Abstract
Purpose
Tissue-based gene expression classifiers (GECs) may assist with management decisions in patients with newly diagnosed prostate cancer. We sought to assess the current use of GEC tests and determine how the test results are associated with primary disease management.
Methods
In this observational study, patients diagnosed with localized prostate cancer were tracked through the Michigan Urological Surgery Improvement Collaborative registry. The utilization and results of three GECs (Decipher Prostate Biopsy, Oncotype DX Prostate, and Prolaris) were prospectively collected. Practice patterns, predictors of GEC use, and effect of GEC results on disease management were investigated.
Results
Of 3,966 newly diagnosed patients, 747 (18.8%) underwent GEC testing. The rate of GEC use in individual practices ranged from 0% to 93%, and patients undergoing GEC testing were more likely to have a lower prostate-specific antigen level, lower Gleason score, lower clinical T stage, and fewer positive cores (all P < .05). Among patients with clinical favorable risk of cancer, the rate of active surveillance (AS) differed significantly among patients with a GEC result above the threshold (46.2%), those with a GEC result below the threshold (75.9%), and those who did not undergo GEC (57.9%; P < .001 for comparison of the three groups). This results in an estimate that, for every nine men with favorable risk of cancer who undergo GEC testing, one additional patient may have their disease initially managed with AS. On multivariable analysis, patients with favorable-risk prostate cancer who were classified as GEC low risk were more likely to be managed on AS than those without testing (odds ratio, 1.84; P = .006).
Conclusion
There is large variability in practice-level use and GEC tests ordered in patients with newly diagnosed, localized prostate cancer. In patients with clinical favorable risk of cancer, GEC testing significantly increased the use of AS. Additional follow-up will help determine whether incorporation of GEC testing into initial patient care favorably affects clinical outcomes.
INTRODUCTION
Accurate risk stratification is critical for patients with localized prostate cancer given the diverse management options available. Although active surveillance (AS) is now recognized as a preferred treatment option for patients with low-risk prostate cancer, more than one third of patients will experience disease progression that requires treatment.1-3 Men with favorable intermediate-risk prostate cancer are rarely entered onto AS, yet growing data suggest that some may be safely treated with AS to avoid immediate radical therapy.4
Multiple new tissue biomarkers have surfaced to aid in risk stratification of newly diagnosed patients, with a goal of improving prognostic accuracy compared with routine clinicopathologic variables. On the basis of multiple retrospective development and validation studies, there are now three widely available tissue-based gene expression classifiers (GECs) for use in this setting: Prolaris (Myriad Genetics, Salt Lake City, UT), Oncotype Dx Prostate (Genomic Health, Redwood City, CA), and Decipher Prostate Biopsy (GenomeDx, Vancouver, Canada).5-12 These tools are now included in the National Comprehensive Cancer Network (NCCN) guidelines as options to aid in decision making for men with newly diagnosed localized prostate cancer, although other national guidelines caution against their routine use.13,14 Each of these tests are RNA-based prognostic biomarkers that analyze a distinct multigene panel that predicts oncologic end points, which range from the likelihood of harboring unrecognized high-grade disease to death as a result of prostate cancer at 10 years. Although published data have demonstrated the prognostic utility of these tests compared with clinicopathologic data alone,7,9,11,12 there is limited understanding of how these tools influence treatment decisions in the contemporary setting.
One unique aspect of these tests is the continuous scale used to report risk, whether for near-term adverse pathology end points or more distant survival end points. Although proposed thresholds that separate low-risk from high-risk patients are provided on each of the test reports, these thresholds have not been prospectively validated. This is in contrast to breast cancer, for which analogous tests have become incorporated into guidelines as a result of multiple prospective trials that validate discrete cut points for management decisions.15-17 Because the emergence of multiple GEC tests—each with various measurement scales, thresholds, outcomes, and reporting tools—is recent, it is unclear how these tests affect real-world management decisions.
To address this complex issue in localized prostate cancer, we sought to perform a prospective clinical utility study by tracking the use of GECs in patients with newly diagnosed prostate cancer and comparing treatment decisions according to GEC result. To capture GEC use and subsequent management across a broad cohort of patients, this study was performed through the Michigan Urological Surgery Improvement Collaborative (MUSIC), a consortium of more than 90% of urology practices in the state of Michigan. We hypothesized that GEC use would be highly variable and that results would correlate closely with subsequent treatment decisions, specifically that rates of AS would be higher in GEC-defined low-risk patients.
METHODS
Study Population
Beginning in January 2017, the use and results of three GEC tests (Prolaris, OncotypeDx Prostate, and Decipher Biopsy) were prospectively tracked within MUSIC. The MUSIC consortium was established in 2012 and is composed of 44 academic and community practices with more than 250 urologists across Michigan. In each participating practice, trained abstractors prospectively enter a standardized set of demographic and pathologic data for every patient with a new prostate cancer diagnosis into an electronic clinical registry. Abstractors also enter data related to treatment—radical prostatectomy, radiotherapy, or other (primary androgen deprivation therapy, high-intensity focused ultrasound, cryotherapy, or other ablative technologies)—and follow-up data. Each site obtains regulatory exemption from local institutional review boards to participate in MUSIC, and funding is provided by Blue Cross Blue Shield of Michigan.
In this analysis, we included all men who were newly diagnosed with clinically localized prostate cancer between January and September 2017. Subsequent treatment decisions (AS, prostatectomy, or radiotherapy) were tracked until the analytic data set was locked on April 1, 2018.
Clinical and Pathologic Variable Definitions
Data elements collected included age, ethnicity, prostate-specific antigen (PSA) level, number of positive cores from biopsy, primary and secondary Gleason patterns, prostate volume, clinical T stage, body mass index (BMI), and comorbidity scores. Prostate magnetic resonance imaging (MRI) was categorized according to maximum prostate imaging and reporting data system (PIRADS) version 2 score as reassuring (PIRADS 1 to 3) or nonreassuring (PIRADS 4 to 5). Using standard clinicopathologic parameters, patients were categorized according to the favorable-risk prostate cancer stratification that has been widely disseminated in the MUSIC consortium as a way to identify patients who may be most appropriate for AS.18 Favorable-risk prostate cancer was defined by a biopsy Gleason score of 6 or low-volume, Gleason 3 + 4 = 7 disease (defined as three or fewer cores positive and no core with > 50% cancer volume). Patients were separately grouped for analysis using the NCCN risk classification system.13
GEC Data
For the three GEC tests, individual thresholds to separate low versus high risk were defined in accordance with published data and test reports for each of these assays.19 A summary of the three assays and cut points used to define GEC low-risk disease are listed below:
Decipher Biopsy (GenomeDx Biosciences): Microarray platform, with score composed of 22 RNA biomarkers reported on a scale of 0 to 1. Threshold for low versus high risk is 0.45.
Oncotype Dx Prostate (Genomic Health): Quantitative real-time polymerase chain reaction platform, with score composed of 17 genes and reported on a scale of 0 to 100. Threshold for low versus high risk is the probability of adverse surgical pathology greater than 20%.
Prolaris (Myriad Genetics): Quantitative real-time polymerase chain reaction platform, with score composed of 46 genes and reported on a scale of 1 to 10. Threshold for low versus high risk is 10-year mortality risk of 3% or greater.
For patients who had more than one test performed (n = 13), only the initial test was included in the analysis.
End Points
The primary study outcome was GEC testing status (did not undergo GEC testing v underwent GEC testing with result below threshold v underwent GEC testing with result above threshold). The secondary outcome was the initial disease management strategy selected (AS v definitive treatment v no treatment). Each patient’s primary disease management strategy was determined 6 months after diagnosis and categorized as definitive treatment or no treatment. Patients whose data were lost to follow-up, deceased, or switched practices for follow-up care were excluded from the evaluation of treatment because MUSIC no longer collected treatment information. Patients who were still missing treatment data at the time of analysis because of a lag in data entry were also excluded.
Statistical Analyses
Patient characteristics were compared between those who did and did not undergo GEC testing using χ2 tests for the overall cohort as well as for patients withfavorable-risk prostate cancer. The rate of GEC testing was summarized and compared at the practice level. The proportion of patients with a test result above the threshold was then summarized by the type of the test used (Prolaris v Oncotype DX v Decipher). The primary management strategy was then compared across three groups defined by the performance and result of the GEC test: no test, GEC test below the threshold, and GEC test above the threshold. The number needed to test calculations are described in the Appendix. A mixed-effects logistic regression model was performed in the favorable-risk subgroup to assess the association between GEC testing and adoption of AS. The model included the performance and result of the GEC test (above or below the threshold) as the primary predictor (with no test as the reference group). The model also adjusted for patient and practice-level characteristics, including patient age, ethnicity, PSA level, biopsy Gleason score, clinical T stage, comorbidity, BMI, practice size, and practice type (academic v private). The model also included random intercepts for practices to account for between-practice variability in the use of AS. All covariates were selected in advance. The statistical analyses were performed in SAS version 9.4 (SAS Institute, Cary, NC), and statistical significance was set at .05.
RESULTS
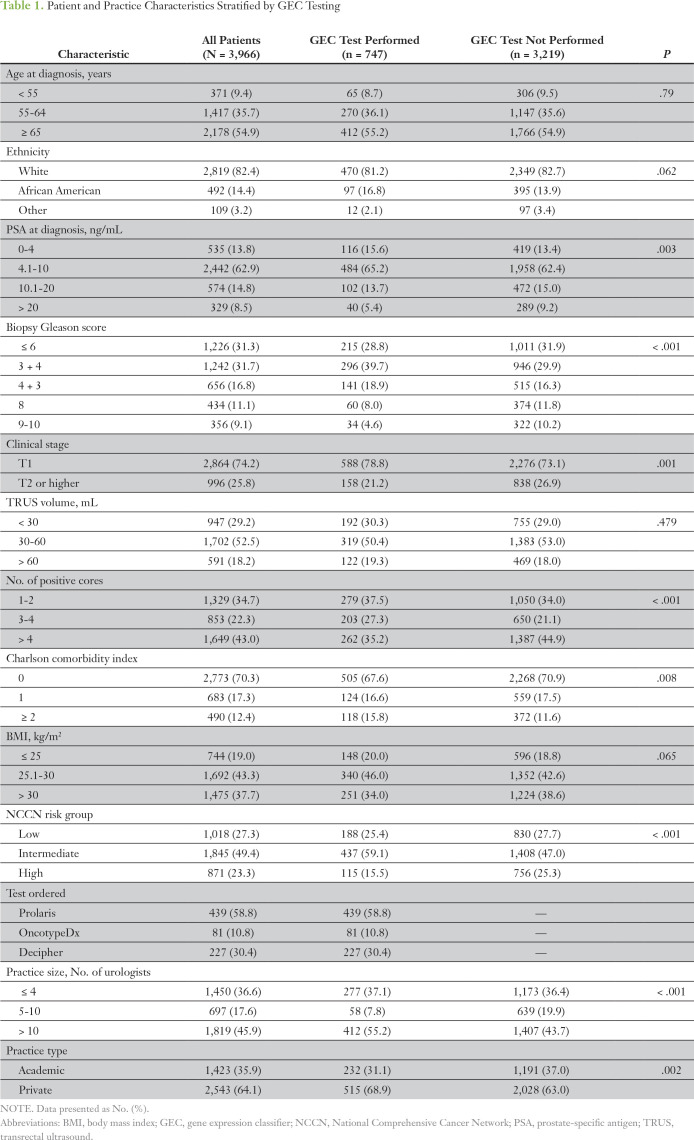
A total of 3,966 men were diagnosed with localized prostate cancer during the study period and were included in the study cohort. Of these, 747 patients (18.8%) underwent biopsy GEC testing. Table 1 lists the patient characteristics stratified by GEC use. Patients who underwent GEC testing were more likely to have a lower PSA level, lower biopsy Gleason score, fewer total positive cores, and a lower clinical T stage (P < .01 for each). In addition, patients with GEC testing had more comorbidities and were more likely to be treated at larger urology practices as well as private rather than academic practices (P < .01 for each). The median time to GEC testing after diagnosis was 0.4 months (interquartile range, 0.2 to 0.9 months), and the median time to MRI after diagnosis was 1.7 months (interquartile range, 1.1 to 2.7 months).
Table 1.
Patient and Practice Characteristics Stratified by GEC Testing
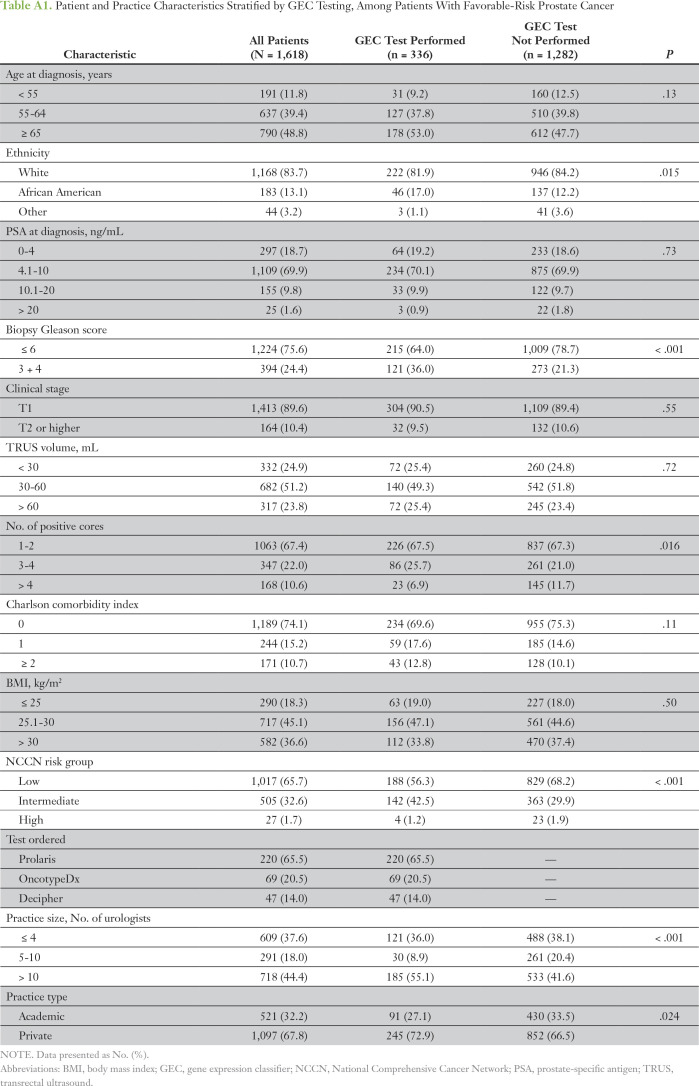
Of the 1,618 patients (40.8%) who were diagnosed with favorable-risk prostate cancer on the basis of MUSIC criteria, 336 (20.8%) underwent testing. Among this subgroup, patients who underwent a GEC test were more likely to be African American and to have a higher biopsy Gleason score (3 + 4 v 6), fewer positive cores, and NCCN intermediate- rather than low-risk cancer (P < .05 for each; Appendix Table A1). Similar to the overall cohort, patients with favorable-risk prostate cancer who underwent a GEC test were more likely to be treated at larger and private urology practices (P < .05 for each).
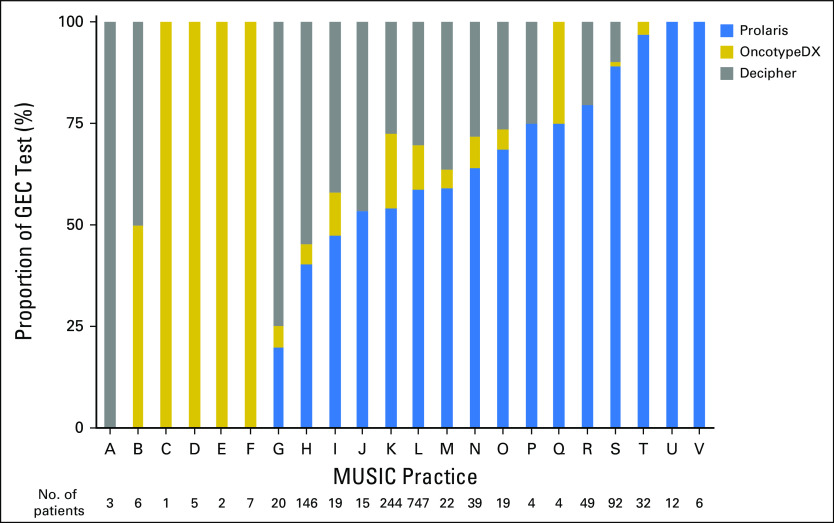
Rates of GEC testing at each of the 44 individual practices were compared (Fig 1). Rates of GEC testing at each practice ranged from 0% to 93% (P < .001). Seven practices ordered a GEC test on more than 50% of patients with newly diagnosed prostate cancer.
Fig 1.
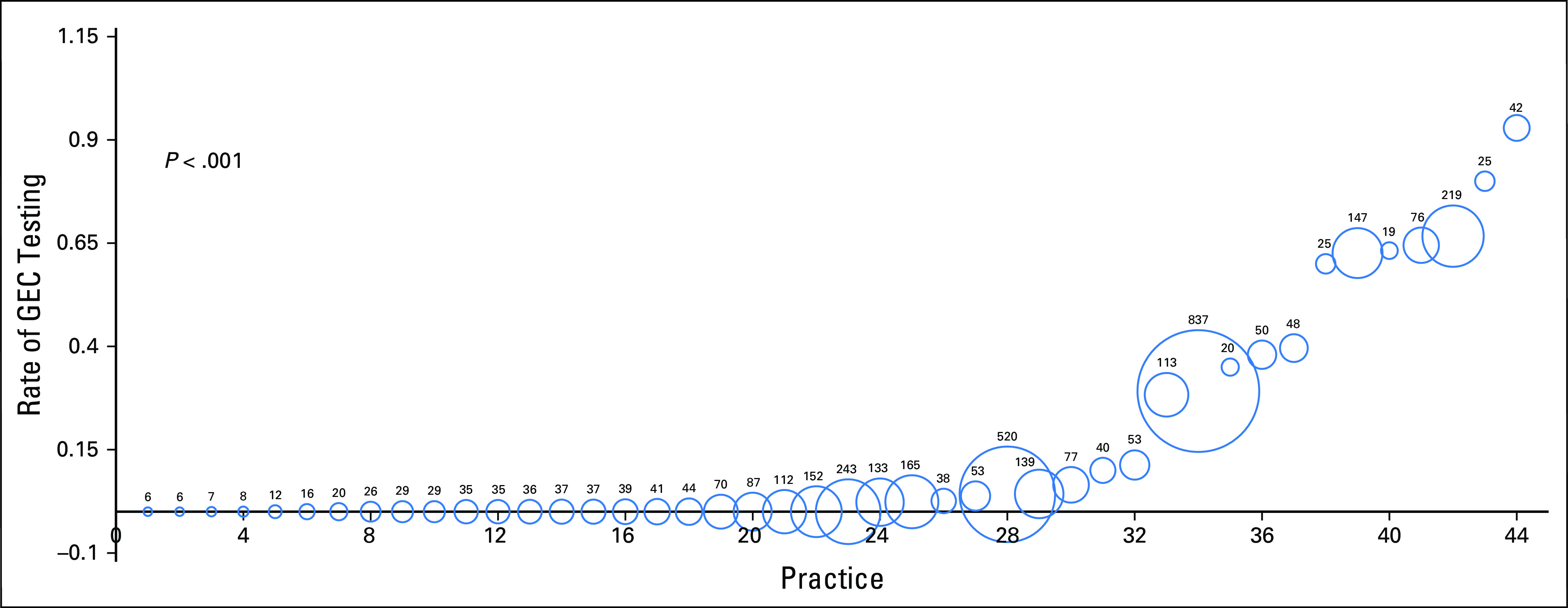
Practice-level rate of genomic test after diagnosis: rate of gene expression classifier (GEC) testing in newly diagnosed men with prostate cancer at each practice (practices are numbered 1 through 44). Area and number inside each sphere represent the total number of diagnosed patients with prostate cancer at each practice during the time period of this study.
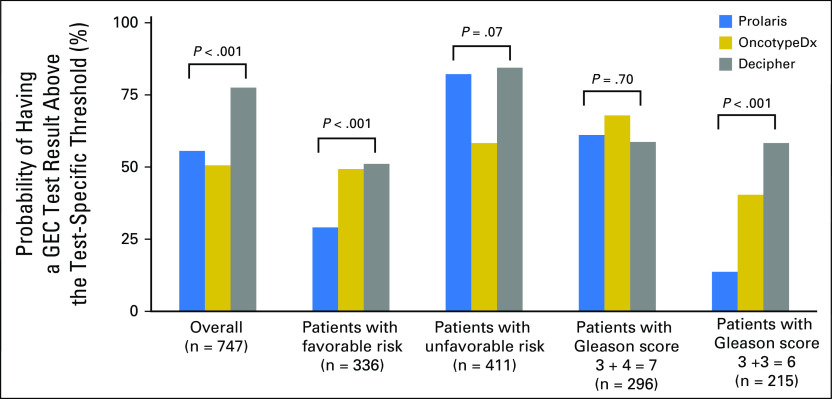
Among the 747 patients who underwent GEC testing, 439 (59%) were tested with Prolaris, 81 (11%) with OncotypeDx, and 227 (30%) with Decipher. The relative use of these three assays for those practices that ordered at least one test is shown in Appendix Fig A1. Using the prespecified thresholds, the probability of a high- versus low-risk test result differed significantly depending on the test (Fig 2). For example, less than 30% of patients with favorable-risk prostate cancer tested with Prolaris had a result above the proposed AS threshold compared with approximately 50% of patients tested with OncotypeDx or Decipher (P < .001). This was even more pronounced in the subgroup with a Gleason score of 6: 14% of patients tested with Prolaris had a GEC score above the low-risk threshold versus 40% and 58% of patients tested with OncotypeDx and Decipher, respectively (P < .001).
Fig 2.
The percentage of patients with a gene expression classifier (GEC) test result above the test-specific threshold is shown according to individual GEC test. The overall rates and rates within patient subgroups are presented separately.
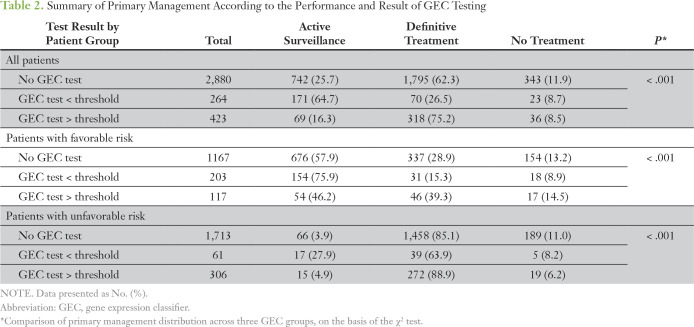
Of the initial 3,966 patients, six patients died before treatment, 16 were lost to follow-up, and 157 switched practices during follow-up care. These patients were missing subsequent treatment data. Another 220 patients were also missing treatment information because of a lag in data entry. Therefore, a total of 3,567 patients were included in the evaluation of treatment decision (Table 2). Of these, 687 (19%) underwent GEC testing, and, overall, rates of AS differed significantly among patients with a GEC result above the threshold (16.3%), those with a GEC test less than or equal to the threshold (64.7%), and those who did not undergo GEC testing (25.7%; P < .001). GEC testing was performed in 367 (17.6%) of 2,080 patients with unfavorable risk; 61 of these men (16.6%) had a low-risk result, and 17 (27.9%) elected AS for initial management.
Table 2.
Summary of Primary Management According to the Performance and Result of GEC Testing
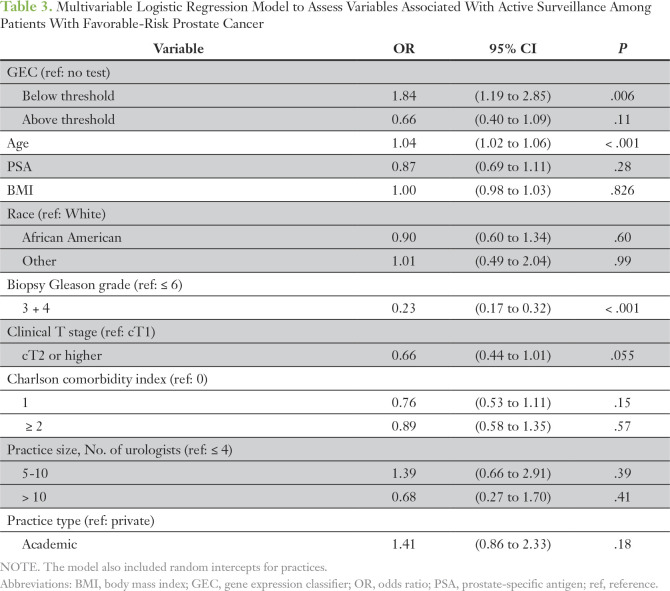
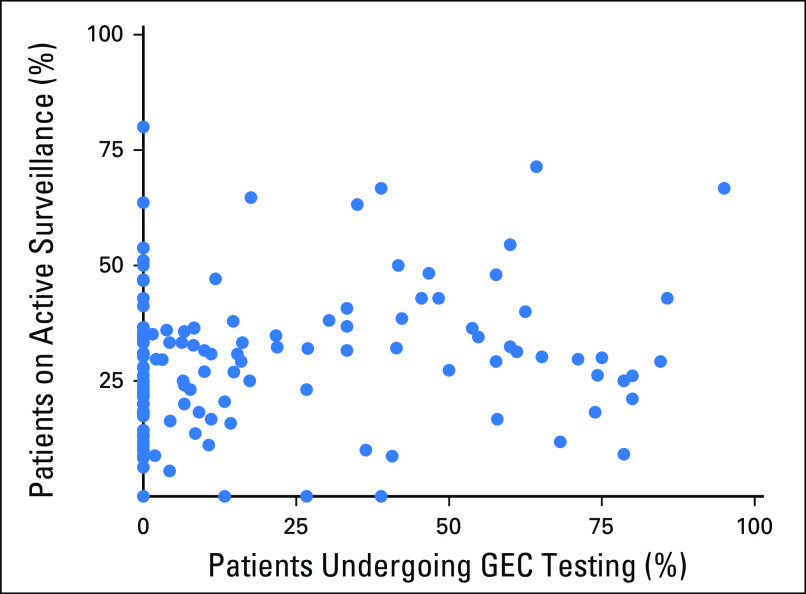
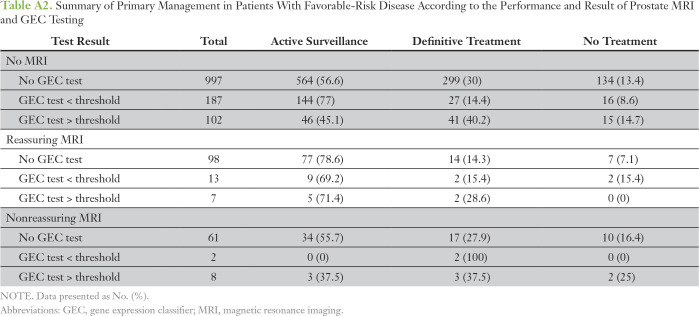
Of the 1,487 patients with favorable-risk disease but with treatment data, 320 (22%) underwent GEC testing. The rate of AS was 57.9% among those without a GEC test compared with 75.9% among those with a GEC result below the threshold and 46.2% among those with a test result above the threshold (P < .001). Of the 1,475 patients favorable-risk disease with evaluable MRI data, 189 (13%) underwent prostate MRI in the first 6 months after diagnosis. Only 30 patients with favorable-risk disease underwent both MRI and GEC testing; this represented 16% of patients who underwent MRI and 9% of patients who underwent GEC testing (Appendix Table A2). When analysis was controlled for patient and practice-level characteristics, patients with a GEC test result below the threshold were significantly more likely than those without a GEC test to receive AS (odds ratio, 1.84; P = .006; Table 3). Rates of GEC testing according to provider (n = 130) also were assessed in patients with favorable-risk disease and ranged from 0% to 95%. GEC testing and use of AS appeared to be independent of each other at a provider level, which indicated the absence of any provider-specific effect (correlation coefficient r = 0.16; Appendix Fig A2).
Table 3.
Multivariable Logistic Regression Model to Assess Variables Associated With Active Surveillance Among Patients With Favorable-Risk Prostate Cancer
With 63% of patients with favorable-risk prostate cancer harboring GEC low-risk disease in this cohort (203 of 320 who underwent GEC testing), these data suggest that, for every nine patients with favorable-risk disease who undergo testing, one patient will receive AS who would have otherwise undergone primary therapy. Conversely, 117 (37%) of 320 GEC-tested patients with favorable-risk prostate cancer were considered molecularly high risk, and 46 (39%) of these 117 patients selected initial definitive management compared with 337 (29%) of 1,167 untested patients. This suggests that, for every 26 patients with favorable-risk prostate cancer who undergo testing, one patient will undergo primary therapy rather than AS on the basis of testing results (Appendix).
DISCUSSION
AS is a standard of care management approach for men with low-risk prostate cancer, and the use of AS has steadily increased during the past decade.20,21 Nonetheless, adverse reclassification is common in patients with low-risk disease, and AS carries increased risks for patients with higher-risk prostate cancer.3,22 Accurate disease risk stratification is critical for patients with newly diagnosed prostate cancer, yet the most commonly used risk classification systems, such as those by D’Amico et al (1998)23 and NCCN,24 include a broad range of risk within defined risk categories. More granular clinical tools to quantify risk, including the Cancer of the Prostate Risk Assessment25 score and Memorial Sloan Kettering Cancer Center26 nomograms, can be affected greatly by variability among pathologists and are still somewhat limited in their prognostic accuracy.27,28 Emerging tools, such as multiparametric MRI (mpMRI), may improve the initial detection of more aggressive tumors, but variability in the performance and interpretation of mpMRI may limit its generalizability for now.29 GEC testing provides an additional objective tool for risk stratification. Prospective data suggest that up to 67% of patients may be reclassified into different risk categories when genomic risk is considered in addition to conventional clinicopathologic parameters of risk.11 Despite this, limited data describe the use of GEC testing in this setting, and the effect of these tools on clinical decision making has not been well established.
A number of unique findings are presented in this study, because this is the first study, to our knowledge, to report on a prospective observational cohort of patients with and without GEC testing. Approximately 20% of patients diagnosed during the study period underwent a GEC test, and there was wide variation in testing patterns across different urology practices in the state of Michigan. Fitting with the current approval details for each test, which are primarily for men with favorable-risk disease, these tests were more likely to be obtained in the setting of clinically less aggressive disease. Patients were more likely to undergo testing if seen at a larger urology practice (with more than 10 providers), which may relate to incorporation of GEC testing into clinical care pathways at some of these practices.
The analysis of the primary end point, clinical utility of GEC testing in favorable-risk prostate cancer, provides key information about how these tests affect treatment decisions. This is one of the key measures used by the Molecular Diagnostic Services program developed by Palmetoto GBA to determine Medicare coverage, and prospective data that compare tested and untested patients. We observed an increase in AS among patients with a GEC test result below threshold compared with untested patients (76% v 58%) in this subgroup. In the favorable-risk subgroup, the data indicate that one additional patient underwent AS instead of surgery or radiation for every nine patients tested and that one additional patient underwent primary therapy rather than AS for every 26 patients tested. In the unfavorable-risk group, utility was less apparent: 17% of tested patients had GEC low risk disease, and only 28% of these men received AS.
Finally, it is important to note that meaningful differences existed among the three tests. Although we did not have sufficient data to compare test results for patients with more than one GEC assay performed, we found substantial variability in the likelihood of GEC high- versus GEC low-risk disease, depending on the test performed. For example, in the subgroup with a Gleason score of 3 + 3 = 6, 13.7% of patients tested with Prolaris had results that were considered above the threshold, whereas 58.3% were above the threshold as reported by Decipher. Importantly, although most of these test-specific thresholds have not been validated, they are included in test reports provided to patients and providers.30 This issue highlights the challenges of applying thresholds to tests developed and validated for assessment along a continuum of risk and also emphasizes the need for research to compare the predictive accuracy of each of these tests. The current thresholds for OncotypeDx and Prolaris could lower AS rates in patients with low-risk disease if strictly followed. Although that was not the case in this cohort, additional study about appropriate and consistent thresholds for these tests is paramount.
It is important note key limitations to this study. Some patients could have undergone GEC testing outside of a MUSIC urology practice, such as by a radiation oncologist, and this would only be captured in the registry if the test results were provided to the primary urologist. However, any contamination of the non-GEC cohort with GEC-tested patients would likely bias these findings toward the null. Also, a full understanding of the clinical utility of these assays requires a prospective, randomized controlled trial, because there may be unmeasured differences in this cohort between patients who did and did not undergo GEC testing. An example of such a trial is the ongoing Genomics in Michigan Impacting Observation or Radiation (G-MINOR) trial in the postprostatectomy setting.31 However, in the absence of a similar trial in patients with newly diagnosed disease, prospective observational data can provide critical insights into how these tests are guiding clinical decisions.
These limitations notwithstanding, strengths of this study include prospective data collection on nearly 4,000 consecutively enrolled patients with newly diagnosed prostate cancer. This cohort represents a broad geographic region with a large degree of heterogeneity in practice size and practice model, including both academic and community settings. GEC testing was prospectively captured, as were the GEC test results and treatment decisions for both tested and untested patients. Although other studies have sought to evaluate treatment decisions related to GEC testing, none included concurrently diagnosed patients without testing, which is vital to understand the clinical utility of these tests.32-35
In conclusion, these data indicate that GEC testing has clinical utility in the favorable-risk setting, and testing of patients with favorable-risk disease is more common in men with higher PSA levels and higher-grade disease who may not typically be considered AS candidates. GEC testing also is being performed in patients with unfavorable intermediate-risk and high-risk disease, for whom the clinical implications are less apparent, and there is marked variability in how GEC tests are being used. The relative accuracy and utility of these tests compared with one other and to other risk stratification tools, including clinical nomograms and mpMRI, must be better understood in the near term to address a key clinical need.
Appendix
For number needed to test calculations, we assumed that the distribution of test results in untested patients would reflect the distribution of test results in tested patients. We first asked the following: of the 1,167 patients with favorable risk of cancer without a gene expression classifier (GEC) test, how many patients with a GEC test result below threshold would likely be reallocated to active surveillance (AS) if they had undergone GEC testing? We calculated this as (203 / 320) × 1,167 = 740, which means that approximately 740 patients with low-risk GEC exist of the 1,167 untested men. Given that 75.9% of patients with a GEC test result below threshold received AS, we assumed that 740 × 75.9%, or 562 of these patients, would receive AS. Similarly, we assumed that, of the 667 untested patients with favorable risk of cancer who initially received AS in this cohort, (203 / 320) × 667, or 429 of these men, are truly GEC low risk. Thus, use of a GEC test should shift 562 − 429, or 133, patients with low-risk GEC to AS and result in a number needed to test of 1,167 / 133, or 8.8 patients with favorable risk of cancer to shift one patient to AS.
Conversely, of the 1,167 patients with favorable risk of cancer without a GEC test, we asked how many patients with a GEC test result above the threshold would likely be reallocated to definitive treatment if they had undergone GEC testing. We assumed that (117 / 320) × 1,167, or 427, would be GEC high risk, using the same approach as for low risk, and that 427 × 39.3%, or 168 patients, would undergo primary therapy. We estimated that (117 / 320) × 337, or 123 of the treated patients without GEC testing, were truly high GEC. Thus, use of a GEC test moves 168 − 123, or 45, patients with a GEC test result above threshold from AS to primary treatment, which results in a number needed to test of 1,167 / 45, or 25.9, patients with favorable risk of cancer to shift one patient to primary therapy.
Fig A1.
Proportion of each gene expression classifier (GEC) test ordered by Michigan Urological Surgery Improvement Collaborative (MUSIC) practice for those practices that have ordered at least one test during the study period.
Fig A2.
Provider-specific rates of gene expression classifier (GEC) testing and rates of active surveillance among patients with favorable risk of cancer are plotted, and they show no correlation between active surveillance use and performance of a GEC test on a per-provider basis (r = 0.16).
Table A1.
Patient and Practice Characteristics Stratified by GEC Testing, Among Patients With Favorable-Risk Prostate Cancer
Table A2.
Summary of Primary Management in Patients With Favorable-Risk Disease According to the Performance and Result of Prostate MRI and GEC Testing
Footnotes
Supported by Blue Cross Blue Shield of Michigan; the Department of Defense Physician Research Training Award No. W81XWH-14-1-0287 (T.M.M.); the Alfred A. Taubman Institute; and the Prostate Cancer Foundation (T.M.M. and D.E.S.).
AUTHOR CONTRIBUTIONS
Conception and design: Jonathan C. Hu, James E. Montie, Kirk Wojno, Frank N. Burks, Daniel E. Spratt, Todd M. Morgan
Collection and assembly of data: Jonathan C. Hu, Deborah Kaye, Anna Johnson, Susan Linsell, James E. Montie, Khurshid R. Ghani, David C. Miller, Kirk Wojno, Frank N. Burks, Todd M. Morgan
Provision of study material or patients: Frank N. Burks, David C. Miller, Kirk Wojno, Daniel E. Spratt, Todd M. Morgan
Data analysis and interpretation: Jonathan C. Hu, Jeffrey J. Tosoian, Ji Qi, Deborah Kaye, James E. Montie, Khurshid R. Ghani, David C. Miller, Kirk Wojno, Frank N. Burks, Daniel E. Spratt, Todd M. Morgan
Administrative support: Susan Linsell, Khurshid R. Ghani, David C. Miller
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Jonathan C. Hu
No relationship to disclose
Jeffrey J. Tosoian
No relationship to disclose
Ji Qi
No relationship to disclose
Deborah Kaye
Research Funding: Blue Cross Blue Shield of Michigan
Travel, Accommodations, Expenses: MedReviews
Anna Johnson
No relationship to disclose
Susan Linsell
No relationship to disclose
James E. Montie
Stock and Other Ownership Interests: Histosonics
Consulting or Advisory Role: Histosonics
Khurshid R. Ghani
Consulting or Advisory Role: Boston Scientific
Research Funding: Boston Scientific
David C. Miller
Research Funding: Blue Cross Blue Shield of Michigan
Kirk Wojno
Employment: Michigan Health Care Professionals, US Medical Management, OncoCellMDx, InformDX
Leadership: OncoCellMDx
Stock and Other Ownership Interests: OncoCellMDx
Honoraria: Strand Diagnostics
Consulting or Advisory Role: Myriad Genetics, Genomic Health, GenomeDx, MDxHealth
Speakers' Bureau: Myriad Genetics, Genomic Health, GenomeDx, MDxHealth, Strand Diagnostics
Research Funding: Myriad Genetics (Inst), Genomic Health (Inst), GenomeDx (Inst), MDxHealth (Inst), NeoGenomics Laboratories (Inst)
Frank N. Burks
No relationship to disclose
Daniel E. Spratt
No relationship to disclose
Todd M. Morgan
Consulting or Advisory Role: Myriad Genetics, TerumoBCT
Research Funding: Myriad Genetics (Inst), MDxHealth (Inst), GenomeDx (Inst)
REFERENCES
- 1.Carroll PR, Parsons JK, Andriole G, et al. NCCN guidelines insights: Prostate cancer early detection, version 2.2016. J Natl Compr Canc Netw. 2016;14:509–519. doi: 10.6004/jnccn.2016.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bokhorst LP, Valdagni R, Rannikko A, et al. A decade of active surveillance in the PRIAS study: An update and evaluation of the criteria used to recommend a switch to active treatment. Eur Urol. 2016;70:954–960. doi: 10.1016/j.eururo.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Newcomb LF, Thompson IM, Jr, Boyer HD, et al. Outcomes of active surveillance for clinically localized prostate cancer in the prospective, multi-institutional Canary PASS cohort. J Urol. 2016;195:313–320. doi: 10.1016/j.juro.2015.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tosoian JJ, Mamawala M, Epstein JI, et al. Intermediate and longer-term outcomes from a prospective active-surveillance program for favorable-risk prostate cancer. J Clin Oncol. 2015;33:3379–3385. doi: 10.1200/JCO.2015.62.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein EA, Haddad Z, Yousefi K, et al. Decipher genomic classifier measured on prostate biopsy predicts metastasis risk. Urology. 2016;90:148–152. doi: 10.1016/j.urology.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 6.Klein EA, Santiago-Jiménez M, Yousefi K, et al. Molecular analysis of low grade prostate cancer using a genomic classifier of metastatic potential. J Urol. 2017;197:122–128. doi: 10.1016/j.juro.2016.08.091. [DOI] [PubMed] [Google Scholar]
- 7.Cuzick J, Stone S, Fisher G, et al. Validation of an RNA cell cycle progression score for predicting death from prostate cancer in a conservatively managed needle biopsy cohort. Br J Cancer. 2015;113:382–389. doi: 10.1038/bjc.2015.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: A retrospective study. Lancet Oncol. 2011;12:245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Den Eeden SK, Lu R, Zhang N, et al. A biopsy-based 17-gene genomic prostate score as a predictor of metastases and prostate cancer death in surgically treated men with clinically localized disease. Eur Urol. 2018;73:129–138. doi: 10.1016/j.eururo.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Klein EA, Cooperberg MR, Magi-Galluzzi C, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol. 2014;66:550–560. doi: 10.1016/j.eururo.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Spratt DE, Zhang J, Santiago-Jiménez M, et al. Development and validation of a novel integrated clinical-genomic risk group classification for localized prostate cancer. J Clin Oncol. 2018;36:581–590. doi: 10.1200/JCO.2017.74.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spratt DE, Yousefi K, Deheshi S, et al. Individual patient-level meta-analysis of the performance of the decipher genomic classifier in high-risk men after prostatectomy to predict development of metastatic disease. J Clin Oncol. 2017;35:1991–1998. doi: 10.1200/JCO.2016.70.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Comprehensive Cancer Network doi: 10.6004/jnccn.2017.0008. https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf NCCN clinical practice guidelines in oncology: Prostate cancer version 1.2017. [DOI] [PubMed]
- 14.Sanda MG, Cadeddu JA, Kirkby E, et al. Clinically localized prostate cancer: AUA/ASTRO/SUO guideline. Part I—Risk stratification, shared decision making, and care options. J Urol. doi: 10.1016/j.juro.2017.11.095. doi: 10.1016/j.juro.2017.11.095 [epub ahead of print on December 15, 2017] [DOI] [PubMed] [Google Scholar]
- 15.Barcenas CH, Sinha AK, Raghavendra AS, et al. Outcomes after chemotherapy in early-stage breast cancer (EBC) patients who underwent a 21-gene expression assay. J Clin Oncol. 2016;34 (suppl; abstr 559) [Google Scholar]
- 16.Sparano JA, Gray RJ, Makower DF, et al. Prospective validation of a 21-gene expression assay in breast cancer. N Engl J Med. 2015;373:2005–2014. doi: 10.1056/NEJMoa1510764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Comprehensive Cancer Network doi: 10.6004/jnccn.2017.0146. https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf NCCN clinical practice guidelines in oncology: Breast cancer, version 2.2017. [DOI] [PMC free article] [PubMed]
- 18.Auffenberg GB, Lane BR, Linsell S, et al. A roadmap for improving the management of favorable risk prostate cancer. J Urol. 2017;198:1220–1222. doi: 10.1016/j.juro.2017.07.085. [DOI] [PubMed] [Google Scholar]
- 19.Cucchiara V, Cooperberg MR, Dall’Era M, et al. Genomic markers in prostate cancer decision making. Eur Urol. 2018;73:572–582. doi: 10.1016/j.eururo.2017.10.036. [DOI] [PubMed] [Google Scholar]
- 20.Womble PR, Montie JE, Ye Z, et al. Contemporary use of initial active surveillance among men in Michigan with low-risk prostate cancer. Eur Urol. 2015;67:44–50. doi: 10.1016/j.eururo.2014.08.024. [DOI] [PubMed] [Google Scholar]
- 21.Cooperberg MR, Broering JM, Kantoff PW, et al. Contemporary trends in low risk prostate cancer: Risk assessment and treatment. J Urol. 2007;178:S14–S19. doi: 10.1016/j.juro.2007.03.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klotz L, Vesprini D, Sethukavalan P, et al. Long-term follow-up of a large active surveillance cohort of patients with prostate cancer. J Clin Oncol. 2015;33:272–277. doi: 10.1200/JCO.2014.55.1192. [DOI] [PubMed] [Google Scholar]
- 23.D’Amico A V, Whittington R, Malkowicz SB, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280:969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 24.Reese AC, Pierorazio PM, Han M, et al. Contemporary evaluation of the National Comprehensive Cancer Network prostate cancer risk classification system. Urology. 2012;80:1075–1079. doi: 10.1016/j.urology.2012.07.040. [DOI] [PubMed] [Google Scholar]
- 25.Cooperberg MR, Pasta DJ, Elkin EP, et al. The University of California, San Francisco, cancer of the prostate risk assessment score: A straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J Urol. 2005;173:1938–1942. doi: 10.1097/01.ju.0000158155.33890.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephenson AJ, Scardino PT, Eastham JA, et al. Preoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Natl Cancer Inst. 2006;98:715–717. doi: 10.1093/jnci/djj190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balachandran VP, Gonen M, Smith JJ, et al. Nomograms in oncology: More than meets the eye. Lancet Oncol. 2015;16:e173–e180. doi: 10.1016/S1470-2045(14)71116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephan C, Siemssen K, Cammann H, et al. Between-method differences in prostate-specific antigen assays affect prostate cancer risk prediction by nomograms. Clin Chem. 2011;57:995–1004. doi: 10.1373/clinchem.2010.151472. [DOI] [PubMed] [Google Scholar]
- 29.Sonn GA, Fan RE, Ghanouni P, et al. Prostate magnetic resonance imaging interpretation varies substantially across radiologists. Eur Urol Focus. doi: 10.1016/j.euf.2017.11.010. doi: 10.1016/j.euf.2017.11.010 [epub ahead of print on December 6, 2017] [DOI] [PubMed] [Google Scholar]
- 30.Lin DW, Crawford ED, Keane T, et al. Identification of men with low-risk biopsy-confirmed prostate cancer as candidates for active surveillance. Urol Oncol. 2018;36:310.e7–310.e13. doi: 10.1016/j.urolonc.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan TM, Miller DC, Dunn R, et al. Prospective randomized trial of genomic classifier impact on treatment decisions in patients at high risk of recurrence following radical prostatectomy (G-MINOR) J Clin Oncol. 2018;36 (suppl; abstr TPS154) [Google Scholar]
- 32.Dall’Era MA, Maddala T, Polychronopoulos L, et al. Utility of the Oncotype DX prostate cancer assay in clinical practice for treatment selection in men newly diagnosed with prostate cancer: A retrospective chart review analysis. Urol Pract. 2015;2:343–348. doi: 10.1016/j.urpr.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Eure G, Germany R, Given R, et al. Use of a 17-gene prognostic assay in contemporary urologic practice: Results of an interim analysis in an observational cohort. Urology. 2017;107:67–75. doi: 10.1016/j.urology.2017.02.052. [DOI] [PubMed] [Google Scholar]
- 34.Shore ND, Kella N, Moran B, et al. Impact of the cell cycle progression test on physician and patient treatment selection for localized prostate cancer. J Urol. 2016;195:612–618. doi: 10.1016/j.juro.2015.09.072. [DOI] [PubMed] [Google Scholar]
- 35.Albala D, Kemeter MJ, Febbo PG, et al. Health economic impact and prospective clinical utility of Oncotype DX genomic prostate score. Rev Urol. 2016;18:123–132. doi: 10.3909/riu0725. [DOI] [PMC free article] [PubMed] [Google Scholar]