Abstract
Our genetic background provides limited information on individual risk of developing vascular complications overtime. New biological layers, namely epigenetic modifications, are now emerging as potent regulators of gene expression thus leading to altered transcriptional programs and vascular disease phenotypes. Such epigenetic modifications, defined as changes to the genome that do not involve changes in DNA sequence, are generally induced by environmental factors and poor lifestyle habits. Of note, adverse epigenetic signals acquired during life can be transmitted to the offspring thus leading to premature alterations of the epigenetic and transcriptional landscape eventually leading to early endothelial dysfunction and vascular senescence. Modifications of the epigenome play a pivotal role in the pathophysiology of cardiometabolic disturbances such as obesity and type 2 diabetes. In these patients, changes of DNA methylation and chromatin structure contribute to alter pathways regulating insulin sensitivity, glucose homeostasis, adipogenesis and vascular function. In this perspective, unveiling the ‘epigenetic landscape’ in cardiometabolic patients may help to identify new players implicated in obesity and diabetes-related vascular dysfunction and may pave the way for personalized therapies in this setting. In the present review, we discuss current knowledge of the epigenetic routes implicated in vascular damage and cardiovascular disease in patients with metabolic alterations.
Keywords: epigenetics, obesity, diabetes, chromatin, vascular risk, precision medicine
Cardiovascular disease burden in people with diabetes
Cardiovascular diseases (CVD) are the main cause of death and disability among patients with obesity and type 2 diabetes mellitus (T2DM). By the year 2035, more than 650 million will have impaired glucose tolerance and 592 million people will have diabetes worldwide (1). A recent study analysing data from 6,264,226 adults (18 years of age or older) who participated in the Behavioral Risk Factor Surveillance System Survey (1993–1994 and 1999–2016) showed that by 2030 nearly 1 in 2 US adults will have obesity with a prevalence ranging from 35 to 50% (2). Moreover, nearly 1 in 4 adults is projected to have severe obesity by 2030 and the prevalence will be higher than 25% in most of US states (2). Taken together, these data indicate that the prevalence of adult obesity and severe obesity will continue to increase in the decades to come. These numbers have strong implications on morbidity and mortality as patients presenting with cardiometabolic alterations are several-fold more likely to develop myocardial infarction and stroke than age-matched subjects without this condition (3). Alterations occurring in patients with obesity and diabetes, namely insulin resistance, defective insulin secretion, low-grade inflammation foster the development of endothelial dysfunction, increased platelet reactivity, and pro-thrombotic states – all factors contributing to atherosclerotic vascular disease and CV events (4). Albeit therapeutic strategies have been significantly implemented over the last decade, residual risk remains high in this patient population. By this background, a better understanding of the intersection between maladaptive metabolic processes and CVD is of paramount importance to identify new targets and design breakthrough therapies in this growing patient population.
Epigenetic regulation of gene expression
Accumulating evidence suggests that the ‘non-genetic’ regulation of vascular function is gaining increasing attention. Studies conducted over the last 2 decades have contributed to unmask the pivotal role of epigenetic changes in the regulation of gene expression (5). Epigenetic modifications are plastic chemical changes of DNA and histone proteins which have the ability to modify the activity of DNA, without affecting the nucleotide sequence (6). This change in DNA activity leads to a fine-tuning regulation of gene transcription. In other words, epigenetics represents an additional layer of biological regulation which, together with the genetic information, modulates gene expression trajectories over the lifetime (7, 8). Several lines of evidence support the fundamental and independent role of epigenetic regulation. For example, genetically identical pluripotent stem cells differentiate towards specific cell lines (i.e, fibroblasts, myocytes) thanks to specific epigenetic changes occurring at given time points (7). Similarly, monozygotic twins – who share an identical genetic makeup - display high discordance rates for common diseases (i.e. asthma), and this phenomenon is explained by epigenetic modifications (9, 10). Epigenetic regulation is a complex process which often requires the cooperation of different signals (DNA methylation, histone modifications, and ncRNAs) acting in concert (5). DNA methylation and histone modifications take place at the level of chromatin, whereas ncRNAs mostly act indirectly at the posttranslational level. However, recent evidence suggests that ncRNAs may directly regulate chromatin accessibility by regulating nucleosome positioning, DNA methylation and activity of chromatin remodelers (11). The mechanism by which epigenetic changes modify DNA activity is via regulation of chromatin structure. The latter may assume two different states: packed in a dense structure which is inaccessible for transcription (heterochromatin) or in a looser, uncondensed and open for transcription state (euchromatin) (12). Dynamic changes of chromatin architecture critically modulate gene transcription by regulating the binding of transcription factors to DNA.
DNA methylation
DNA methylation is a well-studied epigenetic modification which consists in a covalent modification mainly occurring at cytosines followed by guanines (ie, CpG dinucleotides) to form 5-methylcytosine (6). In most cases, methylation of DNA is a process repressing gene transcription. This occurs by hampering the recruitment of transcription factors or by fostering transcriptional silencing (5). The repression of transposons repeats and the X-chromosome inactivation represent valid examples of how DNA methylation changes control gene expression (13). Different families of enzymes, known as DNA methyltransferases (DNMTs), participate in DNA methylation. DNMT1 is responsible for the maintenance of methylation, whereas DNMT3a and DNMT3b mediate de novo methylation of DNA (14). DNA methylation is a reversible modification that can be removed either passively, by reduced activity of DNMT1, or actively, by the ten-eleven translocation enzymes (TETs).
Histone modifications
Together with DNA methylation, chemical modifications of histones (acetylation, methylation, ubiquitination, phosphorylation, sumoylation, citrullination, and ADP-ribosylation) are critically involved in chromatin remodelling and regulation of gene expression (15). The regulation of chromatin accessibility by histone modifications is highly complex due to the diversity of modifications and their different biological function. Indeed, the net effect on gene transcription really depends on the type of modification and on the position of the modified amino acid residue (6). Histone acetylation is mediated by histone acetyltransferases (HATs) and histone deacetylases (HDACs) and is generally associated with an open chromatin and enhanced gene transcription. By contrast, histone methylation may impact differently on gene transcription based on which amino residue is being modified. Histone methyltransferases (HMTs) catalyze the transfer of a methyl group from S-adenosyl-L-methionine to lysine or arginine residues in a highly specific manner (6). Recently, the discovery of histone demethylases (HDMs) increased the complexity of gene regulation by making histone methylation a dynamic rather than a static process. Of note, modifications of histone tail may affect DNA methylation (and vice versa) thus amplifying the complexity of epigenetic regulation (12). A known example of this interaction is represented by the cross-talk between H3K9 methylation and DNA methylation to repress gene expression in a specific chromatin region (16, 17).
Non-coding RNAs
NcRNAs are RNA molecules which are not transcribed into proteins but regulate gene expression at the posttranslational level by interacting with mRNA. Recent evidence indicates that ncRNAs also regulate the expression of enzymes involved in chromatin remodeling (18). Based on size, ncRNAs can be subdivided into two major groups: (i) small ncRNAs (sncRNAs, <200 nucleotides long) including miRNA, PIWI-interacting RNAs, and endogenous short interfering RNAs and (ii) long ncRNAs (lncRNAs), which have a length between 0.2 kb and 2 kb (19).
Epigenetic modifications and cardiovascular damage in obesity and diabetes
Insulin resistance, adiposity and endothelial dysfunction
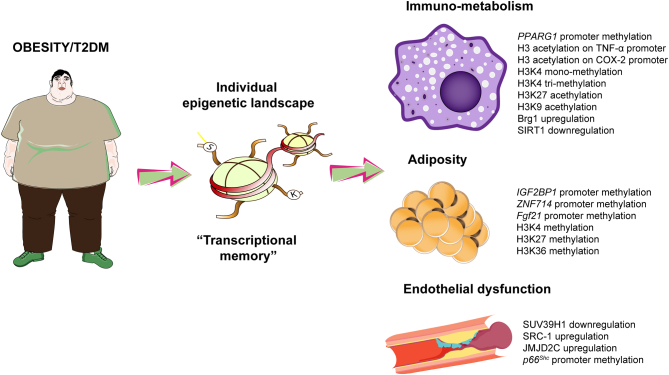
Several studies have shown that epigenetic modifications are responsible for several pathological changes observed in patients with obesity and T2DM (Fig. 1). Environmental factors such as pollution, urban noise and cigarette smoking are potent inducers of epigenetic changes (20). Once acquired, these modifications contribute to derail pathways implicated in the regulation of adipogenesis, low-grade inflammation, liver steatosis and endothelial dysfunction (21). Most importantly, epigenetic alterations are being transmitted across multiple generations thus leading to altered gene expression patterns during childhood and adolescence (22). The transgenerational inheritance of epigenetic signals may certainly contribute to the epidemic of childhood obesity and the increase in CV risk observed in these subjects (23). One out of four obese children shows a detrimental cluster of cardiovascular risk factors which leads to a 10-fold increase in the risk of CVD as compared to non-obese children (24). Epigenetic tags on the promoter of TNF-alpha (TNFα), pyruvate dehydrogenase kinase 4 (PDK4), leptin (LEP), peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) and proopiomelanocortin (POMC) were found to be inherited and causally implicated in the development of metabolic and cardiovascular disease (25, 26).
Figure 1.
Main epigenetic modifications in the cardiometabolic patient. H3, histone 3; K, lysine residue. IL-6, interleukin-6; MCP-1, monocyte chemoattractant protein-1; NF-kB, nuclear factor kappa-B; eNOS, endothelial nitric oxide synthase.
In patients with obesity and T2DM, epigenetic modifications participate to the progressive loss of insulin sensitivity eventually leading to dysglycemia, micro- and macrovascular complications (27). Patients with obesity display altered methylation patterns of genes controlling insulin signalling, namely COL9A1, IGF2BP1, ZNF714, ADCY9 and TBX5 (28). Specifically, ZNF714 was found to be the gene with the largest methylation fold-change among insulin resistant and insulin sensitive patients. Hypomethylation of the zinc finger protein 714 (ZNF714) promoter was associated with higher gene expression in insulin-resistant patients (28). Modifications of the epigenetic landscape also control the expression of genes involved in vascular insulin resistance, a major determinant of systemic loss of insulin sensitivity. DNA methylation and ncRNAs have indeed shown to modulate key pathways, namely the PCKβ/p66Shc, NOX2 and NF-kB, strongly implicated in oxidative stress generation and inflammation eventually fostering the impairment of vascular insulin sensitivity (29, 30). The progressive impairment of endothelial cell function due to ROS accumulation and reduced NO bioavailability also leads to defective capillary recruitment and hampered insulin delivery to hormone sensitive organs. Hence, deregulation of specific vascular pathways by epigenetic modifications may be responsible for systemic metabolic changes thus favouring the development of prediabetes and type 2 diabetes (31). In visceral fat arteries from patients with obesity and insulin resistance, a complex chromatin network characterized by the demethylase SUV39H1, the methyltransferase JMJD2C and the acetyltransferase SRC-1 was found to be causally involved in the generation of vascular oxidative stress and endothelial dysfunction. Targeting these chromatin enzymes was indeed able to rescue obesity-related endothelial dysfunction, thus highlighting the potential relevance of chromatin modifications in the pathogenesis of vascular complications (32).
Epigenetic pathways of vascular damage in diabetes
In patients with diabetes, high blood glucose levels contribute to affect the epigenetic landscape thus leading to persistent deregulation of genes controlling vascular homeostasis. Indeed, hyperglycaemia was found to induce long-lasting transcriptional alterations in the cardiovascular system (4, 33). Such bad legacy of hyperglycemia - mediated by epigenetic changes – may help to explain the lack of benefit of intensive glycemic control on macrovascular complications and mortality in DM patients. Indeed, intensive glycemic control strategies recently failed to reduce the rate of myocardial infarction, stroke and cardiovascular mortality among patients with T2DM (34, 35, 36). This phenomenon – defined ‘hyperglycemic memory’ – indicates a long-term persistence of hyperglycemic stress even after blood glucose normalization (37). Several studies have demonstrated the detrimental impact of persistent transcriptional changes induced by high glucose levels. Fibroblasts isolated from patients with diabetic foot ulcers displayed a persistent demethylation of genes critical to wound repair, angiogenesis, and extracellular matrix assembly. Noteworthy, such gene-related methylation patterns persisted in patient-derived fibroblasts after prolonged passage in normoglycemic conditions, thus demonstrating the existence of a transcriptional memory driven by hyperglycemia (38). Along the same line, experiments in primary human endothelial cells showed that hyperglycemia reduces DNA methylation at the promoter of the mitochondrial adaptor p66Shc, a key protein involved in cytochrome c oxidation and accumulation of free radicals (39). Reduced methylation of p66Shc promoter and increased H3 acetylation – known activating epigenetic marks - were associated with gene upregulation, mitochondrial oxidative stress and impaired availability of nitric oxide, eventually leading to vascular dysfunction. Of note, restoration of normoglycemia was not able to erase the activating marks and this explained persistent transcription of p66Shc and persistent ROS production in the vasculature. Interestingly, blockade of critical enzymes involved in chromatin remodeling such as the acetyltransferase GCN5 reduced H3 acetylation on p66Shc promoter thus restoring physiological p66Shc expression and endothelial function (40). These epigenetic networks were found to be operative also in humans as intensive glycemic control for 6 months was not able to reduce epigenetic changes of p66Shc promoter in patients with T2DM (41). Other epigenetic networks driven by SIRT1 were found to be responsible for the vascular complications of diabetes. Specifically, SIRT1 overexpression prevents endothelial dysfunction by suppressing NF-κB activation and PARP cleavage while restoring hyperglycemia-induced dephosphorylation of LKB1 and AMP kinase (AMPK), two critical regulators of energy balance (42). Interestingly, SIRT1 was also found to modulate p66Shc and p53 in the diabetic vasculature as well as in stem cells from diabetic mice (43, 44, 45). Another activating epigenetic mark, namely acetylation of lysine 9 at histone 3 (H3K9), plays a pivotal role in the pathogenesis of diabetes-related vascular inflammation. Indeed, hyperglycemia-induced H3K9 acetylation was found to promote inflammatory transcriptional programs in vascular smooth muscle cells (VSMCs) isolated from T2DM mice as well as in lymphocytes from type 1 diabetic (T1DM) patients (44, 46). Mono-methylation at lysine 4 of histone 3 (H3K4m), induced by the methyltranfsrease SETD7, also represents a relevant chromatin signature favouring the expression of genes implicated in endothelial inflammation and oxidative stress (47, 48, 49, 50, 51). SETD7 activation is particularly relevant in the setting of diabetes where it is responsible for the upregulation of nuclear factor kappa-B (NF-kB) subunit p65 and subsequent transcription of NF-kB inflammatory genes such as VCAM-1 and ICAM-1 (49). Of clinical relevance, SETD7 is upregulated in peripheral blood monocytes from patients with T2DM, and significantly correlates with NF-kB p65-driven inflammation, oxidative stress and endothelial dysfunction (50). Interestingly, this methyltransferase is amenable of pharmacological intervention, with several compounds already available (52).
microRNAs and cardiovascular damage in obesity and diabetes
Together with DNA methylation and histone marks, ncRNAs are critically involved in obesity and diabetes-related cardiovascular damage. Several miRNAs, including miR-320b, miR-29b, miR-1, miR-133a and miR-499 were found to be involved in the persistence of myocardial damage despite intensive glycemic control (53). Targeting these miRNAs, which are actively involved in myocardial fibrosis, hypertrophic growth, oxidative stress and autophagic response, may represent a potential strategy to prevent diabetic cardiomyopathy and heart failure (53). Along the same line, miR-34 and miR-218 were found to play a pivotal role in the diabetic heart (29). Specifically, miR-34 and miR-218 – respectively targeting deacetylase SIRT1 and methyltransferase DNMT3b - were found to be upstream regulators of chromatin accessibility, leading to persistent upregulation of oxidant genes despite restoration of normoglycemia (29). Zhong et al. have shown that several miRNAs persistently derail transcriptional programs in the vascular endothelium. In particular, miR-125b, miR-146a-5p, and miR-29a-3p – respectively targeting inflammatory cytokines TNF-α-induced protein 3 (TNFAIP3), TNF receptor-associated factor 6 (TRAF6) and IL-1 receptor (IL-1R) – were associated with a persistent impairment of endothelial function and altered expression of proinflammatory genes (54). Direct inhibition of miR-125b or miR-146a-5p overexpression blunted NF-kB-related signals and improved endothelial function (54). A further study showed that restoration of normoglycemia was not able to blunt miR-23b-3p upregulation in retinal endothelial cells, and this contributed to diabetic retinopathy through a SIRT1/NF-kB signalling pathway (55). In patients with T2DM, the expression of miR-326 – a direct modulator of adiponectin and adiponectin receptors (ADIPOR)-1, ADIPOR-2 - was elevated and negatively correlated with adiponectin levels. Of note, miR-326 upregulation was not affected by 12-months of antidiabetic treatment, suggesting its contributions to cardiometabolic disturbances, even in the presence of target HbA1c levels (56).
The emerging role of long non-coding RNAs
A growing body of evidence suggests the involvement of lncRNAs in the pathogenesis of vascular disease in conditions of metabolic stress (57). In a systematic transcriptional screening, Singh and colleagues found that out of 30,586 lncRNAs screened, 100 were upregulated and 186 downregulated in HUVECs cultured under high as compared to normal glucose conditions (58). Similarly, microarray analysis performed in lymphatic endothelial cells from diabetic and non-diabetic patients showed significant variations induced by hyperglycemia, with 31 lncRNAs downregulated and 79 lncRNAs upregulated (59). Among these signatures, several well-known lncRNAs were found, including H19, GAS5, UCA1, CRNDE, GAS5, and LINC00312. Interestingly, network analysis showed that these lncRNAs may be related to insulin reporter signalling pathway and response to insulin stimulus (59).
The lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was found to be elevated during the early stages of high glucose stimulation in HUVECs, and this alteration was associated with a parallel increase in serum amyloid antigen 3 (SAA3), as well as with other pro-inflammatory cytokines such as TNF-α and IL-6. Of note, these alterations were prevented by silencing of MALAT1 (60). A recent study showed that MALAT1 expression was increased in an experimental mouse model of type 2 diabetes and positively correlated with HOMA-IR and resistin levels (61). In line with these findings, MALAT1 siRNA attenuated resistin, Ang II and inflammatory molecules, whereas it enhanced glucose uptake and nitric oxide production (NO). The authors also showed that exercise was able to downregulate MALAT1 with subsequent reduction of resistin levels (61). In line with these findings, Malat1-knockout T1DM animals were protected against hyperglycemia-induced elevations of inflammatory markers (62).
RNCR3 lncRNA has been shown to play a role in T2DM-induced retinal microvascular dysfunction through the RNCR3/KLF2/miR-185-5p axis (63). Activation of the latter pathway was also found to protect against atherosclerotic vascular disease. Indeed, in mouse and human aortic atherosclerotic lesions, as well as cultured HUVECs and VSMCs exposed to high ox-LDL concentrations, RNCR3 depletion accelerated atherosclerosis, aggravated hypercholesterolemia and vascular inflammation while decreasing cell proliferation and migration (64).
The lncRNA called myocardial infarction–associated transcript (MIAT) was also shown to be upregulated in the retina of diabetic rats as well as upon ambient hyperglycemia in cultured endothelial cells. In vitro studies revealed that MIAT knockdown ameliorated diabetes mellitus-induced retinal microvascular dysfunction by inhibiting endothelial cell proliferation, migration, and tube formation (65). LncRNA Maternally expressed gene 3 (MEG3) has been shown to be protective in diabetes-related endothelial dysfunction via activation of PI3k/Akt signalling . MEG3 expression was reduced retinas of STZ-induced diabetic mice and in high glucose-exposed endothelial cells. In experimental mouse models, knockdown of MEG3 was associated with increased number of a cellular capillaries, increased vascular leakage, and increased inflammatory markers such as interleukins, TNF-α, and CCL2 (66). MEG3 knockdown was also associated with exacerbation of inflammatory damage, induction of cell proliferation and suppression of apoptosis in glucose-treated HUVECs. Mechanistic studies revealed that regulation of the apoptotic pathways was associated with upregulation of BCL-2 and downregulation of BAX, caspase-3, and p53, whereas the proliferation processes were related with activation of TGF-β and Wnt/β-catenin pathways (67). In diabetic rats, overexpression of the lncRNA ANRIL promoted Vegf upregulation and increased angiogenesis via activation of NF-κB signalling (68). A recent study also demonstrated that knocking down ANRIL, MALAT1, and ZFAS1 prevented glucose-induced upregulation of ET-1 transcripts (69).
Furthermore, a recent study demonstrated that lncRNA H19 is upregulated in vascular endothelium of patients with T2DM, where it mediates VEGFA overexpression via inhibition of mir-29a. Of note, H19 suppression significantly attenuated high glucose-induced endothelial inflammation and oxidative stress through VEGFA downregulation and AKT/eNOS pathway activation (70).
Toward personalized epigenetic therapies
Several compounds have shown their ability to target chromatin modifications. The unveiling of specific chromatin modifications has enabled the design of specific drugs erasing maladaptive epigenetic signals, thereby restoring vascular homeostasis (Table 1) (27). Activators of histone deacetylases have shown to repress transcription of genes participating to vascular oxidative stress and inflammation. Specifically, the SIRT1 activator resveratrol has demonstrated to prevent eNOS uncoupling and to suppress obesity and hyperglycemia-induced inflammation by inhibition of NF-kB signalling pathway as well as NF-kB-related cytokines such as TNFα (44, 71). SIRT1 activation also modulates adipogenesis and function of perivascular adipose tissue, a major determinant of vascular health (72). The translation of these experimental findings to humans has yielded to conflicting results and ongoing clinical trials will help to clarify whether SIRT1 activators may be employed in the clinical setting to prevent cardiometabolic alterations (73). Other histone deacetylase inhibitors such as vorinostat and thrichostatin A shown beneficial effects in preventing endothelial dysfunction, ischemia/reperfusion injury and diabetic nephropathy (74, 75, 76). Apabetalone (development codes RVX 208, RVX-208, and RVX000222) a small molecule targeting bromodomain and extra terminal domain (BET) proteins (in particular the BET family member), also represent a promising drug given its ability to modulate gene transcription by preventing the interaction of histones with DNA (77). Of interest, Apabetalone was shown to prevent the upregulation of IL-1β, IL-6 and TNF-α in human endothelial cells exposed to high glucose levels as well as in aortic plaques from ApoE-/- mice (78). However, the recent BETonMACE trial failed to show a clear cardiovascular benefit of apabetalone in patients with diabetes and a recent acute coronary syndrome. However, data from secondary endpoints are promising and larger clinical trials are needed in this regard (79). Given the explosion of the new anti-diabetic therapies with glucagon-like peptide-1 receptor agonists (GLP-1 RA) and sodium glucose cotransporter 2 inhibitors (SGLT2i) (80), a plenty of recent studies appearing in rapid succession started to investigate the mechanisms underlying their cardiovascular benefits. Interestingly, GLP-1 RA were shown to modulate the expression of miRNA-338, thus restoring pancreatic β-cell function and glucose homeostasis (81). Other studies showed that GLP1 RA control liver steatosis – a key cardiometabolic risk factor - by regulating the hepatic expression of miRNA-34a, miRNA-21 and miRNA-200b/c (82). Along the same line, the SGLT2i empagliflozin was recently found to modulate hyperglycemia-induced microvascular oxidative stress by modulating the p38 MAPK/miR-21 pathway (83). This finding is interesting as recent work has shown that improvement of microvascular function by SGLT2i reduces myocardial inflammation and oxidative stress (84), important features contributing to heart failure development in people with diabetes (85). Although these studies raise significant interest, more research is needed to appraise how new glucose-lowering drugs induce changes of miRNA landscape, and whether these changes are causally implicated in the disease development.
Table 1.
Available compounds to target chromatin modifications for the prevention of obesity and diabetes-induced vascular dysfunction.
| References | Therapy | Main finding |
|---|---|---|
| Zhang et al. (57, 58), Xia et al. (59) | Resveratrol | SIRT1 inhibition attenuates adipogenesis, inflammation, oxidative stress, and rescues obesity-related endothelial dysfunction |
| Advani et al. (61), Xie et al. (62), Rafehi et al. (63) | Vorinostat | Vorinostat regulates histone acetylation by preventing EP300/CREBBP binding at multiple gene promoters. It prevents eNOS uncoupling, inflammation and insulin signalling in adipose tissue and skeletal muscle from diabetic mice. |
| Hu et al. (64) | Sodium butyrate | Sodium butyrate suppresses NF-kB signalling and NF-kB-dependent inflammatory molecules. Moreover it prevents metabolic alterations in diet-induced obese as well as in aged mice, mainly by enhancing oxidative phosphorylation and beta-oxidation in mitochondria. |
| Zhang et al. (65) | Trichostatin A | Trichostatin A suppresses TNF-α transcription in the context of ischemia-induced left ventricular remodeling. |
| Tsujikawa et al. (67), Ray et al. (68) | Apabetalone | Prevents vascular inflammation by modulating the transcription of VCAM-1, ICAM-1, TNFα and IL1-beta |
Evidence discussed so far suggests that epigenetic therapies are able to interfere with chromatin structure and miRNAs, and may represent a new frontier in cardiovascular medicine. There remain, however, important evidence gaps to be considered before moving to the clinical setting. First, many drugs targeting chromatin modifiers are not specific as they also implicated in the regulation of other pathways. Second, the safety of this approach remains to be proven. Indeed, these compounds may not selectively erase methylation or acetylation signals in specific cells, this may lead to unspecific and undesirable modulation of transcriptional programs. Third, the epigenetic landscape is extremely diverse according to different organs and we currently lack epigenetic biomarkers reflecting chromatin modifications in specific tissues (vasculature, heart). Single-cell RNA sequencing approaches will help to decipher the epigenetic repertoire of different cells with the aim of developing more targeted approaches. The complexity of epigenetic information may significantly contribute to explain why patients with the same disease (obesity) display different vascular risk trajectories overtime. In this respect, understanding the individual epigenetic regulation may open perspectives for personalized therapies to combat obesity and type 2 diabetes in the decades to come.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this work.
Funding
This work did not receive any specific grant from any funding agency in the public, commercial, or not-for-profit sector.
Acknowledgments
F P is the recipient of a Sheikh Khalifa’s Foundation Assistant Professorship at the Faculty of Medicine, University of Zürich. The present work is supported by the Zürich Heart House, the Swiss Heart Foundation, Swiss Life Foundation, the EMDO Stiftung; Kurt und Senta-Hermann Stiftung, and the Schweizerische Diabetes-Stiftung to F P; the Holcim Foundation and the Swiss Heart Foundation (to S C).
References
- 1.Ogurtsova K, da Rocha Fernandes JD, Huang Y, Linnenkamp U, Guariguata L, Cho NH, Cavan D, Shaw JE, Makaroff LE. IDF Diabetes Atlas: global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Research and Clinical Practice 2017. 128 40–50. ( 10.1016/j.diabres.2017.03.024) [DOI] [PubMed] [Google Scholar]
- 2.Ward ZJ, Bleich SN, Cradock AL, Barrett JL, Giles CM, Flax C, Long MW, Gortmaker SL. Projected U.S. State-level prevalence of adult obesity and severe obesity. New England Journal of Medicine 2019. 381 2440–2450. ( 10.1056/NEJMsa1909301) [DOI] [PubMed] [Google Scholar]
- 3.Haffner SM, Lehto S, Ronnemaa T, Pyorala K, Laakso M. Mortality from coronary heart disease in subjects with type 2 diabetes and in nondiabetic subjects with and without prior myocardial infarction. New England Journal of Medicine 1998. 339 229–234. ( 10.1056/NEJM199807233390404) [DOI] [PubMed] [Google Scholar]
- 4.Paneni F, Beckman JA, Creager MA, Cosentino F. Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: part I. European Heart Journal 2013. 34 2436–2443. ( 10.1093/eurheartj/eht149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baccarelli A, Rienstra M, Benjamin EJ. Cardiovascular epigenetics: basic concepts and results from animal and human studies. Circulation: Cardiovascular Genetics 2010. 3 567–573. ( 10.1161/CIRCGENETICS.110.958744) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Handy DE, Castro R, Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation 2011. 123 2145–2156. ( 10.1161/CIRCULATIONAHA.110.956839) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunet A, Berger SL. Epigenetics of aging and aging-related disease. Journals of Gerontology: Series A, Biological Sciences and Medical Sciences 2014. 69 (Supplement 1) S17–S20. ( 10.1093/gerona/glu042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costantino S, Camici GG, Mohammed SA, Volpe M, Luscher TF, Paneni F. Epigenetics and cardiovascular regenerative medicine in the elderly. International Journal of Cardiology 2018. 250 207–214. ( 10.1016/j.ijcard.2017.09.188) [DOI] [PubMed] [Google Scholar]
- 9.Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nature Reviews: Molecular Cell Biology 2015. 16 593–610. ( 10.1038/nrm4048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, et al. Epigenetic differences arise during the lifetime of monozygotic twins. PNAS 2005. 102 10604–10609. ( 10.1073/pnas.0500398102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohmdorfer G, Wierzbicki AT. Control of chromatin structure by long noncoding RNA. Trends in Cell Biology 2015. 25 623–632. ( 10.1016/j.tcb.2015.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swygert SG, Peterson CL. Chromatin dynamics: interplay between remodeling enzymes and histone modifications. Biochimica et Biophysica Acta 2014. 1839 728–736. ( 10.1016/j.bbagrm.2014.02.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. Journal of Cellular Physiology 2007. 213 384–390. ( 10.1002/jcp.21224) [DOI] [PubMed] [Google Scholar]
- 14.Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Frontiers in Oncology 2014. 4 80 ( 10.3389/fonc.2014.00080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenuwein T, Allis CD. Translating the histone code. Science 2001. 293 1074–1080. ( 10.1126/science.1063127) [DOI] [PubMed] [Google Scholar]
- 16.Eskeland R, Eberharter A, Imhof A. HP1 binding to chromatin methylated at H3K9 is enhanced by auxiliary factors. Molecular and Cellular Biology 2007. 27 453–465. ( 10.1128/MCB.01576-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thambirajah AA, Ng MK, Frehlick LJ, Li A, Serpa JJ, Petrotchenko EV, Silva-Moreno B, Missiaen KK, Borchers CH, Adam Hall J, et al. MeCP2 binds to nucleosome free (linker DNA) regions and to H3K9/H3K27 methylated nucleosomes in the brain. Nucleic Acids Research 2012. 40 2884–2897. ( 10.1093/nar/gkr1066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jae N, Heumuller AW, Fouani Y, Dimmeler S. Long non-coding RNAs in vascular biology and disease. Vascular Pharmacology 2019. 114 13–22. ( 10.1016/j.vph.2018.03.003) [DOI] [PubMed] [Google Scholar]
- 19.Poller W, Dimmeler S, Heymans S, Zeller T, Haas J, Karakas M, Leistner DM, Jakob P, Nakagawa S, Blankenberg S, et al. Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. European Heart Journal 2018. 39 2704–2716. ( 10.1093/eurheartj/ehx165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costantino S, Libby P, Kishore R, Tardif JC, El-Osta A, Paneni F. Epigenetics and precision medicine in cardiovascular patients: from basic concepts to the clinical arena. European Heart Journal 2018. 39 4150–4158. ( 10.1093/eurheartj/ehx568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costantino S, Mohammed SA, Ambrosini S, Paneni F. Epigenetic processing in cardiometabolic disease. Atherosclerosis 2019. 281 150–158. ( 10.1016/j.atherosclerosis.2018.09.029) [DOI] [PubMed] [Google Scholar]
- 22.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 2014. 157 95–109. ( 10.1016/j.cell.2014.02.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daxinger L, Whitelaw E. Transgenerational epigenetic inheritance: more questions than answers. Genome Research 2010. 20 1623–1628. ( 10.1101/gr.106138.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayer J, Charakida M, Deanfield JE, Celermajer DS. Lifetime risk: childhood obesity and cardiovascular risk. European Heart Journal 2015. 36 1371–1376. ( 10.1093/eurheartj/ehv089) [DOI] [PubMed] [Google Scholar]
- 25.Lavebratt C, Almgren M, Ekstrom TJ. Epigenetic regulation in obesity. International Journal of Obesity 2012. 36 757–765. ( 10.1038/ijo.2011.178) [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Cardona MC, Huang F, Garcia-Vivas JM, Lopez-Camarillo C, Del Rio Navarro BE, Navarro Olivos E, Hong-Chong E, Bolanos-Jimenez F, Marchat LA. DNA methylation of leptin and adiponectin promoters in children is reduced by the combined presence of obesity and insulin resistance. International Journal of Obesity 2014. 38 1457–1465. ( 10.1038/ijo.2014.30) [DOI] [PubMed] [Google Scholar]
- 27.Costantino S, Ambrosini S, Paneni F. The epigenetic landscape in the cardiovascular complications of diabetes. Journal of Endocrinological Investigation 2019. 42 505–511. ( 10.1007/s40618-018-0956-3) [DOI] [PubMed] [Google Scholar]
- 28.Crujeiras AB, Diaz-Lagares A, Moreno-Navarrete JM, Sandoval J, Hervas D, Gomez A, Ricart W, Casanueva FF, Esteller M, Fernandez-Real JM. Genome-wide DNA methylation pattern in visceral adipose tissue differentiates insulin-resistant from insulin-sensitive obese subjects. Translational Research 2016. 178 13.e15 ( 10.1016/j.trsl.2016.07.002) [DOI] [PubMed] [Google Scholar]
- 29.Costantino S, Paneni F, Mitchell K, Mohammed SA, Hussain S, Gkolfos C, Berrino L, Volpe M, Schwarzwald C, Luscher TF, et al. Hyperglycaemia-induced epigenetic changes drive persistent cardiac dysfunction via the adaptor p66(Shc). International Journal of Cardiology 2018. 268 179–186. ( 10.1016/j.ijcard.2018.04.082) [DOI] [PubMed] [Google Scholar]
- 30.Costantino S, Paneni F, Cosentino F. Targeting chromatin remodeling to prevent cardiovascular disease in diabetes. Current Pharmaceutical Biotechnology 2015. 16 531–543. ( 10.2174/138920101606150407113644) [DOI] [PubMed] [Google Scholar]
- 31.Rask-Madsen C, King GL. Endothelium-dependent delivery of insulin to muscle interstitium. Cell Metabolism 2011. 13 236–238. ( 10.1016/j.cmet.2011.02.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costantino S, Paneni F, Virdis A, Hussain S, Mohammed SA, Capretti G, Akhmedov A, Dalgaard K, Chiandotto S, Pospisilik JA, et al. Interplay among H3K9-editing enzymes SUV39H1, JMJD2C and SRC-1 drives p66Shc transcription and vascular oxidative stress in obesity. European Heart Journal 2019. 40 383–391. ( 10.1093/eurheartj/ehx615) [DOI] [PubMed] [Google Scholar]
- 33.Paneni F, Volpe M, Luscher TF, Cosentino F. SIRT1, p66(Shc), and Set7/9 in vascular hyperglycemic memory: bringing all the strands together. Diabetes 2013. 62 1800–1807. ( 10.2337/db12-1648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boussageon R, Bejan-Angoulvant T, Saadatian-Elahi M, Lafont S, Bergeonneau C, Kassai B, Erpeldinger S, Wright JM, Gueyffier F, Cornu C. Effect of intensive glucose lowering treatment on all cause mortality, cardiovascular death, and microvascular events in type 2 diabetes: meta-analysis of randomised controlled trials. BMJ 2011. 343 d4169 ( 10.1136/bmj.d4169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castagno D, Baird-Gunning J, Jhund PS, Biondi-Zoccai G, MacDonald MR, Petrie MC, Gaita F, McMurray JJ. Intensive glycemic control has no impact on the risk of heart failure in type 2 diabetic patients: evidence from a 37,229 patient meta-analysis. American Heart Journal 2011. 162 938.e932 ( 10.1016/j.ahj.2011.07.030) [DOI] [PubMed] [Google Scholar]
- 36.Ceriello A. Hypothesis: the ‘metabolic memory’, the new challenge of diabetes. Diabetes Research and Clinical Practice 2009. 86 (Supplement 1) S2–S6. ( 10.1016/S0168-8227(09)70002-6) [DOI] [PubMed] [Google Scholar]
- 37.Ceriello A. The emerging challenge in diabetes: the ‘metabolic memory’. Vascular Pharmacology 2012. 57 133–138. ( 10.1016/j.vph.2012.05.005) [DOI] [PubMed] [Google Scholar]
- 38.Park LK, Maione AG, Smith A, Gerami-Naini B, Iyer LK, Mooney DJ, Veves A, Garlick JA. Genome-wide DNA methylation analysis identifies a metabolic memory profile in patient-derived diabetic foot ulcer fibroblasts. Epigenetics 2014. 9 1339–1349. ( 10.4161/15592294.2014.967584) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohammed SA, Ambrosini S, Luscher T, Paneni F, Costantino S. Epigenetic control of mitochondrial function in the vasculature. Frontiers in Cardiovascular Medicine 2020. 7 28 ( 10.3389/fcvm.2020.00028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paneni F, Mocharla P, Akhmedov A, Costantino S, Osto E, Volpe M, Luscher TF, Cosentino F. Gene silencing of the mitochondrial adaptor p66(Shc) suppresses vascular hyperglycemic memory in diabetes. Circulation Research 2012. 111 278–289. ( 10.1161/CIRCRESAHA.112.266593) [DOI] [PubMed] [Google Scholar]
- 41.Costantino S, Paneni F, Battista R, Castello L, Capretti G, Chiandotto S, Tanese L, Russo G, Pitocco D, Lanza GA, et al. Impact of glycemic variability on chromatin remodeling, oxidative stress, and endothelial dysfunction in patients with Type 2 diabetes and with target HbA1c levels. Diabetes 2017. 66 2472–2482. ( 10.2337/db17-0294) [DOI] [PubMed] [Google Scholar]
- 42.Zheng Z, Chen H, Li J, Li T, Zheng B, Zheng Y, Jin H, He Y, Gu Q, Xu X. Sirtuin 1-mediated cellular metabolic memory of high glucose via the LKB1/AMPK/ROS pathway and therapeutic effects of metformin. Diabetes 2012. 61 217–228. ( 10.2337/db11-0416) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou S, Chen HZ, Wan YZ, Zhang QJ, Wei YS, Huang S, Liu JJ, Lu YB, Zhang ZQ, Yang RF, et al. Repression of P66Shc expression by SIRT1 contributes to the prevention of hyperglycemia-induced endothelial dysfunction. Circulation Research 2011. 109 639–648. ( 10.1161/CIRCRESAHA.111.243592) [DOI] [PubMed] [Google Scholar]
- 44.Orimo M, Minamino T, Miyauchi H, Tateno K, Okada S, Moriya J, Komuro I. Protective role of SIRT1 in diabetic vascular dysfunction. Arteriosclerosis, Thrombosis, and Vascular Biology 2009. 29 889–894. ( 10.1161/ATVBAHA.109.185694) [DOI] [PubMed] [Google Scholar]
- 45.Albiero M, Poncina N, Tjwa M, Ciciliot S, Menegazzo L, Ceolotto G, Vigili de Kreutzenberg S, Moura R, Giorgio M, Pelicci P, et al. Diabetes causes bone marrow autonomic neuropathy and impairs stem cell mobilization via dysregulated p66Shc and Sirt1. Diabetes 2014. 63 1353–1365. ( 10.2337/db13-0894) [DOI] [PubMed] [Google Scholar]
- 46.Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. PNAS 2008. 105 9047–9052. ( 10.1073/pnas.0803623105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okabe J, Orlowski C, Balcerczyk A, Tikellis C, Thomas MC, Cooper ME, El-Osta A. Distinguishing hyperglycemic changes by Set7 in vascular endothelial cells. Circulation Research 2012. 110 1067–1076. ( 10.1161/CIRCRESAHA.112.266171) [DOI] [PubMed] [Google Scholar]
- 48.Miao F, Gonzalo IG, Lanting L, Natarajan R. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. Journal of Biological Chemistry 2004. 279 18091–18097. ( 10.1074/jbc.M311786200) [DOI] [PubMed] [Google Scholar]
- 49.El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, Cooper ME, Brownlee M. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. Journal of Experimental Medicine 2008. 205 2409–2417. ( 10.1084/jem.20081188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paneni F, Costantino S, Battista R, Castello L, Capretti G, Chiandotto S, Scavone G, Villano A, Pitocco D, Lanza G, et al. Adverse epigenetic signatures by histone methyltransferase Set7 contribute to vascular dysfunction in patients with type 2 diabetes mellitus. Circulation: Cardiovascular Genetics 2015. 8 150–158. ( 10.1161/CIRCGENETICS.114.000671) [DOI] [PubMed] [Google Scholar]
- 51.Miao F, Wu X, Zhang L, Yuan YC, Riggs AD, Natarajan R. Genome-wide analysis of histone lysine methylation variations caused by diabetic conditions in human monocytes. Journal of Biological Chemistry 2007. 282 13854–13863. ( 10.1074/jbc.M609446200) [DOI] [PubMed] [Google Scholar]
- 52.Barsyte-Lovejoy D, Li F, Oudhoff MJ, Tatlock JH, Dong A, Zeng H, Wu H, Freeman SA, Schapira M, Senisterra GA, et al. (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. PNAS 2014. 111 12853–12858. ( 10.1073/pnas.1407358111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Costantino S, Paneni F, Luscher TF, Cosentino F. MicroRNA profiling unveils hyperglycaemic memory in the diabetic heart. European Heart Journal 2016. 37 572–576. ( 10.1093/eurheartj/ehv599) [DOI] [PubMed] [Google Scholar]
- 54.Zhong X, Liao Y, Chen L, Liu G, Feng Y, Zeng T, Zhang J. The microRNAs in the pathogenesis of metabolic memory. Endocrinology 2015. 156 3157–3168. ( 10.1210/en.2015-1063) [DOI] [PubMed] [Google Scholar]
- 55.Zhao S, Li T, Li J, Lu Q, Han C, Wang N, Qiu Q, Cao H, Xu X, Chen H, et al. miR-23b-3p induces the cellular metabolic memory of high glucose in diabetic retinopathy through a SIRT1-dependent signalling pathway. Diabetologia 2016. 59 644–654. ( 10.1007/s00125-015-3832-0) [DOI] [PubMed] [Google Scholar]
- 56.Santovito D, De Nardis V, Marcantonio P, Mandolini C, Paganelli C, Vitale E, Buttitta F, Bucci M, Mezzetti A, Consoli A, et al. Plasma exosome microRNA profiling unravels a new potential modulator of adiponectin pathway in diabetes: effect of glycemic control. Journal of Clinical Endocrinology and Metabolism 2014. 99 E1681–E1685. ( 10.1210/jc.2013-3843) [DOI] [PubMed] [Google Scholar]
- 57.Zhang HN, Xu QQ, Thakur A, Alfred MO, Chakraborty M, Ghosh A, Yu XB. Endothelial dysfunction in diabetes and hypertension: role of microRNAs and long non-coding RNAs. Life Sciences 2018. 213 258–268. ( 10.1016/j.lfs.2018.10.028) [DOI] [PubMed] [Google Scholar]
- 58.Singh KK, Mantella LE, Pan Y, Quan A, Sabongui S, Sandhu P, Teoh H, Al-Omran M, Verma S. A global profile of glucose-sensitive endothelial-expressed long non-coding RNAs. Canadian Journal of Physiology and Pharmacology 2016. 94 1007–1014. ( 10.1139/cjpp-2015-0585) [DOI] [PubMed] [Google Scholar]
- 59.Qi M, Zhou Q, Zeng W, Shen M, Liu X, Luo C, Long J, Chen W, Zhang J, Yan S. Analysis of long non-coding RNA expression of lymphatic endothelial cells in response to Type 2 diabetes. Cellular Physiology and Biochemistry 2017. 41 466–474. ( 10.1159/000456599) [DOI] [PubMed] [Google Scholar]
- 60.Puthanveetil P, Chen S, Feng B, Gautam A, Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. Journal of Cellular and Molecular Medicine 2015. 19 1418–1425. ( 10.1111/jcmm.12576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu SX, Zheng F, Xie KL, Xie MR, Jiang LJ, Cai Y. Exercise reduces insulin resistance in type 2 diabetes mellitus via mediating the lncRNA MALAT1/MicroRNA-382-3p/Resistin axis. Molecular Therapy: Nucleic Acids 2019. 18 34–44. ( 10.1016/j.omtn.2019.08.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gordon AD, Biswas S, Feng B, Chakrabarti S. MALAT1: a regulator of inflammatory cytokines in diabetic complications. Endocrinology, Diabetes and Metabolism 2018. 1 e00010 ( 10.1002/edm2.10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shan K, Li CP, Liu C, Liu X, Yan B. RNCR3: a regulator of diabetes mellitus-related retinal microvascular dysfunction. Biochemical and Biophysical Research Communications 2017. 482 777–783. ( 10.1016/j.bbrc.2016.11.110) [DOI] [PubMed] [Google Scholar]
- 64.Shan K, Jiang Q, Wang XQ, Wang YN, Yang H, Yao MD, Liu C, Li XM, Yao J, Liu B, et al. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death and Disease 2016. 7 e2248 ( 10.1038/cddis.2016.145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q, Jiang Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circulation Research 2015. 116 1143–1156. ( 10.1161/CIRCRESAHA.116.305510) [DOI] [PubMed] [Google Scholar]
- 66.Qiu GZ, Tian W, Fu HT, Li CP, Liu B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochemical and Biophysical Research Communications 2016. 471 135–141. ( 10.1016/j.bbrc.2016.01.164) [DOI] [PubMed] [Google Scholar]
- 67.Wang Z, Ding L, Zhu J, Su Y, Wang L, Liu L, Ma Q, Yao H. Long non-coding RNA MEG3 mediates high glucose-induced endothelial cell dysfunction. International Journal of Clinical and Experimental Pathology 2018. 11 1088–1100. [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang B, Wang D, Ji TF, Shi L, Yu JL. Overexpression of lncRNA ANRIL up-regulates VEGF expression and promotes angiogenesis of diabetes mellitus combined with cerebral infarction by activating NF-kappaB signaling pathway in a rat model. Oncotarget 2017. 8 17347–17359. ( 10.18632/oncotarget.14468) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Biswas S, Feng B, Thomas A, Chen S, Aref-Eshghi E, Sadikovic B, Chakrabarti S. Endothelin-1 regulation is entangled in a complex web of epigenetic mechanisms in diabetes. Physiological Research 2018. 67 (Supplement 1) S115–S125. ( 10.33549/physiolres.933836) [DOI] [PubMed] [Google Scholar]
- 70.Cheng XW, Chen ZF, Wan YF, Zhou Q, Wang H, Zhu HQ. Long non-coding RNA H19 suppression protects the endothelium against hyperglycemic-induced inflammation via inhibiting expression of miR-29b target gene vascular endothelial growth factor a through activation of the protein kinase B/endothelial nitric oxide synthase pathway. Frontiers in Cell and Developmental Biology 2019. 7 263 ( 10.3389/fcell.2019.00263) H19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng X, Zhu S, Chang S, Cao Y, Dong J, Li J, Long R, Zhou Y. Protective effects of chronic resveratrol treatment on vascular inflammatory injury in streptozotocin-induced type 2 diabetic rats: role of NF-kappa B signaling. European Journal of Pharmacology 2013. 720 147––157.. ( 10.1016/j.ejphar.2013.10.034) [DOI] [PubMed] [Google Scholar]
- 72.Xia N, Forstermann U, Li H. Effects of resveratrol on eNOS in the endothelium and the perivascular adipose tissue. Annals of the New York Academy of Sciences 2017. 1403 132–141. ( 10.1111/nyas.13397) [DOI] [PubMed] [Google Scholar]
- 73.Dyck GJB, Raj P, Zieroth S, Dyck JRB, Ezekowitz JA. The effects of resveratrol in patients with cardiovascular disease and heart failure: a narrative review. International Journal of Molecular Sciences 2019. 20 904 ( 10.3390/ijms20040904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilbert RE, Huang Q, Thai K, Advani SL, Lee K, Yuen DA, Connelly KA, Advani A. Histone deacetylase inhibition attenuates diabetes-associated kidney growth: potential role for epigenetic modification of the epidermal growth factor receptor. Kidney International 2011. 79 1312–1321. ( 10.1038/ki.2011.39) [DOI] [PubMed] [Google Scholar]
- 75.Advani A, Huang Q, Thai K, Advani SL, White KE, Kelly DJ, Yuen DA, Connelly KA, Marsden PA, Gilbert RE. Long-term administration of the histone deacetylase inhibitor vorinostat attenuates renal injury in experimental diabetes through an endothelial nitric oxide synthase-dependent mechanism. American Journal of Pathology 2011. 178 2205–2214. ( 10.1016/j.ajpath.2011.01.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xie M, Kong Y, Tan W, May H, Battiprolu PK, Pedrozo Z, Wang ZV, Morales C, Luo X, Cho G, et al. Histone deacetylase inhibition blunts ischemia/reperfusion injury by inducing cardiomyocyte autophagy. Circulation 2014. 129 1139–1151. ( 10.1161/CIRCULATIONAHA.113.002416) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghosh GC, Bhadra R, Ghosh RK, Banerjee K, Gupta A. RVX 208: a novel BET protein inhibitor, role as an inducer of apo A-I/HDL and beyond. Cardiovascular Therapeutics 2017. 35 e12265. ( 10.1111/1755-5922.12265) [DOI] [PubMed] [Google Scholar]
- 78.Tsujikawa LM, Fu L, Das S, Halliday C, Rakai BD, Stotz SC, Sarsons CD, Gilham D, Daze E, Wasiak S, et al. Apabetalone (RVX-208) reduces vascular inflammation in vitro and in CVD patients by a BET-dependent epigenetic mechanism. Clinical Epigenetics 2019. 11 102 ( 10.1186/s13148-019-0696-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ray KK, Nicholls SJ, Buhr KA, Ginsberg HN, Johansson JO, Kalantar-Zadeh K, Kulikowski E, Toth PP, Wong N, Sweeney M, et al. Effect of apabetalone added to standard therapy on major adverse cardiovascular events in patients with recent acute coronary syndrome and type 2 diabetes: a randomized clinical trial. JAMA 2020. 323 1565–1573. ( 10.1001/jama.2020.3308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paneni F, Luscher TF. Cardiovascular protection in the treatment of Type 2 diabetes: a review of clinical trial results across drug classes. American Journal of Medicine 2017. 130 S18–S29. ( 10.1016/j.amjmed.2017.04.008) [DOI] [PubMed] [Google Scholar]
- 81.Wei J, Ding D, Wang T, Liu Q, Lin Y. MiR-338 controls BPA-triggered pancreatic islet insulin secretory dysfunction from compensation to decompensation by targeting Pdx-1. FASEB Journal 2017. 31 5184–5195. ( 10.1096/fj.201700282R) [DOI] [PubMed] [Google Scholar]
- 82.Capuani B, Pacifici F, Della-Morte D, Lauro D. Glucagon like peptide 1 and microRNA in metabolic diseases: focusing on GLP1 action on miRNAs. Frontiers in Endocrinology 2018. 9 719 ( 10.3389/fendo.2018.00719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Das NA, Carpenter AJ, Belenchia A, Aroor AR, Noda M, Siebenlist U, Chandrasekar B, DeMarco VG. Empagliflozin reduces high glucose-induced oxidative stress and miR-21-dependent TRAF3IP2 induction and RECK suppression, and inhibits human renal proximal tubular epithelial cell migration and epithelial-to-mesenchymal transition. Cellular Signalling 2020. 68 109506 ( 10.1016/j.cellsig.2019.109506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Juni RP, Kuster DWD, Goebel M, Helmes M, Musters RJP, van der Velden J, Koolwijk P, Paulus WJ, van Hinsbergh VWM. Cardiac microvascular endothelial enhancement of cardiomyocyte function is impaired by inflammation and restored by empagliflozin. JACC: Basic to Translational Science 2019. 4 575–591. ( 10.1016/j.jacbts.2019.04.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Waddingham MT, Sonobe T, Tsuchimochi H, Edgley AJ, Sukumaran V, Chen YC, Hansra SS, Schwenke DO, Umetani K, Aoyama K, et al. Diastolic dysfunction is initiated by cardiomyocyte impairment ahead of endothelial dysfunction due to increased oxidative stress and inflammation in an experimental prediabetes model. Journal of Molecular and Cellular Cardiology 2019. 137 119–131. ( 10.1016/j.yjmcc.2019.10.005) [DOI] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a