA, the primer sets were designed to lie within different exons while, particularly, each set of primers for the detection of the IGF-1 isoforms was specific to detect only one specific IGF-1 transcript. Each PCR reaction contained 50 ng of cDNA, 12.5 μl SYBR green master mix, 0.4 μM of each primer, and nuclease free water to a total volume of 20 μl. The real-time PCR parameters were the following: initial denaturation at 95˚C for 5 min followed by 40 cycles of 30 s at 95˚C, 30 s at 62˚C for annealing, and 30 s at 72˚C for extension. Transcript levels of the genes of interest were assessed by automatically calculating the threshold cycle (Ct) as the number of cycles at which the measured fluorescence exceeds the threshold for detection. To normalize the amount of total RNA present in each PCR reaction and the mRNA expression (relative quantification-dCt) of the genes of interest, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as housekeeping gene (internal standard). Each sample was analyzed in duplicate, and the resulting data were averaged. A melting curve (Tm) was also generated by the Bio-Rad iQ5 Real-Time PCR Detection System software after the final cycle for each experimental sample, by continuous monitoring the Bio-Rad SYBR fluorescence throughout the temperature ramp from 70˚C to 95˚C. The specificity of the primers for the corresponding transcript was also confirmed by the melting curve analysis of samples, where there was only one melting curve for each sample and electrophoretic analysis of the real-time PCR products further verified the specificity of the transcript of each gene of interest. Control for specificity included cDNA-free reactions and template-free reactions.
Table I. The sequence of the specific sets of primers used for RT-PCR analyses.
Statistical analysis. One-way analysis of variance (ANOVA) with Dunn's Multiple Comparison post-hoc test was used for statistics, using GraphPad Prism 5. All experiments were performed in triplicate and data are presented as mean±standard error of the mean (S.E.M). The level of statistical significance was set at p<0.05.
Results
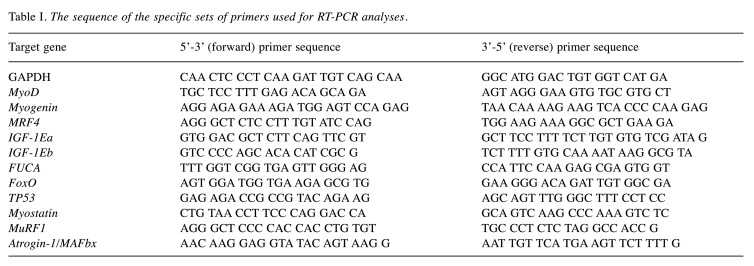
Myogenic regulatory factors. In order to investigate the potential effects of mechanical loading on the myogenic lineage of differentiated myoblasts, we examined the expression levels of both early (MyoD) and late (Myogenin, MRF4) differentiation factors in skeletal myotubes. It was found that only the low frequency (0.25 Hz), low elongation (2%) of longer duration stretching protocols induced significant changes in the expression levels of those MRFs compared to both the control (no stretch) and the higher elongation or frequency conditions (Figure 1A-C). Interestingly, MyoD exhibited differential responses to mechanical loading compared to the late differentiation MRFs; only the 24 h stretching induced a significant up-regulation of MyoD compared to control, in contrast with Myogenin and MRF4 expression, whose expressions were higher in the 12 h stretching condition compared to both the control and the 24 h stretching (Figure 1A-C). Moreover, the 12-h loading protocol was the only that induced significant changes in the expression of MRF4 compared to any other (control or stretching) condition (Figure 1C).
Figure 1. Effects of cyclic mechanical stretch on the expression of MRFs. Quantitative analysis of (A) MyoD, (B) Myogenin and (C) MRF4 mRNA expression in myotubes subjected to mechanical stretch compared to control (non-stretched myotubes); The mRNA values of MRFs in stretched myotubes were normalized to the corresponding GAPDH mRNA and are expressed as fold changes compared to control. Percentages on the columns represent the degree of elongation of the cell culture surface. Note that in the selected stretching protocols as the elongation and frequency (Hz) decrease the duration (Time) increases. *significantly different compared to control; ¤significantly different compared to 15% elongation; &significantly different compared to 10% elongation/1 Hz; #significantly different compared to 10% elongation/0.25 Hz; ¥significantly different compared to 2% elongation/12 h; Mean+SE of 3 independent experiments performed in triplicate; p<0.05.
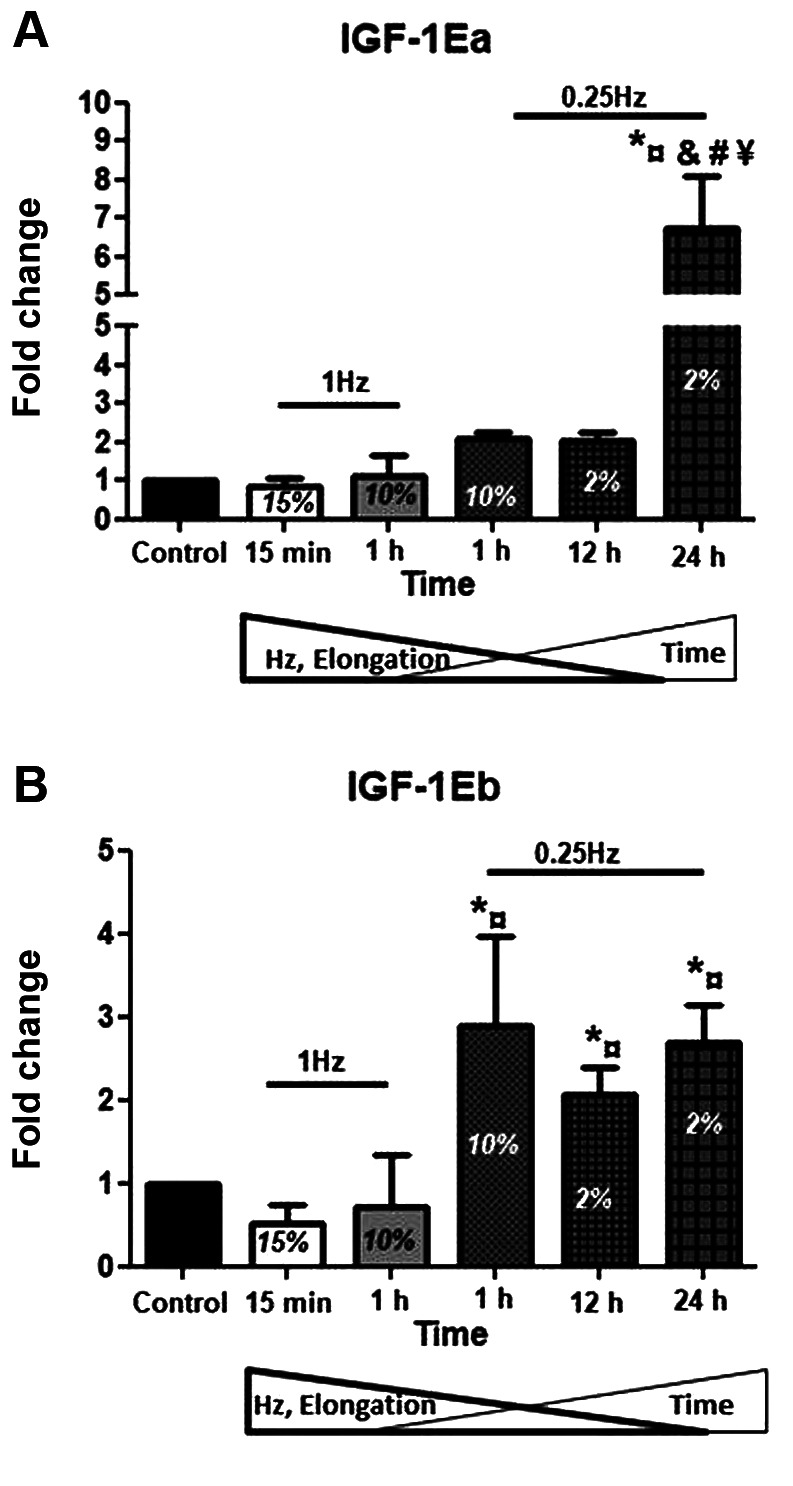
Anabolic factors. As IGF-1 is a major regulator of skeletal muscle development and growth that can induce hypertrophy and block atrophy, we examined the effects of different stretching protocols on the expression of IGF-1 isoforms in skeletal myotubes. For the IGF-1Ea, the most effective protocol was that of 2% strain at 0.25 Hz for 24 h, increasing its expression compared to both the control and the other stretching protocols used (Figure 2A). Interestingly, a differential response was revealed regarding the IGF-1Eb isoform, for which the low frequency protocol (0.25 Hz), regardless of its elongation or duration, resulted in the up-regulation of this isoform (Figure 2B).
Figure 2. Effects of cyclic mechanical stretch on the expression of IGF- 1 isoforms. Quantitative analysis of (A) IGF-1Ea and (B) IGF-1Eb mRNA expression in myotubes subjected to mechanical stretch compared to control (non-stretched myotubes); The mRNA values of IGF-1 isoforms in stretched myotubes were normalized to the corresponding GAPDH mRNA and are expressed as fold changes compared to control. Percentages on the columns represent the degree of elongation of the cell culture surface. Note that in the selected stretching protocols as the elongation and frequency (Hz) decrease the duration (Time) increases. *significantly different compared to control; ¤significantly different compared to 15% elongation; &significantly different compared to 10% elongation/1 Hz; #significantly different compared to 10% elongation/0.25 Hz; ¥significantly different compared to 2% elongation/12 h.
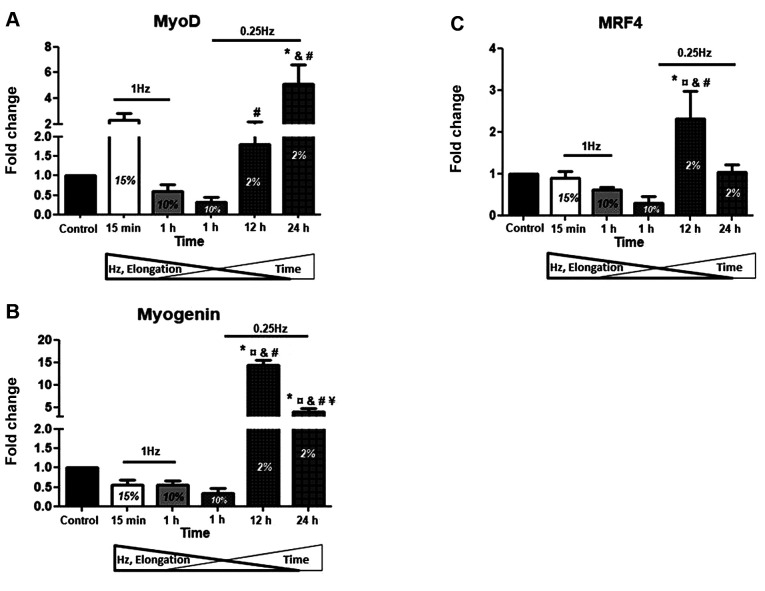
Atrophy factors. In parallel with the effects of mechanical loading on the anabolic factor IGF-1, we also examined the effects of different stretching protocols on the expression of muscle atrophy genes. It is noteworthy that in contrast to the IGF-1 isoforms responses, the stretching protocol with the highest elongation (15%) and frequency (1 Hz) along with the shortest duration (15 min) resulted in significant increases of the atrophy genes expression, particularly of myostatin and Atrogin-1, compared to controls (Figure 3A and C). On the contrary, the low elongation (2%), low frequency (0.25 Hz) for 12 h stretching was the only one to cause a significant decrease in the expression of all three atrophy genes examined (Figure 3A-C). Moreover, the same, 12-hrs low elongation and low frequency protocol resulted in the more pronounced down-regulation of myostatin and Atrogin-1 compared to the high elongation/high frequency protocols (Figure 3A and C).
Figure 3. Effects of cyclic mechanical stretch on the expression of Atrophy Factors. Quantitative analysis of (A) Myostatin, (B) MuRF1 and (C) Atrogin/MAFbx mRNA expression in myotubes subjected to mechanical stretch compared to control (non-stretched myotubes); The mRNA values of atrophy factors in stretched myotubes were normalized to the corresponding GAPDH mRNA and are expressed as fold changes compared to control. Percentages on the columns represent the degree of elongation of the cell culture surface. Note that in the selected stretching protocols as the elongation and frequency (Hz) decrease the duration (Time) increases. *significantly different compared to control; ¤significantly different compared to 15% elongation; &significantly different compared to 10% elongation/1 Hz; #significantly different compared to 10% elongation/0.25 Hz; ¥significantly different compared to 2% elongation/12 h; Mean+SE of 3 independent experiments performed in triplicate; p<0.05.
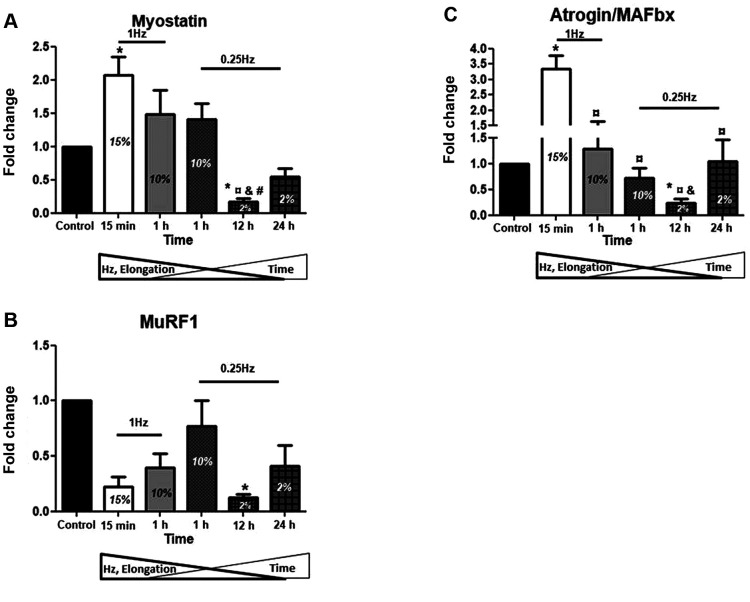
Pro-apoptotic factors. Along with the muscle atrophy genes, we further examined the loading-induced regulation of apoptosis-related factors in the skeletal myotubes. Our results showed that the stretching protocol of 10% elongation at either low (0.25 Hz) or high (1 Hz) frequency for 1 h led to significant up-regulation of the pro-apoptotic factors compared to controls as well as to the 15% elongation for 15 min stretching (Figure 4A-C), indicating that the duration of stretching may determine the particular differences between the high-strain protocols. On the contrary, and similarly to the atrophy genes responses, significant decrease in the expression of those three pro-apoptotic factors was observed in the low elongation (2%), low frequency (0.25 Hz) for 12 h stretching protocol compared to control condition (Figure 4A-C). Moreover, the low elongation, low frequency of the longer durations (12 and 24 h) protocols resulted in more pronounced down-regulation of those factors compared to the high elongation (10%)/short duration (1 h) protocols (Figure 4A-C).
Figure 4. Effects of cyclic mechanical stretch on the expression of proapoptotic factors. Quantitative analysis of (A) FoxO, (B) FUCA and (C) p53 mRNA expression in myotubes subjected to mechanical stretch compared to control (non-stretched myotubes); The mRNA values of apoptotic factors in stretched myotubes were normalized to the corresponding GAPDH mRNA and are expressed as fold changes compared to control. Percentages on the columns represent the degree of elongation of the cell culture surface. Note that in the selected stretching protocols as the elongation and frequency (Hz) decrease the duration (Time) increases. *significantly different compared to control; ¤significantly different compared to 15% elongation; &significantly different compared to 10% elongation/1 Hz; #significantly different compared to 10% elongation/0.25Hz; ¥significantly different compared to 2% elongation/12 h; Mean+SE of 3 independent experiments performed in triplicate; p<0.05.
Discussion
This study examined the effects of various in vitro cell stretching protocols on gene expression responses associated with the myogenic lineage of differentiated C2C12 myotubes, in order to reveal potential loading-specific, detrimental or beneficial effects on the myogenic program in myotubes, depending on the loading characteristics of the protocols used. The expression of myogenic, anabolic, atrophy and pro-apoptotic factors were measured 12 h after the completion of each stretching protocol to determine durable, persistently triggered rather than short transcriptional responses. Our main findings demonstrated that a low strain, low frequency for an intermediate duration mechanical stretching was overall the most effective in inducing beneficial responses in skeletal myotubes, by up-regulating MRFs and IGF-1 while down-regulating atrophy and pro-apoptotic factors. Furthermore, the present study revealed that the late myogenic determination factors, Myogenin and MRF4, exhibited differential responses to mechanical loading compared to the early differentiation factor MyoD. Similarly, a differential response of IGF-1Eb isoform to the cell stretching protocols was revealed compared with the other IGF-1 isoform, IGF-1Ea. Interestingly and in contrast to the responses of the myogenic and anabolic factors, the stretching protocol with the highest strain and frequency along with the shortest duration resulted in increased expression of the atrophy genes. Moreover, the low strain, low frequency and longer duration (12 and 24 h) stretching protocols resulted in the more pronounced down-regulation of the pro-apoptotic factors compared to the high strain, short duration protocols.
The ability of mechanosensitive cells to sense external mechanical stimuli and convert them into electrochemical and biochemical signals (mechanotransduction) is critical for the maintenance of cellular and tissue homeostasis (38-40). In particular, mechanical loading can cause changes in patterns of gene expression in skeletal muscle, influencing protein synthesis and stability and affecting muscle metabolism, while there is a growing body of evidence showing that mechanical cues modulate many aspects of myocyte function, including proliferation and myogenic differentiation (10). The myogenic differentiation program is regulated by MRFs, a family of muscle-specific transcription factors considered myogenic determinants, which share the ability to convert various differentiated cell types to myogenic and direct progenitor cells to establish the skeletal muscle lineage (41). In particular, Myogenin, lying genetically downstream, along with MyoD and MRF4, activate the myogenic differentiation program (8,42). MyoD promotes myoblasts progression to terminal differentiation (43) while it seems, along with Myogenin, to directly or indirectly activate the expression of MRF4 in the later stages of myogenic differentiation (44). Myogenin, as MRF4, is a late differentiation factor stimulating terminal myogenic differentiation and its expression can be induced by IGF-1 (45).
This study showed that only the low frequency/low strain of longer duration stretching conditions induced significant changes in the expression of those MRFs (Figure 1A-C) while, particularly, only the longest duration (24 h) stretching induced a significant up-regulation of MyoD, versus Myogenin and MRF4, whose expressions were higher in the 12 h stretching condition. Τhese findings are similar with those reported by other studies that examined the responses of MyoD after cyclic tensile strain for 24, 48 or 72 h in differentiated myoblasts, or regarding Myogenin and MRF4 responses to cyclic mechanical strain (46-48). Our findings suggest that those MRFs, at least at this (later) stage of myogenic differentiation, are responsive to mechanical loading and more sensitive particularly to long-term, low elongation stretching. Moreover, the time-dependent differential responses of MyoD compared with Myogenin and MRF4 to stretching may reflect a different responsiveness of the early vs late myogenic factors to mechanical stimuli, possibly depending on their distinct role(s) in that stage of the myogenic differentiation program.
Mechanical stretching of skeletal muscle cells both in vivo and in vitro can lead to the up-regulation of many growth factors, such as IGF-1, eventually leading to the enhancement of protein synthesis and muscle cell growth (24-26). Indeed, IGF-1 is a major anabolic factor involved in myogenesis and muscle hypertrophy (11,49-51) and its up-regulation in response to mechanical loading observed in this study is in line with findings of previous studies (51-53). However, to the authors’ best knowledge, this is the first study investigating the distinct expression profiles of IGF-1 isoforms following mechanical loading of myotubes, in vitro. Our data specifically showed that both isoforms appear to be up-regulated by low frequency stretching protocols, while differential responses were also revealed; IGF-1Ea found to be responsive only to the longest duration (24 h) of stretching, while IGF-1Eb exhibited responsiveness to all low-frequency protocols used, regardless of their duration. These findings suggest that IGF-1 isoforms need low frequency loading to be activated, while their distinct, time-dependent responses to stretch may reflect different biological roles in the differentiated myotubes.
In parallel with highlighting the myogenic/anabolic transcriptional profile of the differentiated myotubes in response to different loading conditions, this study also examined the expression responses of muscle atrophy genes to the various mechanical stimuli. Although the defined mechanisms regarding the role of those genes in driving an atrophic phenotype are yet to be fully elucidated, however, it is considered that myostatin has a role in actively inhibiting protein synthesis (27,54), whilst MuRF1 and Atrogin-1 are thought to act through the ubiquitin-proteasome pathway (6,55,56). Specifically, myostatin is a negative regulator of myogenesis by deregulating MyoD activity (57). Similarly, Atrogin-1 appears to be negatively involved in myogenesis by polyubquitination and degradation of Myogenin (24), while MuRF1 plays a major role in regulating skeletal muscle mass by controlling a variety of muscle cell catabolic processes (35).
Our study revealed that, in contrast to the IGF-1 isoforms responses, the stretching protocol with the highest strain/frequency along with the shortest duration resulted in significant increases of myostatin and Atrogin-1. Inversely, the low strain/frequency for 12 h stretching was the only one that led to significant down-regulation of all three atrophy genes examined (Figures 2 and 3). These findings suggest a multiple beneficial effect of the low strain/frequency of intermediate duration mechanical stretching, which simultaneously down-regulates muscle atrophy program and up-regulates myogenic/growth factors in advanced differentiation myotubes.
Various pro-apoptotic factors may potentially be involved in myogenic differentiation; FoxO is a fate decider within the myogenic lineage as opposed to an inducer of the myogenic program (58), p53 suppresses muscle differentiation at the Myogenin step (32), while FUCA inhibits cell growth and induces cell death (59). In our model of applying mechanical stretching on differentiated myoblasts, the loading protocols using a 10% strain for 1 h resulted in the up-regulation of those factors regardless of the stretching frequency; nevertheless, frequency appeared to be the determinant factor for differentiating these increases (Figure 4). Inversely, low strain/low frequency of long duration stretching protocols decreased their expression similarly to the atrophy genes and in contrast to the myogenic factors’ responses (Figures 1, 3 and 4). Our findings indicate that there might be a threshold (or range) of strain and/or duration of mechanical loading for the induction of detrimental effects on myotubes (60-62), such as the up-regulation of those apoptosis-related factors, since they were induced under specific strain and duration of loading.
Overall, our study revealed that a specific mechanical loading protocol (i.e., low strain/frequency of intermediate duration) was beneficial for the differentiated myotubes, not only by up-regulating myogenic and anabolic factors but also by down-regulating atrophy and pro-apoptotic genes.
Conclusion
Muscle cells are mechanosensitive responding in several ways to mechanical signals, produced from native environmental cues, in order to maintain their homeostasis and adapt to external loading. In vitro models of cell mechanical loading can greatly contribute to the improvement of our understanding of cellular mechanotransduction and molecular responses of skeletal muscle cells to loading. This study demonstrated that varying the components of mechanical stretching (i.e., elongation, frequency, duration) applied on myotubes in vitro, resulted in different, beneficial or detrimental, effects on their myogenic lineage, indicating specific features of loading for regulating the myogenic/anabolic program. These findings may be a valuable resource to be utilized as a database for developing more focused in vitro experimental designs to characterize the cellular and molecular mechanisms of mechanical loading-induced muscle adaptations.
Conflicts of Interest
The Authors declare no conflicts of interest in relation to this study.
Authors’ Contributions
AM designed and performed the study, contributed the experimental process, analyzed data and wrote the manuscript; AP wrote and reviewed the manuscript, EZ contributed to the experimental process; OT , contributed to the experimental process; AC analyzed data and reviewed the manuscript; and MK had input in the experimental design, and reviewed the manuscript. The Authors have read and approved the final manuscript.
References
- 1.Passey S, Martin N, Player D, Lewis MP. Stretching skeletal muscle in vitro: Does it replicate in vivo physiology. Biotechnol Lett. 2011;33:1513–1521. doi: 10.1007/s10529-011-0610-z. [DOI] [PubMed] [Google Scholar]
- 2.Tidball JG. Mechanical signal transduction in skeletal muscle growth and adaptation. J Appl Physiol. 2005;(1985)98:1900–1908. doi: 10.1152/japplphysiol.01178.2004. [DOI] [PubMed] [Google Scholar]
- 3.Soltow QA, Zeanah EH, Lira VA, Criswell DS. Cessation of cyclic stretch induces atrophy of c2c12 myotubes. Biochem Biophys Res Commun. 2013;434:316–321. doi: 10.1016/j.bbrc.2013.03.048. [DOI] [PubMed] [Google Scholar]
- 4.Atherton PJ, Szewczyk NJ, Selby A, Rankin D, Hillier K, Smith K, Rennie MJ, Loughna PT. Cyclic stretch reduces myofibrillar protein synthesis despite increases in fak and anabolic signalling in l6 cells. J Physiol. 2009;587:3719–3727. doi: 10.1113/jphysiol.2009.169854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stockdale FE, Miller JB. The cellular basis of myosin heavy chain isoform expression during development of avian skeletal muscles. Dev Biol. 1987;123:1–9. doi: 10.1016/0012-1606(87)90420-9. [DOI] [PubMed] [Google Scholar]
- 6.Glass DJ. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat Cell Biol. 2003;5:87–90. doi: 10.1038/ncb0203-87. [DOI] [PubMed] [Google Scholar]
- 7.Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 8.Karalaki M, Fili S, Philippou A, Koutsilieris M. Muscle regeneration: Cellular and molecular events. In Vivo. 2009;23:779–796. [PubMed] [Google Scholar]
- 9.Kopantseva EE, Belyavsky AV. [key regulators of skeletal myogenesis] Mol Biol (Mosk) 2016;50:195–222. doi: 10.7868/S0026898416010079. [DOI] [PubMed] [Google Scholar]
- 10.Papasani MR, Wang G, Cheguru P, Julien DC, Hofwegen DV, Chapalamadugu KC, Brooking A, Read S. Interactions of muscle regulatory factors and isoforms of insulin growth factor-1 in the muscle regenerative process. J Physiol. 2009;587:2427–2428. doi: 10.1113/jphysiol.2009.169540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philippou A, Halapas A, Maridaki M, Koutsilieris M. Type i insulin-like growth factor receptor signaling in skeletal muscle regeneration and hypertrophy. J Musculoskelet Neuronal Interact. 2007;7:208–218. [PubMed] [Google Scholar]
- 12.Barton ER, Park S, James JK, Makarewich CA, Philippou A, Eletto D, Lei H, Brisson B, Ostrovsky O, Li Z, Argon Y. Deletion of muscle grp94 impairs both muscle and body growth by inhibiting local igf production. FASEB J. 2012;26:3691–3702. doi: 10.1096/fj.11-203026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philippou A, Barton ER. Optimizing igf-i for skeletal muscle therapeutics. Growth Horm IGF Res. 2014;24:157–163. doi: 10.1016/j.ghir.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bikle DD, Tahimic C, Chang W, Wang Y, Philippou A, Barton ER. Role of IGF-i signaling in muscle bone interactions. Bone. 2015;80:79–88. doi: 10.1016/j.bone.2015.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou Y, Dong Y, Meng Q, Zhao Y, Li N. Incorporation of a skeletal muscle-specific enhancer in the regulatory region of igf1 upregulates igf1 expression and induces skeletal muscle hypertrophy. Sci Rep. 2018;8:2781. doi: 10.1038/s41598-018-21122-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durzynska J, Philippou A, Brisson BK, Nguyen-McCarty M, Barton ER. The pro-forms of insulin-like growth factor i (IGF-i) are predominant in skeletal muscle and alter IGF-i receptor activation. Endocrinology. 2013;154:1215–1224. doi: 10.1210/en.2012-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papageorgiou E, Philippou A, Armakolas A, Christopoulos PF, Dimakakos A, Koutsilieris M. The human ec peptide: The active core of a progression growth factor with species-specific mode of action. Hormones (Athens) 2016;15:423–434. doi: 10.14310/horm.2002.1699. [DOI] [PubMed] [Google Scholar]
- 18.Philippou A, Papageorgiou E, Bogdanis G, Halapas A, Sourla A, Maridaki M, Pissimissis N, Koutsilieris M. Expression of igf-1 isoforms after exercise-induced muscle damage in humans: Characterization of the MGF e peptide actions in vitro. In Vivo. 2009;23:567–575. [PubMed] [Google Scholar]
- 19.Vassilakos G, Philippou A, Koutsilieris M. Identification of the igf-1 processing product human ec/rodent eb peptide in various tissues: Evidence for its differential regulation after exercise-induced muscle damage in humans. Growth Horm IGF Res. 2016;32:22–28. doi: 10.1016/j.ghir.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Tsao J, Vernet DA, Gelfand R, Kovanecz I, Nolazco G, Bruhn KW, Gonzalez-Cadavid NF. Myostatin genetic inactivation inhibits myogenesis by muscle-derived stem cells in vitro but not when implanted in the mdx mouse muscle. Stem Cell Res Ther. 2013;4:4. doi: 10.1186/scrt152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 22.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific f-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dapp C, Schmutz S, Hoppeler H, Fluck M. Transcriptional reprogramming and ultrastructure during atrophy and recovery of mouse soleus muscle. Physiol Genomics. 2004;20:97–107. doi: 10.1152/physiolgenomics.00100.2004. [DOI] [PubMed] [Google Scholar]
- 24.Jogo M, Shiraishi S, Tamura TA. Identification of mafbx as a myogenin-engaged f-box protein in scf ubiquitin ligase. FEBS Lett. 2009;583:2715–2719. doi: 10.1016/j.febslet.2009.07.033. [DOI] [PubMed] [Google Scholar]
- 25.Lokireddy S, Wijesoma IW, Sze SK, McFarlane C, Kambadur R, Sharma M. Identification of atrogin-1-targeted proteins during the myostatin-induced skeletal muscle wasting. Am J Physiol Cell Physiol. 2012;303:C512–C529. doi: 10.1152/ajpcell.00402.2011. [DOI] [PubMed] [Google Scholar]
- 26.Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013;280:4294–4314. doi: 10.1111/febs.12253. [DOI] [PubMed] [Google Scholar]
- 27.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new tgf-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 28.Morissette MR, Cook SA, Buranasombati C, Rosenberg MA, Rosenzweig A. Myostatin inhibits igf-i-induced myotube hypertrophy through akt. Am J Physiol Cell Physiol. 2009;297:C1124–C1132. doi: 10.1152/ajpcell.00043.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trendelenburg AU, Meyer A, Rohner D, Boyle J, Hatakeyama S, Glass DJ. Myostatin reduces akt/torc1/p70s6k signaling, inhibiting myoblast differentiation and myotube size. Am J Physiol Cell Physiol. 2009;296:C1258–C1270. doi: 10.1152/ajpcell.00105.2009. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz LM. Atrophy and programmed cell death of skeletal muscle. Cell Death Differ. 2008;15:1163–1169. doi: 10.1038/cdd.2008.68. [DOI] [PubMed] [Google Scholar]
- 31.Wu YJ, Fang YH, Chi HC, Chang LC, Chung SY, Huang WC, Wang XW, Lee KW, Chen SL. Insulin and licl synergistically rescue myogenic differentiation of foxo1 over-expressed myoblasts. PLoS One. 2014;9:e88450. doi: 10.1371/journal.pone.0088450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang ZJ, Broz DK, Noderer WL, Ferreira JP, Overton KW, Spencer SL, Meyer T, Tapscott SJ, Attardi LD, Wang CL. P53 suppresses muscle differentiation at the myogenin step in response to genotoxic stress. Cell Death Differ. 2015;22:560–573. doi: 10.1038/cdd.2014.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Philippou A, Minozzo FC, Spinazzola JM, Smith LR, Lei H, Rassier DE, Barton ER. Masticatory muscles of mouse do not undergo atrophy in space. FASEB J. 2015;29:2769–2779. doi: 10.1096/fj.14-267336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hornberger TA, Mateja RD, Chin ER, Andrews JL, Esser KA. Aging does not alter the mechanosensitivity of the p38, p70s6k, and jnk2 signaling pathways in skeletal muscle. J Appl Physiol. 2005;(1985)98:1562–1566. doi: 10.1152/japplphysiol.00870.2004. [DOI] [PubMed] [Google Scholar]
- 35.Hornberger TA, Armstrong DD, Koh TJ, Burkholder TJ, Esser KA. Intracellular signaling specificity in response to uniaxial vs. Multiaxial stretch: Implications for mechanotransduction. Am J Physiol Cell Physiol. 2005;288:C185–C194. doi: 10.1152/ajpcell.00207.2004. [DOI] [PubMed] [Google Scholar]
- 36.Hua W, Zhang M, Wang Y, Yu L, Zhao T, Qiu X, Wang L. Mechanical stretch regulates microrna expression profile via nf-kappab activation in c2c12 myoblasts. Mol Med Rep. 2016;14:5084–5092. doi: 10.3892/mmr.2016.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moorwood C, Philippou A, Spinazzola J, Keyser B, Macarak EJ, Barton ER. Absence of gamma-sarcoglycan alters the response of p70s6 kinase to mechanical perturbation in murine skeletal muscle. Skelet Muscle. 2014;4:13. doi: 10.1186/2044-5040-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vining KH, Mooney DJ. Mechanical forces direct stem cell behaviour in development and regeneration. Nat Rev Mol Cell Biol. 2015;18:728–742. doi: 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall KL, Lumpkin EA. The molecular basis of mechanosensory transduction. Adv Exp Med Biol. 2012;739:142–155. doi: 10.1007/978-1-4614-1704-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vining KH, Mooney DJ. Mechanical forces direct stem cell behaviour in development and regeneration. Nature reviews. Mol Cell Biol. 2017;18:728–742. doi: 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comai G, Sambasivan R, Gopalakrishnan S, Tajbakhsh S. Variations in the efficiency of lineage marking and ablation confound distinctions between myogenic cell populations. Dev Cell. 2015;31:654–667. doi: 10.1016/j.devcel.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 42.Zammit PS. Function of the myogenic regulatory factors myf5, myod, myogenin and mrf4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev Biol. 2017;72:19–32. doi: 10.1016/j.semcdb.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida N, Yoshida S, Koishi K, Masuda K, Nabeshima Y. Cell heterogeneity upon myogenic differentiation: Down-regulation of myod and myf-5 generates ‘reserve cells’. J Cell Sci. 1998; 111(Pt 6):769–779. doi: 10.1242/jcs.111.6.769. [DOI] [PubMed] [Google Scholar]
- 44.Pavlath GK, Dominov JA, Kegley KM, Miller JB. Regeneration of transgenic skeletal muscles with altered timing of expression of the basic helix-loop-helix muscle regulatory factor mrf4. Am J Pathol. 2003;162:1685–1691. doi: 10.1016/S0002-9440(10)64303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Florini JR, Ewton DZ, Roof SL. Insulin-like growth factor-i stimulates terminal myogenic differentiation by induction of myogenin gene expression. Mol Endocrinol. 1991;5:718–724. doi: 10.1210/mend-5-5-718. [DOI] [PubMed] [Google Scholar]
- 46.Kook SH, Son YO, Choi KC, Lee HJ, Chung WT, Hwang IH, Lee JC. Cyclic mechanical stress suppresses myogenic differentiation of adult bovine satellite cells through activation of extracellular signal-regulated kinase. Mol Cell Biochem. 2008;309:133–141. doi: 10.1007/s11010-007-9651-y. [DOI] [PubMed] [Google Scholar]
- 47.Kumar A, Murphy R, Robinson P, Wei L, Boriek AM. Cyclic mechanical strain inhibits skeletal myogenesis through activation of focal adhesion kinase, rac-1 gtpase, and nf-kappab transcription factor. FASEB J. 2004;18:1524–1535. doi: 10.1096/fj.04-2414com. [DOI] [PubMed] [Google Scholar]
- 48.Zhan M, Jin B, Chen SE, Reecy JM, Li YP. Tace release of tnf-alpha mediates mechanotransduction-induced activation of p38 mapk and myogenesis. J Cell Sci. 2007;120:692–701. doi: 10.1242/jcs.03372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Philippou A, Maridaki M, Halapas A, Koutsilieris M. The role of the insulin-like growth factor 1 (igf-1) in skeletal muscle physiology. In Vivo. 2007;21:45–54. [PubMed] [Google Scholar]
- 50.Goldspink G. Mechanical signals, igf-i gene splicing, and muscle adaptation. Physiology (Bethesda, Md.) 2005;20:232–238. doi: 10.1152/physiol.00004.2005. [DOI] [PubMed] [Google Scholar]
- 51.Juffer P, Bakker AD, Klein-Nulend J, Jaspers RT. Mechanical loading by fluid shear stress of myotube glycocalyx stimulates growth factor expression and nitric oxide production. Cell Biochem Biophys. 2014;69:411–419. doi: 10.1007/s12013-013-9812-4. [DOI] [PubMed] [Google Scholar]
- 52.Rauch C, Loughna PT. Static stretch promotes mef2a nuclear translocation and expression of neonatal myosin heavy chain in c2c12 myocytes in a calcineurin- and p38-dependent manner. Am J Physiol Cell Physiol. 2005;288:C593–C605. doi: 10.1152/ajpcell.00346.2004. [DOI] [PubMed] [Google Scholar]
- 53.Perrone CE, Fenwick-Smith D, Vandenburgh HH. Collagen and stretch modulate autocrine secretion of insulin-like growth factor-1 and insulin-like growth factor binding proteins from differentiated skeletal muscle cells. J Biol Chem. 1995;270:2099–2106. doi: 10.1074/jbc.270.5.2099. [DOI] [PubMed] [Google Scholar]
- 54.Sharples AP, Stewart CE. Myoblast models of skeletal muscle hypertrophy and atrophy. Curr Opin Clin Nutr Metab Care. 2011;14:230–236. doi: 10.1097/MCO.0b013e3283457ade. [DOI] [PubMed] [Google Scholar]
- 55.Gumucio JP, Mendias CL. Atrogin-1, murf-1, and sarcopenia. Endocrine. 2013;43:12–21. doi: 10.1007/s12020-012-9751-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Y, Jemiolo B, Trappe S. Proteolytic mrna expression in response to acute resistance exercise in human single skeletal muscle fibers. J Appl Physiol. 2006;101:1442–1450. doi: 10.1152/japplphysiol.00438.2006. [DOI] [PubMed] [Google Scholar]
- 57.Langley B, Thomas M, Bishop A, Sharma M, Gilmour S, Kambadur R. Myostatin inhibits myoblast differentiation by down-regulating myod expression. J Biol Chem. 2002;277:49831–49840. doi: 10.1074/jbc.M204291200. [DOI] [PubMed] [Google Scholar]
- 58.Kitamura T, Kitamura YI, Funahashi Y, Shawber CJ, Castrillon DH, Kollipara R, DePinho RA, Kitajewski J, Accili D. A foxo/notch pathway controls myogenic differentiation and fiber type specification. J Clin Invest. 2007;117:2477–2485. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ezawa I, Sawai Y, Kawase T, Okabe A, Tsutsumi S, Ichikawa H, Kobayashi Y, Tashiro F, Namiki H, Kondo T, Semba K, Aburatani H, Taya Y, Nakagama H, Ohki R. Novel p53 target gene fuca1 encodes a fucosidase and regulates growth and survival of cancer cells. Cancer Sc. 2016;107:734–745. doi: 10.1111/cas.12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsivitse SK, Mylona E, Peterson JM, Gunning WT, Pizza FX. Mechanical loading and injury induce human myotubes to release neutrophil chemoattractants. Am J Physiol Cell Physiol. 2005;288:C721–C729. doi: 10.1152/ajpcell.00237.2004. [DOI] [PubMed] [Google Scholar]
- 61.Nguyen HX, Lusis AJ, Tidball JG. Null mutation of myeloperoxidase in mice prevents mechanical activation of neutrophil lysis of muscle cell membranes in vitro and in vivo. J Physiol. 2005;565:403–413. doi: 10.1113/jphysiol.2005.085506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sampaolesi M, Yoshida T, Iwata Y, Hanada H, Shigekawa M. Stretch-induced cell damage in sarcoglycan-deficient myotubes. Pflugers Arch. 2001;442:161–170. doi: 10.1007/s004240100516. [DOI] [PubMed] [Google Scholar]