Abstract
We have performed an integrative analysis of SARS-CoV-2 genome sequences from different countries. Apart from mutational analysis, we have predicted host antiviral miRNAs targeting virus genes, PTMs in the virus proteins and antiviral peptides. A comparison of the analyses with other coronavirus genomes has been performed, wherever possible. Our analysis confirms unique features in the SARS-CoV-2 genomes absent in other evolutionarily related coronavirus family genomes, which presumably confer unique infection, transmission and virulence capabilities to the virus. For understanding the crucial factors involved in host-virus interactions, we have performed Bioinformatics aided analysis integrated with experimental data related to other corona viruses. We have identified 42 conserved miRNAs that can potentially target SARS-CoV-2 genomes. Interestingly, out of these, 3 are previously reported to exhibit antiviral activity against other respiratory viruses. Gene expression analysis of known host antiviral factors reveals significant over-expression of IFITM3 and down regulation of cathepsins during SARS-CoV-2 infection, suggesting its active role in pathogenesis and delayed immune response. We also predicted antiviral peptides which can be used in designing peptide based drugs against SARS-CoV-2. Our analysis explores the functional impact of the virus mutations on its proteins and interaction of its genes with host antiviral mechanisms.
Keywords: Bioinformatics, Genetics, Infectious disease, Virology, Coronavirus, Antiviral miRNA, Antiviral peptides
Bioinformatics; Genetics; Infectious Disease; Virology; Coronavirus; antiviral miRNA; antiviral peptides.
1. Introduction
Rapidly spreading Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infections are responsible for the COVID-19 pandemic [1]. The first COVID-19 case was reported in Wuhan in December 2019, the devastating disease quickly spread out across China and many other countries around the globe to eventually turn into a pandemic. SARS-CoV-2 is a single-stranded positive-sense RNA β-coronavirus of the coronaviridae family of viruses. The SARS-CoV-2 genome shares a significant sequence similarity with SARS-CoV, the virus responsible for the 2003 viral outbreak, which comparatively had a much higher lethal rate of 10% [2].
SARS-CoV-2 genomic RNA is translated into two long polypeptides (pp1a/pp1ab), auto-proteolytically processed into 16 non-structural proteins (NSPs), forming the replicase/transcriptase complex (RTC). PP1a codes for 12 NSPs (1–12) including papain-like protease/PLpro domain (NSP3), 3C-like protease (NSP5) and RNA-dependent RNA polymerase/RdRp (NSP12). PP1ab codes for 4 NSPs (13–16), including helicase (NSP13), 3′-to-5′ exoribonuclease (NSP14), followed by structural proteins and downstream ORFs, namely Surface glycoprotein (or Spike), ORF3a, ORF3b, Envelope (E), Membrane (M), ORF6, ORF7a, ORF7b, ORF8, N protein (N), ORF9 and ORF10 [3].
Many coronavirus proteins, including S and M proteins, undergo post-translational modifications (PTMs) necessary for virus receptor binding and replication [4]. The S protein binds to the host cell receptor angiotensin-converting enzyme 2 (ACE2) which mediates its entry and fusion into the host cells [5]. The virus S protein is composed of two functional subunits, namely S1 and S2. The S1 subunit contains the receptor binding domain (RBD), and S2 subunit is responsible for receptor binding and fusion of the virus and cellular membrane. The subunits are cleaved by host proteases like transmembrane protease serine 2 (TMPRSS2) and Cathepsin L [6,7].
The S protein is extensively glycosylated and its binding sites have been reported to serve as an alternative receptor or as an enhancer of the ACE2-mediated infections [4]. Thus, it acts as an important factor to determine the host range and tropism. Hence, the knowledge of different types of S protein PTMs such as O-linked glycosylations, N-linked glycosylations and 3CL-like proteinase cleavage sites have important consequences for development of any therapies targeting the protein.
The virus sequence mutation rate is one of the most fundamental aspects of its evolution in response to selective pressures, which is governed by multiple processes such as polymerase fidelity, 3′ exonuclease activity and post-replicative repair, amongst others [[8], [9]]. Clues from the sequence analysis of the evolving virus genomes in a pandemic have important implications for both strategic planning in the prevention, disease progression and development of vaccines and therapeutic antibodies, even while pandemic is in progression [10].
Owing to the globally alarming COVID-19 outbreak, worldwide efforts are on to mitigate the rapidly spreading viral infection. Bioinformatics aided analysis can enhance these efforts by providing valuable insights into the changes in the evolving virus strains. In the present study, we comprehensively analysed the SARS-CoV-2 genomes from different geographical locations, and identified the crucial factors involved in host-pathogen interactions. Based on the comparative genome analysis, we focused our integrated analysis on different aspects of the evolving SARS-CoV-2, such as mutation analysis and its impact on protein function and stability, identification of host-miRNA targets, host gene expression in response to the viral infections, prediction of antiviral peptides, and correlating it with the literature. The results of the analysis led to a plethora of information regarding the mechanisms involved in evolution and pathogenesis of the virus, which have implications for the COVID-19 related research.
2. Materials and methods
2.1. Retrieval of SARS-CoV-2 genome data
High coverage, complete SARS-CoV-2 genome sequences (10,213 in number), and corresponding metadata were retrieved from the GISAID database, submitted till 15th April 2020 [11]. The SARS-CoV (NC_004718.3) and MERS (KC164505.2) genomes were downloaded from the NCBI genome database and compared with SARS-CoV-2 (NC_045512.2; Wuhan), taken as a reference for the analysis. Gene annotations and protein sequences of the selected SARS-CoV-2 genome were retrieved from the ViPR database [12].
2.2. Genotyping analysis and impact of mutations
The downloaded SARS-CoV-2 genomes were subjected to mutation analysis using Genome Detective Coronavirus Subtyping Tool (version 1.1.3) [13]. In order to remove redundancy, the genome nucleotide sequences that share 99% similarity with the reference genome were excluded from further analysis. To get a consensus prediction of the impact of mutations on protein stability, we employed two machine learning (ML) algorithms, namely I-MUTANT [14] and MuPro [15], using default parameters. Apart from prediction of effect of the identified mutations, we also analysed the effect of previously reported variations in the S protein sequences of the genomes [16].
2.3. Host antiviral miRNAs and its predicted targets
To identify conserved host antiviral miRNAs targeting SARS-CoV-2 genomes, we performed miRNA target predictions. We downloaded a complete list of all the available experimentally validated mature human miRNAs (2654) from the miRBase [17] (Release 22.1), and surveyed the literature to identify experimentally validated human antiviral miRNAs amongst the miRNAs. From the literature survey, we were able to identify 42 miRNAs reported to have antiviral activities against different viruses. These miRNAs were used to identify potential miRNA target sites in the virus genome sequences, using miRanda (3.3 a version) [18], with an energy threshold of -20 kcal/mol, a threshold used in other studies too [19]. We predicted targets of the miRNAs in SARS-CoV (NC_004718.3), MERS (NC_019843.3), and SARS-CoV-2 isolate from Wuhan (NC_045512.2).
2.4. The prediction of post-translational modifications (PTMs) and antiviral peptides
We predicted O and N-linked glycosylation sites in S protein using NetOglyc 4.0 [20] and NetNGly1.0 [21], respectively. Putative 3C-like proteinase sites in SARS-CoV-2 proteins were predicted using the NetCorona1.0 prediction method [22], using default parameters. Palmitoylation of conserved cysteine residues in S protein were predicted using CSS-Palm 2.0 [23]. AVPpred [24] was used for the prediction of antiviral peptides.
2.5. Host gene expression analysis
The gene expression of the host factors previously reported to play a important role in coronavirus infections (ACE2, Cathepsin, TMPRSS11D, IFITM, STAT and few others) [25] were analysed in the SARS-CoV (GSE17400) and SARS-CoV-2 (GSE147507) gene expression datasets downloaded from the NCBI GEO database. The expression levels of selected genes were further examined and compared to get additional insights into similarities or dissimilarities in the host defence mechanisms in SARS-CoV and SARS-CoV-2 infections.
3. Results
3.1. Mutational analysis
Genome mapping analysis revealed that the reference SARS-CoV-2 genome is highly identical to the SARS-CoV genome sequence, at nucleotide as well as protein levels, with 79.57% and 83.36% identities, respectively. The 17% variation in the amino acid level corresponds to mutations in various viral proteins, including the S protein, E, M and NSP of SARS-CoV-2. Wherein, we observed 27, 24 and 17 deletions in ORF1ab, S protein and ORF8.
We observed that the mutations are non-uniformly distributed in all the SARS-CoV-2 proteins as some proteins show a higher number of variations, while others have just one mutation which is not surprising as the virus is evolving, maximum numbers of mutations were observed in SARS-CoV-2 isolates from the USA (n = 1839), China (n = 1511) and Europe (n = 1092) while relatively few mutations were observed in the sequences from Vietnam (n = 56), Japan (n = 78), Australia (n = 103). The high frequency variations that were found to occur maximally in the genomes include L3606F and P4715L (ORF1ab), P323L (RdRp), L37F (NSP6), D614G (S protein), G251V (ORF3a) and L84S (ORF8). Intriguingly, it is observed that few co-mutations such as P323L in RdRp are prevalent and restricted to the countries which are leading in spread and mortality of the infection (including the USA, Spain, Italy, and Germany).
From the entropy calculations, available from GISAID metadata, maximum entropy is observed at the amino-acid position 614 of S, 3606 of ORF1ab, with entropy values of 0.662 and 0.417, respectively.
3.2. Impact of the mutations on protein stability
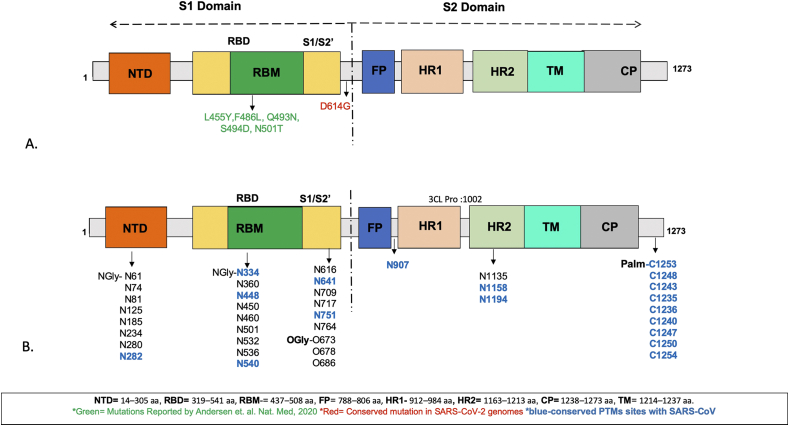
MuPro and I-Mutant servers predict a decrease in the protein stability due to the mutations in S proteins, except P4715L (ORF1ab), P323L (RdRp) (Table1). Out of all the S protein mutations, L455Y, F486L, Q493N, in the receptor binding domain (RBD:319–541), Q493N, S494D, N501T in the receptor binding motif (RBM: 437–508), and D614G are present in S1 subunit (Figure 1A). These findings are in concordance with the previous findings, which suggest S1 to be the highly variable region as compared to the S2 subunit. The mutation D614G was conserved among various countries and was found to be located in S1, in a linking region between S1 and S2.
Table 1.
Predicted effects on the protein stability due to the mutations in the spike glycoproteins in various SARS-CoV-2 genomes, using various prediction methods.
| Mutations | Query Genome | Protein | I-Mutant | MUpro | Study |
|---|---|---|---|---|---|
| L455Y | SARS-CoV | S protein | Decrease | -2.34 | Reported∗ |
| F486L | SARS-CoV | S protein | Decrease | -0.68 | Reported∗ |
| Q493N | SARS-CoV | S protein | Decrease | -0.55 | Reported∗ |
| N501T | SARS-CoV | S protein | Decrease | -1.52 | Reported∗ |
| S494D | SARS-CoV | S protein | Decrease | -0.20 | Reported∗ |
| D614G | SARS-CoV-2 | S protein | Decrease | -1.48 | Present study |
| L3606F | SARS-CoV-2 | ORF 1ab | Decrease | -1.29 | Present study |
| P4715L | SARS-CoV-2 | ORF 1ab | Increase | 0.60 | Present study |
| P323L | SARS-CoV-2 | RdRp | Increase | 0.60 | Present study |
| L37F | SARS-CoV-2 | NSP6 | Decrease | -1.29 | Present study |
| L84S | SARS-CoV-2 | ORF 8 | Decrease | -1.084 | Present study |
| G251V | SARS-CoV-2 | ORF3a | Decrease | -0.45 | Present study |
Andersen et. al. Nat. Med, 2020 [16].
Figure 1.
Schematic domain representation of spike glycoprotein A. Spike glycoprotein mutations B. PTMs identified in the Spike glycoproteins.
3.3. The S protein post-translational modifications (PTMs)
Another important process common in different coronaviruses is the S protein post translational modifications, in which the protein is post-translationally glycosylated [26]. In order to predict the presence and the distribution of PTMs in SARS-CoV-2, we used several prediction methods. The predictions reveal 27 and 23 N-linked glycosylation sites in the S proteins of SARS-CoV-2 and SARS-CoV, respectively. Among the predicted 27 N-linked glycosylations in SARS-CoV-2 S protein, 18 are unique to SARS-CoV-2, whereas 9 are conserved with respect to the SARS-CoV S protein (Figure 1B). Recently, 7 out of the predicted 27 N-linked glycosylation sites in SARS-CoV-2 have been experimentally confirmed by mass spectrometry [27]. We also identified 2 putative O-linked glycosylation sites unique to SARS-CoV-2, in the S1 domain of S protein along with a conserved polybasic site [16]. The conserved cysteine residues in the cytosolic tail of S protein are modified by palmitoylation in SARS-CoV. From the sequence analysis, we predict 9 conserved cysteine residues to undergo palmitoylation that are located in the cytoplasmic domain (1238–1273) of SARS-CoV-2 S protein (Figure 1B). Also, a potential and conserved 3CL proteinase cleavage site TGRLQˆSLQTY is present at the position 1002 in the S protein.
3.4. The prediction of antiviral peptides
AVPpred [24] Antiviral peptides prediction method predicts a peptide KWPWYIWLGFIAGLI to bind S protein with a very high prediction score of 0.98. No antiviral peptides were predicted against N protein and ORF7a. However, NSP7 and NSP10 have a common predicted antiviral peptide VNCLDDRCILHCANF.
3.5. The prediction of antiviral host miRNAs
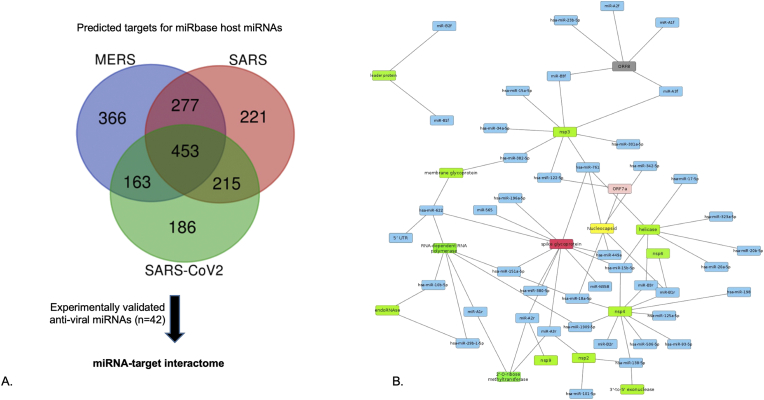
Several host miRNAs are known to activate different defence mechanisms during viral infections [28]. Herein, we predicted the host miRNAs which potentially play a role in the host and the virus interactions by directly targeting the virus genes. We also speculate the impact of mutations identified in SARS-CoV-2 genomes upon miRNA interactions. The analysis of miRNA target predictions revealed that 221, 366 and 186 human miRNAs target SARS-CoV, MERS and SARS-CoV-2 (NC_045512.2) genes, respectively (Figure 2A). We also found 42 conserved antiviral miRNAs predicted to have targets in all the SARS-CoV-2 genomes, irrespective of their geographical location and mutations. These putative antiviral miRNAs are predicted to have target sites specific to NSPs, ORF1ab, ORF8, N protein, 5′UTR and S protein in SARS-CoV-2 (Figure 2B). Interestingly, the maximum number of miRNAs target ORF1ab and S protein genes, two amino acids each of protein products of the genes are found to have maximum entropy change from the mutational analysis (Section 3.1). This shows that despite the higher mutation frequencies in the two genes, the miRNAs targets are conserved, and may be serving as a natural antiviral host-defence mechanism. For instance, hsa-miR-138-5p, hsa-miR-622, hsa-miR-761, miR-A3r have 4 targets each, and hsa-miR-15b-5p, hsa-miR-18a-5p, miR-A2r, miR-B1r have 3 targets each in the SARS CoV-2 genomes. Interestingly, 6 out of these 8 miRNAs are found to target S protein besides other genes, and 7 have 2 or more targets on ORF1ab gene. 12 out of 42 putative antiviral miRNAs have 2 targets each while others have a single target in SARS-CoV-2. We also identified 3 miRNAs, namely, miR-125a, miR-198 and miR-23b, which have been reported to play a crucial antiviral role in respiratory diseases [28].
Figure 2.
miRNA target identification A. Venn diagram showing host-miRNA targets identified in the 3 genomes. B. host-miRNA target network in SARS-CoV-2 protein showing maximum number of miRNAs targeting ORF1ab, followed by spike glycoprotein and nucleocapsid protein.
3.6. Gene expression analysis
Since SARS-CoV-2 shows considerable similarities with SARS-CoV in terms of genotype and phenotype/pathogenesis, we investigated and correlated the host gene expression in SARS-CoV. To understand and compare the changes in gene expression of human genes identified to be induced in the two viral infections, (i.e. IFTIM, Cathespins, STAT, IFN-B, DNM2, GSK3 and others) a comparison of gene expression profiles using publicly available microarray datasets GSE17400 (SARS-CoV) and GSE147507 (SARS-CoV-2) was performed. There were a smaller number of genes found to be differentially expressed in SARS-CoV-2 with respect to SARS-CoV, at 24 h-post infection.
Interestingly, we observed interferon beta (IFN–B), upregulated in SARS-CoV infection, is not differentially expressed in the A549 cell line as well as NHBE treated with SARS-CoV-2, 24hrs post infection. Further, IFITM3 and STAT1 are upregulated in SARS-CoV-2 infection and not found to be the case in SARS-CoV, which is an important modulation of the host factors for facilitation of the virus entry and activation of MAPK pathways [25]. Other host factors such as ACE2 and TMPRSIID remained undetected in both SARS-CoV and SARS-CoV-2, 24 h post infection. The data analysis of SARS-CoV-2 data sets shows unique transcription profiles as compared to other viruses with less expression of interferons and other cytokines, 24 h post infection [29]. However, we observed that cathepsin B and cathepsin H are downregulated in SARS-CoV and SARS-CoV-2 (NHBE cells).
4. Discussion
The identification of genome variation of SARS-CoV-2 and its consequences to its interaction with host factors is crucial for investigating the virus pathogenesis, transmissibility, evolution, designing antiviral therapies, and novel treatment [9]. The maximum number of mutations is observed in SARS-CoV-2 isolates from the USA, Germany and China while relatively few mutations observed in Taiwan, Vietnam, Japan, Australia, Brazil and Hong Kong. The proteins in the descending order of numbers of variations are ORF1ab, NSP3, S protein, N protein, ORF3a, helicase and RdRp, whereas other, including ORF7, ORF10 and E showed very little divergence. The present study findings are in concordance with previous studies reporting ORF1ab as the most variable protein among coronaviruses [30]. NSP3 showed many variations present in different domains which were country specific and had no overlaps. Considering multi-functionality of the NSP3 and the higher frequency of mutations observed, it is speculated that the NSP3 variations may confer important effects on the SARS-CoV-2 pathogenicity. Another important virus enzyme, RdRp (Nsp12), is central to the replication, evolvement and adaptation of coronaviruses [30,31,32]. Additionally, Coronaviruses encode a 3′-to-5′ exoribonuclease activity (ExoN) in NSP14 required for replication fidelity and proofreading, lower efficiency of the enzyme can lead to a 15–20 fold increase in the virus mutation rates [33,34]. This finding has therapeutic implications as it has been shown that coupling nucleoside analog (NA) which targets RdRp with ExoN inhibitors may be a better treatment option reducing viral escape potential [35].
Due to most important interactions in viral pathogenesis [36], S protein serves as one of the main targets for development of antibodies, entry inhibitors, prophylactic vaccines in coronaviruses [37,38]. S1 RBD not only mediates receptor binding and virus entry, but also contains major neutralizing epitopes [[38], [39], [40]]. Further, the S2 domain has heptad repeat 1 and 2 (HR1 and HR2) which tend to form a coiled-coil structure entwined in an antiparallel manner [41,42]. From the present study, we identified that all the mutations lie in the S1 domain while the S2 domain is conserved, suggesting S1 to be the highly variable as compared to the S2 subunit.
PTMs, including glycosylation and proteolytic cleavage, forms an inevitable process in virus pathogenesis [26]. Glycosylation helps spike glycans to mask the protein surface which helps the virus to restrain access to neutralizing antibodies consequently disturbing the humoral immunity. RNA encoded polyproteins are cleaved by proteinases like 3CL pro that cleaves single viral polyprotein in eleven sites in coronavirus [43,44]. In the present study, we predicted the possible 3CL proteinase cleavage site along with the putative glycosylation sites having implications for the viral life cycle and pathogenicity, which were absent in SARS-CoV and may be given due consideration while designing a unique druggable site for SARS-CoV-2. Intriguingly, the N501T mutation, at N-glycosylation site, may affect the SARS-CoV-2 ACE2 binding.
During viral infection, host miRNAs are involved in various signaling pathways, modulation of host-virus interactions, regulation of viral infectivity, transmission and activation of antiviral immune responses [45]. From the host-miRNA target prediction analysis, we identified 42 conserved miRNAs with targets in SARS-CoV-2 genome. Interestingly, these miRNAs are found to target ORF1ab and S protein genes which have maximum entropy change identified from the mutational analysis. Out of these miRNAs, 12 are artificial miRNAs known to efficiently inhibit HIV-1 replication, without any off-targets [46]. Even though, there are genomic differences between HIV and SARS-CoV-2, and hence, it can't be claimed that antiviral mechanism of these miRNAs will be definitely same in SARS-CoV-2. Simply put, prima facie the 12 artificial miRNAs are predicted to have targets in the SARS-CoV-2 genome is an interesting observation, which may be confirmed by experiments. Based on the assertion that these complementary miRNAs are without off-targets, it may be speculated that these can be explored to develop miRNA or RNAi based therapeutics against SARS-CoV-2 infection.
The gene expression analysis of SARS-CoV-2 showed unique transcription profiles at 24 h-post infection, as compared to SARS-CoV. At 24 h, in SARS-CoV-2, no expression of interferons, cytokines, ACE2 and TMPRSIID were identified while IFITM3 was significantly expressed. The increased IFITM3 expression suggests that SARS-CoV-2 uses human IFITM3 during virus entry, facilitating viral infection, and may play a role in pathogenesis [47,48].
Summarily, this study uses integrative data analysis to explore some of the crucial factors involved in host-pathogen interactions, which strongly correlates with the existing literature on related viruses. The novel antiviral miRNAs and antiviral peptides identified in the study can be explored for the development of novel antivirals for COVID-19.
Declarations
Author contribution statement
Rahila Sardar, Dinesh Gupta: Conceived and designed the analysis; Analyzed and interpreted the data; Contributed analysis tools or data; Wrote the paper.
Deepshikha Satish, Shweta Birla: Analyzed and interpreted the data; Contributed analysis tools or data.
Funding statement
Dinesh Gupta was supported by Department of Biotechnology, Ministry of Science and Technology (BT/BI/25/066/2012). Rahila Sardar was supported by Indian Council of Medical Research (2019-5850). Deepshikha Satish was supported by Council of Scientific and Industrial Research, India (IN) (09/0512(0207)/2016/EMR-1). Shweta Birla was supported by Department of Science and Technology, Ministry of Science and Technology (PDF/2017/001326).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of V The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee N., Hui D., Wu A. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348(20):1986–1994. doi: 10.1056/NEJMoa030685. .[published Online First: Epub Date] [DOI] [PubMed] [Google Scholar]
- 3.Ren L.L., Wang Y.M., Wu Z.Q. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin. Med. J. 2020;133(9):1015–1024. doi: 10.1097/CM9.0000000000000722. .[published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han D.P., Lohani M., Cho M.W. Specific asparagine-linked glycosylation sites are critical for DC-SIGN- and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J. Virol. 2007;81(21):12029–12039. doi: 10.1128/JVI.00315-07. .[published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. .[published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuyama S., Nao N., Shirato K. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117(13):7001–7003. doi: 10.1073/pnas.2002589117. .[published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ou X., Liu Y., Lei X. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. .[published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanjuan R., Domingo-Calap P. Mechanisms of viral mutation. Cell. Mol. Life Sci. 2016;73(23):4433–4448. doi: 10.1007/s00018-016-2299-6. .[published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C., Liu Z., Chen Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020;92(6):667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y., Wang Y., Chen Y., Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J. Med. Virol. 2020 doi: 10.1002/jmv.25748. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu Y, McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22(13):30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickett B.E., Sadat E.L., Zhang Y. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40(Database issue):D593–D598. doi: 10.1093/nar/gkr859. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cleemput S., Dumon W., Fonseca V. Genome Detective Coronavirus Typing Tool for rapid identification and characterization of novel coronavirus genomes. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa145. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capriotti E., Fariselli P., Casadio R., I-Mutant2 0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. (Web Server issue) published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng J., Randall A., Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins. 2006;62(4):1125–1132. doi: 10.1002/prot.20810. [published Online First: Epub Date] [DOI] [PubMed] [Google Scholar]
- 16.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–D144. doi: 10.1093/nar/gkj112. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–D153. doi: 10.1093/nar/gkm995. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanna J., Hossain G.S., Kocerha J. The potential for microRNA therapeutics and clinical research. Front. Genet. 2019;10:478. doi: 10.3389/fgene.2019.00478. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steentoft C., Vakhrushev S.Y., Joshi H.J. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32(10):1478–1488. doi: 10.1038/emboj.2013.79. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta R., Brunak S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput. 2002:310–322. [PubMed] [Google Scholar]
- 22.Kiemer L., Lund O., Brunak S., Blom N. Coronavirus 3CLpro proteinase cleavage sites: possible relevance to SARS virus pathology. BMC Bioinf. 2004;5:72. doi: 10.1186/1471-2105-5-72. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 2008;21(11):639–644. doi: 10.1093/protein/gzn039. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thakur N., Qureshi A., Kumar M. AVPpred: collection and prediction of highly effective antiviral peptides. Nucleic Acids Res. 2012;40:W199–204. doi: 10.1093/nar/gks450. (Web Server issue) [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [published Online First: Epub Date] [DOI] [PubMed] [Google Scholar]
- 26.Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13(6):405–430. doi: 10.2217/fvl-2018-0008. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Zhao W., Mao Y. Site-specific N-glycosylation characterization of recombinant SARS-CoV-2 spike proteins using high-resolution mass spectrometry. bioRxiv. 2020 doi: 10.1074/mcp.RA120.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leon-Icaza S.A., Zeng M., Rosas-Taraco A.G. microRNAs in viral acute respiratory infections: immune regulation, biomarkers, therapy, and vaccines. ExRNA. 2019;1(1):1. doi: 10.1186/s41544-018-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanco-Melo D., Nilsson-Payant B., Liu W.-C. SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. BioRxiv. 2020 [Google Scholar]
- 30.Graham R.L., Sparks J.S., Eckerle L.D., Sims A.C., Denison M.R. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 2008;133(1):88–100. doi: 10.1016/j.virusres.2007.02.017. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sexton N.R., Smith E.C., Blanc H., Vignuzzi M., Peersen O.B., Denison M.R. Homology-based identification of a mutation in the coronavirus RNA-dependent RNA polymerase that confers resistance to multiple mutagens. J. Virol. 2016;90(16):7415–7428. doi: 10.1128/JVI.00080-16. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.H.S., Kokic G., Farnung L. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020 doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 33.Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8(2):270–279. doi: 10.4161/rna.8.2.15013. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pachetti M., Marini B., Benedetti F. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shannon A., Le N.T., Selisko B. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antivir. Res. 2020;178:104793. doi: 10.1016/j.antiviral.2020.104793. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [published Online First: Epub Date] [DOI] [PubMed] [Google Scholar]
- 37.Wang Q., Wong G., Lu G., Yan J., Gao G.F. MERS-CoV spike protein: targets for vaccines and therapeutics. Antivir. Res. 2016;133:165–177. doi: 10.1016/j.antiviral.2016.07.015. [published Online First: Epub Date] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tai W., He L., Zhang X. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17(6):613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He Y., Li J., Heck S., Lustigman S., Jiang S. Antigenic and immunogenic characterization of recombinant baculovirus-expressed severe acute respiratory syndrome coronavirus spike protein: implication for vaccine design. J. Virol. 2006;80(12):5757–5767. doi: 10.1128/JVI.00083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses [published correction appears in trends immunol. Trends Immunol. 2020 Apr 24;41(5):355–359. doi: 10.1016/j.it.2020.03.007. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu S., Xiao G., Chen Y. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363(9413):938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wrapp Daniel, Wang Nianshuang, Corbett Kizzmekia S., Goldsmith Jory A., Hsieh Ching-Lin, Abiona Olubukola, Graham Barney S., Mclellan Jason S. Cryo-EM structure of the 2019-ncov spike in the prefusion conformation. Sci. 13 Mar 2020:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muramatsu T., Takemoto C., Kim Y.-T. SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc. Natl. Acad. Sci. Unit. States Am. 2016;113(46):12997–13002. doi: 10.1073/pnas.1601327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L., Lin D., Sun X. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernier A., Sagan S.M. The diverse roles of microRNAs at the host − virus interface. Viruses. 2018;10 doi: 10.3390/v10080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang T., Cheng T., Wei L. Efficient inhibition of HIV-1 replication by an artificial polycistronic miRNA construct. Virol. J. 2012;9(1):118. doi: 10.1186/1743-422X-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao X., Guo F., Liu F. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111(18):6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.X. Zhao, M. Sehgal, Z. Hou, et al., Identification of residues controlling restriction versus enhancing activities of IFITM proteins on entry of human coronaviruses, J Virol 92 (2018). [DOI] [PMC free article] [PubMed]