Abstract
Background:
The presence of at least one MC1R gene variant is associated with a reduction in age at melanoma diagnosis in families with CDKN2A mutations.
Objective:
To describe dermoscopic features of early melanoma in CDKN2A gene mutation-positive Spanish individuals and to evaluate the possibility of a correlation between particular dermatoscopic pattern and MC1R gene variants.
Design:
Patients in whom a melanoma was diagnosed during specific follow up of high-risk individuals carrying CDKN2A mutations (with familial or personal history of previous melanoma) were included in this study. The decision to remove such melanomas was taken on the basis of history, clinical and dermoscopic evaluations including total body photography and digital dermoscopy.
Results:
Of the nine patients included in this study, three were noncarriers of the red hair MC1R polymorphism, three patients had one red hair MC1R polymorphism and three patients had two red hair MC1R polymorphisms. On dermoscopic analysis of suspect melanocytic lesions we found that the mean ABCD Total Dermoscopic Score (TDS) was significantly higher in non-carriers of red-hair MC1R polymorphisms than in carriers of two or more MC1R gene red-hair variants (6.4±0.4 vs. 4.4±0.9 p=0.009).
Conclusions:
Early melanomas in patients with two MC1R red hair variants may be difficult to diagnose definitively by dermoscopy because, in our limited experience, they show fewer colours and structures and have a lower TDS. In such melanomas, subtle atypical vessels and other changes detected by digital image follow up may be useful to confirm the diagnosis of melanoma. An integrated approach including clinical history and dermoscopic data (also considering additional information, such as the presence of atypical vessels) should be utilized in evaluating these high-risk patients. Further studies are necessary to confirm our suggestion.
Keywords: Dermoscopy, Dermatoscopy, Melanocortin 1 receptor, MC1R, melanin, melanoma, CDKN2A, Total Dermoscopy Score (TDS), Red hair
Introduction
The incidence and mortality rates of cutaneous malignant melanoma (CMM) have been increasing steadily in Caucasians worldwide [1, 2]. The principle risk factors for melanoma development are exposure to ultraviolet irradiation and genetic predisposition. Melanoma primarily affects fair-skinned and fair-haired individuals, with poor tanning ability, history of severe sunburns, multiple and especially atypical nevi [3]. As regards the knowledge of the genetic background underyling melanoma development, much progress has been made in recent years. High penetrance genes, such as CDKN2A and CDK4, and low penetrance genes such as the Melanocortin 1 receptor (MC1R) have been demonstrated to contribute to melanoma predisposition. MC1R is the gene responsible for much of the diversity in human pigmentation. Skin and hair colour depends on the relative amount, size and distribution of visible melanin (black/brown eumelanin and red/yellow pheomelanin) pigment, produced within melanosomes of epidermal and follicular melanocytes [4]. MC1R plays an important role in determining the ratio of eumelanin and pheomelanin production. A number of naturally occurring polymorphisms largely determine different skin colours. Some polymorphisms in the MC1R gene result in the increased production of pheomelanin, which is less sun-protective and is largely responsible for red hair colouration. These variants are strongly associated with red hair, pale skin and a poor tanning ability (known as RHC phenotype) are D84E, R151C, R160W, D294H [5, 6, 7, 8].
Recent studies have showed that the presence of at least one MC1R variant is associated with a reduction in age at CMM diagnosis in families with CDKN2A mutations [9, 10, 11]. In individuals with inherited susceptibility for melanoma frequent screening is recommended in order to detect melanoma lesions at the earliest stage possible. We were interested to determine how the presence of an MC1R variant in a patient may effect our ability to diagnose very early melanocytic lesions by dermoscopy. In conditions characterised by lack of pigment in melanocytes, such as albinism, melanoma represents a greater diagnostic challenge because of their hypopigmented appearance [12, 13]. Likewise, in other conditions in which melanocytes produce predominantly red/yellow pheomelanin, as in red headed individuals, melanocytic lesions, including CMM, can be less pigmented and more difficult to identify under clinical examination.
Dermoscopy is currently a pivotal tool for the diagnosis of melanocytic lesions. Dermoscopy is a noninvasive clinical technique that permits the in vivo observation of morphologic structures that are not visible to the naked eye. This technique improves the sensitivity and specificity of CMM diagnosis. It allows the identification of many different lesional structures and colours and correlates them with histopathological findings [14, 15, 16]. However, dermoscopy has limitations in the diagnosis of some very early melanomas [17]. Because colours play an important role in the dermoscopical diagnosis, hypo-or amelanotic melanoma can cause problems in making the correct diagnosis [18]. As far as we are aware this is the first study to report on the dermoscopic features of CMMs associated with genetic changes in MC1R.
The objective of the current work is to describe dermoscopic features of early melanoma associated with MC1R variants in patients under regular screening because they carry CDKN2A mutations.
Material and Methods
Nine patients were included in this study. All of them were treated and followed up at the Melanoma Unit of the Hospital Clínic; Barcelona, Spain.
The inclusion criteria were the following: (i) patient at risk of melanoma in follow up (ii) confirmation of CDKN2A mutation (iii) photographic documentation of the excised melanoma during follow up. The decision to remove such melanomas was taken on the basis of history, clinical and dermoscopic evaluations including total body photography and digital dermoscopy. Written informed consent was obtained from each patient prior to participation.
Mutational analysis
Genomic DNA was obtained from peripheral blood lymphocytes of all patients by standard methods.
Genotyping was conducted at the Molecular Genetics Department of the Hospital Clínic; Barcelona, Spain.
CDKN2A
Primers sequences and protocol for polymerase chain reaction (PCR) amplification of the CDKN2A locus are previously described [19, 20]. Single strand conformation analysis was used to detect mutations. Three μL of denatured PCR product was combined with loading buffer and loaded into GeneGel Excel 12.5 acrylamide gels (Amersham Biosciences, Uppsala, Sweden) and run at 15°C for 2 hours. The gels were silver-stained as previously described [21]. DNA samples with abnormally migrating products were sequenced as following: PCR products were purified using the GFX PCR DNA and Gel Band purification kit (Amersham Biosciences) and automatically sequenced using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA) and an ABI3100 automatic sequencer (Applied Biosystems). The mutations described in this study were designated following the recommendations of den Dunnen and Antonarakis [22].
Sequencing of MC1R
MC1R genotyping was conducted employing PCR amplification of the 1180 bp coding region of MC1R followed by complete direct sequencing of the amplicons. The coding region of MC1R was amplified from 2 μl of total genomic DNA extracted from patient blood samples using PCR primers MC1R NT-F 5’- GCA GCA CCA TGA ACT AAG CA-3’ and MC1R CT-R 5’-CAG GGT CAC ACA GGA ACC A-3’. The reaction mix for PCR amplification contained 15 μl of PCR Master Mix (Promega Catalog # M7502), 2 μl of each Primer F/R (10 pmol/μl), 5% of DMSO (100%) and 7.5 μl of H2O for a final volume of 30 μl. The cycling conditions were set as follows: 5 min activation at 95 °C, followed by 35 cycles of 60 s at 94 °C; 60s at 55 °C; 3 min at 72°C; and finally 10 min at 72 °C. All products from two regions of the PCR were sequenced with the BigDye terminator v1.1/3.1 Cycle Sequencing Kit (Applied Biosystems, Inc.) and an ABI3100 sequence analyzer using sequence primers INT 1F: 5’-TACATCTCCATCTTCTACGC-3’; INT IR: 5’-GTGCTGAAGACGACACTG-3’. All sequences were analyzed and variants were detected.
Dermoscopic analysis
Digital dermoscopic images were captured with a digital camera (Olympus Camedia) equipped with a dermoscopical photo device (Dermlite® Photo) and with a digital epiluminescence microscopic system (Mole Max II; Derma Medical Systems, Vienna, Austria). We studied these digital dermoscopic images to diagnose lesions using the ABCD total dermoscopic score (TDS).
The ABCD rule of dermoscopy, introduced by Stolz and colleagues is a semiquantitative, mathematical approach that scores dermoscopic criteria identified in a lesion to determine the “total dermatoscopy score” (TDS). The dermoscopic criteria found to be significant cofactors for diagnosing melanoma in the ABCD rule are asymmetry (A), borders (B), colours (C), and different structural components (D). The different structural components are the following: structureless areas, pigment network, branched streaks, dots and globules. After identifying all of the criteria, the TDS is calculated by multiplying the points by conversion factors [23, 24]. The total score ranges from 1 to 8.9. A TDS <4.75 is indicative of the benign character of a melanocytic lesion. A TDS of 4.75–5.45 is suggestive but not diagnostic of melanoma. A lesion with a score in this range should be excised or followed up closely. A lesion with a total score of greater than 5.45 should be considered as melanoma.
The digital images provided for ABCD TDS calculation (containing in each case the clinical and dermoscopic images) were converted into JPEG format. These were kept separate from other files containing patient-related information such as gender, age at diagnosis, site of the skin lesion and histopathological diagnosis. The images were coded with unique identification numbers, and only gender, age at diagnosis and the site of the skin lesion were available to the observer. The sets of image files (containing both the clinical and dermoscopic images) were randomly ordered for evaluation. Assessors (F.C. and S.P) were blinded to patient identity and clinical information..
Statistical analysis
Statistical analysis was performed with the T student test to evaluate the differences between the ABCD TDS results. The Levene test was used to determine the equality of variances. Differences were considered to be statistically significant when P ≤ 0.05 (two-sided).
Results
We investigated nine patients, five women and four men, median age 39 years (range 23–53), with histologically confirmed melanoma (five in situ melanomas and four invasive with Clark level II and mean Breslow thickness of 0.5 mm). Eight individuals belonged to six melanoma-prone families and one individual had a personal history of multiple primary CMM without familial background. Table 1 summarizes the clinical features of the nine melanomas together with the histopathological diagnosis and molecular data. Five melanomas were located on the lower limb and four on the trunk. The diagnosis of melanoma and the decision to treat such lesions by surgical excision derived mainly from clinical considerations (history, changes during follow-up assessments) and dermoscopic characteristics.
Table 1.
Clinical, histological and molecular characteristics
| Case # | Gender | Age | Familial MM | Skin color | Hair color | Eye color | MM Localization | Histology | Breslow | Clark | CDKN2A mutation | RHC variants |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 47 | Yes | Very fair | Red | Green/Hazel | Extremities (Lower limb) | in situ | - | I | G101W | R151C, D294H |
| F | 27 | Yes | Fair | Blond | Green/Hazel | Trunk (Back) | Malignant blue nevus | - | - | 358DelG | R160W | |
| M | 39 | No | Fair | Brown | Green/Hazel | Trunk (Back) | SSMM | 0.5 | II | G101W | R151C | |
| F | 35 | Yes | Olive | Brown | Brown | Extremities (Lower limb) | SSMM | 0.5 | II | L65P | - | |
| F | 53 | Yes | Olive | Brown | Green/Hazel | Extremities (Lower limb) | in situ | - | I | G101W | - | |
| M | 46 | Yes | Very Fair | Red | Green/Hazel | Extremities (Lower limb) | SSMM | 0.5 | II | G101W | R151C (homozygous) | |
| M | 52 | Yes | Fair | Brown | Brown | Trunk (Back) | SSMM | 0.5 | II | R87W | - | |
| F | 23 | Yes | Very Fair | Red | Green/Hazel | Extremities (Lower limb) | in situ | - | I | G101W | R160W, R151C | |
| M | 35 | Yes | Fair | Red | Green/Hazel | Trunk (Lateral thorax) | in situ | - | I | 358DelG | R160W |
The most frequent CDKN2A mutation detected in this study was G101W (56%). Other mutations were 358delG (22%), L65P (11%) and R87W (11%) (Table 1). The most frequent MC1R variants found were R151C and R160W (Table 2). As has been previously observed in other studies, there was a strong association between the RHC variants R151C, R160W and D294H and red hair colour. All the subjects with two RHC variants had red hair. In one red-haired patient we found only one MC1R variant (Table 1). Melanomas were subdivided into three groups based on the number of MC1R red hair variants: noncarriers of red hair MC1R polymorphisms (three patients), carriers of one red hair MC1R polymorphism (three patients) and carriers of two red hair MC1R polymorphisms (three patients) (Fig. 1). None of the patients was consensus MC1R gene wild-type carrying at least one nonred hair polymorphism. Table 3 summarizes CDKN2A mutations in relation to MC1R red hair variants. MC1R variants were distributed between all CDKN2A mutations, but were more common for G101W because this is the most common mutation (Table 3).
Table 2.
Classification and prevalence of MC1R polymorphisms in the population studied (No Red hair color= nrhc / Red hair color =RHC)
| Frequency of MC1R variants | ||
|---|---|---|
| Polymorphism | n (%) | |
| V60L | nrhc | 1 (7.1) |
| nrhc | 2 (14.3) | |
| RHC | 4 (28.6) | |
| nrhc | 1 (7.1) | |
| RHC | 3 (21.4) | |
| RHC | 1 (7.1) | |
| nrhc | 2 (14.3) | |
| 14 (100) | ||
Figure 1.
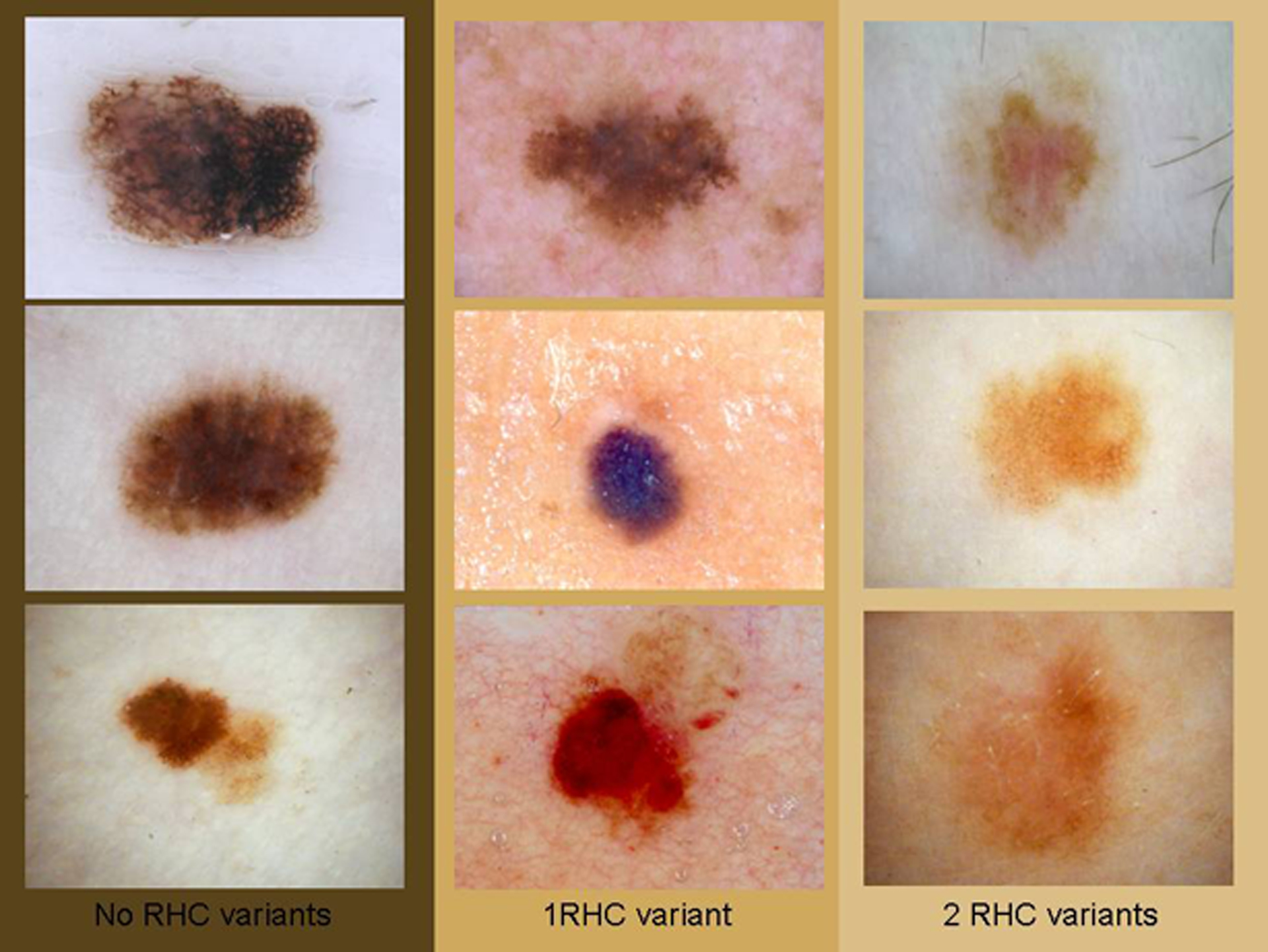
Dermoscopic images of the 9 melanomas classified according to the number of RHC variants in the MC1R gene. On the left, no RHC variants, in the middle, one RHC variant and on the right, 2 RHC variants. The number of colours and the number of structures decrease when the number of RHC variants increases. Upper left case #7, upper middle case #3, upper right case #6, middle left case #4, middle case #2, middle right case #8, lower left case #5, lower middle case #9 and lower right case #1.
Table 3.
Distribution of MC1R variations according to CDKN2A mutation
| 0 RHC n | 1 RHC n | 2 RHC n | TOTAL n (%) | |
|---|---|---|---|---|
| 358delG | 0 | 2 | 0 | 2 (22) |
| 1 | 0 | 0 | 1 (11) | |
| 1 | 1 | 3 | 5 (56) | |
| 1 | 0 | 0 | 1 (11) | |
| 3 (33.3) | 3 (33.3) | 3 (33.3) | 9 (100) |
Dermoscopic classification
The mean ABCD Total Dermoscopic Score (TDS) was significantly higher in non-carriers of red-hair MC1R polymorphisms compared to carriers of two or more MC1R gene red-hair variants (6.8±0.4 vs. 4.4±0.9 p=0.014; Table 4). No statistically significant differences were obtained in asymmetry and borders between the two groups. The groups were mainly distinguished by significant differences in the number of colours (1.8±0.3 vs. 1.0±0.0 p=0.007) and in the number of structures (2.0±0.0 vs. 1.2±0.3 p=0.007).
Table 4.
Mean TDS scores in melanomas from patients according MC1R red hair variants (RHC)
| ABCD TDS | ||||
|---|---|---|---|---|
| 0 RHC (mean) | 1 RHC (mean) | 2 RHC (mean) | p value | |
| Asymmetry | 2.6 | 2.2 | 2.2 | 0.374 |
| 0.4 | 0.2 | 0.03 | 0.157 | |
| 1.8 | 1.8 | 1.0 | 0.007 | |
| 2.0 | 2.0 | 1.2 | 0.007 | |
| 6.8 | 6.2 | 4.4 | 0.014 | |
Interestingly, we observed atypical vessels in two of the three melanomas in carriers of two red hair polymorphisms. The identification of changes during follow-up assessments of the lesions was a very important factor in our diagnosis of melanoma and our decision to remove these three melanomas.
Discussion
The aim of the present study was to examine a possible correlation between MC1R gene variants and particular dermoscopic features during the evaluation of early melanomas among Spanish carriers of CDKN2A gene mutations. To the best of our knowledge this is the first study that correlates dermoscopic findings in melanoma with MC1R variants in carriers of mutations in CDKN2A.
Red hair colour is associated with certain MC1R variants that increase the ratio of pheomelanin/eumelanin produced by melanocytes. As has been previously observed in other studies, we found a strong association between the red hair colour (RHC) variants R151C, R160W, and D294H and red hair colour in our series of Spanish patients. In our study all the subjects with two RHC variants had red hair (three of four red-haired patients). RHC variants of MC1R are necessary but not sufficient for this pigmentary phenotype, and other studies [11] have indicated that not all individuals with two red hair MC1R variants have red hair. In our study, the remaining patient showed just one RHC MC1R variant and has been considered as red haired, although he had primarily blond hair with shades of red.
Melanoma with predominance of light colours is described as hypopigmented. Hypomelanotic melanoma can mimic benign and malignant variants of both melanocytic and non-melanocytic lesions [25]. It is not known whether hypomelanotic melanoma could be more frequent in individuals carrying MC1R variants. We used the ABCD rule algorithm of dermoscopy to evaluate the total dermatoscopy score (TDS) to classify these melanocytic lesions as malignant, suspicious or benign. We found that in Spanish melanoma patients, carriers of a CDKN2A mutation, with two or more MC1R red-hair polymorphisms (R160W, R151C, D294H), the melanomas had a lower ABCD TDS (mean 4.4) than CMMs of non-carriers of RHC variants. An ABCD TDS score <4.75 is suggestive of a benign melanocytic lesion. This means that if we had relied only on the the ABCD rule of dermoscopy, these early melanomas would not have been identified as malignant lesions. In fact all melanomas in carriers of two or more MC1R gene RHC variants in our series were originally misclassified as benign by adherence to the ABCD rule of dermoscopy alone. The main reason for the low TDS was that these melanomas displayed a relatively low number of colours, and consequently a lower number of structures. However in hypopigmented melanomas, it is easier to see atypical vessels, and they were, in fact, present in two from our series.
Diagnostic algorithms can be helpful, particularly for clinicians with less experience in dermoscopy. Those algorithms with highest sensitivity will be the most useful in a primary care setting and will result in fewer melanomas being missed. In our study, all melanomas in carriers of two or more MC1R gene RHC variants were missed by the ABCD rule of dermoscopy. Thus this algorithm, which may be helpful to examiners with limited experience, should not be strictly applied in subjects carrying MC1R variants. These patients may require referral to dermatologists more experience in the diagnosis of melanocytic lesions.
In our center we use a method of digital image follow-up consisting of the combination of total body-photography and digital dermoscopy to assess stability or instability of atypical melanocytic lesions in high-risk patients for melanoma. This method was shown to be useful for the detection of early melanoma in a previously reported study [26]. This digital follow-up examination should be performed every 3–6 months to detect new lesions or any modification in shape, colour or other dermoscopic features in the pre-existing lesions.
The role of clinical history and findings as important cofactors in dermoscopy has been already described in literature [27,28]. It is fundamental to make attention to all signs that could be useful for melanoma diagnosis among patients with MC1R gene RHC variants, considering their high risk of developing a “tricky-to-diagnose” melanomas. The history of changes in a lesion, the presence of vessels under dermoscopy, the presence of regression structures and the evidence of changes in digital follow-up may be crucial in the early identification of melanoma in red hair people.
Despite the small number of patients studied, our findings give interesting results which may have implications for the early detection of melanomas in patients carrying MC1R variants and confirm the important roles of clinical expertise and of an intensive follow up for some lesions in these patients. Additional and larger studies are needed to confirm our findings and to explore the molecular mechanisms that contribute to the relationship between MC1R variants and the morphological appearance of pigmented lesion in these patients.
In conclusion, this study explored a possible correlation between MC1R gene polymorphism status and clinical findings as inferred by dermoscopy algorithms, among patients carrying CDKN2A gene mutations. In patients with two RHC MC1R polymorphisms, CMM may have fewer colours and structures, and this may lead to some false-negative diagnoses if relying strictly on results from the dermoscopic algorithm TDS. For this reason, clinical data, short-term digital image follow up, and the use of complementary dermoscopic information such as the presence of atypical vessels could help expert clinicians in detecting possible difficult-to-diagnose melanoma in these patient subsets. Use of the TDS as a referral tool should be strongly discouraged in high-risk patients and, in any case, they should be referred to expert practitioners.
Acknowledgements
This work was partially funded by grants 03/0019, 06/0265 V2003-REDC03/03 and /07 from Fondo de Investigaciones Sanitarias; grant RO-1 CA 83115 (fund 538226 from National Cancer Institute); and GenoMEL consortium actually founded by CE, Net of Excellence.
This work was developed in the Melanoma Unit of Hospital Clinic I Provincial de Barcelona, IDIBAPS.
Footnotes
All authors declare no conflicts of interest or financial disclosures.
IDIBAPS is Institut de investigacions biomèdiques August Pi Sunyer
References
- 1.Bosetti C, La Vecchia C, Naldi L, Lucchini F, Negri E, Levi F. Mortality from cutaneous malignant melanoma in Europe. Has the epidemic levelled off? Melanoma Res. 2004; 14: 301–9. [DOI] [PubMed] [Google Scholar]
- 2.de Vries and Coebergh 2004 Cutaneous malignant melanoma in Europe. Eur J Cancer. 2004; 40: 2355–66. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein AM, Tucker MA. Etiology, epidemiology, risk factors, and public health issues of melanoma. Curr Opin Oncol. 1993; 5: 358–363. [DOI] [PubMed] [Google Scholar]
- 4.Sturm RA, Box NF and Ramsay M Human pigmentation genetics: the difference is only skin deep. Bioessays. 1998; 20, 712–721. [DOI] [PubMed] [Google Scholar]
- 5.Rees JL. The melanocortin1 receptor (MC1R): more than just reed hair. Pigment Cell Res. 2000; 13: 135–40. [DOI] [PubMed] [Google Scholar]
- 6.Flanagan N, Healy E, Ray A, et al. Pleiotropic effects of the melanocortin 1 receptor (MC1R) gene on human pigmentation. Hum Mol Genet. 2000; 9: 2531–7. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy C, ter Huurne J, Berkhout M, et al. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J Invest Dermatol. 2001; 117: 294–300. [DOI] [PubMed] [Google Scholar]
- 8.Duffy DL, Box NF, Chen W, et al. Interactive effects of MC1R and OCA2 on melanoma risk phenotypes. Hum Mol Genet 2004; 13: 447–61. [DOI] [PubMed] [Google Scholar]
- 9.Box NF, Duffy DL, Chen W, et al. MC1R genotype modifies risk of melanoma in families segregating CDKN2A mutations. Am J Hum Genet 2001; 69: 765–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Velden PA, Sandkuijl LA, Bergman W, et al. Melanocortin-1 receptor variant R151C modifies melanoma risk in Dutch families with melanoma. Am J Hum Genet 2001; 69: 774–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldstein AM, Landi MT, Tsang S, Fraser MC, Munroe DJ, Tucker MA. Association of MC1R variants and risk of melanoma in melanoma-prone families with CDKN2A mutations. Cancer Epidemiol Biomarkers Prev. 2005; 14: 2208–12 [DOI] [PubMed] [Google Scholar]
- 12.Rees JL. Genetics of hair and skin color. Annu Rev Genet. 2003; 37: 67–90. [DOI] [PubMed] [Google Scholar]
- 13.Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet. 2003; 112: 387–399. [DOI] [PubMed] [Google Scholar]
- 14.Benelli C, Roscetti E, Pozzo VD et al. The dermoscopic versus the clinical diagnosis of melanoma. Eur J Dermatol. 1999; 9: 470–6. [PubMed] [Google Scholar]
- 15.Bafounta ML, Beauchet A, Aegerter P et al. Is dermoscopy (epiluminescence microscopy) useful for the diagnosis of melanomas? Results of a meta-analysis using techniques adapted to the evaluation of diagnostic tests. Arch Dermatol 2001; 137: 1343–50. [DOI] [PubMed] [Google Scholar]
- 16.Ascierto PA, Palmieri G, Celentano E, et al. Sensitivity and specificity of epiluminescence microscopy: evaluation on a sample of 2731 excised cutaneous pigmented lesions. Br J Dermatol. 2000; 142: 893–8. [DOI] [PubMed] [Google Scholar]
- 17.Skavara H, Teban L, Fiebiger M, Binder M, Kittler H. Limitations of dermoscopy in the recognition of melanoma. Arch Dermatol. 2005; 141: 155–60. [DOI] [PubMed] [Google Scholar]
- 18.Pizzichetta MA, Talamini R, Stanganelli I, Puddu P, Bono R, Argenziano G, Veronesi A, Trevisan G, Rabinovitz H, Soyer HP. Amelanotic/hypomelanotic melanoma: clinical and dermoscopic features. Br J Dermatol. 2004; 150: 1117–24. [DOI] [PubMed] [Google Scholar]
- 19.Puig S, Ruiz A, Castel T, et al. : Inherited susceptibility to several cancers but absence of linkage between dysplastic nevus syndrome and CDKN2A in a melanoma family with a mutation in the CDKN2A (p16INK4A) gene. Hum Genet. 1997; 101: 359–64. [DOI] [PubMed] [Google Scholar]
- 20.Puig S, Malvehy J, Badenas C, Ruiz A, Jimenez D, Cuellar F, Azon A, Gonzalez U, Castel T, Campoy A, Herrero J, Marti R, Brunet-Vidal J, Mila M. Role of the CDKN2A locus in patients with multiple primary melanomas. J Clin Oncol. 2005; 23: 3043–51 [DOI] [PubMed] [Google Scholar]
- 21.Badenas C, Torra R, Lucero L, et al. : Mutational analysis within the 3_ region of the PKD1 gene. Kidney Int. 1999; 55: 1225–33. [DOI] [PubMed] [Google Scholar]
- 22.den Dunnen JT, Antonarakis E: Mutation nomenclature extensions and suggestions to describe complex mutations: A discusssion. Hum Mutat. 2000; 15: 7–12. [DOI] [PubMed] [Google Scholar]
- 23.Stolz W, Riemann A, Cognetta AB, et al. ABCD rule of dermatoscopy: a new practical method for early recognition of malignant melanoma. Eur J Dermatol. 1994; 4: 521–7. [Google Scholar]
- 24.Nachbar F, Stolz W, Merkle T, et al. The ABCD rule of dermatoscopy: high prospective value in diagnosis of melanocytic lesions. J Am Acad Dermatol. 1994; 30: 551–9. [DOI] [PubMed] [Google Scholar]
- 25.Giuliano AE, Cochran AJ, Morton DL. Melanoma from unknown primary site and amelanotic melanoma. Semin Oncol. 1982; 9: 442–7. [PubMed] [Google Scholar]
- 26.Malvehy J, Puig S. Follow-up of melanocytic skin lesions with digital total-body photography and digital dermoscopy: a two-step method. Clin Dermatol 2002; 20: 297–304. [DOI] [PubMed] [Google Scholar]
- 27.Puig S, Argenziano G, Zalaudek I, Ferrara G, Palou J, Massi D, Hofmann-Wellenhof R, Soyer HP, Malvehy J. Melanomas that failed dermoscopic detection: a combined clinicodermoscopic approach for not missing melanoma. Dermatol Surg. 2007; 33(10):1262–73. [DOI] [PubMed] [Google Scholar]
- 28.Ascierto PA, Palmieri G, Botti G et al. Melanoma Cooperative Group. Early diagnosis of malignant melanoma: proposal of a working formulation for the management of cutaneous pigmented lesions from the Melanoma Cooperative Group. Int J Oncol 2003; 22:1209–15. [PubMed] [Google Scholar]


