Abstract
Tripartite motif (TRIM) proteins are a highly versatile family of host-cell factors that play an integral role in the mammalian defense against pathogens. TRIM proteins regulate either transcription-dependent antiviral responses such as pro-inflammatory cytokine induction, or they modulate other important cell-intrinsic defense pathways like autophagy. Additionally, TRIM proteins exert direct antiviral activity whereby they antagonize specific viral components through diverse mechanisms. Here, we summarize the latest discoveries on the molecular mechanisms of antiviral TRIM proteins and also discuss current and future trends in this fast-evolving field.
Keywords: antiviral response, Innate immunity, TRIM proteins
INTRODUCTION
Tripartite motif (TRIM) and TRIM-like proteins are a highly versatile family of proteins that are known for their modulation of anti-pathogen defenses [1–3]. They are characterized by a conserved N-terminal RING-BBox-coiled-coil (RBCC) motif, and additionally harbor one or more unique C-terminal domains. TRIM-like proteins share the overall domain organization of TRIM proteins but lack parts of the RBCC core components. The RING domain confers E3 ligase activity mediating the ubiquitination (both nondegradative and degradative ubiquitin-linkage types), ISGylation, or SUMOylation of specific substrates. These posttranslational modifications can lead to the degradation of the target protein via the lysosomal or proteasomal route, or they influence the functional activity or interactome of the substrate protein. The RBCC motif also contains one or two zinc-finger BBox domains with largely unknown functions and a coiled-coil domain which is required for oligomerization. Most TRIMs need to at least dimerize to be enzymatically active, and higher-order assemblies have also frequently been observed. The C-terminal domain of TRIM proteins usually confers substrate specificity by facilitating the interaction with specific protein partners in the cell. For some TRIM proteins, the C-terminal domain promotes the binding of nucleic acid or has enzymatic activity. Consequently, human TRIMs are classified according to their C-terminal domain into 11 major groups, comprising a total of ~80 members (Figure 1) [1,2].
Figure 1. TRIM family protein domain structure and groups.
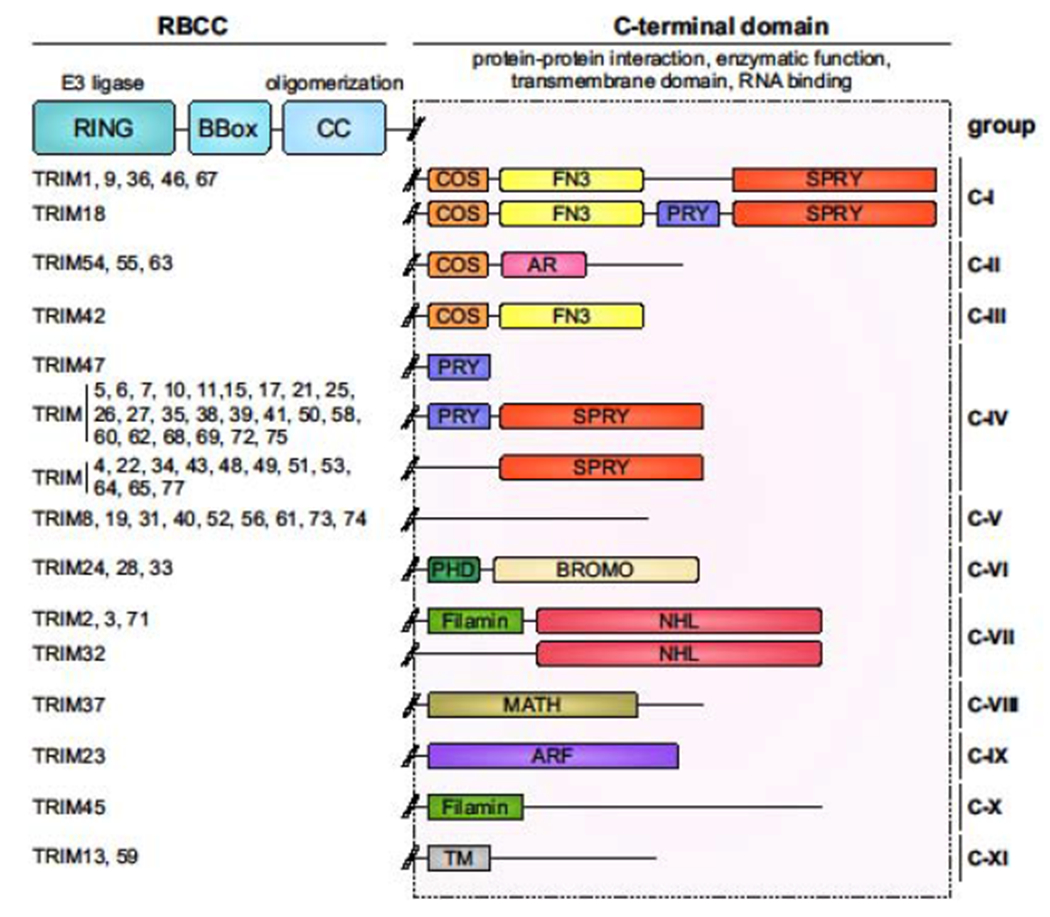
TRIM proteins are characterized by a common RBCC motif. The RBCC motif is comprised of a RING domain conferring E3 ligase activity, one or two BBoxes of mostly unknown function, and a coiled-coil domain that is important for multimerization. The unique C-terminal domains of TRIM proteins have diverse functions including protein-protein interaction, enzymatic activity (e.g. ARF GTPase activity), membrane anchoring, and RNA binding. According to their C-terminal domains, TRIM proteins are classified into 11 major groups. Of note, several TRIM proteins (also called ‘TRIM-like proteins’) that lack the RING or additional domains of the RBCC motif are categorized as ‘unclassified’; these TRIMs are not illustrated for simplicity.
TRIM proteins regulate a variety of cellular processes including transcription, signal transduction, and cell fate and cycle determination (reviewed in [2,4]). Additionally, studies from the past 15 years demonstrated that TRIM proteins are important for mammalian host defenses against viral (and other microbial) pathogens [1,2]. Notably, the gene expression of a large number of the ~80 human TRIM proteins is induced by interferon (IFN) stimulation or upon viral infection as part of an antiviral transcriptional program. At least three major antiviral mechanisms for TRIM proteins have been reported (Figure 2): (I) regulation of cytokine-based innate immune responses; (II) modulation of autophagy-mediated antiviral defense mechanisms; and (III) direct targeting of viral components, which can lead to their degradation or inhibit their functions in key steps of the viral lifecycle through nondegradative mechanisms.
Figure 2. Major antiviral functions of TRIM proteins.
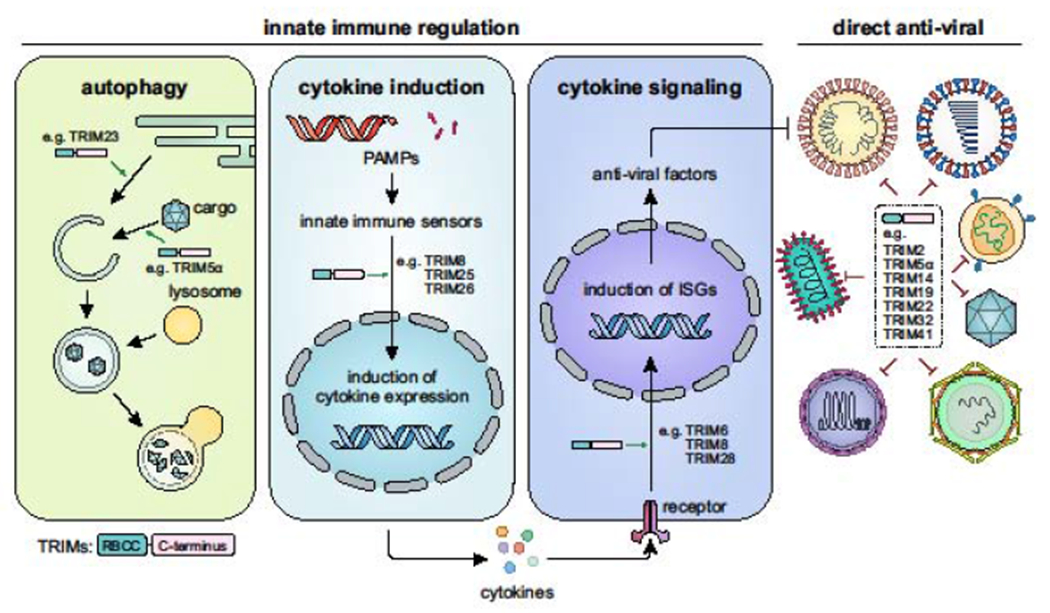
TRIM proteins employ at least three major antiviral mechanisms: (i) modulation of autophagy, a cell-intrinsic autodigestive pathway that limits the replication of certain viruses; (ii) regulation of cytokine induction and cytokine-mediated antiviral innate immune responses; and (iii) direct targeting of viral components, which triggers their degradation or inhibits their functions in key steps of the viral lifecycle. Of note, while some TRIM proteins utilize one of the three antiviral mechanisms, several TRIMs exert immunomodulatory functions and also directly antagonize viral pathogens. Exemplary TRIM proteins involved in specific steps of the antiviral response are indicated. RBCC, RING-BBox-coiled-coil motif.
TRIM proteins modulate cytokine- or autophagy-mediated antiviral defenses
Over the past decade, TRIM proteins were shown to serve important roles in the regulation of innate immune processes, in particular antiviral and proinflammatory cytokine responses [2]. Virus-derived pathogen-associated molecular patterns (PAMPs; predominantly viral RNA or DNA), or cellular danger signals, trigger signaling by specific innate immune receptors that ultimately results in the activation of transcription factors that mediate induction of IFNs (e.g. type-I and -III), proinflammatory cytokines, and chemokines. Released IFN-α/β eventually activates the type-I IFN receptor (IFNAR), thereby promoting the expression of IFN-stimulated genes (ISGs) that create an antiviral milieu. Conceptually, TRIMs can positively or negatively regulate the signal transduction pathways that lead to type-I and/or - III IFN expression, or they can modulate gene expression downstream of IFNAR and other cytokine receptors (e.g. TRIM6, TRIM8 and TRIM28 [1,2]) (Figure 2). Some TRIM proteins induce classical K48-linked ubiquitination of innate immune proteins, leading to their proteasomal degradation, which ultimately dampens innate immunity [1,2]. For example, TRIM21 induces the turnover of activated IRF3, while TRIM13 negatively regulates signaling by the cytoplasmic dsRNA sensor MDA5. TRIM29 was identified to trigger the degradation of the adaptor protein STING during certain DNA virus infections [2]. The suppression of innate immune signaling by TRIM proteins prevents excessive or accidental immune responses, or it may be usurped by viruses to benefit viral replication. On the other hand, many TRIMs catalyze nondegradative polyubiquitin-linkage types, most prominently K63-linked polyubiquitin, which can promote the multimerization of specific innate signaling proteins, or facilitate protein-protein interactions, thereby boosting innate immune responses. Well-characterized TRIM proteins in cytokine-mediated antiviral immunity include TRIM25, which activates the viral RNA sensor RIG-I via K63-linked polyubiquitin, or TRIM56 that regulates the cGAS-STING axis to mediate immune responses to immunostimulatory DNA [2,5].
The functions of TRIM proteins within the complex network of innate immunity have recently been extensively reviewed elsewhere [1–3]. Therefore, only recent developments in this field are briefly summarized here. An emerging concept is that several TRIM proteins have nucleic acid binding activity; these include TRIM25, TRIM28, and TRIM56 [6–8]. In the case of TRIM25, RNA binding to TRIM25 is critical for its enzymatic activity to ubiquitinate RIG-I and to mediate antiviral responses [6], and the long noncoding RNA, Lnczc3h7a, was identified as a physiological RNA ligand for TRIM25 in mouse cells [9]. Furthermore, while most TRIM proteins are widely expressed, several TRIMs were recently identified to exert distinct functions in specific cells of the immune system. For example, TRIM7 was reported to promote cytokine responses mediated by Toll-like receptor 4 in macrophages [10]. TRIM8 was identified to be required for optimal antiviral IFN responses in plasmacytoid dendritic cells [11]. This indicates that TRIM proteins may have tissue- or cell type-specific functions. In the past few years, it also has become clear that many TRIM proteins have more than one antiviral function; for example, several TRIMs that regulate antiviral host responses (e.g. cytokine induction or autophagy) also directly antagonize viral pathogens. Moreover, recent studies discovered specific virus-encoded TRIM antagonists (reviewed in detail in [1,2]), strengthening the notion that TRIM proteins are important antiviral molecules that the virus needs to neutralize in order to overcome TRIM-mediated restriction.
Autophagy is an emerging antiviral process that is induced upon infection with certain viruses (e.g. herpesviruses) and can target viral components for lysosomal degradation (Figure 2). However, autophagy can also degrade signaling components of the cytokine response [3], thereby dampening antiviral gene expression. TRIM proteins were recently identified to regulate virus-triggered autophagy on multiple levels [3]. TRIM5α was reported to target the capsid protein of human immunodeficiency virus-1 (HIV-1) for autophagic degradation [12]. TRIM16 was found to promote antiviral autophagy by facilitating activation of the p62-NRF2 axis [13]. TRIM23 regulates autophagy in response to multiple viruses including herpes simplex virus type 1 (HSV-1) by activating the TBKl-p62 signaling axis [14]; in turn, one of the HSV-1-encoded proteins, Us11, prevents TRIM23 from binding to TBK1 and thereby antagonizes TRIM23-mediated autophagy [15].
TRIM proteins that directly antagonize viral components
Besides modulating cellular signaling cascades of the antiviral innate immune response, TRIM proteins also directly target viral components (Table 1). Only a few TRIM proteins are known to restrict viruses from multiple families by a common mechanism. For example, TRIM21 targets intracellular antibody-opsonized viruses for degradation, seemingly regardless of viral species [16–18]. TRIM25 acts as a co-factor of zinc-finger antiviral protein (ZAP) to target CpG-rich sites in the genomes of Sindbis virus (SINV) and HIV-1 [19–21]. On the other hand, viral restriction by many TRIM proteins seems to be virus species- or family-specific, whereby multiple TRIM proteins contribute to the effective restriction of a particular virus. Below, we summarize in detail the mechanisms utilized by TRIM proteins to block the lifecycle of specific viruses or virus families.
TABLE 1.
TRIM proteins directly antagonizing viruses.
| Virus family | Virus | TRIM | Molecular mechanism | Ref. |
|---|---|---|---|---|
| Retroviruses | HIV-1 | TRIM5α | premature uncoating of the capsid | [27] |
| TRIM5α | enhanced recognition by the immune system | [12,28] | ||
| TRIM11 | premature uncoating of the capsid | [24] | ||
| TRIM19 | stabilizes Daxx to inhibit reverse transcription | [25] | ||
| TRIM22 | blocks binding of SP1 to the HIV-1 LTR | [22] | ||
| TRIM25 | co-factor of ZAP | [19–21] | ||
| TRIM33 | targets HIV-1 IN for degradation | [26] | ||
| TRIM37 | inhibits viral DNA synthesis | [23] | ||
| HIV-1 & SIV | TRIM34 | targets the capsid | [32] | |
| MLV | TRIM5α | premature uncoating of the capsid | [30,31] | |
| Hepadnaviruses | HBV | TRIM5α | inhibits viral transcription | [33] |
| TRIM6 | inhibits viral transcription | [33] | ||
| TRIM14 | inhibits viral transcription | [33] | ||
| TRIM21 | targets HBV Pol for degradation | [34] | ||
| TRIM22 | represses the HBV core promoter | [35] | ||
| TRIM25 | inhibits viral transcription | [33] | ||
| TRIM25 | restricts replication | [36] | ||
| TRIM26 | inhibits viral transcription | [33] | ||
| TRIM31 | inhibits viral transcription | [33] | ||
| TRIM41 | targets the HBV promoter to inhibit transcription | [33] | ||
| Herpesviruses | HSV-1 | TRIM19 | restricts replication | [37] |
| TRIM19 | co-sensor for viral DNA | [38] | ||
| EBV | TRIM5α | targets EBV Rta for degradation | [40] | |
| EBV & hCMV | TRIM28 | promotes unintended reactivation | [41] | |
| KSHV | TRIM28 | represses lytic gene expression | [42] | |
| VZV | TRIM19 | entraps the forming nucleocapsids | [39] | |
| Multiple Herpesviruses | TRIM43 | targets the centrosomal protein pericentrin for degradation to represses active viral chromatin states | [43] | |
| Negative-strand RNA viruses | JUNV & TCRV | TRIM2 | inhibits virus entry | [51] |
| IAV | TRIM14 | targets IAV NP for degradation | [49] | |
| TRIM22 | target IAV NP for degradation | [48] | ||
| TRIM32 | targets IAV PB1 for degradation | [50] | ||
| TRIM41 | targets IAV NP for degradation | [47] | ||
| IAV & IBV | TRIM56 | restricts RNA synthesis | [46] | |
| VSV | TRIM41 | targets VSV N for degradation | [45] | |
| TRIM69 | sequesters VSV P | [44] | ||
| Flaviviruses and other positive-strand viruses | HCV | TRIM14 | targets HCV NS5A for degradation | [55] |
| TRIM22 | targets HCV NS5A for degradation | [56] | ||
| JEV | TRIM52 | targets JEV NS2A for degradation | [57] | |
| ZIKV | TRIM56 | restricts replication | [52] | |
| DENV | TRIM69 | targets DENV NS3 for degradation | [54] | |
| DENV & YFV | TRIM56 | restricts early stages of replication | [53] | |
| SINV | TRIM25 | co-factor of ZAP | [19–21] | |
| PRRSV | TRIM22 | interacts with PRRSV N | [58] | |
| Various virus species | TRIM21 | Targets antibody-opsonized viruses for degradation | [16–18] | |
| TRIM25 | co-factor of antiviral ZAP | [19–21] | ||
Retroviruses
At least 6 TRIM proteins restrict the replication of retroviruses (Table 1). TRIM22 prevents the transcription factor Sp1 from binding to the viral promotor long terminal repeats (LTRs) of HIV-1, thus decreasing viral gene expression and replication [22]. TRIM37 inhibits retroviral DNA synthesis and genomic integration, and is incorporated into the HIV virion [23]. One of the major anti-retroviral targets of TRIM proteins are the capsids. The HIV-1 capsid is prematurely uncoated by TRIM11 [24], leading to blockage of reverse transcription. In addition to targeting the viral capsid upon entry, TRIM11 also affects the release of HIV-1 particles. TRIM19 targets HIV-1 by inhibiting reverse transcription in a cell type-dependent manner via the stabilization of Daxx [25]. TRIM33 targets the viral integrase (IN), thereby inhibiting genomic integration of the provirus [26]. The best-studied anti-retroviral TRIM is TRIM5α, which limits retroviral replication by premature disassembly of the capsid [27] and enhanced recognition by the immune system [12,28]. The ability of TRIM5α to catalyze K63-linked ubiquitination is required for HIV inhibition [29]. Interestingly, only old-world monkey-derived, but not human, TRIM5α can antagonize HIV-1. Human TRIM5α, however, is still active against other retroviruses like murine leukemia virus (MLV) [30,31]. TRIM34 was recently identified to restrict HIV-1 and simian immunodeficiency virus (SIV) in a TRIM5α-dependent manner [32], also by targeting the capsid.
Hepadnaviruses
Several TRIM proteins were reported to counteract hepatitis B virus (HBV) transcription from its core promotor: TRIM5α, TRIM6, TRIM14, TRIM25, TRIM26, TRIM31 and TRIM41 [33]. Mechanistic analysis of TRIM41 revealed that both the C-terminal SPRY and the N-terminal RING domains are required for binding and targeting a 150-nucleotide long motif in the HBV promotor. TRIM21 was reported to target the DNA polymerase of HBV for degradation via direct ubiquitination [34]. TRIM22 represses the HBV core promoter in a RING-dependent manner [35] while TRIM25, which is upregulated by type I IFN and interleukin-27, restricts HBV replication presumably by facilitating innate immune responses [36].
Herpesviruses
TRIM19/PML is well-known for its ability to restrict herpesvirus infection. It was first identified to tightly control the establishment of latency of HSV-1 and human cytomegalovirus (hCMV) via epigenetic silencing [37]. In the absence of the viral TRIM19-antagonist ICP0, HSV-1 DNA is captured and trapped, effectively blocking viral replication [38]. This study demonstrated that removal of a virus-encoded TRIM antagonist reveals the potent antiviral activity of a TRIM protein which may otherwise be obscured. In a second function, TRIM19 reportedly facilitates IFI16-dependent innate immune responses by presumably acting as a (co)-sensor for HSV-1 dsDNA [38]. The incoming capsid of varicella zoster virus (VZV) is targeted by TRIM19 [39]. TRIM5α targets the replication and transcription activator (Rta) protein of Epstein-Barr Virus (EBV) for ubiquitination, attenuating EBV lytic replication [40]. Phosphorylation of TRIM28 (also known as KAP-1) induced by chloroquine treatment promotes unintended latency reactivation of multiple herpesviruses, including EBV and hCMV [41]. Additionally, TRIM28 represses lytic gene expression during the early stage of infection of Kaposi sarcoma-associated herpesvirus (KSHV) [42]. TRIM43 inhibits the lytic replication or reactivation of a broad range of herpesviruses, including HSV-1, EBV and KSHV, by modulating the nuclear lamina architecture via degradation of the centrosomal protein pericentrin. TRIM43 expression is profoundly induced following herpesviral infection via DUX4, a germline transcription factor [43] (Table 1).
Negative-strand RNA viruses
RNA synthesis of the prototypic negative-strand RNA virus vesicular stomatitis virus (VSV) is disturbed by TRIM69 that sequesters the polymerase co-factor P [44]. The nucleoprotein of VSV is ubiquitinated by TRIM41 and targeted for degradation by the proteasome, limiting viral replication [45]. TRIM56 decreases RNA synthesis of both influenza A virus (IAV) and influenza B virus (IBV) [46]. For IAV, one of the major targets of TRIM proteins seems to be the nucleoprotein NP. Multiple TRIM proteins, including TRIM14, TRIM41 and TRIM22, mediate the ubiquitination and subsequent degradation of NP [47–49]. TRIM32 senses the presence of IAV polymerase basic protein 1 (PB1) and targets it for proteasomal destruction via K48-linked polyubiquitination [50]. New World arenaviruses, such as Junin virus (JUNV) and Tacaribe virus (TCRV), are inhibited by TRIM2 at the stage of virus entry, whereby the activity of TRIM2 to regulate phagocytosis independent of its E3 ligase activity is believed to play a role [51] (Table 1).
Flaviviruses and other positive-strand RNA viruses
Positive-strand RNA viruses such as Flaviviridae family members are restricted by multiple TRIM proteins (Table 1). TRIM56 blocks the replication of dengue virus (DENV), Zika virus (ZIKV) and Yellow fever virus (YFV) [52,53], and the RNA-binding activity of TRIM56 was reported to be required for restriction [52]. The DENV non-structural protein 3 (NS3) is directly targeted for degradation by TRIM69 [54]. The hepatitis C virus (HCV) NS5A protein is sent for proteasomal destruction by both TRIM14 [55] and TRIM22 [56] via K48-linked polyubiquitination. The replication of Japanese encephalitis virus (JEV) is inhibited by TRIM52, which targets the NS2A protein for ubiquitination and degradation [57]. The nucleoprotein of porcine reproductive and respiratory syndrome virus (PRRSV) is targeted by TRIM22, thereby restricting virus replication [58].
CONCLUDING REMARKS
Recent years have identified TRIM proteins as potent antagonists of viral replication that target viruses either directly, or indirectly by regulating antiviral innate immune responses. In turn, many TRIM proteins are antagonized by viruses that block their antiviral properties. However, mechanistic insights into the antiviral function of many TRIM proteins as well as viral countermeasures remain elusive. Notably, some TRIMs promote virus replication by ubiquitinating and degrading specific innate immune signaling proteins to dampen cytokine responses, or by facilitating specific viral replication steps. The TRIM-mediated degradation of cellular components of the innate immune system can also function to avoid the overzealous activation or accidental misfiring of the antiviral response.
Recent studies also identified novel functions for some TRIM proteins whose mechanisms were thought to be well understood, highlighting that TRIMs are versatile multifunctional proteins. One prominent example is TRIM25, whose role in the activation of the RNA sensor RIG-I is well demonstrated [1,2]. More recently, TRIM25 was found to also function as a co-factor of the nuclease ZAP which degrades viral RNA [19,20]. Overall, despite detailed mechanistic research over the past several years, we may not have fully understood the complete functional repertoire of many TRIM proteins.
All major virus families are restricted by TRIM proteins using a wide variety of mechanisms. Common strategies of TRIMs that target multiple virus species or families are just beginning to emerge. It remains to be determined whether TRIM proteins recognize unique motifs or three-dimensional structural elements in specific viral proteins, such as the viral nucleoproteins or capsid proteins [12,25,27,48]. Along these lines, several TRIMs target the same viral component, prompting the question of whether these TRIM proteins act redundantly, or are required only in specific cell types or at different stages in the viral lifecycle. Future studies should determine the relevance of antiviral TRIM proteins in different tissues, organs, and host species. Finally, since the function of a large majority of TRIMs depends on their enzymatic activities, TRIMs may be potential drug targets for therapeutic intervention. Future research therefore needs to unravel the detailed mechanisms of TRIM proteins and explore their therapeutic impact.
HIGHLIGHTS.
TRIM proteins are E3 ubiquitin ligases that target specific substrates.
TRIM proteins regulate antiviral cytokine induction or autophagy.
Several TRIM proteins directly target and antagonize viral components.
New antiviral mechanisms of TRIM proteins are still emerging.
ACKNOWLEDGMENTS
We apologize for not being able to cite and discuss all relevant literature due to space limitations. Research in Dr. Gack’s laboratory is funded by U.S. National Institutes of Health (NIH) grants (R01 AI127774, R01 AI087846, and R21 AI148082) and an award from the ClayCo Foundation. Dr. Sparrer’s lab is supported by the German Society for Research (DFG) grants (SP1600/4-1, SPP1923, and CRC1279). L.K. is supported by Ulm University Medical Center intramural funding (L.SBN.0150).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTEREST
The authors have no conflict of interest to declare.
REFERENCES
- *1.van Tol S, Hage A, Giraldo MI, Bharaj P, Rajsbaum R: The TRIMendous Role of TRIMs in Virus-Host Interactions. Vaccines 2017, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *2.van Gent M, Sparrer KMJ, Gack MU: TRIM Proteins and Their Roles in Antiviral Host Defenses. Annu Rev Virol 2018, 5:385–405. [DOI] [PMC free article] [PubMed] [Google Scholar]; Van Tol et al. and van Gent et al. provide a well-structured and comprehensive overview of the diverse anti-viral functions and innate immune regulation by TRIM proteins.
- 3.Sparrer KMJ, Gack MU: TRIM proteins: New players in virus-induced autophagy. PLoS Pathog 2018, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venuto S, Merla G: E3 Ubiquitin Ligase TRIM Proteins, Cell Cycle and Mitosis. Cells 2019, 8:510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seo GJ, Kim C, Shin WJ, Sklan EH, Eoh H, Jung JU: TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat Commun 2018, doi: 10.1038/s41467-018-02936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *6.Sanchez JG, Sparrer KMJ, Chiang C, Reis RA, Chiang JJ, Zurenski MA, Wan Y, Gack MU, Pornillos O: TRIM25 Binds RNA to Modulate Cellular Anti-viral Defense. J Mol Biol 2018, doi: 10.1016/j.jmb.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *7.Choudhury NR, Heikel G, Trubitsyna M, Kubik P, Nowak JS, Webb S, Granneman S, Spanos C, Rappsilber J, Castello A, et al. : RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. BMC Biol 2017, doi: 10.1186/s12915-017-0444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Sanchez et al, Choudhury et al. establishes an important role for RNA-binding by TRIM25 in activating innate immune signaling.
- 8.Williams FP, Haubrich K, Perez-Borrajero C, Hennig J: Emerging RNA-binding roles in the TRIM family of ubiquitin ligases. Biol Chem 2019, doi: 10.1515/hsz-2019-0158. [DOI] [PubMed] [Google Scholar]
- **9.Lin H, Jiang M, Liu L, Yang Z, Ma Z, Liu S, Ma Y, Zhang L, Cao X: The long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat Immunol 2019, doi: 10.1038/s41590-019-0379-0. [DOI] [PubMed] [Google Scholar]; This study is the first description of a long non-coding RNA acting as a scaffold to promote TRIM25 and RIG-I mediated antiviral innate immune responses.
- 10.Lu M, Zhu X, Yang Z, Zhang W, Sun Z, Ji Q, Chen X, Zhu J, Wang C, Nie S: E3 ubiquitin ligase tripartite motif 7 positively regulates the TLR4-mediated immune response via its E3 ligase domain in macrophages. Mol Immunol 2019, 109:126–133. [DOI] [PubMed] [Google Scholar]
- *11.Maarifi G, Smith N, Maillet S, Moncorgé O, Chamontin C, Edouard J, Sohm F, Blanchet FP, Herbeuval JP, Lutfalla G, et al. : TRIM8 is required for virus-induced IFN response in human plasmacytoid dendritic cells. Sci Adv 2019, 5:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]; This publication shows that certain TRIM proteins regulate innate signal transduction only in specific cell types but are dispensable for signaling in other cell types.
- 12.Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C, Silvestri G, Münch J, Kirchhoff F, Simonsen A, et al. : TRIM Proteins Regulate Autophagy and Can Target Autophagic Substrates by Direct Recognition. Dev Cell 2014, 30:394–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jena KK, Kolapalli SP, Mehto S, Nath P, Das B, Sahoo PK, Ahad A, Syed GH, Raghav SK, Senapati S, et al. : TRIM16 controls assembly and degradation of protein aggregates by modulating the p62- NRF2 axis and autophagy. EMBO J 2018, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sparrer KMJ, Gableske S, Zurenski MA, Parker ZM, Full F, Baumgart GJ, Kato J, Pacheco-Rodriguez G, Liang C, Pornillos O, et al. : TRIM23 mediates virus-induced autophagy via activation of TBK1. Nat Microbiol 2017, 2:1543–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *15.Liu X, Matrenec R, Gack MU, He B: Disassembly of the TRIM23-TBK1 Complex by the Us11 Protein of Herpes Simplex Virus 1 Impairs Autophagy. J Virol 2019, doi: 10.1128/jvi.00497-19. [DOI] [PMC free article] [PubMed] [Google Scholar]; This exemplary study (and many others) demonstrate that the antiviral functions of many TRIM proteins, here TRIM23, are antagonized by viral proteins, highlighting their importance as antiviral restriction factors.
- 16.McEwan WA, Tam JCH, Watkinson RE, Bidgood SR, Mallery DL, James LC: Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol 2013, 14:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foss S, Bottermann M, Jonsson A, Sandlie I, James LC, Andersen JT: TRIM21—From intracellular immunity to therapy. Front Immunol 2019, 10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bottermann M, Foss S, Van Tienen LM, Vaysburd M, Cruickshank J, O’Connell K, Clark J, Mayes K, Higginson K, Hirst JC, et al. : TRIM21 mediates antibody inhibition of adenovirus-based gene delivery and vaccination. Proc Natl Acad Sci U S A 2018, 115:10440–10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li MMH, Lau Z, Cheung P, Aguilar EG, Schneider WM, Bozzacco L, Molina H, Buehler E, Takaoka A, Rice CM, et al. : TRIM25 Enhances the Antiviral Action of Zinc-Finger Antiviral Protein (ZAP). PLOS Pathog 2017, 13:el006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *20.Kmiec D, Nchioua R, Sherrill-Mix S, Stürzel CM, Heusinger E, Braun E, Gondim MVP, Hotter D, Sparrer KMJ, Hahn BH, et al. : CpG frequency in the 5’ third of the env gene determines sensitivity of primary HIV-1 strains to the zinc-finger antiviral protein. MBio 2020, 11:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *21.Ficarelli M, Antzin-Anduetza I, Hugh-White R, Firth AE, Sertkaya H, Wilson H, Neil SJD, Schulz R, Swanson CM: CpG Dinucleotides Inhibit HIV-1 Replication through Zinc Finger Antiviral Protein (ZAP)-Dependent and -Independent Mechanisms. J Virol 2019, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kmiec et al. and Ficarelli et al. show that TRIM25 as a co-factor of ZAP, targets CpG dinucleotide-rich regions in HIV and that this may cause suppression of CpGs in HIV strains and possibly other viruses.
- 22.Turrini F, Marelli S, Kajaste-Rudnitski A, Lusic M, Van Lint C, Das AT, Harwig A, Berkhout B, Vicenzi E: HIV-1 transcriptional silencing caused by TRIM22 inhibition of Sp1 binding to the viral promoter. Retrovirology 2015, 12:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabah AA, Tardif K, Mansky LM: Anti-HIV-1 activity of Trim 37. J Gen Virol 2014, 95:960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan T, Yao W, Tokunaga K, Yang R, Sun B: An HIV-1 capsid binding protein TRIM11 accelerates viral uncoating. Retrovirology 2016, 13:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutrieux J, Maarifi G, Portilho DM, Arhel NJ, Chelbi-Alix MK, Nisole S: PML/TRIM19-Dependent Inhibition of Retroviral Reverse-Transcription by Daxx. PLOS Pathog 2015, 11:e1005280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ali H, Mano M, Braga L, Naseem A, Marini B, Vu DM, Collesi C, Meroni G, Lusic M, Giacca M: Cellular TRIM33 restrains HIV-1 infection by targeting viral integrase for proteasomal degradation. Nat Commun 2019, doi: 10.1038/s41467-019-08810-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J: The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 2004, 427:848–853. [DOI] [PubMed] [Google Scholar]
- 28.Jimenez-Moyano E, Ruiz A, Kløverpris HN, Rodriguez-Plata MT, Peña R, Blondeau C, Selwood DL, Izquierdo-Useros N, Moris A, Clotet B, et al. : Nonhuman TRIM5 Variants Enhance Recognition of HIV-1-Infected Cells by CD8 + T Cells. J Virol 2016, 90:8552–8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **29.Imam S, Kömürlü S, Mattick J, Selyutina A, Talley S, Eddins A, Diaz-Griffero F, Campbell EM: K63-Linked Ubiquitin Is Required for Restriction of HIV-1 Reverse Transcription and Capsid Destabilization by Rhesus TRIM5α. J Virol 2019, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provided new mechanistic insights for TRIM5α, whose function is well-known, by showing that K63-linked ubiquitin chains are required for HIV capsid disassembly and subsequent association with autophagosomal membranes.
- 30.Yap MW, Nisole S, Lynch C, Stoye JP: Trim5 protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci 2004, 101:10786–10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yap MW, Nisole S, Stoye JP: A Single Amino Acid Change in the SPRY Domain of Human Trim5α Leads to HIV-1 Restriction. Curr Biol 2005, 15:73–78. [DOI] [PubMed] [Google Scholar]
- 32.Id MO, Id KK, Keceli SK, Felton A, Campbell E, Id JL, Id ME: TRIM34 restricts HIV-1 and SIV capsids in a TRIM5 α -dependent manner. 2020, doi: 10.1371/journal.ppat.1008507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang S, Guo J-T, Wu JZ, Yang G: Identification and Characterization of Multiple TRIM Proteins That Inhibit Hepatitis B Virus Transcription. PLoS One 2013, 8:e70001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mu T, Zhao X, Zhu Y, Fan H, Tang H: The E3 ubiquitin ligase TRIM21 promotes HBV DNA polymerase degradation. Viruses 2020, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao B, Duan Z, Xu W, Xiong S: Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located RING domain. Hepatology 2009, 50:424–433. [DOI] [PubMed] [Google Scholar]
- 36.Tan G, Xiao Q, Song H, Ma F, Xu F, Peng D, Li N, Wang X, Niu J, Gao P, et al. : Type i IFN augments IL-27-dependent TRIM25 expression to inhibit HBV replication. Cell Mol Immunol 2018, doi: 10.1038/cmi.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *37.Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A: PML Contributes to a Cellular Mechanism of Repression of Herpes Simplex Virus Type 1 Infection That Is Inactivated by ICP0. J Virol 2006, 80:7995–8005. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study highlights that the antiviral activity of many TRIM proteins may be masked by viral antagonists.
- 38.Alandijany T, Roberts APE, Conn KL, Loney C, McFarlane S, Orr A, Boutell C: Distinct temporal roles for the promyelocytic leukaemia (PML) protein in the sequential regulation of intracellular host immunity to HSV-1 infection. PLoS Pathog 2018, doi: 10.1371/journal.ppat.1006769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reichelt M, Wang L, Sommer M, Perrino J, Nour AM, Sen N, Baiker A, Zerboni L, Arvin AM: Entrapment of viral capsids in nuclear PML cages is an intrinsic antiviral host defense against Varicella-zoster virus. PLoS Pathog 2011, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang H-H, Chen C-S, Wang W-H, Hsu S-W, Tsai H-H, Liu S-T, Chang L-K: TRIM5α Promotes Ubiquitination of Rta from Epstein–Barr Virus to Attenuate Lytic Progression. Front Microbiol 2017, 7:2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Burton EM, Bhaduri-McIntosh S: Chloroquine triggers Epstein-Barr virus replication through phosphorylation of KAP1/TRIM28 in Burkitt lymphoma cells. PLOS Pathog 2017, 13:e1006249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun R, Liang D, Gao Y, Lan K: Kaposi’s Sarcoma-Associated Herpesvirus-Encoded LANA Interacts with Host KAP1 To Facilitate Establishment of Viral Latency. J Virol 2014, doi: 10.1128/jvi.00596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **43.Full F, van Gent M, Sparrer KMJ, Chiang C, Zurenski MA, Scherer M, Brockmeyer NH, Heinzerling L, Stürzl M, Korn K, et al. : Centrosomal protein TRIM43 restricts herpesvirus infection by regulating nuclear lamina integrity. Nat Microbiol 2019, 4:164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified the antiviral function and molecular mechanism of TRIM43, which targets not only a single virus but rather several members of the herpesviridae family, suggesting that some TRIM proteins may broadly inhibit viruses by a shared mechanism.
- 44.Kueck T, Bloyet L-M, Cassella E, Zang T, Schmidt F, Brusic V, Tekes G, Pornillos O, Whelan SPJ, Bieniasz PD: Vesicular Stomatitis Virus Transcription Is Inhibited by TRIM69 in the Interferon-Induced Antiviral State. J Virol 2019, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patil G, Xu L, Wu Y, Song K, Hao W, Hua F, Wang L, Li S: TRIM41-Mediated Ubiquitination of Nucleoprotein Limits Vesicular Stomatitis Virus Infection. Viruses 2020, 12:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu B, Li NL, Shen Y, Bao X, Fabrizio T, Elbahesh H, Webby RJ, Li K: The C-Terminal Tail of TRIM56 Dictates Antiviral Restriction of Influenza A and B Viruses by Impeding Viral RNA Synthesis. J Virol 2016, 90:4369–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patil G, Zhao M, Song K, Hao W, Bouchereau D, Wang L, Li S: TRIM41-Mediated Ubiquitination of Nucleoprotein Limits Influenza A Virus Infection. J Virol 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Pietro A, Kajaste-Rudnitski A, Oteiza A, Nicora L, Towers GJ, Mechti N, Vicenzi E: TRIM22 Inhibits Influenza A Virus Infection by Targeting the Viral Nucleoprotein for Degradation. J Virol 2013, 87:4523–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu X, Wang J, Wang S, Wu F, Chen Z, Li C, Cheng G, Qin FX-F: Inhibition of Influenza A Virus Replication by TRIM14 via Its Multifaceted Protein-Protein Interaction With NP. Front Microbiol 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu B, Wang L, Ding H, Schwamborn JC, Li S, Dorf ME: TRIM32 Senses and Restricts Influenza A Virus by Ubiquitination of PB1 Polymerase. PLOS Pathog 2015, 11:e1004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *51.Sarute N, Ibrahim N, Medegan Fagla B, Lavanya M, Cuevas C, Stavrou S, Otkiran-Clare G, Tyynismaa H, Henao-Mejia J, Ross SR: TRIM2, a novel member of the antiviral family, limits New World arenavirus entry. PLOS Biol 2019, 17:e3000137. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study highlights that some TRIM proteins restrict virus infection independent of the enzymatic function of their RING domain.
- 52.Yang D, Li NL, Wei D, Liu B, Guo F, Elbahesh H, Zhang Y, Zhou Z, Chen G-Y, Li K: The E3 ligase TRIM56 is a host restriction factor of Zika virus and depends on its RNA-binding activity but not miRNA regulation, for antiviral function. PLoS Negl Trop Dis 2019, 13:e0007537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu B, Li NL, Wang J, Shi P-Y, Wang T, Miller MA, Li K: Overlapping and Distinct Molecular Determinants Dictating the Antiviral Activities of TRIM56 against Flaviviruses and Coronavirus. J Virol 2014, 88:13821–13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang K, Zou C, Wang X, Huang C, Feng T, Pan W, Wu Q, Wang P, Dai J: Interferon-stimulated TRIM69 interrupts dengue virus replication by ubiquitinating viral nonstructural protein 3. PLOS Pathog 2018, 14:e1007287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S, Chen Y, Li C, Wu Y, Guo L, Peng C, Huang Y, Cheng G, Qin FX-F: TRIM14 inhibits hepatitis C virus infection by SPRY domain-dependent targeted degradation of the viral NS5A protein. Sci Rep 2016, 6:32336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang C, Zhao X, Sun D, Yang L, Chong C, Pan Y, Chi X, Gao Y, Wang M, Shi X, et al. : Interferon alpha (IFNα)-induced TRIM22 interrupts HCV replication by ubiquitinating NS5A. Cell Mol Immunol 2016, 13:94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan W, Wu M, Qian S, Zhou Y, Chen H, Li X, Qian P: TRIM52 inhibits Japanese Encephalitis Virus replication by degrading the viral NS2A. Sci Rep 2016, 6:33698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jing H, Tao R, Dong N, Cao S, Sun Y, Ke W, Li Y, Wang J, Zhang Y, Huang H, et al. : Nuclear localization signal in TRIM22 is essential for inhibition of type 2 porcine reproductive and respiratory syndrome virus replication in MARC-145 cells. Virus Gems 2019, 55:660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]


