Supplemental Digital Content is available in the text.
Keywords: genomics, congenital heart disease, heart transplantation, mortality, survival
Abstract
Background:
De novo genic and copy number variants are enriched in patients with congenital heart disease, particularly those with extra-cardiac anomalies. The impact of de novo damaging variants on outcomes following cardiac repair is unknown.
Methods:
We studied 2517 patients with congenital heart disease who had undergone whole-exome sequencing as part of the CHD GENES study (Congenital Heart Disease Genetic Network).
Results:
Two hundred ninety-four patients (11.7%) had clinically significant de novo variants. Patients with de novo damaging variants were 2.4 times more likely to have extra-cardiac anomalies (P=5.63×10−12). In 1268 patients (50.4%) who had surgical data available and underwent open-heart surgery exclusive of heart transplantation as their first operation, we analyzed transplant-free survival following the first operation. Median follow-up was 2.65 years. De novo variants were associated with worse transplant-free survival (hazard ratio, 3.51; P=5.33×10−04) and longer times to final extubation (hazard ratio, 0.74; P=0.005). As de novo variants had a significant interaction with extra-cardiac anomalies for transplant-free survival (P=0.003), de novo variants conveyed no additional risk for transplant-free survival for patients with these anomalies (adjusted hazard ratio, 1.96; P=0.06). By contrast, de novo variants in patients without extra-cardiac anomalies were associated with worse transplant-free survival during follow-up (hazard ratio, 11.21; P=1.61×10−05) than that of patients with no de novo variants. Using agnostic machine-learning algorithms, we identified de novo copy number variants at 15q25.2 and 15q11.2 as being associated with worse transplant-free survival and 15q25.2, 22q11.21, and 3p25.2 as being associated with prolonged time to final extubation.
Conclusions:
In patients with congenital heart disease undergoing open-heart surgery, de novo variants were associated with worse transplant-free survival and longer times on the ventilator. De novo variants were most strongly associated with adverse outcomes among patients without extra-cardiac anomalies, suggesting a benefit for preoperative genetic testing even when genetic abnormalities are not suspected during routine clinical practice.
Registration:
URL: https://www.clinicaltrials.gov. Unique identifier: NCT01196182.
Congenital heart disease (CHD) is the most common birth defect, occurring in nearly 1% of live births.1–3 Despite advances in diagnosis, cardiac surgery, and perioperative management, CHD remains a leading cause of infant and child mortality. Recent discoveries have identified de novo damaging variants in ≈10% of patients with diverse CHD malformations.4–19 These variants include de novo single nucleotide variants and small insertions or deletions that are predicted to adversely impact the function or expression of one protein (denoted damaging genic variants [DGVs]), as well as de novo copy number variants (CNVs) that delete or duplicate contiguous genes. Damaging de novo variants in genes that are expressed in the developing heart and other organs are significantly more prevalent in CHD patients with extra-cardiac anomalies (ECAs), compared with patients with isolated CHD.4,5,18,19 Given the potential pleiotropic effects of de novo variants, an emerging question is whether genotype also influences broader clinical outcomes in patients with CHD.
A few prior studies have investigated large CNVs (>300 kb) discovered using genotype arrays in small cohorts. These studies demonstrated worse linear growth in CHD patients with single ventricle20; >3-fold increase in risk of death or transplant among nonsyndromic CHD patients21 and longer surgical bypass times, more re-operations, and increased intensive care stays in patients with chromosome 22q11.2 deletions and tetralogy of Fallot, truncus arteriosus, or interrupted aortic arch.22,23 To date, no studies have comprehensively examined the impact of de novo variants on postoperative outcomes in CHD patients with and without ECAs.
To better elucidate the influence of de novo genotype on clinical characteristics and postoperative outcomes after open-heart surgery, we analyzed 2517 CHD trios (patients and biological parents) enrolled by the Pediatric Cardiac Genomics Consortium. Using exome sequences, we identified clinically significant de novo variants, both CNVs and DGVs18 and analyzed their associations with ECAs. Among patients undergoing open-heart surgery, we then investigated the association of de novo variants with transplant-free survival and postoperative respiratory support, and their interaction with ECAs.
Methods
TOP Guidelines Statement
All supporting data are available within the article and the Data Supplement. Details methods used in this study are provided in the Data Supplement. The study protocol was approved by the Institutional Review Boards of Boston Children’s Hospital, Brigham and Women’s Hospital, Great Ormond St Hospital, Children’s Hospital of Los Angeles, Children’s Hospital of Philadelphia, Columbia University Medical Center, Icahn School of Medicine at Mount Sinai, Rochester School of Medicine and Dentistry, Steven and Alexandra Cohen Children’s Medical Center of New York, and Yale School of Medicine. Written informed consent was obtained from participating subjects or their parents.
Results
De Novo Variants Are Associated With ECA
Of 2517 patients with CHD in our cohort, 294 (11.7%) carried de novo variants: 131 (5.2%) had CNVs (3.2-fold more frequent than controls) and 169 (6.7%) had DGVs in multi-hit genes (denoted throughout as clinically significant; Methods and Tables IV through VIII in the Data Supplement). Patients with de novo variants were 2.4 times more likely to have ECAs (P=5.63×10−12). This association persisted when considering only patients with de novo CNVs, novel de novo CNVs not commonly associated with CHD (Table IX in the Data Supplement), de novo DGVs, and de novo DGVs in genes highly expressed in the heart (Table 1 and Methods in the Data Supplement).
Table 1.
Frequency of ECA in Patients With De Novo Damaging Variants

De Novo Variants Are Associated With Worse Transplant-Free Survival and Time to Extubation After Open-Heart Surgery
Surgical data were available in 1413 patients with CHD (Figure 1A) and all who had open-heart surgery exclusive of those with heart transplantation as their first surgery (n=1268 patients) were studied for postoperative transplant-free survival. Among these patients, 947 patients also had ventilator data for analyses of respiratory outcomes. Surgical patients, compared with those in whom surgical data were not available, were younger, had more severe CHD, and had more ECAs (Figure 1B).
Figure 1.
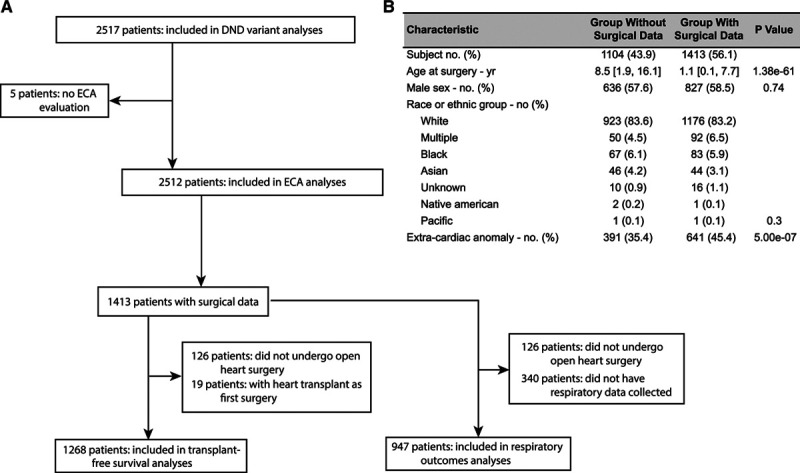
Description of study population. A, Flow chart depicting the derivation of the study population. B, Comparison of patients with and without available surgical data. Surgical data were not available for patients who underwent surgery before the treatment center began collecting data. Damaging genic variants (DGVs) include only those that are clinically significant (occur in multi-hit genes; see Methods in the Data Supplement). DND indicates de novo damaging.
Among the 1268 open-heart surgery patients, 143 (11.3%) had de novo CNVs and clinically significant DGVs (Figure 1B). ECAs occurred more frequently among surgical CHD patients with de novo variants. There was a difference in STAT (Society of Thoracic Surgeons-European Association for Cardio-Thoracic Surgery) mortality categories (reflecting increasing risk and severity of surgery) between those with and without de novo variants, primarily due to differences in categories 4 and 5. The remaining demographic and operative factors, as well as follow-up times and cardiac diagnoses, were similar (Table 2 and Table X in the Data Supplement).
Table 2.
Characteristics of Patients With and Without De Novo Damaging Variants

De novo variants were associated with significantly worse transplant-free survival during a median follow-up of 2.65 years after the first open-heart surgery in unadjusted and adjusted analyses taking into account age, total cardiopulmonary support time, STAT mortality category, center where the surgery was performed and preoperative factors (adjusted hazard ratio [HR], 3.51 [95% CI, 1.96–6.07], P=5.33×10−05; Methods and Table XI in the Data Supplement). This association was robust in sensitivity analyses that excluded individual STAT categories (Table XII in the Data Supplement). Subgroup analyses showed worse transplant-free survival in patients carrying de novo CNVs, novel de novo CNVs and de novo DGVs (Table 3, Figure 2, and Figure I in the Data Supplement). Atrial and ventricular septal defects were the most common malformations encountered, and de novo variants were also associated with worse transplant-free survival in this cohort (adjusted HR, 3.79 [95% CI, 1.67–8.23], P=0.002), as well as in patients with left-sided lesions (adjusted HR, 3.40, P=0.003; Table XIII in the Data Supplement).
Table 3.
Transplant-Free Survival and Time to Extubation in Patients With De Novo Variants

Figure 2.

Kaplan-Meier curves depicting (A) time to death or heart transplantation following first open-heart surgery and (B) time to final extubation following open-heart surgery for congenital heart disease patients with and without de novo damaging variants (includes only clinically significant damaging genic variants [DGVs]). Adjusted hazard ratios (HR) are shown.
De novo variants were also associated with significantly longer time to final extubation following open-heart surgery in unadjusted and adjusted analyses accounting for age, total cardiopulmonary support time, STAT mortality category, center where surgery was performed and preoperative factors (adjusted HR, 0.74 [95% CI, 0.60–0.91], P=0.005; Table XIV in the Data Supplement). This association was robust in sensitivity analyses that excluded individual STAT categories (Table XV in the Data Supplement). The time to final extubation was most prolonged in patients with septal defects and de novo variants (adjusted HR, 0.62 [95% CI, 0.47–0.82], P=7.45×10−04; Table XVI in the Data Supplement). Subgroup analyses showed longer time to final extubation in patients with de novo CNVs, novel de novo CNVs, but not de novo DGVs (Table 3, Figure 2, and Figure I in the Data Supplement).
De Novo Variants, ECA, and Clinical Outcomes
Since de novo variants are associated with ECAs, and ECAs have been reported to lead to worse outcomes,22,23 we investigated the association between the 2 and clinical outcomes. De novo variants and ECAs had a significant interaction (P=0.003) in their association with transplant-free survival; that is, the magnitude of association between de novo variants and death or transplant varied according to the presence or absence of ECAs. In patients with ECAs, there was no additional hazard associated with the presence of de novo variants on transplant-free survival (HR, 1.96 [95% CI, 0.96–3.79], P=0.06). In contrast, among patients without ECAs, de novo variants were associated with worse transplant-free survival (HR, 11.21 [95% CI, 4.12–29.13], P=1.61×10−05; Figure 3). Moreover, transplant-free survival of patients with versus without variants in nonsyndromic genes (adjusted HR, 3.84 [95% CI, 2.12–6.94]; P=8.77×10−06) was worse than similar comparisons of variants in syndromic genes (adjusted HR, 1.21 [95% CI, 0.28–5.31]; P=0.80).
Figure 3.

Interaction between de novo damaging variants and extra-cardiac anomalies (ECA) in their association with transplant-free survival and time to final extubation. A, Models for transplant-free survival and time to final extubation that include interaction between DND variants and ECAs. Analyses include only clinically significant damaging genic variants (DGVs). There was a significant interaction between the 2 for transplant-free survival. Among patients without ECAs, de novo damaging variants were associated with worse transplant-free survival, but there was no significant association among patients with ECAs. For time to final extubation, both de novo damaging variants and ECAs were independently associated with worse outcome without interaction between the 2. B and C, Kaplan-Meier curves depicting time to death or heart transplantation following first open-heart surgery for patients with and without ECAs. Adjusted hazard ratios (HR) are shown. STAT indicates Society of Thoracic Surgeons-European Association for Cardio-Thoracic Surgery.
Both de novo variants (HR, 0.75 [95% CI, 0.61–0.94], P=0.01) and ECAs (HR, 0.86 [95% CI, 0.76–0.99], P=0.03) were independently associated with longer time to final extubation. However, there was no significant interaction between de novo variants and ECAs with regard to time to final extubation (P=0.89).
Machine-Learning Insights Into High-Risk De Novo Variants Associated With Adverse Clinical Outcomes
We used agnostic, model-free, machine-learning methods24–26 to determine if some de novo variants were associated with worse clinical outcomes (detailed in Methods in the Data Supplement). Analyses were limited to the 1268 CHD patients with surgical data, who were randomly assigned into discovery and validation cohorts. For each of one thousand pervariants, we sought to identify de novo variants that were associated with worse transplant-free survival and prolonged time to final extubation (Figure 4). For transplant-free survival de novo CNVs at 2 loci, 15q25.2 and 15q11.2, were identified in both the discovery and validation cohorts at substantial frequencies (48% and 43%, respectively; Figure 4A). Patients with CHD harboring these high-risk CNVs had significantly worse transplant-free survival compared with CHD patients with no de novo variants (HR, 17.44; P=3.53×10−04) and those with other de novo variants (Figure 4C). The remaining de novo variants were still associated with worse transplant-free survival but at a lower hazard (HR, 2.68 [95% CI, 1.47–4.63], P=0.002).
Figure 4.
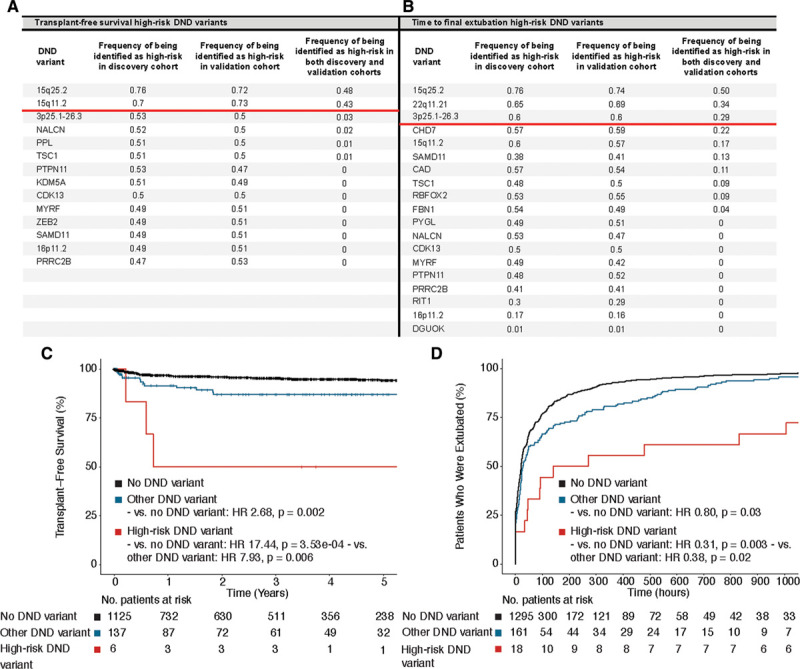
Identification and validation of high-risk de novo damaging variants. Permutation analyses identified de novo copy number variants at 15q25.2 and 15q11.2 as high-risk for transplant-free survival (A) and 15q25.2, 22q11.21, and 3p25.1-26.3 as high-risk for prolonged time to final extubation (B). C, Kaplan-Meier curves depicting time to death or heart transplantation after first open-heart surgery in congenital heart disease (CHD) patients with high-risk de novo damaging variants (15q25.2 and 15q11.2), any other de novo damaging variants, and no de novo variants. D, Kaplan-Meier curves depicting time to final extubation after open-heart surgery in patients with high-risk de novo damaging variants (15q25.2, 22q11.21, and 3p25.1-26.3), other de novo damaging variants and no de novo variants. Adjusted hazard ratios (HR) are shown.
De novo CNVs affecting chromosomes 15q25.2, 22q11.21 and 3p25.1-26.3 were most frequently identified in both the discovery and validation cohorts (50%, 34%, and 29% of pervariants, respectively) in association with prolonged time to final extubation. These de novo variants were also associated with significantly prolonged times to final extubation compared with the remaining de novo variants (HR, 0.38 [95% CI, 0.17–0.86]), although the remaining de novo variants also had a residual hazard for prolonged intubation (HR, 0.80 [95% CI, 0.65–0.98]).
Discussion
Genomic analyses of 2517 patients with CHD and their parents revealed clinically significant de novo variants in 11.7% of patients. Patients with de novo variants were more likely to have ECAs, reinforcing prior evidence that de novo variants are more prevalent in syndromic than in isolated CHD.4,19 In a subset of 1268 patients with CHD who had undergone open-heart surgery and for whom we had surgical data, we also found that de novo variants were associated with worse transplant-free survival, as well as worse postoperative respiratory outcomes, including a greater likelihood of postoperative re-intubation, and longer times to first and final extubation. Importantly, de novo variants were strongly associated with adverse outcomes among patients without ECAs, in whom genetic abnormalities might not have been suspected during routine clinical practice.
Smaller studies have previously reported associations between CNVs and clinical outcomes,20–23 but this is the first large scale study to examine the overall impact of all de novo variants, including DGVs, on postoperative outcomes. Our findings are consistent with prior studies that have demonstrated worse transplant-free survival in patients with large CNVs, as well as in patients with 22q11.2 deletions. Notably, subgroup analyses that excluded CNVs commonly associated with CHD, including 22q11.2 deletions, still demonstrated significantly worse outcomes, suggesting that the observed associations are not solely driven by patients with well-established CNVs. Rare, not commonly reported de novo CNVs were equally important determinants of postoperative outcomes.
DGVs that occurred 2 or more times within the same gene had significant associations with ECAs, transplant-free survival, and respiratory outcomes. This observation supports 2 conclusions. First, some DGVs, particularly those identified only once in the study cohort, were likely incidental mutations that did not account for CHD and associated phenotypes. As larger cohorts are studied, we expect that a more complete repertoire of genes that are critical for heart and organ development will emerge. Second, because DGVs affect a single gene, while the majority of CNVs in this cohort spanned multiple genes, the observed associations likely reflect the pleiotropic effects of individual genes. Further elucidation of the developmental function of variants in these genes may inform mechanisms for these clinically important associations.
Several prior studies demonstrated a significant enrichment of de novo variants among CHD patients with ECAs.4,18,19 These findings indicate that some de novo variants cause pleiotropic effects that are responsible for both cardiac and extra-cardiac defects. Moreover, because de novo variants are associated with ECAs, and ECAs are associated with worse outcomes,22,23 ECAs are in the causal pathway between de novo mutations and worse surgical outcomes. Consistent with this interpretation, our interaction analyses demonstrated that the magnitude of the association between de novo variants and death/transplant was different for patients with versus those without ECAs. Within the subgroup of patients with ECAs, the association of de novo variants with adverse outcome was modest and did not reach statistical significance.
CHD patients without ECAs have a lower burden of de novo variants.4,18,19 But among those without ECAs who do have de novo variants, our study provided the first and strong evidence that these genotypes were associated with a statistically significantly greater hazard of mortality or cardiac transplantation during a median follow-up of 2.65 years. This finding may reflect the impact of de novo variants on cardiopulmonary demands, resilience after open-heart surgery, or other effects independent of overt ECAs. The effects of unrecognized de novo variants might contribute to increased, early mortality of patients with simple (isolated, uncomplicated septal defects and patent ductus arteriosus) CHD.27 The mechanisms and genetic pathways through which de novo variants and ECAs act warrant further study.
Currently, the majority of patients with CHD and ECAs undergo genetic testing, which leads to early diagnosis, further diagnostic testing as dictated by the specific genetic diagnosis, and anticipatory guidance over time. Our data suggest that genetic testing is also important in CHD patients without clinically overt ECAs for 2 reasons. First, these patients also have significantly worse outcomes, and second, although we excluded from sequencing patients with clinically diagnosed syndromic CHD, a number of syndromic variants were nevertheless discovered by exome sequence analyses. This suggests that even at high-volume centers, many syndromic patients remain under-diagnosed.
To explore the relationships between specific genotypes and open-heart surgery outcomes in patients with CHD, we leveraged model-free machine-learning predictive tools. Among all CHD subjects with relevant postoperative data, we found that specific de novo variants were associated with the highest risks of mortality, heart transplantation, or prolonged time to extubation. Because we had only one CHD cohort with both genetic and clinical data, we used permutation analyses to confirm these associations. Our finding that de novo CNVs on chromosome 22q11.2 conveyed high risk for prolonged time to extubation provides support for this approach, as independent data associate this locus with respiratory dysfunction.23,28 Overall, we identified de novo CNVs at 2 loci (15q25.2 and 15q11.2) associated with markedly worse transplant-free survival and 3 loci (15q25.2, 22q11.2, and 3p25.1-26.3) associated with longer time to final extubation. Patients with these variants had worse outcomes than those with other de novo variants, who in turn had worse outcomes than patients with no de novo variants. Together, these data suggest that different de novo variants confer varying amounts of risk, presumably due to the gene functions that are perturbed.
There are limitations in our study. We had surgical data for about half of the cohort that had undergone genetic sequencing. However, to our knowledge, this is the largest study that has investigated the relationship between de novo genotype and postoperative outcomes in CHD.
The absence of surgical data resulted largely because participating centers in our study began standardized surgical data collection at different times between 1991 and 2015, sometimes after patients had undergone open-heart surgery. Consistent with this, the mean age of patients with surgical data was 1.1 years, whereas that of patients without surgical data was 8.5 years, creating some differences between the groups. Patients without surgical data had fewer ECAs and were more likely to have been enrolled in this study, years after cardiac surgery. However, de novo variants occurred with comparable prevalence among patients with and without surgical data, suggesting that our inability to capture surgical cases for administrative reasons was unlikely to have biased our findings. Some patients with CHD might have had unrecognized subclinical ECAs, as phenotypes were only identified as part of clinical care, and misclassification of such subjects as ECA-negative could have contributed risk that we ascribed to genotype. Whereas all study patients received care in tertiary clinical centers from physicians with considerable expertise in recognizing syndromic CHD, future re-evaluation of these patients may improve phenotyping. Finally, our analyses examined exome sequences, which enabled broad detection of DGVs and CNVs spanning ≥3 exons,15 but excluded most intronic de novo variants and would not capture epigenetic modifiers.
We conclude that in addition to providing insights into CHD causes, de novo genetic variants are associated with noncardiac phenotypes and negative outcomes after cardiac surgery. CHD patients with de novo variants have more ECA, require longer postoperative ventilator support and have decreased transplant-free survival. With the increased availability and reduced costs of genomic sequencing, clinical application of this technology may be warranted to identify high-risk patients with CHD before cardiac surgical interventions.
Acknowledgments
We are grateful to the patients and families who participated in this research. We thank the following team members for outstanding contributions to patient recruitment: A. Julian, M. Mac Neal, Y. Mendez, T. Mendiz-Ramdeen, C. Mintz (Icahn School of Medicine at Mount Sinai); N. Cross (Yale School of Medicine); K. Sadamistu and N. Tran (Children’s Hospital of Los Angeles); B. McDonough, A. Monafo, J. Stryker, J. Geva (Harvard Medical School), K. Flack, L. Panesar, N. Taylor (University College London); E. Taillie (University of Rochester School of Medicine and Dentistry); S. Edman, J. Garbarini, J. Tusi, S. Woyciechowski, (Children’s Hospital of Philadelphia); D. Awad, C. Breton, K. Celia, C. Duarte, D. Etwaru, N. Fishman, M. Kaspakoval, J. Kline, R. Korsin, A. Lanz, E. Marquez, D. Queen, A. Rodriguez, J. Rose, J.K. Sond, D. Warburton, A. Wilpers, and R. Yee (Columbia Medical School).
Sources of Funding
This work was supported by the U01-HL098153 and grant UL1TR000003 from the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health (NIH), grants to the Pediatric Cardiac Genomics Consortium (U01-HL098188, U01-HL098147, U01-HL098153, U01-HL098163, U01-HL098123, and U01-HL098162), the NIH Centers for Mendelian Genomics (5U54HG006504), the Howard Hughes Medical Institute (CES) and the Simons Foundation (WKC). Dr Boskovski was supported by 5T32HL007572-33 from the National Heart, Lung, and Blood Institute. Dr Homsy was supported by the John S. LaDue Fellowship at Harvard Medical School and is a recipient of the Alan Lerner Research Award at the Brigham and Women’s Hospital. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Heart, Lung, and Blood Institute, the National Center for Research Resources, or the NIH.
Disclosure
None.
Supplementary Material
Footnotes
Nonstandard Abbreviations and Acronyms
- CHD
- congenital heart disease
- CNV
- copy number variants
- DGV
- damaging genic variant
- DND
- de novo damaging
- ECA
- extra-cardiac anomaly
- HR
- hazard ratio
- NDD
- neurodevelopmental delay
Drs Newburger and Seidman are joint senior authors.
For Sources of Funding and Disclosures, see page 192 and 193.
The Data Supplement is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCGEN.119.002836.
References
- 1.Reller MD, Strickland MJ, Riehle-Colarusso T, Mahle WT, Correa A. Prevalence of congenital heart defects in metropolitan Atlanta, 1998-2005. J Pediatr. 2008;153:807–813. doi: 10.1016/j.jpeds.2008.05.059. doi: 10.1016/j.jpeds.2008.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ, Roos-Hesselink JW. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58:2241–2247. doi: 10.1016/j.jacc.2011.08.025. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 3.Matthews TJ, MacDorman MF. Infant mortality statistics from the 2010 period linked birth/infant death data set. Natl Vital Stat Rep. 2013;62:1–26. [PubMed] [Google Scholar]
- 4.Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erdogan F, Larsen LA, Zhang L, Tümer Z, Tommerup N, Chen W, Jacobsen JR, Schubert M, Jurkatis J, Tzschach A, et al. High frequency of submicroscopic genomic aberrations detected by tiling path array comparative genome hybridisation in patients with isolated congenital heart disease. J Med Genet. 2008;45:704–709. doi: 10.1136/jmg.2008.058776. doi: 10.1136/jmg.2008.058776. [DOI] [PubMed] [Google Scholar]
- 7.Soemedi R, Wilson IJ, Bentham J, Darlay R, Töpf A, Zelenika D, Cosgrove C, Setchfield K, Thornborough C, Granados-Riveron J, et al. Contribution of global rare copy-number variants to the risk of sporadic congenital heart disease. Am J Hum Genet. 2012;91:489–501. doi: 10.1016/j.ajhg.2012.08.003. doi: 10.1016/j.ajhg.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenway SC, Pereira AC, Lin JC, DePalma SR, Israel SJ, Mesquita SM, Ergul E, Conta JH, Korn JM, McCarroll SA, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41:931–935. doi: 10.1038/ng.415. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci USA. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warburton D, Ronemus M, Kline J, Jobanputra V, Williams I, Anyane-Yeboa K, Chung W, Yu L, Wong N, Awad D, et al. The contribution of de novo and rare inherited copy number changes to congenital heart disease in an unselected sample of children with conotruncal defects or hypoplastic left heart disease. Hum Genet. 2014;133:11–27. doi: 10.1007/s00439-013-1353-9. doi: 10.1007/s00439-013-1353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silversides CK, Lionel AC, Costain G, Merico D, Migita O, Liu B, Yuen T, Rickaby J, Thiruvahindrapuram B, Marshall CR, et al. Rare copy number variations in adults with tetralogy of Fallot implicate novel risk gene pathways. PLoS Genet. 2012;8:e1002843. doi: 10.1371/journal.pgen.1002843. doi: 10.1371/journal.pgen.1002843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldmuntz E, Paluru P, Glessner J, Hakonarson H, Biegel JA, White PS, Gai X, Shaikh TH. Microdeletions and microduplications in patients with congenital heart disease and multiple congenital anomalies. Congenit Heart Dis. 2011;6:592–602. doi: 10.1111/j.1747-0803.2011.00582.x. doi: 10.1111/j.1747-0803.2011.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thienpont B, Mertens L, de Ravel T, Eyskens B, Boshoff D, Maas N, Fryns JP, Gewillig M, Vermeesch JR, Devriendt K. Submicroscopic chromosomal imbalances detected by array-CGH are a frequent cause of congenital heart defects in selected patients. Eur Heart J. 2007;28:2778–2784. doi: 10.1093/eurheartj/ehl560. doi: 10.1093/eurheartj/ehl560. [DOI] [PubMed] [Google Scholar]
- 14.Hitz MP, Lemieux-Perreault LP, Marshall C, Feroz-Zada Y, Davies R, Yang SW, Lionel AC, D’Amours G, Lemyre E, Cullum R, et al. Rare copy number variants contribute to congenital left-sided heart disease. PLoS Genet. 2012;8:e1002903. doi: 10.1371/journal.pgen.1002903. doi: 10.1371/journal.pgen.1002903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glessner JT, Bick AG, Ito K, Homsy J, Rodriguez-Murillo L, Fromer M, Mazaika E, Vardarajan B, Italia M, Leipzig J, et al. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ Res. 2014;115:884–896. doi: 10.1161/CIRCRESAHA.115.304458. doi: 10.1161/CIRCRESAHA.115.304458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards AA, Santos LJ, Nichols HA, Crider BP, Elder FF, Hauser NS, Zinn AR, Garg V. Cryptic chromosomal abnormalities identified in children with congenital heart disease. Pediatr Res. 2008;64:358–363. doi: 10.1203/PDR.0b013e31818095d0. doi: 10.1203/PDR.0b013e31818095d0. [DOI] [PubMed] [Google Scholar]
- 17.Fahed AC, Gelb BD, Seidman JG, Seidman CE. Genetics of congenital heart disease: the glass half empty. Circ Res. 2013;112:707–720. doi: 10.1161/CIRCRESAHA.112.300853. doi: 10.1161/CIRCRESAHA.112.300853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, Zeng X, Qi H, Chang W, Sierant MC, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49:1593–1601. doi: 10.1038/ng.3970. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sifrim A, Hitz MP, Wilsdon A, Breckpot J, Turki SH, Thienpont B, McRae J, Fitzgerald TW, Singh T, Swaminathan GJ, et al. INTERVAL Study; UK10K Consortium; Deciphering Developmental Disorders Study. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016;48:1060–1065. doi: 10.1038/ng.3627. doi: 10.1038/ng.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carey AS, Liang L, Edwards J, Brandt T, Mei H, Sharp AJ, Hsu DT, Newburger JW, Ohye RG, Chung WK, et al. Effect of copy number variants on outcomes for infants with single ventricle heart defects. Circ Cardiovasc Genet. 2013;6:444–451. doi: 10.1161/CIRCGENETICS.113.000189. doi: 10.1161/CIRCGENETICS.113.000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim DS, Kim JH, Burt AA, Crosslin DR, Burnham N, Kim CE, McDonald-McGinn DM, Zackai EH, Nicolson SC, Spray TL, et al. Burden of potentially pathologic copy number variants is higher in children with isolated congenital heart disease and significantly impairs covariate-adjusted transplant-free survival. J Thorac Cardiovasc Surg. 2016;151:1147.e4–1151.e4. doi: 10.1016/j.jtcvs.2015.09.136. doi: 10.1016/j.jtcvs.2015.09.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mercer-Rosa L, Pinto N, Yang W, Tanel R, Goldmuntz E. 22q11.2 Deletion syndrome is associated with perioperative outcome in tetralogy of Fallot. J Thorac Cardiovasc Surg. 2013;146:868–873. doi: 10.1016/j.jtcvs.2012.12.028. doi: 10.1016/j.jtcvs.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Byrne ML, Yang W, Mercer-Rosa L, Parnell AS, Oster ME, Levenbrown Y, Tanel RE, Goldmuntz E. 22q11.2 Deletion syndrome is associated with increased perioperative events and more complicated postoperative course in infants undergoing infant operative correction of truncus arteriosus communis or interrupted aortic arch. J Thorac Cardiovasc Surg. 2014;148:1597–1605. doi: 10.1016/j.jtcvs.2014.02.011. doi: 10.1016/j.jtcvs.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshizato T, Dumitriu B, Hosokawa K, Makishima H, Yoshida K, Townsley D, Sato-Otsubo A, Sato Y, Liu D, Suzuki H, et al. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med. 2015;373:35–47. doi: 10.1056/NEJMoa1414799. doi: 10.1056/NEJMoa1414799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. Ann Appl Stat. 2008;2:841–860. [Google Scholar]
- 26.Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011;39:1–13. doi: 10.18637/jss.v039.i05. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Videbæk J, Laursen HB, Olsen M, Høfsten DE, Johnsen SP. Long-term nationwide follow-up study of simple congenital heart disease diagnosed in otherwise healthy children. Circulation. 2016;133:474–483. doi: 10.1161/CIRCULATIONAHA.115.017226. doi: 10.1161/CIRCULATIONAHA.115.017226. [DOI] [PubMed] [Google Scholar]
- 28.Mercer-Rosa L, Paridon SM, Fogel MA, Rychik J, Tanel RE, Zhao H, Zhang X, Yang W, Shults J, Goldmuntz E. 22q11.2 deletion status and disease burden in children and adolescents with tetralogy of Fallot. Circ Cardiovasc Genet. 2015;8:74–81. doi: 10.1161/CIRCGENETICS.114.000819. doi: 10.1161/CIRCGENETICS.114.000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


