Abstract
The COVID-19 pandemic has rapidly spread around the world with significant morbidity and mortality in a subset of patients including the elderly. The poorer outcomes are associated with ‘cytokine storm-like’ immune responses, otherwise referred to as ‘hyperinflammation’. While most of the infected individuals show minimal or no symptoms and recover spontaneously, a small proportion of the patients exhibit severe symptoms characterized by extreme dyspnea and low tissue oxygen levels, with extensive damage to the lungs referred to as acute respiratory distress symptom (ARDS). The consensus is that the hyperinflammatory response of the host is akin to the cytokine storm observed during sepsis and is the major cause of death. Uncertainties remain on the factors that lead to hyperinflammatory response in some but not all individuals. Hyperinflammation is a common feature in different viral infections such as dengue where existing low-titer antibodies to the virus enhances the infection in immune cells through a process called antibody-dependent enhancement or ADE. ADE has been reported following vaccination or secondary infections with other corona, Ebola and dengue virus. Detailed analysis has shown that antibodies to any viral epitope can induce ADE when present in sub-optimal titers or is of low affinity. In this review we will discuss ADE in the context of dengue and coronavirus infections including Covid-19.
Keywords: Covid-19, Cytokine storm, SARS-CoV2, ADE
Abbreviations: ACE2, Angiotensin I converting enzyme 2; ADE, antibody dependent enhancement; DENV, dengue virus; DHF, dengue hemorrhagic fever; FcγR, Fc gamma receptor; FCoV, feline corona virus; FECV, feline enteric corona virus; FIPV, feline infectious peritonitits virus; HIV, human immunodeficiency virus; IHC, immunohistochemistry; MERS, Middle East respiratory syndrome virus; MLN, mesenteric lymph nodes; RBD, receptor binding domain; SARS, severe acute respiratory syndrome; TMPRSS2, Transmembrane serine protease 2; YFV, yellow fever virus; ZIKV, Zika virus
1. Introduction
SARS-CoV2 infection is asymptomatic in about 80% of the infected individuals at the population level [1]. Epidemiology suggests that SARS-CoV2 infections may be similar to influenza rather than SARS-CoV or MERS-CoV infections as the transmission is observed during the prodromal phase thereby accounting for the high rates of infection [2], [3]. The virus infects the upper respiratory tract by binding to its receptor ACE2 aided by TMPRSS2 protease to permit the infection of epithelial cells of the respiratory tract [4], [5]. Mild form of the infection observed in a majority of adults and children, is localized to nasal and upper respiratory tract and is usually resolved without any intervention and in the absence of overt immune responses [6], [7], [8]. Even though some of the asymptomatic individuals exhibit seropositivity to the SARS viruses [9], [10], [11], [12], it is not known yet if they generate high titers of neutralizing antibodies and long-lasting immunity. Greatest susceptibility to the infection is seen in older individuals and in those with underlying health conditions [13], [14]. Some of the reasons for the susceptibility of the elderly population to lower respiratory tract infections such as Covid-19 include weakened immune system, co-morbidities, reduced ability to restore epithelium after damage and age-related reduction in mucociliary functions [6], [8], [15].
Certain patterns are beginning to emerge from the analysis of anti-SARS-CoV2 antibodies in Covid-19 patients. In general IgM antibodies specific to SARS-CoV2 are observed from 7 days to 1 month after the first detection of the virus by RT-PCR while IgG responses are observed from about 7 days but persist at higher levels over longer period of time [16], [17], [18], [19]. Additionally, antibodies to the nucleocapsid protein appears before antibodies to the spike protein [18], [20]. The titer of IgG anti-SARS-CoV2 antibodies appear to be higher in patients with severe form of the disease and may be related to the commercial ELISA kits used [18], [21], [22]. However, analyses of convalescent sera obtained 39 days after infection suggest that the neutralizing titers generated are generally low [23]. Nonetheless, the antibody titers positively correlated with virus-specific T cells [24], indicating that T cell responses play an important role in the development of protective immunity.
Studies during the past 6 months have characterized in detail the cytokine storm, that is observed in Covid-19 patients, but the underlying reasons have not been completely elucidated. In certain other infections, severity of the infection has been shown to be increased by a process, referred to as antibody-mediated enhancement (ADE), that can be induced by low titers of neutralizing antibodies. Rather than neutralizing the virus, the antibodies promotes ADE in immune cells as observed in certain viral infections [25]. In this review we will describe dengue hemorrhagic fever (DHF), feline infectious peritonitis (FIP) and SARS-CoV infections to assess the role of ADE in Covid-19 and discuss the cytokine storm observed to date in Covid-19 patients.
2. Antibody dependent enhancement (ADE) of infection
In general, vaccination strategies elicit high affinity antibodies directed towards the proteins on the surface of viral particles thereby contributing to the elimination of the pathogen. These antibodies reduce the infectivity by different mechanisms. For example, antibodies can interfere with the interaction of the viruses with their receptors thereby preventing infection. As the epitopes on the surface of the virus are usually repetitive, multiple molecules of the antibody can bind to a single virion and induce the formation of immune complexes that are eliminated by phagocytosis. Alternately, antibodies can interfere with the release of the viral genome in endosomes following phagocytosis [26]. However sub-optimal concentration of antibodies or cross-reactive antibodies promote ADE. ADE has been extensively studied in various viral infections [27]. Both IgG and IgM antibodies are capable of promoting ADE. Extrinsic ADE refers to the increase in the number of cells that are infected due to the binding of the virus-Ab complex to the complement receptors or FcγR, while intrinsic ADE refers to the increased production of the virus particles per cell as a consequence of suppression of the inflammatory responses [27], [28], [29]. Complement-mediated ADE has been observed in Human immunodeficiency virus (HIV) and West Nile virus infections [30], [31]. At sub-optimal concentrations of neutralizing antibodies, the virus-antibody complex fixes complement and subsequently bind to complement receptors expressed on different cell types. As complement receptors are expressed in a wide variety of cells, it can lead to infection of non-target cells and contribute to chronicity [32].
FcγR-mediated ADE is observed in situations where IgG antibodies to the virus from a previous infection that are present at low titers or, cross-reactive antibodies to a related heterologous virus promote the uptake of the virus through type I FcγR-mediated phagocytosis [27], [33]. Studies in different infection models have shown that all the type I FcγR subtypes can promote ADE. FcγRI (CD64) binds monomeric IgG with high affinity and plays an important role in priming adaptive immune responses [34], [35], [36]. Increased surface expression of FcγRI by inflammatory cytokines induces aggregation and potentiates the binding of immune complexes [37]. FcγRII (CD32) and FcγRIII (CD16) are low affinity receptors for IgG but are activated by immune complexes. With the exception of FcγRIIb that contains an ITIM motif in the cytoplasmic tail, engagement of the other FcγR activates signaling through the ITAM motifs that they express or present in the associated proteins [38]. Type I FcγR are expressed by myeloid, polymorphonuclear, dendritic, B and NK cells [33]. However ADE is primarily observed in monocytes, macrophages and dendritic cells [39], [40], [41], [42], even though FcγR are expressed by B cells.
2.1. ADE in Flaviviridae infections
Dengue fever is caused by single-stranded, positive-sense RNA virus of the Flaviviridae family. Dengue virus consists of 4 closely related serotypes (DENV1-4) that show trophism for monocytes, macrophages and dendritic cells [43]. A vast majority of the infections are subclinical, while clinical presentation of the disease is associated fever and malaise that resolves after 7–10 days. Adaptive immune responses contribute to long-term protection against the same DENV serotype and afford variable degree of cross-protection against the other three serotypes that wanes over time. A small percentage of patients exhibit dengue hemorrhagic fever (DHF) with severe symptoms such as spontaneous bleeding and vascular leakage akin to sepsis. These patients show characteristics of cytokine storm, with elevated levels of cytokines, chemokines and hepatic transaminases [44]. As individuals can be infected with any of the four serotypes over their life span, increased disease severity is observed in a fraction of patients following infection with a different serotype. High viremia and vascular leakage are also observed in infants of seropositive mothers who have low anti-dengue titers and were infected with a heterologous DENV strain [45]. ADE resulting from the antibodies generated during previous infections that are cross-reactive with the strain causing the current infection has been proposed to be the underlying cause for DHF. Recently two studies analyzed the anti-DENV antibodies and severity of infection in a large cohort of adults and pediatric population from dengue infected endemic areas [46], [47] and reached similar conclusions. They observed that DHF correlated with a narrow range (low titers) of pre-existing anti-DENV antibodies while high titers of anti-DENV antibodies were protective irrespective of the DENV subtype. These observations highlight the importance of the ability of a primary immune response (or vaccination) to induce high-titer antibodies to confer protection and avoid ADE in humans. As a corollary, the immunogenicity of live attenuated yellow fever vaccination was increased by ADE in a proportion of individuals who had received inactivated Japanese encephalomyelitis vaccine previously [48].
A role for anti-DENV antibodies in promoting ADE during the outbreak of Zika (ZIKV) infections and modulating its severity has been postulated [49], [50]. However, two reports on the analysis of adult and pediatric populations from these endemic areas suggest otherwise. High titers of anti-DENV antibodies reduced the severity of subsequent ZIKV infections [51], [52]. It may be important to analyze in vivo immune responses to heterologous infections before proposing a role for ADE based purely on in vitro analyses. Immune responses to ZIKV in non-human primates previously infected with DENV or yellow fever virus (YFV) were compared to naïve animals [53]. While the ZIKV infection of cell lines were enhanced by sera from DENV-infected animals, there was no significant difference between the various groups in vivo following ZIKV infection.
In addition to the efficacy of high titer neutralizing antibodies, T cell-mediated immune responses play an important role in preventing subsequent infections. In non-human primates, T cell-mediated immunity, but not antibodies to DENV protects efficiently from subsequent ZIKV infection [54]. The importance of promoting T cell responses to DENV (or other flavivirus infections) is highlighted by the fact that of the various anti-DENV vaccines developed over the past 30 years using different platforms only one candidate has reached phase 3 clinical trials [55]. CYD-TDV (chimeric yellow fever dengue-tetravalent dengue vaccine, Dengvaxia), a tetravalent vaccine on YFV backbone that incorporates antigens from the four DENV viruses induces a robust T and antibody responses [56]. In seropositive individuals, the vaccine reduced hospitalization due to DHF, but in seronegative individuals the incidence of hospitalization was increased [57]. It is possible that in seropositive individuals the efficacy of the vaccination was higher as the vaccine could have infected greater number of cells accompanied by higher ‘viremia’ due to ADE [58]. On the other hand, Dengvaxia sensitized seronegative individuals to subsequent dengue infections as the vaccine ‘behaved’ like an infection with one of the 4 DENV strains [59], [60]. Alternately, the YFP backbone in the Dengvaxia may contribute to sensitizing the vaccinated individuals to subsequent dengue infections by enhancing ADE. Head-to-head comparison of Dengvaxia, a mixture of the reference DENV1-4 strains and tetravalent sub-unit vaccine (DSV4) in murine models of dengue and Zika infection suggest that the tetravalent sub-unit vaccine does not induce ADE while maintaining vaccine efficacy against dengue and zika infections [61], [62].
2.2. ADE in Coronaviridae
Pathogenic members of the Coronavirus family infect primarily the epithelial cells in the respiratory and gastrointestinal tracts [63]. Among the various members, immune responses have been well characterized in vivo in cats following natural infection with feline corona virus (FCoV; an alphacoronavirus, previously referred to as feline enteric corona virus (FECV) and/ or feline infectious peritonitis virus (FIPV). It is a relatively new disease that is believed to have jumped species sometime during the mid-20th century [64]. Feline infectious peritonitis (FIP) is the pathogenic manifestation of otherwise non-pathogenic infection by FCoV that is a ubiquitous pathogen present in cats throughout the world [65]. FCoV infects enterocytes, is shed in the feces and causes mild diarrhea. Kittens get infected through the fecal-oral route at 2 months of age [65]. Even if the infected cats do not show any symptoms, they can be seropositive and cats with high titers show fecal shedding of the virus, while those with low titers do not [66]. Extensive expansion of the virus during the initial period of replication (enteric FCoV) leads to mutations in the spike protein and in ORFs coding for non-structural proteins that promote enhanced systemic spread (systemic FCoV) [67], [68]. Systemic dissemination per se is not associated with disease severity. FIP is manifested in 2 distinct forms. The most common form is the development of effusive inflammatory condition with visceral serosa and omentum with exudates in the abdomen. The dry form of the disease is characterized by granulomatous inflammatory lesions in paranchymatous tissues such as kidneys and other organs [64]. Despite the extensive presence of FCoV infections in cat populations around the world, the morbidity is less than 5% [69]. Genetic factors may play an important role in susceptibility as FIP prevalence is high in certain breeds of cats but not others [70]. Follow-up of 820 household cats over 6 years suggested that despite seropositivity for FCoV the incidence of FIP was not different from groups which were seronegative [69]. The incidence of FIP was decreased with age suggesting that the cats develop protective long-lasting immunity to FCoV [69].
Mutations in the Spike protein of FCoV is believed to contribute to the infection of monocytes and macrophages that result in the dissemination of the virus to the mesenteric lymph nodes (MLN) [68]. In healthy cats, FCoV was detected by immunohistochemistry (IHC) in the epithelial cells lining the intestine, sinusoidal macrophages in the MLN and in lung alveolar macrophages [71]. Systemic spread of FCoV is associated with an increase in viremia but does not necessarily induce FIP. Despite the detection of the virus by PCR in the MLN of non-FIP animals, the low viral load precludes its detection by IHC [71]. To better understand the pathology of FIP, Malbon et al. [72] compared the expression of transcripts for various cytokines, chemokines and TLRs in mesenteric lymph node (MLN) samples from cats with systemic infection that developed or not FIP. They observed that the expression of IL-1β, TNFα, IL-6, IFNγ, IL-15, CXCL10 and CCL8 and IFNβ was increased in cats with FIP when compared to those without FIP, indicating that FIP is associated with hyperinflammation. Additionally, there was no significant differences in the expression of IL-10 and TGFβ between the two groups. Nonetheless, non-symptomatic systemic viral load was associated with a modest increase in the expression of some of the above-mentioned inflammatory mediators, pointing to an ongoing immune response [72], [73], [74]. Decrease in the numbers of circulating lymphocytes is a characteristic of FIP and may be a consequence of viremia and cytokine responses, akin to cytokine storm [75]. High viremia leads to the deposition of immune complexes and viral particles containing macrophages around small venules, otherwise referred to as Arthus-type vasculitis [76].
Similar to dengue infections, titer of the neutralizing antibodies plays an important role in protection. Live attenuated vaccine generated by deleting orf3abc or orf7ab induced high titers of neutralizing antibodies and protected cats from lethal challenge [77], but no follow up studies were published. However, in one study, low titers of antibodies present from enteric presence of the virus does not protect from challenge with the virulent virus and it is not known whether it was associated with augmented infections through ADE [66], [69]. Immunization with vaccinia virus expression the Spike protein generated low titer of antibodies in kittens and accelerated the disease following challenge with the virulent virus by ADE [78]. Again, transfer of antibodies from FIP or seropositive healthy cats accelerated the disease process following infection of recipients [79], [80], [81]. The role of pre-existing antibodies is not clear as vaccination with low virulent with ORF3abc truncated strain of FCoV induced comparable neutralizing antibodies in SPF-reared and conventional cats [82]. However, protection from subsequent lethal challenge was seen only in cats reared in SPF but not in conventional facilities. As the 2 strains of cats were different in this study it is not possible to arrive at conclusions by comparing the 2 groups. Nonetheless, cats that showed high titers following vaccination succumbed at later timepoints to fatal disease supporting a protective role of high titer neutralizing antibodies in keeping viremia under check.
Various lines of evidence suggest that a robust T cell mediated immunity is required to confer long-lasting immunity. Immunization of cats with avirulent or sublethal dose of the virus induced robust DTH responses and elicited partial protection [83]. Granulomatous lesions are made up of virus containing macrophages surrounded by plasma cells and T cells [64]. Animals that survived acute infection were characterized by robust T cell responses during the later stages of the infection [84]. Furthermore, these survivors were resistant to subsequent pathogenic FCoV virus challenge. This study further reiterated the observation that the titers of anti-spike antibodies did not correlate with the development of FIP or the viral RNA concentrations. Even though these experiments were done with cats from specific pathogen free (SPF) facility, the observation that household cats older than 5 years are resistant to the development of FIP [64], [85] suggests that despite the presence of antibodies that promote ADE during FCoV infections, long-lasting immunity is conferred by T cell mediated protection.
2.3. SARS-CoV infection
Humans have been exposed to different coronaviruses before the arrival of the three unwanted guests during the past 2 decades. At least 4 other coronaviruses, namely 229E, HKU1, NL63 and OC43 cause respiratory tract infections and have different rates of prevalence [86]. During the SARS epidemic in 2003, sera of SARS patients were analyzed for cross-reactive antibodies to NL63, 229E and OC43 [87]. Seropositivity to SARS-CoV was detected only in patients who tested positive for the virus by RNA indicating that SARS-CoV does not seem to be recognized by antibodies generated in response to previous coronavirus infections. As the number of individuals infected with SARS-CoV and MERS-CoV was quite low, it is possible that the absence of cross-reactivity cannot be concluded definitely. Analysis of samples from a hospital in Canada in 2003 during the SARS outbreak suggested that antibodies to N protein of HCoV-OC43 can cross-react with SARS-CoV [88]. However, analysis of antibody responses in a large panel of pre-Covid-19 sera to a microarray of SARS-CoV2 antigens indicate low level of cross-reactivity to S2 domain of spike protein and N protein [89]. It is not known whether ADE can be promoted by antibodies to any epitope of SARS-CoV2 virus and whether the prevalence of these cross-reactive antibodies are high in the older population.
While infection through receptor-mediated uptake as a consequence of binding of spike protein to ACE2 expressed by epithelial cells is the classical mode of infection that contributes to the range of classical pathology in Covid-19, monocytes and macrophages show minimal expression of ACE2 [90] and hence may not be infected directly by the virus. Furthermore, SARS-CoV replicates poorly in monocytes and macrophages, similar to OC43 [91]. However, antibody mediated uptake of the virus has been observed in human monocytes even though they do not replicate in these cells [92]. In Covid-19 patients with a severe pathology, increase in the presence of plasma cells were observed in bronchoalveolar lavage fluid (BALF) [93]. Extrafollicular B cell activation have been observed in critically ill patients [94]. Analysis of Covid-19 patients with x-linked agammaglobulinemia (lack of B cells and circulating immunoglobulins) and common variable immune deficiencies (CVID, lack Ig but have dysfunctional B cells) showed increased disease severity in the latter group [95], [96].
Convalescent sera has been used to control the severity of the infection and bring down the viral load during the previous pandemics [97], [98], [99]. Even though the clinical trial carried out in China was underpowered due to absence of new infections [100], treatment with convalescent sera that had high titers of neutralizing antibodies was observed to be beneficial in Covid-19 patients with moderate disease severity, and there was no evidence for ADE. Additional trials are ongoing in different countries. Even though monoclonal antibodies developed against the receptor-binding domain (RBD) of SARS-CoV do not recognize RBD of SARS-CoV2, data from convalescent sera suggest that cross-reactive antibodies are possible in vivo [101]. Comparative analyses of responses of convalescent sera to SARS-CoV, MERS-CoV, SARS-CoV2 and other coronaviruses indicate that sera from SARS-CoV and MERS-CoV infected patients show some cross-reactivity to S2 and RBD (receptor binding domain) of the spike protein but neutralization is not observed [102], [103]. Vaccines for SARS and MERS have been developed and tested in different pre-clinical models (reviewed in [104]) and some of the vaccines exhibited ADE. The characteristics, the animal models and the nature of the vaccines are detailed in the review [104]. In hamsters, vaccination with recombinant trimeric spike protein elicited neutralizing antibodies and were protected from subsequent SARS-CoV challenge [105]. Vaccination of non-human primates with inactivated SARS-CoV conferred protection during subsequent challenge and did not induce ADE [106]. On the other hand, ADE was observed in monkeys following immunization with one of the 4 peptides derived from spike protein [107]. However, the titer of the antibodies elicited by the vaccines and the ability of the vaccines to induce humoral and cell-mediated immune responses are not known in the above studies. Analyses of ADE in dengue infections (discussed above) suggest that generation of high titers of the neutralizing antibodies is one of the important factors that needs to be considered to prevent ADE. In animal models where SARS-CoV2 infections have been established [104], [108], [109], [110] it may be important to assess ADE using graded titers of convalescent sera. Similarly, follow up of recovered individuals for anti-viral titers and reinfection will help determine the susceptibility and severity to re-infection, if any. However, confirming the absence of cross-reactivity of sera from older age groups who have been exposed to different coronaviruses during their lifetime and characterization of the antigens that can induce ADE, is required to rule out ADE in older patients. Thus, the role of ADE in promoting infections in older patients has not been completely ruled out, even though there is no supportive evidence to date either.
3. Overview of the cytokine storm in SARS-CoV2 infection
The pathological characteristics of Covid-19 greatly resemble those of SARS (2002–2003) and MERS (2012) [111]. Despite years of research, the specific factors contributing to the morbidity associated with CoV are yet to be completely understood [112]. Nevertheless, studies in humans who died from SARS and more recent studies done in animal models show a dysregulated immune response associated with uncontrolled inflammation [112]. In the present pandemic, most severe cases of Covid-19 present with acute respiratory distress syndrome (ARDS) which is the main cause of death in Covid-19 patients as in SARS and MERS patients [113], [114]. In patients with severe infection, the virus replicates in the alveolar epithelial cells leading to diffuse alveolar damage, oedema, hyaline membrane formation, immune cell infiltration in the lungs and desquamation of pneumocytes that are characteristics of ARDS [114]. Most of the above pathologies were also observed before the onset of pneumonia in 2 patients undergoing surgery for lung adenocarcinoma and were incidentally diagnosed with COVID-19 [115], [116]. Additional studies indicate that the patients with ARDS exhibit symptoms of cytokine storm [117]. However, unlike DHF and FIP where ADE contributes to the initiation of the severe pathology, we do not yet know the factors that trigger ARDS in Covid-19 patients.
Impaired type 1 IFN signature appears to be an important factor that distinguishes moderate and severe infections [113], [118]. In animal models of SARS infections, delayed induction of type 1 IFNs has been shown to contribute to the severity [119], [120], [121]. Nonetheless, IFN I responsiveness was intact in the patients suggesting that IFN 1 therapy can reduce the viral load [113], [118]. In support, exogenous IFN I was capable of reducing the viral load in infected epithelial cells [118]. As SARS-CoV2 can induce Type 1 IFN response [120] it is possible that interferon-inducible transmembrane (IFITM) proteins suppress viral replication as has been shown for SARS-CoV virus [122], [123]. Equally SARS-CoV-derived proteins can suppress type 1 IFN responses [118], [124], [125].
Sera of SARS-CoV infected patients showed early induction of CXCL10 (before any lung damage) followed by overproduction of proinflammatory cytokines such as IL-6 together with a lack of anti-inflammatory cytokine IL-10 [126], [127], [128]. Similarly, bronchoalveolar lavage fluid showed higher levels of TNFα and IL-6 as well as IL-8, RANTES and CCL2 [129]. SARS-CoV infected respiratory tract epithelial cells have been shown to be a major source of chemokines and inflammatory cytokines [130]. Similar results were obtained in severe case of SARS-CoV2 patients where high levels of proinflammatory cytokines IL-6 and TNFα were observed [131]. Moreover, more severe cases were associated with higher concentration of G-CSF, CXCL10, CCL2 and CCL3 [132]. Contrary to SARS-CoV, higher levels of anti-inflammatory cytokine IL-10 were found in severe cases of Covid-19 [131]. Following infection with SARS-CoV2 in ferrets, CCL8 and CXCL9 were detected in the upper respiratory tract as early as 3 days post infection [118]. Over time, expression of other chemokines was upregulated. In patients with severe SARS-CoV2, circulating levels of HRP, ferritin, lactate dehydrogenase, alanine aminotransferase, IL-6, IL-8, TNFα, IL-10 and IL-1RA were elevated with a concomitant lymphopenia [131]. The increase in IL-10 and IL-1RA may be pointing to a hyperinflammation process that is out of control. Presence of IL-6, IL-8 and TNFα in the sera correlated with the severity of the infection [132], [133], [134].
The source of the cytokines in Covid-19-associated inflammation can point towards the mechanisms involved. CAR-T induced cytokine storm is characterized by the presence of T cell-associated cytokines such as IL-3, IFNγ and GM-CSF that subsequently activate macrophages to produce IL-6, TNFα and IL-1 [135], [136]. Excessive immune response generated by PRR activation initiates the cytokine storm in sepsis and involves IL-6, TNFα, IL-12, IL-1β and IL-18 [137], [138]. Unlike in sepsis, CAR-T cell therapy, DHF and FIP, where myeloid and lymphoid cells are the source of inflammatory cytokines, in SARS infections, the epithelial cells of the respiratory tract appear to be important players. In Covid-19 patients who exhibit ARDS and cytokine storm, the expression of chemokines, IL-6 and TNFα is elevated with minimal implication for IL-1, IFNγ or IL-17 [113], [133], [139]. Elevated serum IL-6 and TNFα levels in the patients with severe disease correlated with signaling pathways induced by these cytokines in PBMC, but was not reflected in the RNA transcripts from PBMC [113], [139]. While IL1B transcripts were higher, circulating IL-1β protein was low indicating towards minimal inflammasome activation. These observations suggest that source of IL-6 and TNFα may be from epithelial/endothelial and other cells in the respiratory tract. The macrophage composition of BALF was distinct between patients with moderate and severe disease and differed from that of healthy controls [140]. Severe disease was characterized by high neutrophil infiltration. Thus it is possible that the airway epithelial cells are the primary source of inflammatory mediators in severely infected patients, as SARS-CoV infects epithelial cells in the lung and the hematopoietic cells poorly support viral infection and replication, as seen from the viral loads and production of pro-inflammatory mediators [97], [112], [141]. These observations raise the question as to why some individuals mount an exaggerated immune response in the airway epithelium while others do not.
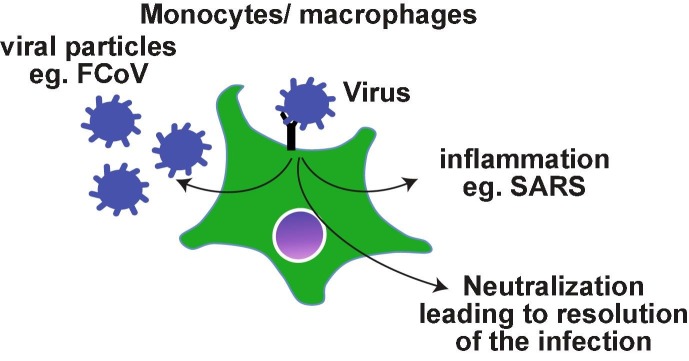
Even though SARS-CoV does not target monocyte lineage primarily, the role of antibodies and the immune complexes is not as well-defined as in DHF or FIP. Nonetheless there are evidences to suggest a role for antibodies in the lung pathology. Severe disease and higher death rate were associated with older patients who had high titers of antibody within 2 weeks after SARS infection, while seroconversion was observed in most of the SARS patients between 3 and 5 weeks after infection [142]. In this cohort, lower neutralizing titers were associated with prolonged illness. The antibody levels were higher and long-lasting in health-care workers with severe MERS pathology, while patients with milder course of the disease had variable presence of antibodies [143]. It is possible that the severe disease may be associated with robust long-lasting immune responses. Reflecting these observations, antibodies generated in New Zealand white rabbits following primary MERS-CoV infection exacerbated the pulmonary pathology during re-infection despite reduced viremia [144]. Passive transfer of serum from previously infected animals also induced similar pathology in naïve infected animals. Again, re-infected animals developed robust immunity. Th2 immunopathology with eosinophilic infiltrations were uniformly observed following immunization of mice with 4 different SARS-CoV vaccine candidates [145] suggesting that acute lung pathology may be part of the natural immune responses to SARS infections. In SARS-CoV infected or Spike DNA vaccinated macaques, anti-Spike antibodies skewed the monocyte/macrophage responses towards pro-inflammatory and wound healing phenotype simultaneously, resembling the pathology observed in SARS patients [146]. Thus, the antibodies generated during infection with pathogenic corona viruses may activate monocytes/macrophages and promote lung pathology rather than enhancing the viral infection as observed with macrophage tropic viruses such as dengue and FCoV (Fig. 1 ).
Fig. 1.
Possible outcomes of antibody-mediated uptake of viral particles by monocytes/macrophages. Antibody binding to the viral particles ideally leads to the neutralization. In the case of monocyte/macrophage tropic viruses such as FCoV, the viremia ensues if the neutralization is not achieved. As SARS-CoV viruses replicate poorly in monocytes/macrophages, it is possible that the virus induces an exaggerated inflammatory response.
4. Concluding remarks
The inflammatory response of the airway epithelium appears to play an important role in the progression of SARS-CoV2 infection. Antibodies can exacerbate the lung pathology by activating macrophages even though it may not necessarily enhance viral load and dissemination, as seen in classical ADE. It may be important that vaccines promote robust T cell immunity in addition to antibody responses [147], [148], [149], [150]. Knowledge about the mechanisms involved in activating the innate immune responses and the priming the adaptive immune response will contribute to the identification of parameters that can be used to evaluate protective immune responses generated by SARS-CoV2 vaccines.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Availability of data and materials
Not applicable
Authors' contributions
MC, MN, SR and SI designed the content and wrote the manuscript. HAC and AUK contributed to the discussions and specific sub- sections in the manuscript. All authors reviewed and corrected the manuscript.
Funding
This work was supported by NSERC Discovery grant to SI and SR. MN is a recipient of FRQS post-doctoral fellowship.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Wu Z., McGoogan J.M. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 2.Wilder-Smith A., Chiew C.J., Lee V.J. Can we contain the COVID-19 outbreak with the same measures as for SARS? Lancet Infect. Dis. 2020;20(5):e102–e107. doi: 10.1016/S1473-3099(20)30129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceccarelli M., Berretta M., Venanzi Rullo E., Nunnari G., Cacopardo B. Differences and similarities between Severe Acute Respiratory Syndrome (SARS)-CoronaVirus (CoV) and SARS-CoV-2. Would a rose by another name smell as sweet? Eur. Rev. Med. Pharmacol. Sci. 2020;24(5) doi: 10.26355/eurrev_202003_20551. pp. 2781–2783 10.26355/eurrev_202003_20551. [DOI] [PubMed] [Google Scholar]
- 4.M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Kruger, T. Herrler, S. Erichsen, T.S. Schiergens, G. Herrler, N.H. Wu, A. Nitsche, M.A. Muller, C. Drosten, S. Pohlmann, SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell 181(2) (2020) 271-280 e8 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed]
- 5.S. Lukassen, R.L. Chua, T. Trefzer, N.C. Kahn, M.A. Schneider, T. Muley, H. Winter, M. Meister, C. Veith, A.W. Boots, B.P. Hennig, M. Kreuter, C. Conrad, R. Eils, SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells, EMBO J 39(10) (2020) e105114 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed]
- 6.P. Pavone, M. Ceccarelli, R. Taibi, G. La Rocca, G. Nunnari, Outbreak of COVID-19 infection in children: fear and serenity, Eur Rev Med Pharmacol Sci 24(8) (2020) 4572-4575 10.26355/eurrev_202004_21043. [DOI] [PubMed]
- 7.D. Korean Society of Infectious, D. Korean Society of Pediatric Infectious, E. Korean Society of, T. Korean Society for Antimicrobial, C. Korean Society for Healthcare-associated Infection, Prevention, C. Korea Centers for Disease, Prevention, Report on the Epidemiological Features of Coronavirus Disease 2019 (COVID-19) Outbreak in the Republic of Korea from January 19 to March 2, 2020, J Korean Med Sci 35(10) (2020) e112 10.3346/jkms.2020.35.e112. [DOI] [PMC free article] [PubMed]
- 8.Mason R.J. Pathogenesis of COVID-19 from a cell biology perspective. Eur. Respir. J. 2020;55(4) doi: 10.1183/13993003.00607-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hains D.S., Schwaderer A.L., Carroll A.E., Starr M.C., Wilson A.C., Amanat F., Krammer F. Asymptomatic seroconversion of immunoglobulins to SARS-CoV-2 in a pediatric dialysis unit. JAMA. 2020 doi: 10.1001/jama.2020.8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kliger A.S., Silberzweig J. Mitigating risk of COVID-19 in dialysis facilities. Clin. J. Am. Soc. Nephrol. 2020;15(5):707–709. doi: 10.2215/CJN.03340320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung D.T., van Maren W.W., Chan F.K., Chan W.S., Lo A.W., Ma C.H., Tam F.C., To K.F., Chan P.K., Sung J.J., Lim P.L. Extremely low exposure of a community to severe acute respiratory syndrome coronavirus: false seropositivity due to use of bacterially derived antigens. J. Virol. 2006;80(18):8920–8928. doi: 10.1128/JVI.00649-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su K., Ma Y., Wang Y., Song Y., Lv X., Wei Z., Shi M., Ding G., Shen B., Wang H. How we mitigate and contain COVID-19 outbreak in hemodialysis center (HD): lessons and experiences. Infect. Control Hosp. Epidemiol. 2020:1–6. doi: 10.1017/ice.2020.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nile S.H., Nile A., Qiu J., Li L., Jia X., Kai G. COVID-19: Pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev. 2020;53:66–70. doi: 10.1016/j.cytogfr.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho J.C., Chan K.N., Hu W.H., Lam W.K., Zheng L., Tipoe G.L., Sun J., Leung R., Tsang K.W. The effect of aging on nasal mucociliary clearance, beat frequency, and ultrastructure of respiratory cilia. Am. J. Respir. Crit. Care Med. 2001;163(4):983–988. doi: 10.1164/ajrccm.163.4.9909121. [DOI] [PubMed] [Google Scholar]
- 16.Qu J., Wu C., Li X., Zhang G., Jiang Z., Li X., Zhu Q., Liu L. Profile of IgG and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao A.T., Gao C., Zhang S. Profile of specific antibodies to SARS-CoV-2: the first report. J. Infect. 2020 doi: 10.1016/j.jinf.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang G., Nie S., Zhang Z., Zhang Z. Longitudinal change of SARS-Cov2 antibodies in patients with COVID-19. J. Infect. Dis. 2020 doi: 10.1093/infdis/jiaa229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., Wang D.Q., Hu Y., Ren J.H., Tang N., Xu Y.Y., Yu L.H., Mo Z., Gong F., Zhang X.L., Tian W.G., Hu L., Zhang X.X., Xiang J.L., Du H.X., Liu H.W., Lang C.H., Luo X.H., Wu S.B., Cui X.P., Zhou Z., Zhu M.M., Wang J., Xue C.J., Li X.F., Wang L., Li Z.J., Wang K., Niu C.C., Yang Q.J., Tang X.J., Zhang Y., Liu X.M., Li J.J., Zhang D.C., Zhang F., Liu P., Yuan J., Li Q., Hu J.L., Chen J., Huang A.L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 20.Guo L., Ren L., Yang S., Xiao M., Chang, Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q., Xu S., Zhu H., Xu Y., Jin Q., Sharma L., Wang L., Wang J. Profiling early humoral response to diagnose novel Coronavirus Disease (COVID-19) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okba N.M.A., Muller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C., Bosch B.J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerg. Infect. Dis. 2020;26(7) doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., Hagglof T., Oliveira T.Y., Viant C., Hurley A., Hoffmann H.H., Millard K.G., Kost R.G., Cipolla M., Gordon K., Bianchini F., Chen S.T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A.W., Waltari E., Pak J.E., Huey-Tubman K.E., Koranda N., Hoffman P.R., West A.P., Jr., Rice C.M., Hatziioannou T., Bjorkman P.J., Bieniasz P.D., Caskey M., Nussenzweig M.C. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020 doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., Wei P., Ge J., Gou M., Li X., Sun L., Cao T., Wang P., Zhou C., Zhang R., Liang P., Guo H., Wang X., Qin C.F., Chen F., Dong C. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020 doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tetro J.A. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020;22(2):72–73. doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klasse P.J., Sattentau Q.J. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 2002;83(Pt 9):2091–2108. doi: 10.1099/0022-1317-83-9-2091. [DOI] [PubMed] [Google Scholar]
- 27.Taylor A., Foo S.S., Bruzzone R., Dinh L.V., King N.J., Mahalingam S. Fc receptors in antibody-dependent enhancement of viral infections. Immunol. Rev. 2015;268(1):340–364. doi: 10.1111/imr.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flipse J., Wilschut J., Smit J.M. Molecular mechanisms involved in antibody-dependent enhancement of dengue virus infection in humans. Traffic. 2013;14(1):25–35. doi: 10.1111/tra.12012. [DOI] [PubMed] [Google Scholar]
- 29.Halstead S.B., Mahalingam S., Marovich M.A., Ubol S., Mosser D.M. Intrinsic antibody-dependent enhancement of microbial infection in macrophages: disease regulation by immune complexes. Lancet Infect. Dis. 2010;10(10):712–722. doi: 10.1016/S1473-3099(10)70166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cardosa M.J., Porterfield J.S., Gordon S. Complement receptor mediates enhanced flavivirus replication in macrophages. J. Exp. Med. 1983;158(1):258–263. doi: 10.1084/jem.158.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fust G., Toth F.D., Kiss J., Ujhelyi E., Nagy I., Banhegyi D. Neutralizing and enhancing antibodies measured in complement-restored serum samples from HIV-1-infected individuals correlate with immunosuppression and disease. AIDS. 1994;8(5):603–609. doi: 10.1097/00002030-199405000-00005. [DOI] [PubMed] [Google Scholar]
- 32.von Kietzell K., Pozzuto T., Heilbronn R., Grossl T., Fechner H., Weger S. Antibody-mediated enhancement of parvovirus B19 uptake into endothelial cells mediated by a receptor for complement factor C1q. J. Virol. 2014;88(14):8102–8115. doi: 10.1128/JVI.00649-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pincetic A., Bournazos S., DiLillo D.J., Maamary J., Wang T.T., Dahan R., Fiebiger B.M., Ravetch J.V. Type I and type II Fc receptors regulate innate and adaptive immunity. Nat. Immunol. 2014;15(8):707–716. doi: 10.1038/ni.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barnes N., Gavin A.L., Tan P.S., Mottram P., Koentgen F., Hogarth P.M. FcgammaRI-deficient mice show multiple alterations to inflammatory and immune responses. Immunity. 2002;16(3):379–389. doi: 10.1016/s1074-7613(02)00287-x. [DOI] [PubMed] [Google Scholar]
- 35.Ioan-Facsinay A., de Kimpe S.J., Hellwig S.M., van Lent P.L., Hofhuis F.M., van Ojik H.H., Sedlik C., da Silveira S.A., Gerber J., de Jong Y.F., Roozendaal R., Aarden L.A., van den Berg W.B., Saito T., Mosser D., Amigorena S., Izui S., van Ommen G.J., van Vugt M., van de Winkel J.G., Verbeek J.S. FcgammaRI (CD64) contributes substantially to severity of arthritis, hypersensitivity responses, and protection from bacterial infection. Immunity. 2002;16(3):391–402. doi: 10.1016/s1074-7613(02)00294-7. [DOI] [PubMed] [Google Scholar]
- 36.van der Poel C.E., Spaapen R.M., van de Winkel J.G., Leusen J.H. Functional characteristics of the high affinity IgG receptor, FcgammaRI. J. Immunol. 2011;186(5):2699–2704. doi: 10.4049/jimmunol.1003526. [DOI] [PubMed] [Google Scholar]
- 37.Brandsma A.M., Schwartz S.L., Wester M.J., Valley C.C., Blezer G.L.A., Vidarsson G., Lidke K.A., Ten Broeke T., Lidke D.S., Leusen J.H.W. Mechanisms of inside-out signaling of the high-affinity IgG receptor FcgammaRI. Sci. Signal. 2018;11(540) doi: 10.1126/scisignal.aaq0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bournazos S., Ravetch J.V. Anti-retroviral antibody FcgammaR-mediated effector functions. Immunol. Rev. 2017;275(1):285–295. doi: 10.1111/imr.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M. Li, L. Zhao, C. Zhang, X. Wang, W. Hong, J. Sun, R. Liu, L. Yu, J. Wang, F. Zhang, X. Jin, Dengue immune sera enhance Zika virus infection in human peripheral blood monocytes through Fc gamma receptors, PLoS One 13(7) (2018) e0200478 10.1371/journal.pone.0200478. [DOI] [PMC free article] [PubMed]
- 40.Kou Z., Quinn M., Chen H., Rodrigo W.W., Rose R.C., Schlesinger J.J., Jin X. Monocytes, but not T or B cells, are the principal target cells for dengue virus (DV) infection among human peripheral blood mononuclear cells. J. Med. Virol. 2008;80(1):134–146. doi: 10.1002/jmv.21051. [DOI] [PubMed] [Google Scholar]
- 41.Sun P., Bauza K., Pal S., Liang Z., Wu S.J., Beckett C., Burgess T., Porter K. Infection and activation of human peripheral blood monocytes by dengue viruses through the mechanism of antibody-dependent enhancement. Virology. 2011;421(2):245–252. doi: 10.1016/j.virol.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 42.Flipse J., Diosa-Toro M.A., Hoornweg T.E., van de Pol D.P., Urcuqui-Inchima S., Smit J.M. Antibody-dependent enhancement of dengue virus infection in primary human macrophages; balancing higher fusion against antiviral responses. Sci. Rep. 2016;6:29201. doi: 10.1038/srep29201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothman A.L. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat. Rev. Immunol. 2011;11(8):532–543. doi: 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- 44.Srikiatkhachorn A., Green S. Markers of dengue disease severity. Curr. Top. Microbiol. Immunol. 2010;338:67–82. doi: 10.1007/978-3-642-02215-9_6. [DOI] [PubMed] [Google Scholar]
- 45.Halstead S.B. Immune enhancement of viral infection. Prog. Allergy. 1982;31:301–364. [PubMed] [Google Scholar]
- 46.Katzelnick L.C., Gresh L., Halloran M.E., Mercado J.C., Kuan G., Gordon A., Balmaseda A., Harris E. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358(6365):929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salje H., Cummings D.A.T., Rodriguez-Barraquer I., Katzelnick L.C., Lessler J., Klungthong C., Thaisomboonsuk B., Nisalak A., Weg A., Ellison D., Macareo L., Yoon I.K., Jarman R., Thomas S., Rothman A.L., Endy T., Cauchemez S. Reconstruction of antibody dynamics and infection histories to evaluate dengue risk. Nature. 2018;557(7707):719–723. doi: 10.1038/s41586-018-0157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan K.R., Wang X., Saron W.A.A., Gan E.S., Tan H.C., Mok D.Z.L., Zhang S.L., Lee Y.H., Liang C., Wijaya L., Ghosh S., Cheung Y.B., Tannenbaum S.R., Abraham S.N., St John A.L., Low J.G.H., Ooi E.E. Cross-reactive antibodies enhance live attenuated virus infection for increased immunogenicity. Nat. Microbiol. 2016;1:16164. doi: 10.1038/nmicrobiol.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahalingam S., Teixeira M.M., Halstead S.B. Zika enhancement: a reality check. Lancet Infect. Dis. 2017;17(7):686–688. doi: 10.1016/S1473-3099(17)30340-7. [DOI] [PubMed] [Google Scholar]
- 50.Fischer C., de Oliveira-Filho E.F., Drexler J.F. Viral emergence and immune interplay in flavivirus vaccines. Lancet Infect. Dis. 2020;20(1):15–17. doi: 10.1016/S1473-3099(19)30697-8. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-Barraquer I., Costa F., Nascimento E.J.M., Nery N.J., Castanha P.M.S., Sacramento G.A., Cruz J., Carvalho M., De Olivera D., Hagan J.E., Adhikarla H., Wunder E.A., Jr., Coelho D.F., Azar S.R., Rossi S.L., Vasilakis N., Weaver S.C., Ribeiro G.S., Balmaseda A., Harris E., Nogueira M.L., Reis M.G., Marques E.T.A., Cummings D.A.T., Ko A.I. Impact of preexisting dengue immunity on Zika virus emergence in a dengue endemic region. Science. 2019;363(6427):607–610. doi: 10.1126/science.aav6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.A. Gordon, L. Gresh, S. Ojeda, L.C. Katzelnick, N. Sanchez, J.C. Mercado, G. Chowell, B. Lopez, D. Elizondo, J. Coloma, R. Burger-Calderon, G. Kuan, A. Balmaseda, E. Harris, Prior dengue virus infection and risk of Zika: a pediatric cohort in Nicaragua, PLoS Med 16(1) (2019) e1002726 10.1371/journal.pmed.1002726. [DOI] [PMC free article] [PubMed]
- 53.M.K. McCracken, G.D. Gromowski, H.L. Friberg, X. Lin, P. Abbink, R. De La Barrera, K.H. Eckles, L.S. Garver, M. Boyd, D. Jetton, D.H. Barouch, M.C. Wise, B.S. Lewis, J.R. Currier, K. Modjarrad, M. Milazzo, M. Liu, A.B. Mullins, J.R. Putnak, N.L. Michael, R.G. Jarman, S.J. Thomas, Impact of prior flavivirus immunity on Zika virus infection in rhesus macaques, PLoS Pathog 13(8) (2017) e1006487 10.1371/journal.ppat.1006487. [DOI] [PMC free article] [PubMed]
- 54.C. Serrano-Collazo, E.X. Perez-Guzman, P. Pantoja, M.A. Hassert, I.V. Rodriguez, L. Giavedoni, V. Hodara, L. Parodi, L. Cruz, T. Arana, M.I. Martinez, L. White, J.D. Brien, A. de Silva, A.K. Pinto, C.A. Sariol, Effective control of early Zika virus replication by Dengue immunity is associated to the length of time between the 2 infections but not mediated by antibodies, PLoS Negl Trop Dis 14(5) (2020) e0008285 10.1371/journal.pntd.0008285. [DOI] [PMC free article] [PubMed]
- 55.Wilder-Smith A. Dengue vaccine development: status and future. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2020;63(1):40–44. doi: 10.1007/s00103-019-03060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.H. Friberg, L.J. Martinez, L. Lin, J.M. Blaylock, R.A. De La Barrera, A.L. Rothman, J.R. Putnak, K.H. Eckels, S.J. Thomas, R.G. Jarman, J.R. Currier, Cell-Mediated Immunity Generated in Response to a Purified Inactivated Vaccine for Dengue Virus Type 1, mSphere 5(1) (2020) 10.1128/mSphere.00671-19. [DOI] [PMC free article] [PubMed]
- 57.S.R. Hadinegoro, J.L. Arredondo-Garcia, M.R. Capeding, C. Deseda, T. Chotpitayasunondh, R. Dietze, H.I. Muhammad Ismail, H. Reynales, K. Limkittikul, D.M. Rivera-Medina, H.N. Tran, A. Bouckenooghe, D. Chansinghakul, M. Cortes, K. Fanouillere, R. Forrat, C. Frago, S. Gailhardou, N. Jackson, F. Noriega, E. Plennevaux, T.A. Wartel, B. Zambrano, M. Saville, C.-T.D.V.W. Group, Efficacy and Long-Term Safety of a Dengue Vaccine in Regions of Endemic Disease, N Engl J Med 373(13) (2015) 1195-206 10.1056/NEJMoa1506223. [DOI] [PubMed]
- 58.D.Z.L. Mok, K.R. Chan, The Effects of Pre-Existing Antibodies on Live-Attenuated Viral Vaccines, Viruses 12(5) (2020) 10.3390/v12050520. [DOI] [PMC free article] [PubMed]
- 59.Halstead S.B. Dengvaxia sensitizes seronegatives to vaccine enhanced disease regardless of age. Vaccine. 2017;35(47):6355–6358. doi: 10.1016/j.vaccine.2017.09.089. [DOI] [PubMed] [Google Scholar]
- 60.Sridhar S., Luedtke A., Langevin E., Zhu M., Bonaparte M., Machabert T., Savarino S., Zambrano B., Moureau A., Khromava A., Moodie Z., Westling T., Mascarenas C., Frago C., Cortes M., Chansinghakul D., Noriega F., Bouckenooghe A., Chen J., Ng S.P., Gilbert P.B., Gurunathan S., DiazGranados C.A. Effect of dengue serostatus on dengue vaccine safety and efficacy. N. Engl. J. Med. 2018;379(4):327–340. doi: 10.1056/NEJMoa1800820. [DOI] [PubMed] [Google Scholar]
- 61.V. Ramasamy, U. Arora, R. Shukla, A. Poddar, R.K. Shanmugam, L.J. White, M.M. Mattocks, R. Raut, A. Perween, P. Tyagi, A.M. de Silva, S.K. Bhaumik, M.K. Kaja, F. Villinger, R. Ahmed, R.E. Johnston, S. Swaminathan, N. Khanna, A tetravalent virus-like particle vaccine designed to display domain III of dengue envelope proteins induces multi-serotype neutralizing antibodies in mice and macaques which confer protection against antibody dependent enhancement in AG129 mice, PLoS Negl Trop Dis 12(1) (2018) e0006191 10.1371/journal.pntd.0006191. [DOI] [PMC free article] [PubMed]
- 62.R. Shukla, J.A. Brown, H. Beesetti, R. Ahuja, V. Ramasamy, R.K. Shanmugam, A. Poddar, G. Batra, F. Krammer, J.K. Lim, S. Kale, A. Lal, S. Swaminathan, N. Khanna, Dengue and Zika Virus Infections are enhanced by Live Attenuated Dengue. [DOI] [PMC free article] [PubMed]
- 63.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pedersen N.C. A review of feline infectious peritonitis virus infection: 1963–2008. J. Feline Med. Surg. 2009;11(4):225–258. doi: 10.1016/j.jfms.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pedersen N.C. An update on feline infectious peritonitis: diagnostics and therapeutics. Vet. J. 2014;201(2):133–141. doi: 10.1016/j.tvjl.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pedersen N.C., Allen C.E., Lyons L.A. Pathogenesis of feline enteric coronavirus infection. J. Feline Med. Surg. 2008;10(6):529–541. doi: 10.1016/j.jfms.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vennema H., Poland A., Foley J., Pedersen N.C. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology. 1998;243(1):150–157. doi: 10.1006/viro.1998.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Porter E., Tasker S., Day M.J., Harley R., Kipar A., Siddell S.G., Helps C.R. Amino acid changes in the spike protein of feline coronavirus correlate with systemic spread of virus from the intestine and not with feline infectious peritonitis. Vet. Res. 2014;45:49. doi: 10.1186/1297-9716-45-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Addie D.D., Toth S., Murray G.D., Jarrett O. Risk of feline infectious peritonitis in cats naturally infected with feline coronavirus. Am. J. Vet. Res. 1995;56(4):429–434. [PubMed] [Google Scholar]
- 70.Pesteanu-Somogyi L.D., Radzai C., Pressler B.M. Prevalence of feline infectious peritonitis in specific cat breeds. J. Feline Med. Surg. 2006;8(1):1–5. doi: 10.1016/j.jfms.2005.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kipar A., Meli M.L., Baptiste K.E., Bowker L.J., Lutz H. Sites of feline coronavirus persistence in healthy cats. J. Gen. Virol. 2010;91(Pt 7):1698–1707. doi: 10.1099/vir.0.020214-0. [DOI] [PubMed] [Google Scholar]
- 72.Malbon A.J., Meli M.L., Barker E.N., Davidson A.D., Tasker S., Kipar A. Inflammatory mediators in the mesenteric lymph nodes, site of a possible intermediate phase in the immune response to feline coronavirus and the pathogenesis of feline infectious peritonitis? J. Comp. Pathol. 2019;166:69–86. doi: 10.1016/j.jcpa.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gelain M.E., Meli M., Paltrinieri S. Whole blood cytokine profiles in cats infected by feline coronavirus and healthy non-FCoV infected specific pathogen-free cats. J. Feline Med. Surg. 2006;8(6):389–399. doi: 10.1016/j.jfms.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.N. Safi, A. Haghani, S.W. Ng, G.T. Selvarajah, F. Mustaffa-Kamal, A.R. Omar, Expression profiles of immune mediators in feline Coronavirus-infected cells and clinical samples of feline Coronavirus-positive cats, BMC Vet Res 13(1) (2017) 92 10.1186/s12917-017-1019-2. [DOI] [PMC free article] [PubMed]
- 75.Paltrinieri S., Ponti W., Comazzi S., Giordano A., Poli G. Shifts in circulating lymphocyte subsets in cats with feline infectious peritonitis (FIP): pathogenic role and diagnostic relevance. Vet. Immunol. Immunopathol. 2003;96(3–4):141–148. doi: 10.1016/s0165-2427(03)00156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paltrinieri S., Cammarata M.P., Cammarata G., Comazzi S. Some aspects of humoral and cellular immunity in naturally occuring feline infectious peritonitis. Vet. Immunol. Immunopathol. 1998;65(2–4):205–220. doi: 10.1016/s0165-2427(98)00155-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haijema B.J., Volders H., Rottier P.J. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 2004;78(8):3863–3871. doi: 10.1128/jvi.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vennema H., de Groot R.J., Harbour D.A., Dalderup M., Gruffydd-Jones T., Horzinek M.C., Spaan W.J. Early death after feline infectious peritonitis virus challenge due to recombinant vaccinia virus immunization. J. Virol. 1990;64(3):1407–1409. doi: 10.1128/jvi.64.3.1407-1409.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weiss R.C., Scott F.W. Antibody-mediated enhancement of disease in feline infectious peritonitis: comparisons with dengue hemorrhagic fever. Comp. Immunol. Microbiol. Infect. Dis. 1981;4(2):175–189. doi: 10.1016/0147-9571(81)90003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hardy W.D., Jr. Immunopathology induced by the feline leukemia virus. Springer Semin. Immunopathol. 1982;5(1):75–106. doi: 10.1007/BF00201958. [DOI] [PubMed] [Google Scholar]
- 81.Takano T., Yamada S., Doki T., Hohdatsu T. Pathogenesis of oral type I feline infectious peritonitis virus (FIPV) infection: antibody-dependent enhancement infection of cats with type I FIPV via the oral route. J. Vet. Med. Sci. 2019;81(6):911–915. doi: 10.1292/jvms.18-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Balint A., Farsang A., Szeredi L., Zadori Z., Belak S. Recombinant feline coronaviruses as vaccine candidates confer protection in SPF but not in conventional cats. Vet. Microbiol. 2014;169(3–4):154–162. doi: 10.1016/j.vetmic.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pedersen N.C., Black J.W. Attempted immunization of cats against feline infectious peritonitis, using avirulent live virus or sublethal amounts of virulent virus. Am. J. Vet. Res. 1983;44(2):229–234. [PubMed] [Google Scholar]
- 84.Mustaffa-Kamal F., Liu H., Pedersen N.C., Sparger E.E. Characterization of antiviral T cell responses during primary and secondary challenge of laboratory cats with feline infectious peritonitis virus (FIPV) BMC Vet Res. 2019;15(1):165. doi: 10.1186/s12917-019-1909-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Foley J.E., Poland A., Carlson J., Pedersen N.C. Risk factors for feline infectious peritonitis among cats in multiple-cat environments with endemic feline enteric coronavirus. J. Am. Vet. Med. Assoc. 1997;210(9):1313–1318. [PubMed] [Google Scholar]
- 86.Gaunt E.R., Hardie A., Claas E.C., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48(8):2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chan K.H., Cheng V.C., Woo P.C., Lau S.K., Poon L.L., Guan Y., Seto W.H., Yuen K.Y., Peiris J.S. Serological responses in patients with severe acute respiratory syndrome coronavirus infection and cross-reactivity with human coronaviruses 229E, OC43, and NL63. Clin. Diagn. Lab. Immunol. 2005;12(11):1317–1321. doi: 10.1128/CDLI.12.11.1317-1321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Patrick D.M., Petric M., Skowronski D.M., Guasparini R., Booth T.F., Krajden M., McGeer P., Bastien N., Gustafson L., Dubord J., Macdonald D., David S.T., Srour L.F., Parker R., Andonov A., Isaac-Renton J., Loewen N., McNabb G., McNabb A., Goh S.H., Henwick S., Astell C., Guo J.P., Drebot M., Tellier R., Plummer F., Brunham R.C. An outbreak of human coronavirus OC43 infection and serological cross-reactivity with SARS coronavirus. Can. J. Infect. Dis. Med. Microbiol. 2006;17(6):330–336. doi: 10.1155/2006/152612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.S. Khan, R. Nakajima, A. Jain, R.R. de Assis, A. Jasinskas, J.M. Obiero, O. Adenaiye, S. Tai, F. Hong, D.K. Milton, H. Davies, P.L. Felgner, Analysis of serologic cross-reactivity between common human coronaviruses and SARS-CoV-2 using coronavirus antigen microarray, bioRxiv (2020) 2020.03.24.006544 10.1101/2020.03.24.006544.
- 90.Bost P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H., Deczkowska A., Zhang S., Schwikowski B., Zhang Z., Amit I. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020 doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yilla M., Harcourt B.H., Hickman C.J., McGrew M., Tamin A., Goldsmith C.S., Bellini W.J., Anderson L.J. SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res. 2005;107(1):93–101. doi: 10.1016/j.virusres.2004.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yip M.S., Leung N.H., Cheung C.Y., Li P.H., Lee H.H., Daeron M., Peiris J.S., Bruzzone R., Jaume M. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol J. 2014;11:82. doi: 10.1186/1743-422X-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Giani M., Seminati D., Lucchini A., Foti G., Pagni F. Exuberant plasmocytosis in bronchoalveolar lavage specimen of the first patient requiring extracorporeal membrane oxygenation for SARS-CoV-2 in Europe. J. Thorac. Oncol. 2020;15(5):e65–e66. doi: 10.1016/j.jtho.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.M. Woodruff, R. Ramonell, K. Cashman, D. Nguyen, A. Ley, S. Kyu, A. Saini, N. Haddad, W. Chen, J.C. Howell, T. Ozturk, S. Lee, J. Estrada, A. Morrison-Porter, A. Derrico, F. Anam, H. Wu, S. Le, S. Jenks, W. Hu, F.E. Lee, I. Sanz, Critically ill SARS-CoV-2 patients display lupus-like hallmarks of extrafollicular B cell activation, medRxiv 10.1101/2020.04.29.20083717 (2020) 10.1101/2020.04.29.20083717.
- 95.I. Quinti, V. Lougaris, C. Milito, F. Cinetto, A. Pecoraro, I. Mezzaroma, C.M. Mastroianni, O. Turriziani, M.P. Bondioni, M. Filippini, A. Soresina, G. Spadaro, C. Agostini, R. Carsetti, A. Plebani, A possible role for B cells in COVID-19? Lesson from patients with agammaglobulinemia, J. Allergy Clin. Immunol. 10.1016/j.jaci.2020.04.013 (2020) 10.1016/j.jaci.2020.04.013. [DOI] [PMC free article] [PubMed]
- 96.A. Soresina, D. Moratto, M. Chiarini, C. Paolillo, G. Baresi, E. Foca, M. Bezzi, B. Baronio, M. Giacomelli, R. Badolato, Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover, Pediatr. Allergy Immunol. (2020) 10.1111/pai.13263. [DOI] [PMC free article] [PubMed]
- 97.Cheng Y., Wong R., Soo Y.O., Wong W.S., Lee C.K., Ng M.H., Chan P., Wong K.C., Leung C.B., Cheng G. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24(1):44–46. doi: 10.1007/s10096-004-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hung I.F., To K.K., Lee C.K., Lee K.L., Chan K., Yan W.W., Liu R., Watt C.L., Chan W.M., Lai K.Y., Koo C.K., Buckley T., Chow F.L., Wong K.K., Chan H.S., Ching C.K., Tang B.S., Lau C.C., Li I.W., Liu S.H., Chan K.H., Lin C.K., Yuen K.Y. Convalescent plasma treatment reduced mortality in patients with severe pandemic influenza A (H1N1) 2009 virus infection. Clin. Infect. Dis. 2011;52(4):447–456. doi: 10.1093/cid/ciq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Luke T.C., Kilbane E.M., Jackson J.L., Hoffman S.L. Meta-analysis: convalescent blood products for Spanish influenza pneumonia: a future H5N1 treatment? Ann. Intern. Med. 2006;145(8):599–609. doi: 10.7326/0003-4819-145-8-200610170-00139. [DOI] [PubMed] [Google Scholar]
- 100.L. Li, W. Zhang, Y. Hu, X. Tong, S. Zheng, J. Yang, Y. Kong, L. Ren, Q. Wei, H. Mei, C. Hu, C. Tao, R. Yang, J. Wang, Y. Yu, Y. Guo, X. Wu, Z. Xu, L. Zeng, N. Xiong, L. Chen, J. Wang, N. Man, Y. Liu, H. Xu, E. Deng, X. Zhang, C. Li, C. Wang, S. Su, L. Zhang, J. Wang, Y. Wu, Z. Liu, Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life-threatening COVID-19: a randomized clinical trial, JAMA 10.1001/jama.2020.10044 (2020) 10.1001/jama.2020.10044. [DOI] [PMC free article] [PubMed]
- 101.X. Tian, C. Li, A. Huang, S. Xia, S. Lu, Z. Shi, L. Lu, S. Jiang, Z. Yang, Y. Wu, T. Ying, Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody, Emerg Microbes Infect 9(1) (2020) 382-385 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed]
- 102.B. Ju, Q. Zhang, J. Ge, R. Wang, J. Sun, X. Ge, J. Yu, S. Shan, B. Zhou, S. Song, X. Tang, J. Yu, J. Lan, J. Yuan, H. Wang, J. Zhao, S. Zhang, Y. Wang, X. Shi, L. Liu, J. Zhao, X. Wang, Z. Zhang, L. Zhang, Human neutralizing antibodies elicited by SARS-CoV-2 infection, Nature 10.1038/s41586-020-2380-z (2020) 10.1038/s41586-020-2380-z. [DOI] [PubMed]
- 103.H. Lv, N.C. Wu, O.T. Tsang, M. Yuan, R. Perera, W.S. Leung, R.T.Y. So, J.M.C. Chan, G.K. Yip, T.S.H. Chik, Y. Wang, C.Y.C. Choi, Y. Lin, W.W. Ng, J. Zhao, L.L.M. Poon, J.S.M. Peiris, I.A. Wilson, C.K.P. Mok, Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections, Cell Rep. 31(9) (2020) 107725 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed]
- 104.Lambert P.H., Ambrosino D.M., Andersen S.R., Baric R.S., Black S.B., Chen R.T., Dekker C.L., Didierlaurent A.M., Graham B.S., Martin S.D., Molrine D.C., Perlman S., Picard-Fraser P.A., Pollard A.J., Qin C., Subbarao K., Cramer J.P. Consensus summary report for CEPI/BC March 12–13, 2020 meeting: assessment of risk of disease enhancement with COVID-19 vaccines. Vaccine. 2020;38(31):4783–4791. doi: 10.1016/j.vaccine.2020.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kam Y.W., Kien F., Roberts A., Cheung Y.C., Lamirande E.W., Vogel L., Chu S.L., Tse J., Guarner J., Zaki S.R., Subbarao K., Peiris M., Nal B., Altmeyer R. Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcgammaRII-dependent entry into B cells in vitro. Vaccine. 2007;25(4):729–740. doi: 10.1016/j.vaccine.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qin E., Shi H., Tang L., Wang C., Chang G., Ding Z., Zhao K., Wang J., Chen Z., Yu M., Si B., Liu J., Wu D., Cheng X., Yang B., Peng W., Meng Q., Liu B., Han W., Yin X., Duan H., Zhan D., Tian L., Li S., Wu J., Tan G., Li Y., Li Y., Liu Y., Liu H., Lv F., Zhang Y., Kong X., Fan B., Jiang T., Xu S., Wang X., Li C., Wu X., Deng Y., Zhao M., Zhu Q. Immunogenicity and protective efficacy in monkeys of purified inactivated Vero-cell SARS vaccine. Vaccine. 2006;24(7):1028–1034. doi: 10.1016/j.vaccine.2005.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang Q., Zhang L., Kuwahara K., Li L., Liu Z., Li T., Zhu H., Liu J., Xu Y., Xie J., Morioka H., Sakaguchi N., Qin C., Liu G. Correction: immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates. ACS Infect. Dis. 2020;6(5):1284–1285. doi: 10.1021/acsinfecdis.0c00148. [DOI] [PubMed] [Google Scholar]
- 108.L. Bao, W. Deng, B. Huang, H. Gao, J. Liu, L. Ren, Q. Wei, P. Yu, Y. Xu, F. Qi, Y. Qu, F. Li, Q. Lv, W. Wang, J. Xue, S. Gong, M. Liu, G. Wang, S. Wang, Z. Song, L. Zhao, P. Liu, L. Zhao, F. Ye, H. Wang, W. Zhou, N. Zhu, W. Zhen, H. Yu, X. Zhang, L. Guo, L. Chen, C. Wang, Y. Wang, X. Wang, Y. Xiao, Q. Sun, H. Liu, F. Zhu, C. Ma, L. Yan, M. Yang, J. Han, W. Xu, W. Tan, X. Peng, Q. Jin, G. Wu, C. Qin, The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice, Nature (2020) 10.1038/s41586-020-2312-y. [DOI] [PubMed]
- 109.Y.I. Kim, S.G. Kim, S.M. Kim, E.H. Kim, S.J. Park, K.M. Yu, J.H. Chang, E.J. Kim, S. Lee, M.A.B. Casel, J. Um, M.S. Song, H.W. Jeong, V.D. Lai, Y. Kim, B.S. Chin, J.S. Park, K.H. Chung, S.S. Foo, H. Poo, I.P. Mo, O.J. Lee, R.J. Webby, J.U. Jung, Y.K. Choi, Infection and Rapid Transmission of SARS-CoV-2 in Ferrets, Cell Host Microbe 27(5) (2020) 704-709 e2 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed]
- 110.L. Yuan, Q. Tang, T. Cheng, N. Xia, Animal models for emerging coronavirus: progress and new insights, Emerg Microbes Infect 9(1) (2020) 949-961 10.1080/22221751.2020.1764871. [DOI] [PMC free article] [PubMed]
- 111.Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., Xia H., Tang J., Zhang K., Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26(4):502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.J. Hadjadj, N. Yatim, L. Barnabei, A. Corneau, J. Boussier, H. Pere, B. Charbit, V. Bondet, C. Chenevier-Gobeaux, P. Breillat, N. Carlier, R. Gauzit, C. Morbieu, F. Pene, N. Marin, N. Roche, T.-A. Szwebel, N. Smith, S. Merkling, J.-M. Treluyer, D. Veyer, L. Mouthon, C. Blanc, P.-L. Tharaux, F. Rozenberg, A. Fischer, D. Duffy, F. Rieux-Laucat, S. Kerneis, B. Terrier, Impaired type I interferon activity and exacerbated inflammatory responses in severe Covid-19 patients, medRxiv (2020) 10.1101/2020.04.19.20068015.
- 114.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tian S., Hu W., Niu L., Liu H., Xu H., Xiao S.Y. Pulmonary pathology of early-phase 2019 Novel Coronavirus (COVID-19) pneumonia in two patients with lung cancer. J. Thorac. Oncol. 2020;15(5):700–704. doi: 10.1016/j.jtho.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hanley B., Lucas S.B., Youd E., Swift B., Osborn M. Autopsy in suspected COVID-19 cases. J. Clin. Pathol. 2020;73(5):239–242. doi: 10.1136/jclinpath-2020-206522. [DOI] [PubMed] [Google Scholar]
- 117.P. Mehta, D.F. McAuley, M. Brown, E. Sanchez, R.S. Tattersall, J.J. Manson, U.K. Hlh Across Speciality Collaboration, COVID-19: consider cytokine storm syndromes and immunosuppression, Lancet 395(10229) (2020) 1033-1034 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed]
- 118.D. Blanco-Melo, B.E. Nilsson-Payant, W.C. Liu, S. Uhl, D. Hoagland, R. Moller, T.X. Jordan, K. Oishi, M. Panis, D. Sachs, T.T. Wang, R.E. Schwartz, J.K. Lim, R.A. Albrecht, B.R. tenOever, Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19, Cell 181(5) (2020) 1036-1045 e9 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed]
- 119.Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated Type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19(2):181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Z. Zhou, L. Ren, L. Zhang, J. Zhong, Y. Xiao, Z. Jia, L. Guo, J. Yang, C. Wang, S. Jiang, D. Yang, G. Zhang, H. Li, F. Chen, Y. Xu, M. Chen, Z. Gao, J. Yang, J. Dong, B. Liu, X. Zhang, W. Wang, K. He, Q. Jin, M. Li, J. Wang, Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients, Cell Host Microbe 10.1016/j.chom.2020.04.017 (2020) 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed]
- 121.Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., Sompallae R., McCray P.B., Jr., Meyerholz D.K., Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;129(9):3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.I.C. Huang, C.C. Bailey, J.L. Weyer, S.R. Radoshitzky, M.M. Becker, J.J. Chiang, A.L. Brass, A.A. Ahmed, X. Chi, L. Dong, L.E. Longobardi, D. Boltz, J.H. Kuhn, S.J. Elledge, S. Bavari, M.R. Denison, H. Choe, M. Farzan, Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus, PLoS Pathog 7(1) (2011) e1001258 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed]
- 123.Wrensch F., Winkler M., Pohlmann S. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses. 2014;6(9):3683–3698. doi: 10.3390/v6093683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hu Y., Li W., Gao T., Cui Y., Jin Y., Li P., Ma Q., Liu X., Cao C. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits Type I interferon production by interfering with TRIM25-mediated RIG-I Ubiquitination. J. Virol. 2017;91(8):e02143–e2216. doi: 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Minakshi R., Padhan K., Rani M., Khan N., Ahmad F., Jameel S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS ONE. 2009;4(12):e8342. doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chien J.Y., Hsueh P.R., Cheng W.C., Yu C.J., Yang P.C. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology. 2006;11(6):715–722. doi: 10.1111/j.1440-1843.2006.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H., Lit L.C., Hui D.S., Chan M.H., Chung S.S., Sung J.J. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136(1):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang Y., Li J., Zhan Y., Wu L., Yu X., Zhang W., Ye L., Xu S., Sun R., Wang Y., Lou J. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect. Immun. 2004;72(8):4410–4415. doi: 10.1128/IAI.72.8.4410-4415.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang C.H., Liu C.Y., Wan Y.L., Chou C.L., Huang K.H., Lin H.C., Lin S.M., Lin T.Y., Chung K.F., Kuo H.P. Persistence of lung inflammation and lung cytokines with high-resolution CT abnormalities during recovery from SARS. Respir. Res. 2005;6:42. doi: 10.1186/1465-9921-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Qian Z., Travanty E.A., Oko L., Edeen K., Berglund A., Wang J., Ito Y., Holmes K.V., Mason R.J. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am. J. Respir. Cell Mol. Biol. 2013;48(6):742–748. doi: 10.1165/rcmb.2012-0339OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.D.M. Del Valle, S. Kim-schulze, H. Hsin-hui, N.D. Beckmann, S. Nirenberg, B. Wang, Y. Lavin, T. Swartz, D. Madduri, A. Stock, T. Marron, H. Xie, M.K. Patel, O. van Oekelen, A. Rahman, P. Kovatch, J. Aberg, E. Schadt, S. Jagannath, M. Mazumdar, A. Charney, A. Firpo-Betancourt, D.R. Mendu, J. Jhang, D. Reich, K. Sigel, C. Cordon-Cardo, M. Feldmann, S. Parekh, M. Merad, S. Gnjatic, An inflammatory cytokine signature helps predict COVID-19 severity and death, medRxiv (2020) 10.1101/2020.05.28.20115758.
- 134.J. Wang, M. Jiang, X. Chen, L.J. Montaner, Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: Review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts, J Leukoc Biol 10.1002/JLB.3COVR0520-272R (2020) 10.1002/JLB.3COVR0520-272R. [DOI] [PMC free article] [PubMed]
- 135.Giavridis T., van der Stegen S.J.C., Eyquem J., Hamieh M., Piersigilli A., Sadelain M. CAR T cell-induced cytokine release syndrome is mediated by macrophages and abated by IL-1 blockade. Nat. Med. 2018;24(6):731–738. doi: 10.1038/s41591-018-0041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Norelli M., Camisa B., Barbiera G., Falcone L., Purevdorj A., Genua M., Sanvito F., Ponzoni M., Doglioni C., Cristofori P., Traversari C., Bordignon C., Ciceri F., Ostuni R., Bonini C., Casucci M., Bondanza A. Monocyte-derived IL-1 and IL-6 are differentially required for cytokine-release syndrome and neurotoxicity due to CAR T cells. Nat. Med. 2018;24(6):739–748. doi: 10.1038/s41591-018-0036-4. [DOI] [PubMed] [Google Scholar]
- 137.Chousterman B.G., Swirski F.K., Weber G.F. Cytokine storm and sepsis disease pathogenesis. Semin Immunopathol. 2017;39(5):517–528. doi: 10.1007/s00281-017-0639-8. [DOI] [PubMed] [Google Scholar]
- 138.van der Poll T., van de Veerdonk F.L., Scicluna B.P., Netea M.G. The immunopathology of sepsis and potential therapeutic targets. Nat. Rev. Immunol. 2017;17(7):407–420. doi: 10.1038/nri.2017.36. [DOI] [PubMed] [Google Scholar]
- 139.A.J. Wilk, A. Rustagi, N.Q. Zhao, J. Roque, G.J. Martinez-Colon, J.L. McKechnie, G.T. Ivison, T. Ranganath, R. Vergara, T. Hollis, L.J. Simpson, P. Grant, A. Subramanian, A.J. Rogers, C.A. Blish, A single-cell atlas of the peripheral immune response in patients with severe COVID-19, Nat. Med. (2020) 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed]
- 140.M. Liao, Y. Liu, J. Yuan, Y. Wen, G. Xu, J. Zhao, L. Cheng, J. Li, X. Wang, F. Wang, L. Liu, I. Amit, S. Zhang, Z. Zhang, Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19, Nat. Med. (2020) 10.1038/s41591-020-0901-9. [DOI] [PubMed]
- 141.Law H.K., Cheung C.Y., Ng H.Y., Sia S.F., Chan Y.O., Luk W., Nicholls J.M., Peiris J.S., Lau Y.L. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood. 2005;106(7):2366–2374. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ho M.S., Chen W.J., Chen H.Y., Lin S.F., Wang M.C., Di J., Lu Y.T., Liu C.L., Chang S.C., Chao C.L., King C.C., Chiou J.M., Su I.J., Yang J.Y. Neutralizing antibody response and SARS severity. Emerg. Infect. Dis. 2005;11(11):1730–1737. doi: 10.3201/eid1111.040659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Alshukairi A.N., Khalid I., Ahmed W.A., Dada A.M., Bayumi D.T., Malic L.S., Althawadi S., Ignacio K., Alsalmi H.S., Al-Abdely H.M., Wali G.Y., Qushmaq I.A., Alraddadi B.M., Perlman S. Antibody response and disease severity in healthcare worker MERS survivors. Emerg. Infect. Dis. 2016;22(6):1113–1115. doi: 10.3201/eid2206.160010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.K.V. Houser, A.J. Broadbent, L. Gretebeck, L. Vogel, E.W. Lamirande, T. Sutton, K.W. Bock, M. Minai, M. Orandle, I.N. Moore, K. Subbarao, Enhanced inflammation in New Zealand white rabbits when MERS-CoV reinfection occurs in the absence of neutralizing antibody, PLoS Pathog 13(8) (2017) e1006565 10.1371/journal.ppat.1006565. [DOI] [PMC free article] [PubMed]
- 145.C.T. Tseng, E. Sbrana, N. Iwata-Yoshikawa, P.C. Newman, T. Garron, R.L. Atmar, C.J. Peters, R.B. Couch, Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus, PLoS One 7(4) (2012) e35421 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed]
- 146.L. Liu, Q. Wei, Q. Lin, J. Fang, H. Wang, H. Kwok, H. Tang, K. Nishiura, J. Peng, Z. Tan, T. Wu, K.W. Cheung, K.H. Chan, X. Alvarez, C. Qin, A. Lackner, S. Perlman, K.Y. Yuen, Z. Chen, Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection, JCI Insight 4(4) (2019) e123158 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed]
- 147.K. Garber, Coronavirus vaccine developers wary of errant antibodies, Nat. Biotechnol. 10.1038/d41587-020-00016-w (2020) 10.1038/d41587-020-00016-w. [DOI] [PubMed]
- 148.Diamond M.S., Pierson T.C. The challenges of vaccine development against a new virus during a pandemic. Cell Host Microbe. 2020;27(5):699–703. doi: 10.1016/j.chom.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hotez P.J., Corry D.B., Bottazzi M.E. COVID-19 vaccine design: the Janus face of immune enhancement. Nat. Rev. Immunol. 2020;20(6):347–348. doi: 10.1038/s41577-020-0323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chen Z., John Wherry E. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable