Abstract
Triple-negative breast cancer (TNBC) is an extremely diverse group of breast tumors, with aggressive clinical behavior, higher rates of distant recurrence and worse overall survival compared to other types of breast cancers. The genetic, transcriptional histological and clinical heterogeneity of this disease has been an obstacle in the progression of targeted therapeutic approaches, as a ubiquitous TNBC marker has not yet been discerned. In terms of that, current studies focus on the classification of TNBC tumors in subgroups with similar characteristics in order to develop a treatment specialized for each group of patients. To date, a series of gene expression profiles analysis in order to identify the different molecular subtypes have been used. Complementary DNA microarrays, PAM50 assays, DNA and RNA sequencing as well as immunohistochemical analysis are some of the methods utilized to classify TNBC tumors. In 2012, the Cancer Genome Atlas (TCGA) Research Network conducted a major analysis of breast cancers using six different platforms, the genomic DNA copy number arrays, DNA methylation, exome sequencing, messenger RNA arrays, microRNA sequencing and reverse-phase protein arrays, in order to assort the tumors in homogenous subgroups. Since then, an increasing number of breast cancer data sets are being examined in an attempt to distinguish the classification with biological interpretation and clinical implementation. In this review, the progress in molecular subtyping of TNBC is discussed, providing a brief insight in novel TNBC biomarkers and therapeutic strategies.
Keywords: Triple-negative breast cancer, molecular, classification, targeted, therapies, review
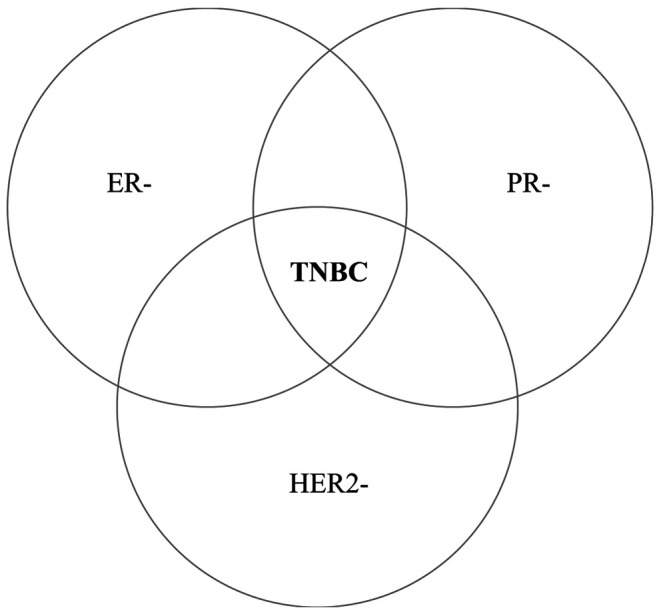
In 2005, Brenton et al., mentioned a group of basal-like breast cancers with “triple-negative receptor status”, not responding to conventional targeted therapies (hormonal or anti-HER2), which is commonly named as Triple-Negative Breast Cancer (TNBC) (1). TNBC is a diverse and heterogeneous type of carcinoma characterized by the absence of estrogen and progesterone receptors (ER and PR respectively), as well as lack of HER2 gene amplification (Figure 1) (2). TNBC represent 10-20% of all types of breast cancers (3). These carcinomas occur more frequently in younger women (<50 years old), in women harboring mutations in BRCA1 gene (more than 75% of BRCA1 carcinomas are triple-negative) and women with specific extraction or economic status (more common to African-American and Hispanic origin) (4-7). The median time to death for patients with TNBC is estimated at 4.2 years in comparison with the 6 years of other breast cancers, while survival after diagnosis of TNBC patients is 10 years at most, compared to 18 years in patients with other breast cancers (8). Furthermore, tumor’s distant recurrence risk in patients with TNBC is maximized in 1-3 years after surgery and remains steady afterwards, while the likelihood of metastasis within 5 years of diagnosis is significantly increased in TNBC compared to other types of cancers (9). The median survival time to death after relapse is only 9 months, although disease-free patients for more than 8 years are less likely to die because of breast cancer (10). Importantly, women with TNBC have higher rates of pathological complete response (pCR) following neoadjuvant chemotherapy, with favorable lasting outcome and lower percentages of recurrence. However, in the case of remaining disease after the therapy, TNBC patients display decreased overall survival (11,12).
Figure 1. Definition of triple-negative breast cancer (TNBC). TNBC is characterized by the absence of estrogen and progesterone receptors (ER and PR respectively), as well as lack of HER2 gene amplification. HER2: Human epidermal growth factor receptor 2.
Histologically, TNBC are mainly high-grade invasive carcinomas of no special type (approximately 90% of the cases), also referred as invasive ductal carcinomas-not otherwise specified (IDC-NOS) (13,14). However, numerous high-grade TNBC have special histological features and can be classified as apocrine, medullary or metaplastic breast carcinomas (MBC) (15). IDCs-NOS are large and highly malignant tumors (16), with pushing invasive border configuration, dense lymphocytic infiltration, nuclear pleomorphism and high mitotic rates (8,17). Apocrine carcinomas are extremely rare (<1%), exhibit nuclear pleomorphism and are characterized by a lymphocytic environment as well as presence of androgen receptors (AR) (18-20). The presence of AR could imply prognostic value, but so far, the data are rather uncertain and not conclusive (21,22). Medullary breast carcinomas usually display basal-like characteristics, with well-defined borders, syncytial growth and brisk lymphocytic infiltrate and have been related to better prognosis (23). In fact, the lymphocytic infiltrate of these carcinomas displays prognostic value for chemotherapy-treated women with TNBC (24,25). MBC consist a group of high-grade, heterogeneous lesions with poor prognosis, including carcinomas with either claudin-low (CLDN-l) or basal-like characteristics (26).
On the other hand, a few TNBC are low-grade, less aggressive carcinomas and can be divided into two subgroups, the salivary gland-like breast tumors with unique genetic alterations and the low-grade triple-negative breast neoplasia family whose expression patterns resemble those of high-grade TNBC (27). Nevertheless, it is important to mention that all subsets of low-grade TNBC can progress to high-grade tumors and during this process, the salivary gland-like subset maintains the differences from the conventional high-grade TNBC (28-30), whereas the low-grade TNBC family is rather similar to conventional high-grade TNBC (31).
The salivary gland-like tumors do not harbor recurrent TP53 mutations, carry specific genes’ expression profile and they represent mostly two subsets; the rare adenoid cystic carcinomas (AdCC) (less than 0.1% of all breast cancers) and the secretory carcinomas (less than 0.15% of all breast cancers) (32). The former show favorable prognosis, have basal-like features and harbor the MYB-NFIB fusion gene due to the t(6;9)q(22-23;p23-24) genetic translocation (33), whereas the latter display excellent clinical outcome, abundant intra- and extracellular secretion of eosinophilic material and harbor the ETV6-NTRK3 fusion gene as a result of the t(12;15)(p13;q25) translocation (34). As far as the low-grade TN breast neoplasia family is concerned, it includes microglandular adenosis (MGA), atypical MGA (AMGA) and acinic cell carcinoma (ACC) (35). MGA/AMGA are clonal neoplastic lesions expressing the S100 protein, display specific genetic alterations (e.g. 5q losses or 8q gains) and carry mutations of TP53 (80%) and other genes (BRCA1, PI3K pathway genes, receptor tyrosine kinases genes) (36,37). ACCs show good prognosis with rare local or distant recurrences, different genomic alterations as well as TP53 (80% of cases) and PIK3CA (10% of cases) mutations (38).
To date, there are considerably restricted prognostic biomarkers and Food and Drug Administration (FDA)-approved therapies for TNBC cases (39,40). The complexity of histopathological and molecular features of these carcinomas along with the diverse clinical response, point out the necessity of the TNBC classification in subgroups with similar characteristics so as to develop new targeted treatments.
Materials and Methods
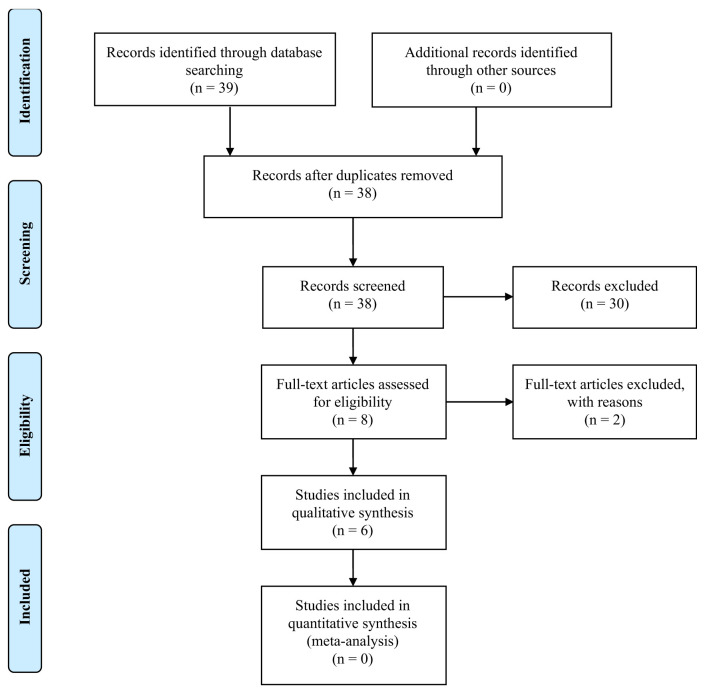
A review of the literature was performed through the PubMed database and Cochrane library aiming to identify articles regarding molecular classification of TNBC. Specifically, our research used keywords, separately and in numerous combinations, such as triple-negative breast cancer, classification, molecular, treatment and prognosis. Furthermore, we double-checked the references from all articles in order to include any other eligible studies. The initial research identified 39 articles. No duplicates were found. These articles were screened and 30 were excluded for various reasons. Some of them were only abstracts, whereas others were not completely relevant to the topic. It should be mentioned that publications in non-English language were also excluded. The full-text articles assessed for eligibility were 8 and 2 of them were excluded because they were reviews of the classifications. Finally, 6 articles were included. The inclusion process is shown in Figure 2.
Figure 2. PRISMA flow diagram for the current study.
Results
Molecular subtypes. In 2000, Perou et al. analyzed the gene expression patterns of 65 human breast tumors and distinguished four clusters, the ER+/luminal-like, basal-like, Erb-B2+ (or HER2+) and normal breast (2). Subsequently, another intrinsic subtype of breast cancer, the CLDN-l tumors, with low expression of luminal markers, high expression of genes related to the epithelial to mesenchymal transition, immune response markers and cancer stem-cell like characteristics, was described. Clinically, most CLDN-l tumors are triple-negative IDCs with poor prognosis and different response to chemotherapy if compared to luminal or basal-like tumors. However, the majority (50-75%) of TNBC exhibit basal-like characteristics and a smaller part can be correlated with any of the aforementioned subgroups (41). Furthermore, basal-like breast carcinomas display histological and immunohistochemical similarities to myoepithelial cells of the breast (42). The identification of basal-like carcinomas requires molecular analysis of genes, however, more commonly, it is used the immunohistochemical expression profile of specific markers such as cytokeratins 5/6,14 and 17, cyclin E, P-cadherin, epidermal growth factor receptor (EGFR) and receptor tyrosine kinase (C-kit) (43,44). Particularly, EGFR and C-kit are overexpressed in 77% of basal-like breast cancers (45). Importantly, 80% of basal-like tumors harbor mutated TP53, whereas there is a 9% having PI3KC mutations (46).
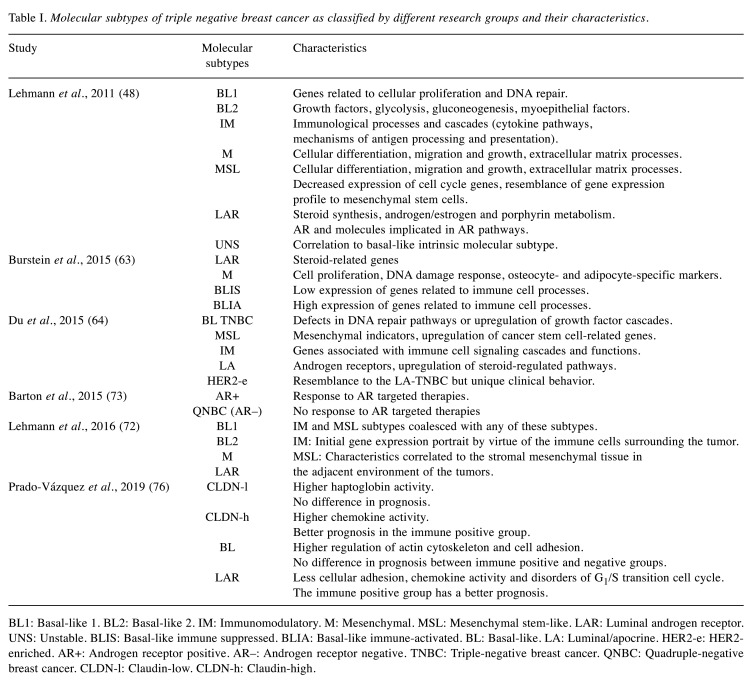
The molecular heterogeneity of TNBC, the poor prognosis and the absence of targeted therapeutic strategies highlight the requirement to classify this diverse disease. Recently, a number of studies concentrated on the investigation of the gene expression profile of TNBC with the use of high-throughput technology tools (47). In 2011, a gene expression database analysis led Lehmann et al., to the identification of six reproducible subtypes of TNBC: Basal-like 1 (BL1), basal-like 2 (BL2), immunomodulatory (IM), mesenchymal (M), mesenchymal stem-like (MSL) and luminal androgen receptor (LAR) subtype. Another type of unstable carcinomas (UNS), was also proposed (48).
The BL1 subtype consists of genes involved in cellular proliferation as well as DNA repair. The database analysis revealed an augmented expression of genes related to cell cycle and DNA damage response [neuroblastoma-RAS (NRAS), MYC, polo-like kinase 1 (PLK1), checkpoint kinase 1 (CHEK1), metalloprotease/disintegrin-like 1 (MDC1), breast cancer gene (BRAC) etc.] (48,49). On the other hand, in subtype BL2 there are genes involved in the signaling pathways of growth factors (EGFR, nerve growth factor (NGF), receptor tyrosine kinase MET, Wnt/β-catenin and insulin-like growth factor 1 (IGF1R) pathways etc.), glycolysis, gluconeogenesis and in the expression of myoepithelial factors [tumor protein 63 (TP63) and membrane metalloendopeptidase (ΜΜΕ) etc.] (50). The IM subtype includes genes associated to immunological processes and cascades (cytokine pathways, mechanisms of antigen processing, nuclear factor-ĸB, tumor necrosis factor (TNF) and JAK/STAT signaling etc.) (51). The other two subtypes, M and MSL, are described by genes related to cellular differentiation, migration and growth, as well as extracellular matrix processes. However, the MSL subtype differs because of the decreased expression of cell-cycle genes and the resemblance of its gene expression profile to that of mesenchymal stem cells [HOX, NGF receptor (NGFR), vascular cell adhesion protein 1 (VCAM1) etc.] (52,53). Finally, the LAR subtype displays a luminal-like expression profile with increased presence of AR and molecules involved in AR pathways (54). It is, also, enriched with genes involved in steroid synthesis, androgen/estrogen and porphyrin metabolism (48). The LAR TNBC subtype has been also linked to molecular apocrine type of breast cancer (55). Cytokeratins can be also criteria for breast cancer characterization as basal-like or luminal (56). Particularly, TNBC subtypes either highly express basal cytokeratins (UNS, BL1, BL2 and M), or luminal cytokeratins (LAR) (57,58).
As far as the intrinsic molecular classification is concerned, the majority of TNBC have basal-like molecular subtype, whereas the M and MSL subtype are mostly normal breast-like. However, it is still controversial if the intrinsic classification is appropriate for TNBC, considering the variety of results in BL2, M and MSL analysis. On the other hand, the vast percentage of LAR TNBC (82%) are luminal A or B (48). A later study conducted to investigate the correlation among PAM50 breast cancer intrinsic subtypes (2) and TNBC subtypes (59), analyzed data from 374 TNBC cases. The results indicated a high percentage of BL1 (99%), BL2 (95%), IM (84%) and M (97%) subtypes exhibiting basal-like phenotype, verifying previous studies (60). On the other hand, half of the MSL TNBC resemble to the basal-like intrinsic subclass, while 28% is normal-like and 14% luminal B. Finally, LAR molecular subtype mostly pertains to HER2 (74%) group, whereas a 14% is luminal B (61). Intriguingly, a cluster of TNBC lacked luminal-specific markers and expressed CLDN-l-related genes (62).
Recently, Burstein et al., proposed 4 stable subtypes of the TNBC with differential prognoses, through mRNA expression analysis and DNA profiling. These subtypes are the luminal/androgen receptor (LAR), the mesenchymal (MES), the basal-like immune suppressed (BLIS) and the basal-like immune-activated subtype (BLIA). The aforementioned subtypes are described by distinguishing RNA and DNA indicators that could be used in treatment strategies. The LAR subtype expresses steroid-related genes (androgen receptor (AR), progesterone receptor (PGR), GATA3 etc.). Interestingly, gene expression patterns of the LAR TNBC subgroup showed that in 1% of these ER-negative tumors (as shown by immunohistochemistry) occurs ER activation, as a result of ER low expression. The MES subtype is enriched with genes implicated in cell proliferation, DNA damage response as well as with osteocyte- and adipocyte-specific markers [osteoglycin (OGN), adiponectin-C1Q and collagen domain containing (ADIPOQ) etc.]. The basal-like immune suppressed subgroup of TNBC, as implied by its name, displays low expression of genes related to immune cell processes. On the contrary, basal-like immune activated cluster is characterized by augmentation of immune cell-specific pathways (63).
Furthermore, a comprehensive study conducted by Le Du et al. gathered published data about TNBC and suggested 5 potential clinically applicable molecular subtypes of TNBC. These are: 1) the basal-like TNBC (BL-TNBC), which appears to be the most common group (25-80% based on the characterization), 2) the mesenchymal-like TNBC (ML-TNBC) 3) the immunomodulatory TNBC (I-TNBC), 4) the luminal/apocrine TNBC (LA-TNBC) and 5) the HER2-enriched TNBC (HER2e-TNBC) (64). The first cluster of TNBC is described by the defects in DNA repair pathways or by the up-regulation of growth factor cascades, as previously reported, mentioning the Lehmann’s division to BL1 and BL2 subtypes (48). The BL-TNBC display higher rates of post-chemotherapy pCR, (65), but it seems that these rates refer to Lehman’s BL1 subtype (51% compared to BL2 with 0%), indicating the possibility of grouping BL2 cluster with other molecular subtypes (66). The ML-TNBC (that is mesenchymal, mesenchymal stem-like, CLDN-l carcinomas) display elevated expression of mesenchymal indicators as well as up-regulation of cancer stem cell-related genes (48,62). PAM50 analysis, in which CLDN-l cluster is not included, assorts MSL to normal-like tumors and the M to basal-like (67). I-TNBC highly express genes associated with immune cell signaling cascades and functions. Despite the diversity of tumor characterization, it is certain that there is a cluster of cancers with increased immune cells infiltration (CLDN-l tumors and Burstein’s BLIA molecular subtype) (68,69). In the LA-TNBC subtype (LAR, luminal A-B, molecular apocrine) there are high levels of androgen receptors leading to up-regulation of steroid-regulated pathways. Finally, the HER2-enriched (low HER2 expression-no HER2 amplification) TNBC subgroup (6-8% of TNBC), shows quite resemblance to the LA TNBC and its separation on account of a unique clinical behavior is yet controversial (2,10,70,71).
Reassessment of previous TNBC grouping by Lehmann et al. led to lessening the number of subtypes from 6 to 4, based on the cell types ambulating in the tumor environment. The redefined subgroups are the BL1, BL2, M and LAR TNBC, whereas the other two (IM and MSL) subtypes were merged with the above, as their initial classification proved to be rather the result of the tumors’ neighboring diverse cellular composition. Particularly, the gene expression portrait of the former subtype referred as IM was proposed to be by virtue of the immune cells surrounding the tumor, rather than a tumor-specific indicator. Likewise, the MSL-cluster characteristics were correlated to the stromal mesenchymal tissue in the adjacent environment of the tumors (72).
Interestingly, a different classification of the TNBC based on the presence or absence of AR, has been recently proposed. Therefore, the two major subclasses are the AR+ TNBC, expressing AR, and the quadruple-negative breast cancer (QNBC) which lacks the androgen receptor (73). Several ongoing clinical trials with AR-targeting agents indicate the significance of the above grouping (74). On the other hand, a comparative study about the inflammatory and non-inflammatory TNBC, revealed no correlation between the molecular subtype of the TNBC and its inflammatory status (75).
In 2019, Prado-Vázquez et al. described two different molecular classifications which were based to cellular type and immune activity. The initial classification included 4 subgroups: 1) CLDN-low, 2) CLDN-high, 3) basal-like and 4) LAR group and was performed according to the stem cell cancer hypothesis (76,77). This classification is related to the differentiation process and especially the initiation of carcinogenesis. This means that the LAR and the CLDN-low type will have the higher and lower cell differentiation rate respectively. As far as the clinical presentation is concerned, the CLDN-low and LAR tumors had a bigger size and higher infiltration rate than the other two groups. The basal-like group had a small size and did not demonstrate lymph node infiltration. The CLDN-high type was large in size but did not show node infiltration. The CLDN-low tumors were related more to haptoglobin, whereas the CLDN-high subtype was related to chemokine. The LAR type demonstrated less cellular adhesion, chemokine activity and disorders of G1/S transition cell cycle. The basal-like type had higher regulation of actin cytoskeleton and cell adhesion (76).
An immune classification was added in the cellular aiming to associate the cellular types to prognosis. The tumors with positive immune activity had better prognosis than the ones with negative immune activity. This was significantly important to the LAR subtype tumors in which the immune positive group demonstrated better survival rates. The CLDN-high tumors with positive immune activity had also a better prognosis. However, these results did not appear in the other two groups.
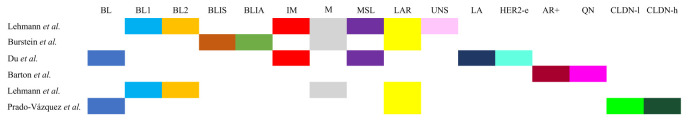
Table I summarizes all the proposed classifications of TNBC and Figure 3 describes their similarities and their differences.
Table I. Molecular subtypes of triple negative breast cancer as classified by different research groups and their characteristics.
BL1: Basal-like 1. BL2: Basal-like 2. IM: Immunomodulatory. M: Mesenchymal. MSL: Mesenchymal stem-like. LAR: Luminal androgen receptor. UNS: Unstable. BLIS: Basal-like immune suppressed. BLIA: Basal-like immune-activated. BL: Basal-like. LA: Luminal/apocrine. HER2-e: HER2- enriched. AR+: Androgen receptor positive. AR–: Androgen receptor negative. TNBC: Triple-negative breast cancer. QNBC: Quadruple-negative breast cancer. CLDN-l: Claudin-low. CLDN-h: Claudin-high
Figure 3. Similarities and differences between the molecular classification of triple-negative breast cancer. BL: Basal-like. BL1: Basal-like 1. BL2: Basal-like 2. BLIS: Basal-like immune suppressed. BLIA: Basal-like immune-activated. IM: Immunomodulatory. M: Mesenchymal. MSL: Mesenchymal stem-like. LAR: Luminal androgen receptor. UNS: Unstable. LA: Luminal/apocrine. HER2-e: HER2-enriched. AR+: Androgen receptor positive. QN: Quadruple-negative breast cancer. CLDN-l: Claudin-low. CLDN-h: Claudin-high.
Therapeutic strategies. Therapeutically, the absence of specific targets in TNBC makes chemotherapy (anthracyclines, taxanes) the only established approach in TNBC treatment, usually with the addition of platinum agents (66,78). The pathological complete response (pCR) rates, differ amongst TNBC subtypes, with BL1 having the highest rate, while BL2 and LAR displaying the lowest (66). Unfortunately, women who do not achieve pCR have worse outcomes compared to patient with other types of breast cancers (79). Generally, non-metastatic TNBC develop better response to neoadjuvant chemotherapy, in contrast to other types of breast cancers. However, there is a diverse response among the different molecular subtypes. During the last decade, a growing number of different therapeutic approaches is under research so as to develop subtype-specific treatments.
Approximately 75% of breast tumors containing the BRCA1 mutation are triple-negative and sensitive to double-stranded DNA breaks. As a result, platinum agents (carboplatin, cisplatin, epirubicin) are widely used in clinical trials, as the DNA damage they cause cannot be repaired by the endogenous homologous recombination mechanism (80). Platinum-based chemotherapy has been shown to increase pCR rate in TNBC patients, whereas patients with metastatic TNBC and BRCA mutations have a better response to carboplatin and docetaxel (12,81,82). However, the results are contradictive when it comes to disease-free survival (DFS), overall survival (OS), as well as metastatic progression (83). Furthermore, ongoing pre-clinical data and clinical trials on PARP inhibitors seem to be promising in basal-like TNBC (84-86). Regarding the mesenchymal TNBC, there are no data from clinical trials and so far, there is little progress in specific targeting (64).
At the same time, immune-based therapy, is also being investigated by several groups (87,88). The immune cells that infiltrate the tumors (TILs) are of great importance in TNBC clinical features as well as in the tumor’s response to treatments (89). The immune-based subtype in the classification of Prado-Vázquez can possibly provide information concerning response to immunotherapy (76). Paradoxically, higher TILs are correlated to worse clinical characteristics, but improved adjuvant and neoadjuvant chemotherapy response (66). Notably, IM TNBC display promising response to pembrolizumab, an immune checkpoint inhibitor (90).
Furthermore, next-generation sequencing revealed some of the most frequent mutations in metastatic TNBC [TP53, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), phosphatase and tensin homolog (PTEN)] as well as an AR protein amplification (AR+ TNBC), indicating new promising therapeutic strategies (91). Currently, it is estimated that approximately 30% of TNBC, including non-LAR subtypes, are AR-dependent, highlighting enzalutamide as a potential targeted chemotherapy in diverse TNBC subtypes (73). Moreover, clinical trials using bicalutamide and enobosarm (AR-inhibitors) report relatively high rates of clinical benefits in AR+ TNBC (92-94). Notably, Garmpis et al., suggested that histone deacetylase inhibitors (HDACi) display some promising in vitro and in vivo results, but clinical trials are preliminary (95).
Several signaling pathways play an important role in different TNBC subtypes, suggesting potential targets (64). Damaskos et al., reported the PI3K/AKT/mTOR pathway, poly ADP-ribose polymerase-1 (PARP-1) inhibitors, tyrosine kinase inhibitors, anti-androgens and anti-angiogenetic factors as possible therapeutic tools against TNBC (96). One particular target is EGFR, which is overexpressed in ML, BL and HER2e TNBC and has been shown to have beneficial results when combined with chemotherapy (97,98). Recently, it has been proposed that the circulating tumor DNA (ctDNA) could be either used as a biomarker for early-phase TNBC diagnosis, or as a way of «monitoring» the tumor clones resistant to chemotherapy in order to redefine the treatment (99).
On the basis of these data, novel targeted approaches should be developed for TNBC. There is no known particular target for TNBC up to now. However, TNBC is a type of breast cancer with challenging biological features and the scientific community’s increasing understanding of these features leads us to improve clinical outcomes. A promising future goal is the application of valuable biomarkers that can predict therapies for TNBC. Concerning the chemosensitive TNBC seem to respond to cytoxic agents such as anthracyclines and taxanes. Moreover, platinum derivatives lead to promising results. Preliminary results with AR inhibitors and PARP inhibitors show hopeful results and can be potential therapeutic tools against TNBC. Anti-angiogenic drugs and EGFR inhibitors are also promising therapeutic tools and up to date results indicate that they may play a significant role TNBC therapy. Recently, clinical studies among patients with breast matrix-producing carcinomas (MPCs), which are rare and usually triple-negative, have been reported (100). Patients with TNBC received anthracycline- and taxane-based neoadjuvant chemotherapy (NAC), but they showed a poor response to standard NAC. A future target could be further studies in order to improve the treatment of this rare but aggressive tumor. At the same time, the investigation of the prognostic value of tumor-infiltrating lymphocytes (TILs) in systemically untreated and early-stage TNBC led to promising results (101). More specifically, the clinical studies suggest that a low TIL level (≤10%) was correlated to beneficial results in patients with T1a/b or N0 disease. Therefore, patients with a low TIL level, even in the T1a/bN0 subset display promising response to adjuvant chemotherapy regardless of the status or the tumor size. Finally, it is known that the radiation therapy (RT) represents a therapeutic strategy in breast cancer. It has been proposed that there are cell lines at a transcriptional level which demonstrate resistance to RT (102). Taking all these into consideration, we are able to better investigate all the mechanisms involved in RT as a therapeutic tool in TNBC in order to develop upgraded targeted therapies.
Conclusion
TNBC are undoubtedly one of the most diverse breast cancer categories. New technologies have facilitated researchers to analyze a large amount of data sets so as to compare different TNBC and categorize them in stable subgroups based on their similarities. The ongoing molecular classification of TNBC is crucial for the understanding of the genetic targets characterizing these tumors and provides potential prognostic and therapeutic approaches. Obviously, the identification of unique subtype-specific genetic targets is the highest ambition for a successful TNBC therapy.
Conflicts of Interest
All the Authors declare that there are no conflicts of interest regarding this study.
Authors’ Contributions
NG, CD and ΑΝ designed the study. NG, CD, AG and KN wrote the article. NG, CD, AG, KN, ED, PF, AP, EV, AS and CGZ collected the data. NN and KK offered scientific advice. DD, EAA, AK and DS revised the manuscript. AN was the supervisor.
References
- 1.Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: Ready for clinical application. J Clin Oncol. 2005;23(29):7350–7360. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Plasilova ML, Hayse B, Killelea BK, Horowitz NR, Chagpar AB, Lannin DR. Features of triple-negative breast cancer: Analysis of 38,813 cases from the national cancer database. Medicine (Baltimore) 2016;95(35):e4614. doi: 10.1097/MD.0000000000004614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrés R, Pajares I, Balmaña J, Llort G, Ramón Y Cajal T, Chirivella I, Aguirre E, Robles L, Lastra E, Pérez-Segura P, Bosch N, Yagüe C, Lerma E, Godino J, Miramar MD, Moros M, Astier P, Saez B, Vidal MJ, Arcusa A, Ramón y Cajal S, Calvo MT, Tres A. Association of BRCA1 germline mutations in young onset triple-negative breast cancer (TNBC) Clin Transl Oncol. 2014;16(3):280–284. doi: 10.1007/s12094-013-1070-9. [DOI] [PubMed] [Google Scholar]
- 5.Siegel R, Ma J, Zou Z, Jemal A. Cancer statisticσ. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 6.Morris GJ, Naidu S, Topham AK, Guiles F, Xu Y, McCue P, Schwartz GF, Park PK, Rosenberg AL, Brill K, Mitchell EP. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: A single-institution compilation compared with the National Cancer Institute’s Surveillance, Epidemiology, and End Results database. Cancer. 2007;110(4):876–884. doi: 10.1002/cncr.22836. [DOI] [PubMed] [Google Scholar]
- 7.Brewster AM, Chavez-MacGregor M, Brown P. Epidemiology, biology, and treatment of triple-negative breast cancer in women of African ancestry. Lancet Oncol. 2014;15(13):e625–e634. doi: 10.1016/S1470-2045(14)70364-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin Cancer Res . 2007;13(15 Pt1):4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 9.Dent R, Hanna WM, Trudeau M, Rawlinson E, Sun P, Narod SA. Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res Treat. 2009;115(2):423–428. doi: 10.1007/s10549-008-0086-2. [DOI] [PubMed] [Google Scholar]
- 10.Haffty BG, Yang Q, Reiss M, Kearney T, Higgins SA, Weidhaas J, Harris L, Hait W, Toppmeyer D. Locoregional relapse and distant metastasis in conservatively managed triple-negative early-stage breast cancer. J Clin Oncol. 2006;24(36):5652–5657. doi: 10.1200/JCO.2006.06.5664. [DOI] [PubMed] [Google Scholar]
- 11.von Minckwitz G, Untch M, Blohmer J-U, Costa SD, Eidtmann H, Fasching PA, Gerber B, Eiermann W, Hilfrich J, Huober J, Jackisch C, Kaufmann M, Konecny GE, Denkert C, Nekljudova V, Mehta K, Loibl S. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol. 2012;30(15):1796–1804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 12.von Minckwitz G, Schneeweiss A, Loibl S, Salat C, Denkert C, Rezai M, Blohmer JU, Jackisch C, Paepke S, Gerber B, Zahm DM, Kümmel S, Eidtmann H, Klare P, Huober J, Costa S, Tesch H, Hanusch C, Hilfrich J, Khandan F, Fasching PA, Sinn BV, Engels K, Mehta K, Nekljudova V, Untch M. Neoadjuvant carboplatin in patients with triple-negative and HER2-positive early breast cancer (GeparSixto; GBG 66): A randomised phase 2 trial. Lancet Oncol. 2014;15(7):747–756. doi: 10.1016/S1470-2045(14)70160-3. [DOI] [PubMed] [Google Scholar]
- 13.Bose S. Triple-negative breast carcinoma: morphologic and molecular subtypes. Adv Anat Pathol. 2015;22(5):306–313. doi: 10.1097/PAP.0000000000000084. [DOI] [PubMed] [Google Scholar]
- 14.Geyer FC, Pareja F, Weigelt B, Rakha E, Ellis IO, Schnitt SJ, Reis-Filho JS. The spectrum of triple-negative breast disease: High- and low-grade lesions. Am J Pathol. 2017;187(10):2139–2151. doi: 10.1016/j.ajpath.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rakha EA, Reis-Filho JS, Baehner F, Dabbs DJ, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, Palacios J, Richardson AL, Schnitt SJ, Schmitt FC, Tan PH, Tse GM, Badve S, Ellis IO. Breast cancer prognostic classification in the molecular era: The role of histological grade. Breast Cancer Res. 2010;12(4):207. doi: 10.1186/bcr2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Ding A. Comparison of invasive micropapillary and triple-negative invasive ductal carcinoma of the breast. Breast. 2015;24(6):723–731. doi: 10.1016/j.breast.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Krizmanich-Conniff KM, Paramagul C, Patterson SK, Helvie MA, Roubidoux MA, Myles JD, Jiang K, Sabel M. Triple receptor-negative breast cancer: Imaging and clinical characteristics. AJR Am J Roentgenol. 2012;199(2):458–464. doi: 10.2214/AJR.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasahara M, Matsui A, Ichimura Y, Hirakata Y, Murata Y, Marui E. Overexpression of androgen receptor and forkhead-box A1 protein in apocrine breast carcinoma. Anticancer Res. 2014;34(3):1261–1267. [PubMed] [Google Scholar]
- 19.Durham JR, Fechner RE. The histologic spectrum of apocrine lesions of the breast. Am J Clin Pathol . 2000;113(5 Suppl 1):S3–S18. doi: 10.1309/7A2P-YMWJ-B1PD-UDN9. [DOI] [PubMed] [Google Scholar]
- 20.Tsutsumi Y. Apocrine carcinoma as triple-negative breast cancer: Novel definition of apocrine-type carcinoma as estrogen/progesterone receptor-negative and androgen receptor-positive invasive ductal carcinoma. Jpn J Clin Oncol. 2012;42(5):375–386. doi: 10.1093/jjco/hys034. [DOI] [PubMed] [Google Scholar]
- 21.Choi JE, Kang SH, Lee SJ, Bae YK. Androgen receptor expression predicts decreased survival in early stage triple-negative breast cancer. Ann Surg Oncol. 2015;22(1):82–99. doi: 10.1245/s10434-014-3984-z. [DOI] [PubMed] [Google Scholar]
- 22.Vera-Badillo FE, Templeton AJ, de Gouveia P, Diaz-Padilla I, Bedard PL, Al-Mubarak M, Seruga B, Tannock IF, Ocana A, Amir E. Androgen receptor expression and outcomes in early breast cancer: A systematic review and meta-analysis. J Natl Cancer Inst. 2013;106(1):djt319. doi: 10.1093/jnci/djt319. [DOI] [PubMed] [Google Scholar]
- 23.Kleer CG. Carcinoma of the breast with medullary-like features: Diagnostic challenges and relationship with BRCA1 and EZH2 functions. Arch Pathol Lab Med. 2009;133(11):1822–1825. doi: 10.1043/1543-2165-133.11.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, Rouas G, Francis P, Crown JP, Hitre E, de Azambuja E, Quinaux E, Di Leo A, Michiels S, Piccart MJ, Sotiriou C. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol. 2013;31(7):860–867. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 25.Adams S, Gray RJ, Demaria S, Goldstein L, Perez EA, Shulman LN, Martino S, Wang M, Jones VE, Saphner TJ, Wolff AC, Wood WC, Davidson NE, Sledge GW, Sparano JA, Badve SS. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol. 2014;32(27):2959–2966. doi: 10.1200/JCO.2013.55.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weigelt B, Ng CK, Shen R, Popova T, Schizas M, Natrajan R, Mariani O, Stern MH, Norton L, Vincent-Salomon A, Reis-Filho JS. Metaplastic breast carcinomas display genomic and transcriptomic heterogeneity. Mod Pathol. 2015;28(3):340–351. doi: 10.1038/modpathol.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinn HP, Kreipe H. A brief overview of the WHO classification of breast tumors, 4th Edition, focusing on issues and updates from the 3rd Edition. Breast Care (Basel) 2013;8(2):149–154. doi: 10.1159/000350774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fusco N, Geyer FC, De Filippo MR, Martelotto LG, Ng CKY, Piscuoglio S, Guerini-Rocco E, Schultheis AM, Fuhrmann L, Wang L, Jungbluth AA, Burke KA, Lim RS, Vincent-Salomon A, Bamba M, Moritani S, Badve SS, Ichihara S, Ellis IO, Reis-Filho JS, Weigelt B. Genetic events in the progression of adenoid cystic carcinoma of the breast to high-grade triple-negative breast cancer. Mod Pathol. 2016;29(11):1292–1305. doi: 10.1038/modpathol.2016.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seethala RR, Cieply K, Barnes EL, Dacic S. Progressive genetic alterations of adenoid cystic carcinoma with high-grade transformation. Arch Pathol Lab Med. 2011;135(1):123–130. doi: 10.1043/2010-0048-OAR.1. [DOI] [PubMed] [Google Scholar]
- 30.Del Castillo M, Chibon F, Arnould L, Croce S, Ribeiro A, Perot G, Hostein I, Geha S, Bozon C, Garnier A, Lae M, Vincent-Salomon A, MacGrogan G. Secretory breast carcinoma: A histopathologic and genomic spectrum characterized by a joint specific ETV6-NTRK3 gene fusion. Am J Surg Pathol. 2015;39(11):1458–1467. doi: 10.1097/PAS.0000000000000487. [DOI] [PubMed] [Google Scholar]
- 31.Khalifeh IM, Albarracin C, Diaz LK, Symmans FW, Edgerton ME, Hwang RF, Sneige N. Clinical, histopathologic, and immunohistochemical features of microglandular adenosis and transition into in situ and invasive carcinoma. Am J Surg Pathol. 2008;32(4):544–552. doi: 10.1097/PAS.0b013e31815a87e2. [DOI] [PubMed] [Google Scholar]
- 32.Pareja F, Geyer FC, Marchiò C, Burke KA, Weigelt B, Reis-Filho JS. Triple-negative breast cancer: The importance of molecular and histologic subtyping, and recognition of low-grade variants. NPJ Breast Cancer. 2016;2(1):16036. doi: 10.1038/npjbcancer.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Persson M, Andren Y, Mark J, Horlings HM, Persson F, Stenman G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci USA. 2009;106(44):18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, Becker L, Carneiro F, MacPherson N, Horsman D, Poremba C, Sorensen PH. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2(5):367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 35.Geyer FC, Berman SH, Marchio C, Burke KA, Guerini-Rocco E, Piscuoglio S, Ng CK, Pareja F, Wen HY, Hodi Z, Schnitt SJ, Rakha EA, Ellis IO, Norton L, Weigelt B, Reis-Filho JS. Genetic analysis of microglandular adenosis and acinic cell carcinomas of the breast provides evidence for the existence of a low-grade triple-negative breast neoplasia family. Mod Pathol. 2017;30(1):69–84. doi: 10.1038/modpathol.2016.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guerini-Rocco E, Piscuoglio S, Ng CKY, Geyer FC, De Filippo MR, Eberle CA, Akram M, Fusco N, Ichihara S, Sakr RA, Yatabe Y, Vincent-Salomon A, Rakha EA, Ellis IO, Wen YH, Weigelt B, Schnitt SJ, Reis-Filho JS. Microglandular adenosis associated with triple-negative breast cancer is a neoplastic lesion of triple-negative phenotype harbouring TP53 somatic mutations. J Pathol. 2016;238(5):677–688. doi: 10.1002/path.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geyer FC, Lacroix-Triki M, Colombo P-E, Patani N, Gauthier A, Natrajan R, Lambros MB, Khalifeh I, Albarracin C, Orru S, Marchiò C, Sapino A, Mackay A, Weigelt B, Schmitt FC, Wesseling J, Sneige N, Reis-Filho JS. Molecular evidence in support of the neoplastic and precursor nature of microglandular adenosis. Histopathology. 2012;60(6B):E115–E130. doi: 10.1111/j.1365-2559.2012.04207.x. [DOI] [PubMed] [Google Scholar]
- 38.Ripamonti CB, Colombo M, Mondini P, Siranoush M, Peissel B, Bernard L, Radice P, Carcangiu ML. First description of an acinic cell carcinoma of the breast in a BRCA1 mutation carrier: A case report. BMC Cancer. 2013;13(1):46. doi: 10.1186/1471-2407-13-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saha P, Nanda R. Concepts and targets in triple-negative breast cancer: Recent results and clinical implications. Ther Adv Med Oncol. 2016;8(5):351–359. doi: 10.1177/1758834016657071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fleisher B, Clarke C, Ait-Oudhia S. Current advances in biomarkers for targeted therapy in triple-negative breast cancer. Breast Cancer (Dove Med Press) 2016;8:183–197. doi: 10.2147/BCTT.S114659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma CX, Luo J, Ellis MJ. Molecular profiling of triple-negative breast cancer. Breast Dis. 2010;32(1-2):73–84. doi: 10.3233/BD-2010-0309. [DOI] [PubMed] [Google Scholar]
- 42.Venkitaraman R. Triple-negative/basal-like breast cancer: Clinical, pathologic and molecular features. Expert Rev Anticancer Ther. 2010;10(2):199–207. doi: 10.1586/era.09.189. [DOI] [PubMed] [Google Scholar]
- 43.Arnes JB, Brunet J-S, Stefansson I, Begin LR, Wong N, Chappuis PO, Akslen LA, Foulkes WD. Placental cadherin and the basal epithelial phenotype of BRCA1-related breast cancer. Clin Cancer Res. 2005;11(11):4003–4311. doi: 10.1158/1078-0432.CCR-04-2064. [DOI] [PubMed] [Google Scholar]
- 44.Bori R, Cserni G. Basal phenotype in breast carcinoma occurring in women aged 35 or younger. Pathol Oncol Res. 2009;15(1):41–45. doi: 10.1007/s12253-008-9090-4. [DOI] [PubMed] [Google Scholar]
- 45.Lerma E, Peiro G, Ramon T, Fernandez S, Martinez D, Pons C, Muñoz F, Sabate JM, Alonso C, Ojeda B, Prat J, Barnadas A. Immunohistochemical heterogeneity of breast carcinomas negative for estrogen receptors, progesterone receptors and Her2/neu (basal-like breast carcinomas) Mod Pathol. 2007;20(11):1200–1207. doi: 10.1038/modpathol.3800961. [DOI] [PubMed] [Google Scholar]
- 46.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, Burleigh A, Yap D, Bernard V, McPherson A, Shumansky K, Crisan A, Giuliany R, Heravi-Moussavi A, Rosner J, Lai D, Birol I, Varhol R, Tam A, Dhalla N, Zeng T, Ma K, Chan SK, Griffith M, Moradian A, Cheng SW, Morin GB, Watson P, Gelmon K, Chia S, Chin SF, Curtis C, Rueda OM, Pharoah PD, Damaraju S, Mackey J, Hoon K, Harkins T, Tadigotla V, Sigaroudinia M, Gascard P, Tlsty T, Costello JF, Meyer IM, Eaves CJ, Wasserman WW, Jones S, Huntsman D, Hirst M, Caldas C, Marra MA, Aparicio S. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Judes G, Rifai K, Daures M, Dubois L, Bignon Y-J, Penault-Llorca F, Bernard-Gallon D. High-throughput «Omics» technologies: New tools for the study of triple-negative breast cancer. Cancer Lett. 2016;382(1):77–85. doi: 10.1016/j.canlet.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zugazagoitia J, Perez-Segura P, Manzano A, Blanco I, Vega A, Custodio A, Teulé A, Fachal L, Martínez B, González-Sarmiento R, Cruz-Hernández JJ, Chirivella I, Garcés V, Garre P, Romero A, Caldés T, Díaz-Rubio E, de la Hoya M. Limited family structure and triple-negative breast cancer (TNBC) subtype as predictors of BRCA mutations in a genetic counseling cohort of early-onset sporadic breast cancers. Breast Cancer Res Treat. 2014;148(2):415–421. doi: 10.1007/s10549-014-3167-4. [DOI] [PubMed] [Google Scholar]
- 50.Orzol P, Nekulova M, Holcakova J, Muller P, Votesek B, Coates PJ. DeltaNp63 regulates cell proliferation, differentiation, adhesion, and migration in the BL2 subtype of basal-like breast cancer. Tumour Biol. 2016;37(8):10133–10140. doi: 10.1007/s13277-016-4880-x. [DOI] [PubMed] [Google Scholar]
- 51.Bertucci F, Finetti P, Cervera N, Charafe-Jauffret E, Mamessier E, Adélaïde J, Debono S, Houvenaeghel G, Maraninchi D, Viens P, Charpin C, Jacquemier J, Birnbaum D. Gene expression profiling shows medullary breast cancer is a subgroup of basal breast cancers. Cancer Res. 2006;66(9):4636–4644. doi: 10.1158/0008-5472.CAN-06-0031. [DOI] [PubMed] [Google Scholar]
- 52.Zhu A, Li Y, Song W, Xu Y, Yang F, Zhang W, Yin Y, Guan X. Antiproliferative effect of androgen receptor inhibition in mesenchymal stem-like triple-negative breast cancer. Cell Physiol Biochem. 2016;38(3):1003–1014. doi: 10.1159/000443052. [DOI] [PubMed] [Google Scholar]
- 53.Wahdan-Alaswad R, Harrell JC, Fan Z, Edgerton SM, Liu B, Thor AD. Metformin attenuates transforming growth factor beta (TGF-beta) mediated oncogenesis in mesenchymal stem-like/claudin-low triple negative breast cancer. Cell Cycle. 2016;15(8):1046–1059. doi: 10.1080/15384101.2016.1152432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rahim B, O’Regan R. AR Signaling in breast cancer. Cancers (Basel) 2017;9(3):E21. doi: 10.3390/cancers9030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, Macgrogan G, Bergh J, Cameron D, Goldstein D, Duss S, Nicoulaz AL, Brisken C, Fiche M, Delorenzi M, Iggo R. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24(29):4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 56.Valentin MD, da Silva SD, Privat M, Alaoui-Jamali M, Bignon YJ. Molecular insights on basal-like breast cancer. Breast Cancer Res Treat. 2012;134(1):21–30. doi: 10.1007/s10549-011-1934-z. [DOI] [PubMed] [Google Scholar]
- 57.Yang XR, Chang-Claude J, Goode EL, Couch FJ, Nevanlinna H, Milne RL, Gaudet M, Schmidt MK, Broeks A, Cox A, Fasching PA, Hein R, Spurdle AB, Blows F, Driver K, Flesch-Janys D, Heinz J, Sinn P, Vrieling A, Heikkinen T, Aittomäki K, Heikkilä P, Blomqvist C, Lissowska J, Peplonska B, Chanock S, Figueroa J, Brinton L, Hall P, Czene K, Humphreys K, Darabi H, Liu J, Van’t Veer LJ, van Leeuwen FE, Andrulis IL, Glendon G, Knight JA, Mulligan AM, O'Malley FP, Weerasooriya N, John EM, Beckmann MW, Hartmann A, Weihbrecht SB, Wachter DL, Jud SM, Loehberg CR, Baglietto L, English DR, Giles GG, McLean CA, Severi G, Lambrechts D, Vandorpe T, Weltens C, Paridaens R, Smeets A, Neven P, Wildiers H, Wang X, Olson JE, Cafourek V, Fredericksen Z, Kosel M, Vachon C, Cramp HE, Connley D, Cross SS, Balasubramanian SP, Reed MW, Dörk T, Bremer M, Meyer A, Karstens JH, Ay A, Park-Simon TW, Hillemanns P, Arias Pérez JI, Menéndez Rodríguez P, Zamora P, Benítez J, Ko YD, Fischer HP, Hamann U, Pesch B, Brüning T, Justenhoven C, Brauch H, Eccles DM, Tapper WJ, Gerty SM, Sawyer EJ, Tomlinson IP, Jones A, Kerin M, Miller N, McInerney N, Anton-Culver H, Ziogas A, Shen CY, Hsiung CN, Wu PE, Yang SL, Yu JC, Chen ST, Hsu GC, Haiman CA, Henderson BE, Le Marchand L, Kolonel LN, Lindblom A, Margolin S, Jakubowska A, Lubiński J, Huzarski T, Byrski T, Górski B, Gronwald J, Hooning MJ, Hollestelle A, van den Ouweland AM, Jager A, Kriege M, Tilanus-Linthorst MM, Collée M, Wang-Gohrke S, Pylkäs K, Jukkola-Vuorinen A, Mononen K, Grip M, Hirvikoski P, Winqvist R, Mannermaa A, Kosma VM, Kauppinen J, Kataja V, Auvinen P, Soini Y, Sironen R, Bojesen SE, Ørsted DD, Kaur-Knudsen D, Flyger H, Nordestgaard BG, Holland H, Chenevix-Trench G, Manoukian S, Barile M, Radice P, Hankinson SE, Hunter DJ, Tamimi R, Sangrajrang S, Brennan P, McKay J, Odefrey F, Gaborieau V, Devilee P, Huijts PE, Tollenaar RA, Seynaeve C, Dite GS, Apicella C, Hopper JL, Hammet F, Tsimiklis H, Smith LD, Southey MC, Humphreys MK, Easton D, Pharoah P, Sherman ME, Garcia-Closas M. Associations of breast cancer risk factors with tumor subtypes: A pooled analysis from the Breast Cancer Association Consortium Studies. J Natl Cancer Inst. 2011;103(3):250–263. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, Nobel A, Parker J, Ewend MG, Sawyer LR, Wu J, Liu Y, Nanda R, Tretiakova M, Ruiz Orrico A, Dreher D, Palazzo JP, Perreard L, Nelson E, Mone M, Hansen H, Mullins M, Quackenbush JF, Ellis MJ, Olopade OI, Bernard PS, Perou CM. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7(1):96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen X, Li J, Gray WH, Lehmann BD, Bauer JA, Shyr Y, Pietenpol JA. TNBCtype: A Subtyping tool for triple-negative breast cancer. Cancer Inform. 2012;11:147–156. doi: 10.4137/CIN.S9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prat A, Adamo B, Cheang MCU, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18(2):123–133. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol. 2014;232(2):142–150. doi: 10.1002/path.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Fuqua SAW, Savage MI, Osborne CK, Hilsenbeck SG, Chang JC, Mills GB, Lau CC, Brown PH. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21(7):1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du F Le, Eckhardt BL, Lim B, Litton JK, Moulder S, Meric-Bernstam F, Gonzalez-Angulo AM, Ueno NT. Is the future of personalized therapy in triple-negative breast cancer based on molecular subtype. Oncotarget. 2015;6(15):12890–12908. doi: 10.18632/oncotarget.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, Hess KR, Stec J, Ayers M, Wagner P, Morandi P, Fan C, Rabiul I, Ross JS, Hortobagyi GN, Pusztai L. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11(16):5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 66.Masuda H, Baggerly KA, Wang Y, Zhang Y, Gonzalez-Angulo AM, Meric-Bernstam F, Valero V, Lehmann BD, Pietenpol JA, Hortobagyi GN, Symmans WF, Ueno NT. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res. 2013;19(19):5533–5540. doi: 10.1158/1078-0432.CCR-13-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mayer IA, Abramson VG, Lehmann BD, Pietenpol JA. New strategies for triple-negative breast cancer-deciphering the heterogeneity. Clin Cancer Res. 2014;20(4):782–790. doi: 10.1158/1078-0432.CCR-13-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, Park M. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14(5):518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 69.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK, Hilsenbeck SG, Chang JC, Mills GB, Lau CC, Brown PH. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21(7):1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dalenc F. Medical treatment of breast cancer: Chemotherapy and tailored therapy. Rev Prat. 2013;63(10):1408–1414. [PubMed] [Google Scholar]
- 71.Cancer Genome Atlas Network, Collaborators Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, Moses HL, Sanders ME, Pietenpol Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadjuvant chemotherapy selection. PLoS One. 2016;11(6):e0157368. doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barton VN, D’Amato NC, Gordon MA, Christenson JL, Elias A, Richer JK. Androgen receptor biology in triple negative breast cancer: A case for classification as AR+ or quadruple negative disease. Horm Cancer. 2015;6(5-6):206–213. doi: 10.1007/s12672-015-0232-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hon JDC, Singh B, Sahin A, Du G, Wang J, Wang VY, Deng FM, Zhang DY, Monaco ME, Lee P. Breast cancer molecular subtypes: From TNBC to QNBC. Am J Cancer Res. 2016;6(9):1864–1872. [PMC free article] [PubMed] [Google Scholar]
- 75.Masuda H, Baggerly KA, Wang Y, Iwamoto T, Brewer T, Pusztai L, Kai K, Kogawa T, Finetti P, Birnbaum D, Dirix L, Woodward WA, Reuben JM, Krishnamurthy S, Symmans W, Van Laere SJ, Bertucci F, Hortobagyi GN, Ueno NT. Comparison of molecular subtype distribution in triple-negative inflammatory and non-inflammatory breast cancers. Breast Cancer Res. 2013;15(6):R112. doi: 10.1186/bcr3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prado-Vázquez G, Gámez-Pozo A, Trilla-Fuertes L, Arevalillo JM, Zapater-Moros A, Ferrer-Gómez M, Díaz-Almirón M, López-Vacas R, Navarro H, Maín P, Feliú J, Zamora P, Espinosa E, Fresno Vara JÁ. A novel approach to triple-negative breast cancer molecular classification reveals a luminal immune-positive subgroup with good prognoses. Sci Rep. 2019;9(1):1538. doi: 10.1038/s41598-018-38364-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shipitsin M, Polyak K. The cancer stem cell hypothesis: In search of definitions, markers, and relevance. Lab Invest. 2008;88(5):459–463. doi: 10.1038/labinvest.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petrelli F, Coinu A, Borgonovo K, Cabiddu M, Ghilardi M, Lonati V, Barni S. The value of platinum agents as neoadjuvant chemotherapy in triple-negative breast cancers: A systematic review and meta-analysis. Breast Cancer Res Treat. 2014;144(2):223–232. doi: 10.1007/s10549-014-2876-z. [DOI] [PubMed] [Google Scholar]
- 79.Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, Cristofanilli M, Hortobagyi GN, Pusztai L. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26(8):1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 80.Villarreal-Garza C, Khalaf D, Bouganim N, Clemons M, Pena-Curiel O, Baez-Revueltas B, Kiss A, Kassam F, Enright K, Verma S, Pritchard K, Myers J, Dent R. Platinum-based chemotherapy in triple-negative advanced breast cancer. Breast Cancer Res Treat. 2014;146(3):567–572. doi: 10.1007/s10549-014-3033-4. [DOI] [PubMed] [Google Scholar]
- 81.Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, Fatima A, Gelman RS, Ryan PD, Tung NM, De Nicolo A, Ganesan S, Miron A, Colin C, Sgroi DC, Ellisen LW, Winer EP, Garber JE. Efficacy of neoadjuvant cisplatin in triple-negative breast cancer. J Clin Oncol. 2010;28(7):1145–1153. doi: 10.1200/JCO.2009.22.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sikov WM, Berry DA, Perou CM, Singh B, Cirrincione CT, Tolaney SM, Kuzma CS, Pluard TJ, Somlo G, Port ER, Golshan M, Bellon JR, Collyar D, Hahn OM, Carey LA, Hudis CA, Winer EP. Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple-negative breast cancer: CALGB 40603 (Alliance) J Clin Oncol. 2015;33(1):13–21. doi: 10.1200/JCO.2014.57.0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.La Belle A, Khatib J, Schiemann WP, Vinayak S. Role of platinum in early-stage triple-negative breast cancer. Curr Treat Options Oncol. 2017;18(11):68. doi: 10.1007/s11864-017-0506-9. [DOI] [PubMed] [Google Scholar]
- 84.Shen Y, Rehman FL, Feng Y, Boshuizen J, Bajrami I, Elliott R, Wang B, Lord CJ, Post LE, Ashworth A. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin Cancer Res. 2013;19(18):5003–5015. doi: 10.1158/1078-0432.CCR-13-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mateo J, Ong M, Tan DSP, Gonzalez MA, de Bono JS. Appraising iniparib, the PARP inhibitor that never was--what must we learn. Nat Rev Clin Oncol. 2013;10(12):688–696. doi: 10.1038/nrclinonc.2013.177. [DOI] [PubMed] [Google Scholar]
- 86.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, Wardley A, Mitchell G, Earl H, Wickens M, Carmichael J. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet. 2010;376(9737):235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 87.Emens LA. Breast cancer immunotherapy: Facts and hopes. Clin Cancer Res. 2017;24(3):511–520. doi: 10.1158/1078-0432.CCR-16-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van Rooijen JM, Stutvoet TS, Schroder CP, de Vries EGE. Immunotherapeutic options on the horizon in breast cancer treatment. Pharmacol Ther. 2015;156:90–101. doi: 10.1016/j.pharmthera.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 89.Ibrahim EM, Al-Foheidi ME, Al-Mansour MM, Kazkaz GA. The prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancer: A meta-analysis. Breast Cancer Res Treat. 2014;148(3):467–476. doi: 10.1007/s10549-014-3185-2. [DOI] [PubMed] [Google Scholar]
- 90.Nanda R, Chow LQM, Dees EC, Berger R, Gupta S, Geva R, Pathiraja K, Aktan G, Cheng JD, Karantza V, Buisseret L. Pembrolizumab in patients with advanced triple-negative breast cancer: Phase Ib KEYNOTE-012 study. J Clin Oncol. 2016;34(21):2460–2467. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Millis SZ, Gatalica Z, Winkler J, Vranic S, Kimbrough J, Reddy S, O'Shaughnessy JA. Predictive biomarker profiling of >6000 breast cancer patients shows heterogeneity in TNBC, with treatment implications. Clin Breast Cancer. 2015;15(6):473–481. doi: 10.1016/j.clbc.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 92.Cochrane DR, Bernales S, Jacobsen BM, Cittelly DM, Howe EN, D’Amato NC, Spoelstra NS, Edgerton SM, Jean A, Guerrero J, Gómez F, Medicherla S, Alfaro IE, McCullagh E, Jedlicka P, Torkko KC, Thor AD, Elias AD, Protter AA, Richer JK. Role of the androgen receptor in breast cancer and preclinical analysis of enzalutamide. Breast Cancer Res. 2014;16(1):R7. doi: 10.1186/bcr3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cortes Castan J, Schmid P, Awada A, Uppal H, Tudor IC, Blaney ME, Steinberg JL, Yardley DA, Hudis C, Traina TA. Stage 1 results from MDV3100-11: A 2-stage study of enzalutamide (ENZA), an androgen receptor (AR) inhibitor, in advanced AR+ triple-negative breast cancer (TNBC) Ann Oncol. 2015;26(Suppl 3):iii6. doi: 10.1093/annonc/mdv115.01. [DOI] [Google Scholar]
- 94.Overmoyer B, Sanz-Altamira P, Taylor RP, Hancock ML, Dalton JT, Johnston MA, Steiner MS. Enobosarm: A targeted therapy for metastatic, androgen receptor positive, breast cancer. J Clin Oncol. 2014;32(15 Suppl):568. doi: 10.1200/jco.2014.32.15_suppl.568. [DOI] [Google Scholar]
- 95.Garmpis N, Damaskos C, Garmpi A, Kalampokas E, Kalampokas T, Spartalis E, Daskalopoulou A, Valsami S, Kontos M, Nonni A, Kontzoglou K, Perrea D, Nikiteas N, Dimitroulis D. Histone deacetylases as new therapeutic targets in triple-negative breast cancer: Progress and promises. Cancer Genomics Proteomics. 2017;14(5):299–313. doi: 10.21873/cgp.20041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Damaskos C, Garmpi A, Nikolettos K, Vavourakis M, Diamantis E, Patsouras A, Farmaki P, Nonni A, Dimitroulis D, Mantas D, Antoniou EA, Nikolettos N, Kontzoglou K, Garmpis N. Triple-negative breast cancer: The progress of targeted therapies and future tendencies. Anticancer. 2019;Res39(10):5285–5296. doi: 10.21873/anticanres.13722. [DOI] [PubMed] [Google Scholar]
- 97.Baselga J, Gomez P, Greil R, Braga S, Climent MA, Wardley AM, Kaufman B, Stemmer SM, Pêgo A, Chan A, Goeminne JC, Graas MP, Kennedy MJ, Ciruelos Gil EM, Schneeweiss A, Zubel A, Groos J, Melezínková H, Awada A. Randomized phase II study of the anti-epidermal growth factor receptor monoclonal antibody cetuximab with cisplatin versus cisplatin alone in patients with metastatic triple-negative breast cancer. J Clin Oncol. 2013;31(20):2586–2592. doi: 10.1200/JCO.2012.46.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nabholtz J, Weber B, Mouret-Reynier M, Gligorov J, Coudert BP, Vanlemmens L, Petit T, Tredan O, Van Praagh-Doreau I, Dubray-Longeras P, Ferriere J, Nayl B, Tubiana-Mathieu N, Jouannaud C, Devaud H, Abrial C, Planchat E, Chalabi N, Penault-Llorca FM, Chollet PJM. Panitumumab in combination with FEC 100 (5-fluorouracil, epidoxorubicin, cyclophosphamide) followed by docetaxel (T) in patients with operable, triple-negative breast cancer (TNBC): Preliminary results of a multicenter neoadjuvant pilot phase II study. J Clin Oncol. 2011;29(15 Suppl):e11574. doi: 10.1200/jco.2011.29.15_suppl.e11574. [DOI] [Google Scholar]
- 99.Saliou A, Bidard FC, Lantz O, Stern MH, Vincent-Salomon A, Proudhon C, Pierga JY. Circulating tumor DNA for triple-negative breast cancer diagnosis and treatment decisions. Expert Rev Mol Diagn. 2016;16(1):39–50. doi: 10.1586/14737159.2016.1121100. [DOI] [PubMed] [Google Scholar]
- 100.Shimada K, Ishikawa T, Yamada A, Sugae S, Narui K, Shimizu D, Chishima T, Endo I. Matrix-producing carcinoma as an aggressive triple-negative breast cancer: Clinicopathological features and response to neoadjuvant chemotherapy. Anticancer Res. 2019;39(7):3863–3869. doi: 10.21873/anticanres.13536. [DOI] [PubMed] [Google Scholar]
- 101.Park JH, Lee HJ, Lee SB, Ahn JH, Kim JE, Jung KH, Gong G, Son BH, Ahn SH, Kim SB. Intrinsic prognostic impact of tumor-infiltrating lymphocytes in systemically untreated patients with early-stage triple-negative breast cancer. Anticancer Res. 2019;39(6):3111–3119. doi: 10.21873/anticanres.13447. [DOI] [PubMed] [Google Scholar]
- 102.Bravatà V, Cammarata FP, Minafra L, Musso R, Pucci G, Spada M, Fazio I, Russo G, Forte GI. Gene expression profiles induced by high-dose ionizing radiation in MDA-MB-231 triple-negative breast cancer cell line. Cancer Genomics Proteomics. 2019;16(4):257–266. doi: 10.21873/cgp.20130. [DOI] [PMC free article] [PubMed] [Google Scholar]