Abstract
PROM1 has played a pivotal role in the identification and isolation of tumor stem cells. This study aimed to assess the association between PROM1 promoter methylation and head and neck squamous cell carcinoma (HNSCC), and its diagnostic and prognostic value.
Bioinformatic analysis was performed using data from the Cancer Genome Atlas-HNSC and Gene Expression Omnibus datasets.
The results showed that PROM1 promoter was hypermethylated in HNSCCs compared with normal head and neck tissues (P = 4.58E-37). The area under the receiver-operating characteristic curve based on methylated PROM1 data was 0.799. In addition, PROM1 hypermethylation independently predicted poor overall survival (hazard ratio [HR]: 1.459, 95% confidence interval [CI]: 1.071–1.987, P = .016) and recurrence-free survival (HR: 1.729, 95% CI: 1.088–2.749, P = .021) in HNSCC patients. Moreover, PROM1 methylation was weakly negatively correlated with its mRNA expression (Pearson r = –0.148, P < .001).
In summary, our study reveals that methylated PROM1 might serve as a valuable diagnostic biomarker and predictor of poor survival for HNSCC patients. PROM1 hypermethylation might partially contribute to its downregulation in HNSCC.
Keywords: CD133, diagnosis, HNSCC, methylation, prognosis, PROM1
1. Introduction
Head and neck cancers represent the sixth most common cancer and fifth leading cause of cancer-related death worldwide.[1] More than 90% of these tumors are head and neck squamous cell carcinomas (HNSCCs), which develop in the mucosal linings of the upper aerodigestive tract (lip, oral cavity, pharynx, and larynx) and exhibit microscopic evidence of squamous differentiation.[2] According to the latest report of the International Agency for Research on Cancer, >1 million new patients were diagnosed with HNSCC in 2018, representing 5.5% of all newly diagnosed cancers and accounting for 5.7% of cancer-related deaths worldwide.[1] Recently, the American Cancer Society has estimated that there will be 63,030 new cases of and 13,360 deaths from HNSCC in the United States alone in 2019.[3] HNSCCs develop either by exogenous carcinogen exposure (smoking, alcohol drinking) or by human papillomavirus (HPV) infection (mainly associated with HPV-16 and HPV-18 types), particularly those in the oral cavity and oropharynx.[4] Despite the recent multimodal therapeutic strategies that have yielded some significant improvements, including surgical resection, chemotherapy, targeted therapy, and radiotherapy, the 5-year survival rate for HNSCC patients over the last decade has remained at approximately 50%.[5] The therapeutic strategy and prognosis differ substantially between early and advanced stage HNSCCs. Because of the lack of symptoms and effective screening tests in the early stage, the majority of HNSCC patients are diagnosed at an advanced stage with high local recurrence and/or distant metastatic rates.[6,7] Therefore, the identification of potential biomarkers for early diagnosis, cancer risk assessment, molecular classification, and targeted treatment will be essential for the advancement of HNSCC patient outcomes.
With the establishment of precision medicine, modalities such as anti-EGFR antibody (cetuximab) and checkpoint inhibitors (pembrolizumab, nivolumab) that have recently been approved for the treatment of advanced and metastatic HNSCC, are promising options for the management of high-risk patients.[8,9] Tumor node metastasis (TNM) staging classification is still a vital tool in the prediction of tumor patient outcomes.[10] However, HNSCC is characterized as a heterogeneous and molecularly complex set of cancers that are caused by a variety of genetic and epigenetic aberrations.[11] DNA methylation, a heritable modification known to alter gene expression that is not mediated by changes in the DNA sequence, is a major form of epigenetic modification.[12] Aberrant methylation in the promoter region of tumor suppressor genes (TSGs) is associated with expression silencing and is correlated with cancer initiation, progress, invasion, and metastasis.[13–15] Recent studies have revealed that DNA methylation shows promise for improving the characterization of malignancy to predict treatment response and prognosis.[16] Moreover, aberrant methylation is a relatively early molecular change in carcinogenesis[17,18] and has been proposed as a diagnostic biomarker for a wide range of cancers.[19,20] Thus, the identification of HNSCC-specific methylation biomarkers has emerged as one of the most promising approaches to improve HNSCC diagnosis and prognostic prediction, as it presents several advantages compared to other markers.
Recent evidence unveiled a small population of cancer cells that are highly tumorigenic, capable of self-renewal and that behave as tumor progenitor cells in HNSCC and are responsible for tumor recurrence and metastatic metastasis.[21] Then the further studies affirmed that this subpopulation of cancer stem cells (CSCs) exhibits properties of normal stem cells.[22] Therefore, great efforts exerted on the identification and isolation of biomarkers for this subpopulation tumor cells. Prominin 1 (PROM1), namely CD133, is one of characterized potential marker for CSCs, and CD133+ cells were found to have increased in clonality when compared to CD133–cells.[23]Prominin 1 (PROM1), located on chromosome 4p15.32, encodes CD133, which is a pentaspan membrane glycoprotein first identified in humans as a hematopoietic stem cell marker[24] and is currently used as a marker to isolate CSCs from several tissues and cancer types.[25,26] Understanding the molecular biology of CD133+ cancer cells is now essential for developing more effective cancer treatments, including drugs targeting organelles, such as mitochondria or lysosomes, using highly efficient and selective inducers of apoptosis.[27] Aberrant CD133 expression is found in cancer-initiating cells in many cancers, including stomach cancer,[28] hepatocellular cancer,[29] and pancreatic carcinomas.[30] CD133 expression has also been correlated with adverse cancer properties, such as metastasis, recurrence, and therapy resistance.[31,32] The complex transcription of PROM1 is controlled in a tissue-specific manner by 5 alternative promoters generating at least 16 alternative splicing patterns of the 5’-UTR.[33] Three of the 5 promoters responsible for PROM1 transcription are located in a CpG island, indicating a possible epigenetic regulation of PROM1 expression through DNA methylation.[34] Several studies have suggested that promoter methylation regulates PROM1 expression in various cancers, such as gastrointestinal stromal tumors,[35] gliomas,[36] and colorectal adenocarcinoma.[37] However, until now, the association between PROM1 promoter methylation status and HNSCC, as well as its clinical value, has not been clearly delineated.
In the present study, using bioinformatics analysis, we explored PROM1 promoter methylation in HNSCC and its clinical value. In addition, we investigated the association between PROM1 promoter methylation and its expression.
2. Materials and methods
2.1. Data mining in the cancer genome atlas-head and neck squamous cell carcinoma (TCGA-HNSC)
The level-3 TCGA-HNSC methylation and expression data were obtained by using the University of California Santa Cruz Xena browser (https://xenabrowser.net/). The methylation data were globally normalized using β values (methylation ratio). The RNA-seq data were normalized based on the RPKM (reads per kilobase per million mapped reads) values. The average β values of CpG probes mapping 200 bp downstream of the transcription start sites (TSS200) of PROM1 were defined as the PROM1 promoter methylation level. The clinicopathological data, including sample type, age at initial pathologic diagnosis, sex, alcohol history, tobacco smoking history, anatomic neoplasm subdivision, HPV status by p16 testing, neoplasm histologic grade, pathologic T, pathologic N, pathological stage, overall survival (OS) status, OS time, recurrence-free survival (RFS) status, and RFS time were collected and downloaded for secondary analysis. A total of 525 primary HNSCC cases had both methylation and OS data recorded. A total of 445 primary HNSCC cases had both methylation and RFS data recorded. Our research was performed based on public database and did not involve ethnic issue.
2.2. Data mining Gene Expression Omnibus database
PROM1 mRNA expression data were obtained from published databases (GSE38823)[38] and downloaded from Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo). We focused on the expression changes of PROM1 in 4 HNSCC cell lines (OC3, SAS, SCC-15, and HSC3) with and without 5’-aza-2’-deoxycytidine treatment.
2.3. Statistical analysis
All statistical analyses were performed using Statistical Program for Social Sciences (SPSS) 20.0 software (SPSS Inc, Chicago, IL) and R 3.1.2 software (https://www.r-project.org/), which was also used to generate figures. Data are presented as the means and standard deviations. Welch t test was performed to compare the differences between 2 groups. A receiver-operating characteristic (ROC) curve was used to assess the diagnostic value of PROM1 methylation for HNSCC. Then the value of Youden index was calculated. HNSCC patients with integrated survival data were divided into high and low PROM1 methylation groups according to the maximum Youden index based on ROC curves for death and recurrence detection in HNSCC patients. Kaplan–Meier curves of OS and RFS after initial therapy were generated, and log-rank tests were performed to identify the significance of the difference between the survival curves. Cox regression models were used to evaluate the independent prognostic value of PROM1 methylation in terms of OS and RFS in HNSCC patients. Only the characteristics that had a significant association with OS and RFS in the univariate analysis were included in the multivariate analysis. The correlation between PROM1 methylation and expression was tested using Pearson rank correlation coefficient. A P value <.05 was considered statistically significant.
3. Results
3.1. PROM1 promoter methylation was significantly elevated in HNSCC tissues
In this study, we downloaded the methylation profiles of 528 HNSCC and 50 normal tissues from the TCGA data portal to investigate the association of PROM1 promoter methylation with HNSCC. Three CpG sites (cg10630155, cg04203238, and cg26260038) of Illumina Human Methylation 450K in the TSS200 region of PROM1 are shown in Figure 1 using the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu/). Due to the significant correlations between the 3 CpGs (r > 0.8, P < .001), the average methylation level of 3 CpG sites was applied for the following analyses. The results showed that HNSCC tissues exhibited significantly elevated PROM1 promoter methylation levels compared with normal tissues (P = 4.58E-37; Fig. 2).
Figure 1.

Genomic position of 3 available CpG probes (cg10630155, cg04203238, and cg26260038 in the Illumina Human Methylation 450K) in the PROM1 promoter is shown in the UCSC genome browser (human 2009 assembly; GRCh37/hg19).
Figure 2.
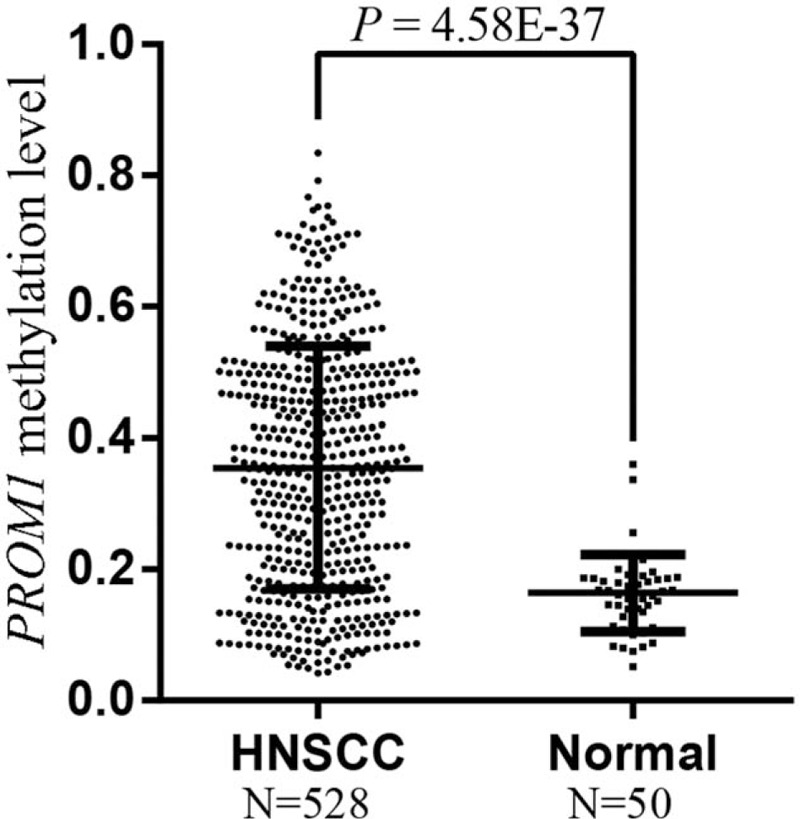
HNSCC tissues have significantly elevated PROM1 promoter methylation levels compared with nontumor tissues. HNSCC = head and neck squamous cell carcinoma, N = sample number.
3.2. Association of PROM1 promoter methylation with clinicopathological characteristics of HNSCC patients
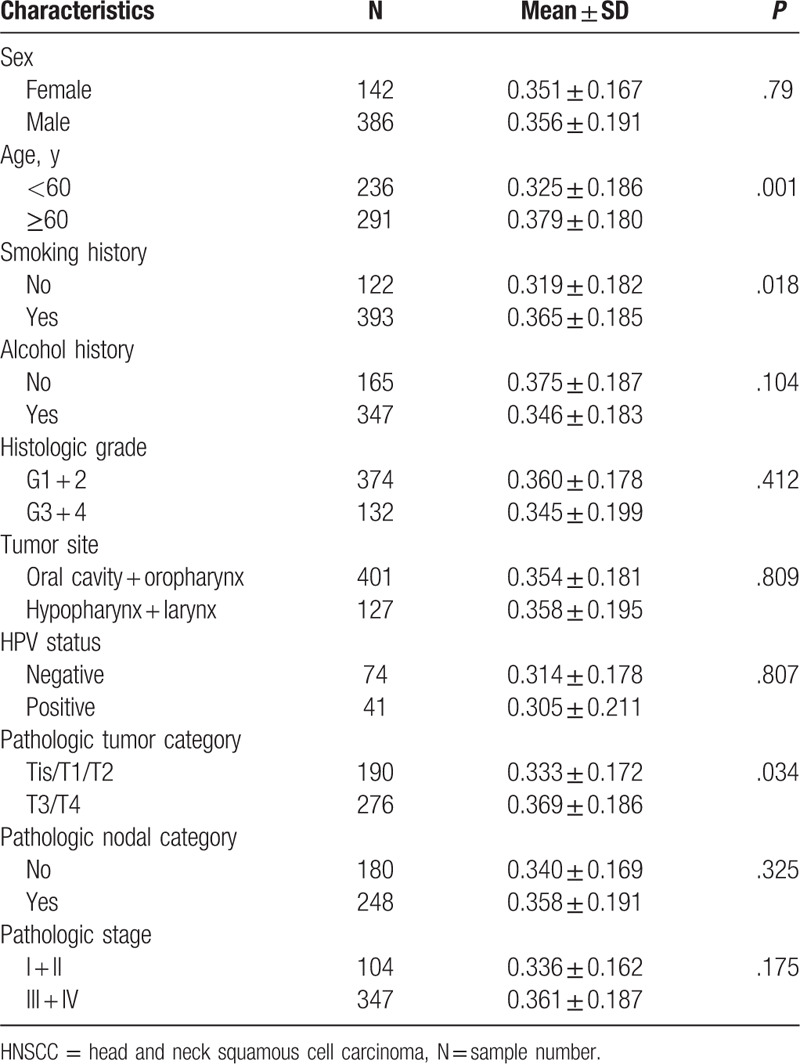
Subsequently, we examined the association between PROM1 promoter methylation levels and the clinicopathological characteristics of HNSCC patients, including sex, age, smoking history, alcohol history, histological grade, tumor site, HPV status, pathologic tumor classification, lymph node metastasis, and pathological stage. In the present study, PROM1 promoter methylation levels were significantly associated with age (P = .001), smoking history (P = .018), and tumor classification (P = 0.034). However, no statistically significant correlation was found with other clinicopathological characteristics (Table 1).
Table 1.
Association between CD133 promoter methylation and clinicopathological features of HNSCC patients.
3.3. Diagnostic value of PROM1 promoter methylation for HNSCC
The ROC curve was used to evaluate the diagnostic value of PROM1 promoter methylation for HNSCC. An area under the ROC curve (AUC) closer to 1.0 signifies that the test exhibits higher diagnostic accuracy. The maximum Youden index was used as a cutoff point. The result showed that PROM1 promoter methylation yielded an AUC of 0.799 at a cutoff value of 0.2256 (Fig. 3). The sensitivity and specificity were 0.706 and 0.940, respectively.
Figure 3.
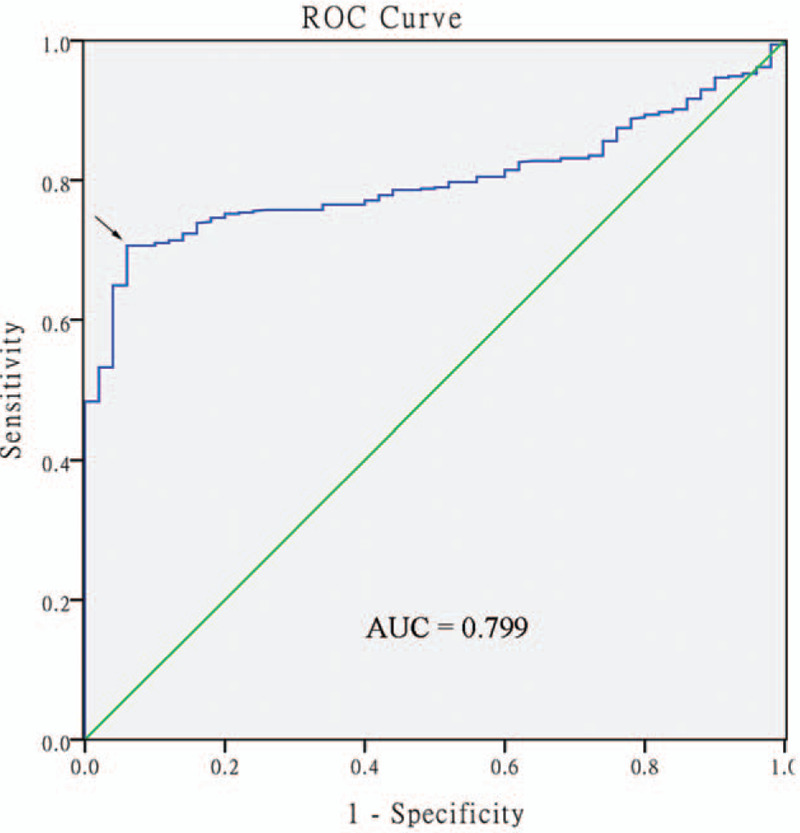
ROC curves to assess the diagnostic value of PROM1 promoter methylation in head and neck squamous cell carcinoma patients. The AUC was 0.799. The arrow indicates the cutoff point. ROC = receiver-operating characteristic, AUC = area under the curve.
3.4. The hypermethylated PROM1 promoter was an independent predictor of unfavorable OS and RFS in HNSCC patients
Using the maximum Youden index as the cutoff point (0.355), 525 HNSCC patients with integrated OS data were divided into high PROM1 promoter methylation (N = 261) and low PROM1 promoter methylation groups (N = 264). The median survival time of high PROM1 promoter methylation group and low PROM1 promoter methylation group were 2.99 and 8.35 years, respectively. Kaplan–Meier curves and log-rank tests revealed that PROM1 promoter hypermethylation was significantly associated with poor OS in HNSCC (Fig. 4A, P = 1.7E-4). Moreover, we classified 445 HNSCC patients with integrated RFS data into high (N = 248) and low (N = 197) PROM1 promoter methylation groups according to a cutoff value of 0.308. Kaplan–Meier curves and log-rank tests confirmed that HNSCC patients in the high PROM1 promoter methylation group exhibited significantly worse RFS (Fig. 4B, P = .017).
Figure 4.
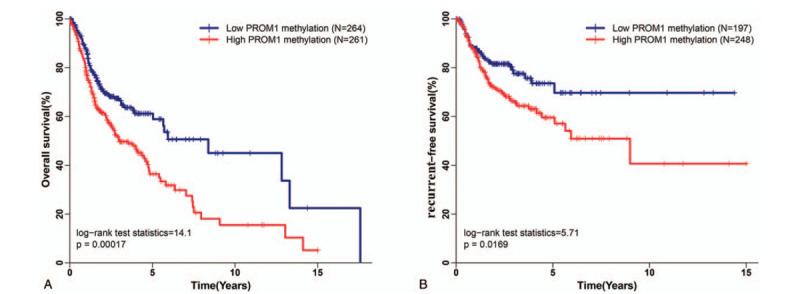
Association between PROM1 promoter methylation and head and neck squamous cell carcinoma (HNSCC) prognosis. (A) PROM1 promoter hypermethylation is associated with poor overall survival in HNSCC patients. (B) PROM1 promoter hypermethylation is associated with poor recurrence-free survival in HNSCC patients.
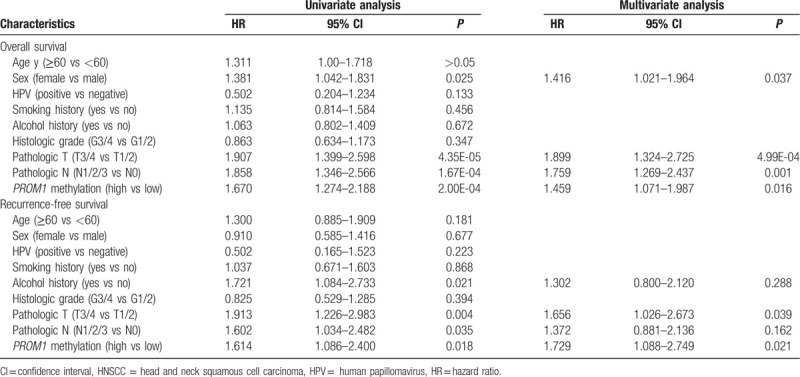
As shown in Table 2, univariate Cox proportional hazards analysis revealed a significantly increased risk of death for HNSCC patients who were female (hazard ratio [HR]: 1.381, 95% confidence interval [CI]: 1.042–1.831, P = .025), patients with advanced tumor classification (HR: 1.907, 95% CI: 1.399–2.598, P = 4.35E-05), lymph node metastasis (HR: 1.858, 95% CI: 1.346–2.566, P = 1.67E-04), and hypermethylated PROM1 promoter (HR: 1.670, 95% CI: 1.274–2.188, P = 2.00E-04). Additionally, we revealed that alcohol consumption history (HR: 1.721, 95% CI: 1.084–2.733, P = .021), advanced tumor classification (HR: 1.913, 95% CI: 1.226–2.983, P = .004), lymph node metastasis (HR: 1.602, 95% CI: 1.034–2.482, P = .035), and PROM1 promoter hypermethylation (HR: 1.614, 95% CI: 1.086–2.400, P = .018) were significantly correlated with shorter RFS. Multivariate Cox proportional hazard analysis was conducted to investigate the independent prognostic factors in terms of OS and RFS in HNSCC patients by adjusting only variables that exhibited significance in a univariate analysis. The results showed that being female (HR: 1.416, 95% CI: 1.021–1.964, P = .037) and lymph node metastasis (HR: 1.759, 95% CI: 1.269–2.437, P = .001) were independent unfavorable OS predictors for HNSCC patients. Moreover, we confirmed that both advanced tumor classification (OS: HR: 1.899, 95% CI: 1.324–2.725, P = 4.99E-04; RFS: HR: 1.656, 95% CI: 1.026–2.673, P = .039) and PROM1 promoter hypermethylation (OS: HR: 1.459, 95% CI: 1.071–1.987, P = .016; RFS: HR: 1.729, 95% CI: 1.088–2.749, P = .021) were independent unfavorable prognostic factors in terms of both OS and RFS in HNSCC patients.
Table 2.
Univariate and multivariate analysis of overall survival and recurrence-free survival in HNSCC patients.
3.5. PROM1 promoter methylation level was negatively correlated with its expression in HNSCC
We examined the association of the PROM1 promoter methylation level with its expression using 483 HNSCC patients with integrated methylation and expression data in the TCGA database. PROM1 was downregulated in the HNSCC tissue (Fig. 5). Regression analysis revealed a negative correlation (Pearson r=–0.148, P < .001) between PROM1 promoter methylation level and its expression level in HNSCC samples (Fig. 6A).
Figure 5.
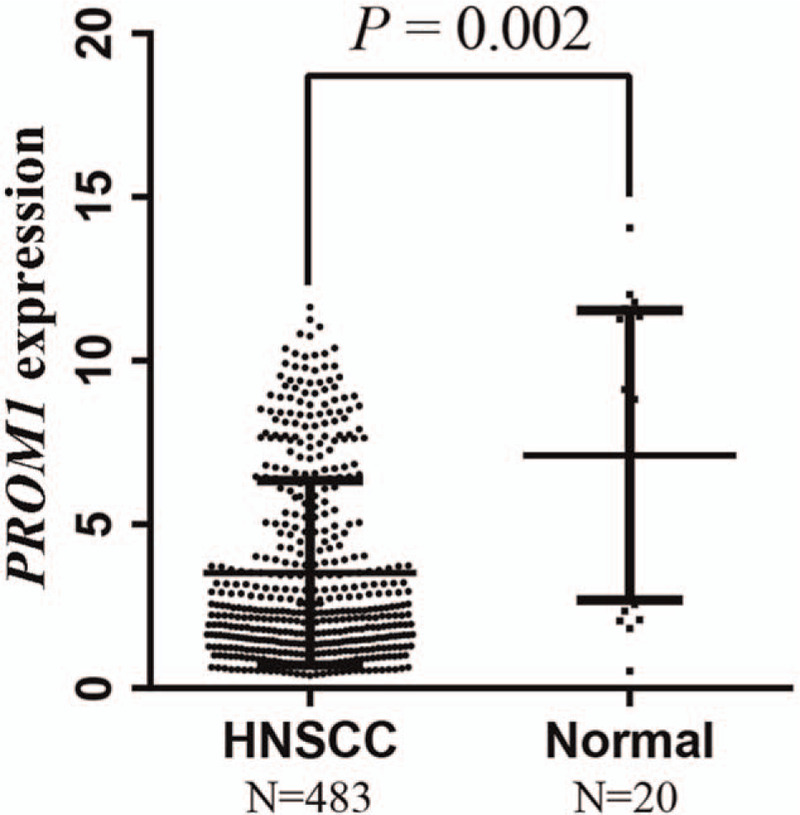
PROM1 expression levels are significantly reduced in HNSCC tissues compared with normal tissues.
Figure 6.
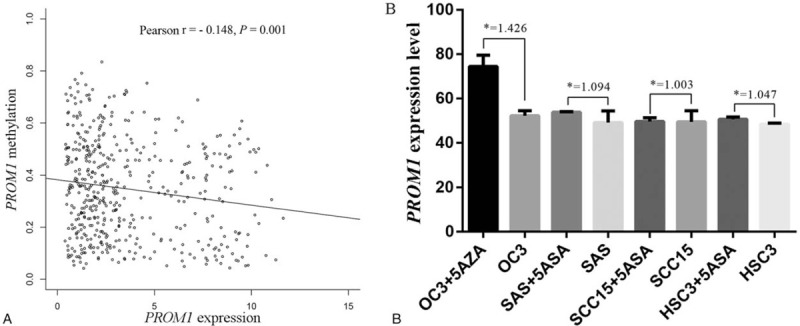
The correlation between PROM1 expression and its promoter methylation in HNSCC tissues and cell lines. (A) PROM1 expression was negatively correlated with its promoter methylation in HNSCC. (B) PROM1 expression increased in HNSCC cell lines after 5’-aza-2’-deoxycytidine treatment (5-AZA) treatment. ∗: fold change = with 5-AZA treatment/without 5-AZA treatment. HNSCC = head and neck squamous cell carcinoma, N = sample number.
In addition, using the GEO database (GSE38823), we found that PROM1 expression in 4 HNSCC cell lines (OC3, SAS, SCC-15, and HSC3) all increased after 5’-aza-2’-deoxycytidine (5-AZA) treatment (Fig. 6B). Therefore, PROM1 was likely hypermethylated in HNSCC cell lines, which potentially suppressed PROM1 expression.
4. Discussion
DNA methylation is one of the most widely studied epigenetic modifications.[39] Previous studies have shown that the inactivation of TSGs in numerous cancers, including HNSCC, may be attributed to the hypermethylation of CpG islands in the promoter region.[40,41]PROM1 encodes CD133, which plays a pivotal role in the identification and isolation of CSCs. CD133-positive CSCs were also reported to possess enhanced chemoresistance and radioresistance, resulting in tumor progression and recurrence.[42,43] Promoter methylation is one epigenetic modification mechanism that is associated with downregulation of PROM1 in several cancers, including glioblastoma and colon cancer.[44,45] However, the association between PROM1 promoter methylation and HNSCC remains unclear. In the present study, using public data from TCGA-HNSC, we revealed that the PROM1 promoter is highly methylated in HNSCC tissues.
Subsequently, we also determined the association between PROM1 promoter methylation and the clinicopathological characteristics of patients with HNSCC. Smoking behavior is a crucial factor for increasing the risk of HNSCC.[46] In addition, tobacco smoking has long-lasting effects on DNA methylation patterns, which play an integral role in tumorigenesis.[47] In this study, using RNA-seq data in TCGA-HNSC, we found that increased PROM1 promoter methylation levels were significantly correlated with smoking history, suggesting that smoking might contribute to HNSCC by inducing PROM1 methylation. Besides, our result showed higher methylation level of PROM1 in elder patients when compared with youngers. Aging is a complex process characterized by a global decline in physiological functions and is associated with an increased risk for several diseases, including cancer. Previous epigenome-wide association studies report that CpG island, mainly placed within genes promoter regions, is hypermethylated in the elderly. Hypermethylation of PROM1 promoter was observed in advanced tumor stages compared with early tumor stages in HNSCC, suggesting the involvement of PROM1 promoter methylation in the invasion progression of HNSCC.
One of the most important issues concerning cancer patients is how to screen and diagnose at an early stage.[48] Early-stage HNSCC patients may only receive minimally invasive surgery or radiation therapy alone, with good outcomes, whereas late-stage patients receive aggressive therapy, such as expanded surgery and/or concomitant chemoradiotherapy, resulting in dismal survival rates and poor quality of life.[49] Screening for HNSCC depends on clinical symptoms and imaging examinations, and a definite diagnosis depends on biopsy and histopathological examination.[50] However, given the nonspecificity of symptoms in the early stage and lack of effective biomarkers, the early detection of HNSCC remains unsatisfactory.[51] Due to occurring early in carcinogenesis and having other advantageous characteristics, methylation markers represent potential clinical applications for the early detection of cancer.[20] In the present study, we constructed ROC curves and calculated the AUC to determine the diagnostic value of PROM1 promoter methylation for HNSCC. The sensitivity, specificity, and AUC values were 0.706, 0.940, and 0.799, respectively, signifying greater diagnostic power compared with conventional cancer-related biomarkers for HNSCC patients, such as squamous cell carcinoma antigen, carcinoembryonic antigen, tissue polypeptide-specific antigen, and Cyfra 21-1.[52] However, considering the relatively low sensitivity, PROM1 methylation might not be useful for clinical application alone. Recent evidence has shown that the combination of several epigenetic biomarkers can improve the diagnostic accuracy for cancers.[53,54] It would be logical that a panel of epigenetic biomarkers, including PROM1, represents a potential diagnostic test for HNSCC. Future rigorous studies using larger sample sizes will be essential to validate this approach.[8–11] In the present study, using the large TCGA dataset, the log-rank test demonstrated that HNSCC patients with PROM1 hypermethylation had remarkably shorter OS and RFS. By performing univariate Cox proportional hazard and multivariate Cox proportional hazard analysis, we further showed that PROM1 hypermethylation was an independent poor prognosis factor for OS and RFS in HNSCC patients. Besides, our finding revealed a significantly increased risk of death for HNSCC patients who were female. Previous studies have demonstrated that the initial of HNSCC is the consequence of persistent infection with high-risk HPV in combination with other aetiological factors (such as smoking behaviors). The epidemiology of HNSCC showed that the incidence rates of HNSCC have remained or decreased, whereas the rates of non-HPV HNSCCs among females have increased which largely reflect changes in sex-specific smoking rate. Therefore, female patients without HPV infection might be induced in alternative pathway characterized with highly invasiveness. These results suggest that PROM1 hypermethylation may be of great value for tailoring individual therapies and risk stratification of recurrence and subsequent death in HNSCC patients, which might help these patients benefit from intensified first-line treatment and surveillance. In this study, we found that PROM1 is significantly downregulated in HNSCC tissues compared with normal head and neck tissues. Many studies show that hypermethylation in the promoter area of TSGs results in loss of gene expression, which plays an important role in the initiation and progression of HNSCC.[41,55] In the present study, by comparing PROM1 promoter methylation and gene expression in HNSCC tissues, we observed a negative correlation between PROM1 promoter methylation and its expression level. Moreover, GEO data showed that PROM1 expression in 4 HNSCC cell lines increased after demethylation treatment. The above evidence suggests that PROM1 hypermethylation might contribute to the development of HNSCC by at least partly downregulating PROM1 expression.
5. Conclusions
PROM1 promoter methylation is significantly elevated in HNSCC tissues; this methylation is partly related to its transcriptional inactivation and may be involved in the invasion of HNSCC. Additionally, PROM1 hypermethylation is a potential epigenetic biomarker for the early diagnosis and prognosis of HNSCC. However, well-designed prospective studies with a large sample size and cellular experiments are urgently needed to support our findings.
Author contributions
Conceptualization: Zele Hu, Huigao Liu, Zhenhua Wu, Qun Li, Chongchang Zhou.
Data curation: Huigao Liu.
Formal analysis: Huigao Liu.
Supervision: Chongchang Zhou.
Validation: Xinrong Zhang, Bin Hong.
Writing – original draft: Zele Hu, Xinrong Zhang, Bin Hong.
Writing – review & editing: Zele Hu.
Chongchang Zhou orcid: 0000-0002-8728-6819.
Footnotes
Abbreviations: AUC = area under the receiver-operating characteristic curve, CSC = cancer stem cell, GEO = Gene Expression Omnibus, HNSCC = head and neck squamous cell carcinoma, HPV = human papillomavirus, OS = overall survival, RFS = recurrence-free survival, ROC = receiver-operating characteristic, TNM = tumor node metastasis, TSG = tumor suppressor gene.
How to cite this article: Hu Z, Liu H, Zhang X, Hong B, Wu Z, Li Q, Zhou C. Promoter hypermethylation of CD133/PROM1 is an independent poor prognosis factor for head and neck squamous cell carcinoma. Medicine. 2020;99:11(e19491).
The authors report no conflicts of interest.
This work was supported by grants from the Zhejiang Provincial Natural Science Foundation of China (No. LY19H160090), Ningbo Natural Science Foundation (No. 2015A610221 and No. 2017A610236), Clinical research funding from Zhejiang Medical Association (No. 2015ZYC-A49), and the Ningbo Health Branding Subject Fund (PPXK2018-02).
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [2].Pai SI, Westra WH. Molecular pathology of head and neck cancer: implications for diagnosis, prognosis, and treatment. Ann Rev Pathol 2009;4:49–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- [4].Kindt N, Descamps G, Lechien JR, et al. Involvement of HPV infection in the release of macrophage migration inhibitory factor in head and neck squamous cell carcinoma. J Clin Med 2019;8:pii: E75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Shield KD, Ferlay J, Jemal A, et al. The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA Cancer J Clin 2017;67:51–64. [DOI] [PubMed] [Google Scholar]
- [6].De Felice F, Thomas C, Barrington S, et al. Analysis of loco-regional failures in head and neck cancer after radical radiation therapy. Oral Oncol 2015;51:1051–5. [DOI] [PubMed] [Google Scholar]
- [7].Marur S, Forastiere AA. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc 2016;91:386–96. [DOI] [PubMed] [Google Scholar]
- [8].Alfieri S, Cavalieri S, Licitra L. Immunotherapy for recurrent/metastatic head and neck cancer. Curr Opin Otolaryngol Head Neck Surg 2018;26:152–6. [DOI] [PubMed] [Google Scholar]
- [9].Kaidar-Person O, Gil Z, Billan S. Precision medicine in head and neck cancer. Drug Resist Updat 2018;40:13–6. [DOI] [PubMed] [Google Scholar]
- [10].Chen K, Chen H, Yang F, et al. Validation of the eighth edition of the TNM staging system for lung cancer in 2043 surgically treated patients with non-small-cell lung cancer. Clin Lung Cancer 2017;18:e457–66. [DOI] [PubMed] [Google Scholar]
- [11].Stadler ME, Patel MR, Couch ME, et al. Molecular biology of head and neck cancer: risks and pathways. Hematol Oncol Clin North Am 2008;22:1099–124. vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bird AP. CpG-rich islands and the function of DNA methylation. Nature 1986;321:209–13. [DOI] [PubMed] [Google Scholar]
- [13].Lin S, Liu Y, Goldin LR, et al. Sex-related DNA methylation differences in B cell chronic lymphocytic leukemia. Biol Sex Differ 2019;10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liu J, Li H, Sun L, et al. Epigenetic alternations of MicroRNAs and DNA methylation contribute to liver metastasis of colorectal cancer. Digest Dis Sci 2019;64:1523–34. [DOI] [PubMed] [Google Scholar]
- [15].Liu S, Hausmann S, Carlson SM, et al. METTL13 methylation of eEF1A increases translational output to promote tumorigenesis. Cell 2019;176:491–504. e421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hao X, Luo H, Krawczyk M, et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci U S A 2017;114:7414–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Azzollini J, Pesenti C, Pizzamiglio S, et al. Constitutive BRCA1 promoter hypermethylation can be a predisposing event in isolated early-onset breast cancer. Cancers 2019;11: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Trujillo KA, Jones AC, Griffith JK, et al. Markers of field cancerization: proposed clinical applications in prostate biopsies. Prostate Cancer 2012;2012:302894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Conway K, Edmiston SN, Parker JS, et al. Identification of a robust methylation classifier for cutaneous melanoma diagnosis. J Invest Dermatol 2019;139:1349–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pan Y, Liu G, Zhou F, et al. DNA methylation profiles in cancer diagnosis and therapeutics. Clin Exp Med 2018;18:1–4. [DOI] [PubMed] [Google Scholar]
- [21].Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A 2007;104:973–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- [23].Wei XD, Zhou L, Cheng L, et al. In vivo investigation of CD133 as a putative marker of cancer stem cells in Hep-2 cell line. Head Neck 2009;31:94–101. [DOI] [PubMed] [Google Scholar]
- [24].Yin AH, Miraglia S, Zanjani ED, et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood 1997;90:5002–12. [PubMed] [Google Scholar]
- [25].Mia-Jan K, Jung SY, Kim IY, et al. CD133 expression is not an independent prognostic factor in stage II and III colorectal cancer but may predict the better outcome in patients with adjuvant therapy. BMC Cancer 2013;13:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shmelkov SV, St Clair R, Lyden D, et al. AC133/CD133/Prominin-1. Int J Biochem Cell Biology 2005;37:715–9. [DOI] [PubMed] [Google Scholar]
- [27].Neuzil J, Stantic M, Zobalova R, et al. Tumour-initiating cells vs. cancer 'stem’ cells and CD133: what's in the name? Biochem Biophys Res Commun 2007;355:855–9. [DOI] [PubMed] [Google Scholar]
- [28].Yu JW, Zhang P, Wu JG, et al. Expressions and clinical significances of CD133 protein and CD133 mRNA in primary lesion of gastric adenocacinoma. J Exp Clin Cancer Res 2010;29:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Smith LM, Nesterova A, Ryan MC, et al. CD133/prominin-1 is a potential therapeutic target for antibody-drug conjugates in hepatocellular and gastric cancers. Br J Cancer 2008;99:100–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shimizu K, Itoh T, Shimizu M, et al. CD133 expression pattern distinguishes intraductal papillary mucinous neoplasms from ductal adenocarcinomas of the pancreas. Pancreas 2009;38:e207–14. [DOI] [PubMed] [Google Scholar]
- [31].Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature 2007;445:111–5. [DOI] [PubMed] [Google Scholar]
- [32].Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature 2004;432:396–401. [DOI] [PubMed] [Google Scholar]
- [33].Shmelkov SV, Jun L, St Clair R, et al. Alternative promoters regulate transcription of the gene that encodes stem cell surface protein AC133. Blood 2004;103:2055–61. [DOI] [PubMed] [Google Scholar]
- [34].Grosse-Gehling P, Fargeas CA, Dittfeld C, et al. CD133 as a biomarker for putative cancer stem cells in solid tumours: limitations, problems and challenges. J Pathol 2013;229:355–78. [DOI] [PubMed] [Google Scholar]
- [35].Geddert H, Braun A, Kayser C, et al. Epigenetic regulation of CD133 in gastrointestinal stromal tumors. Am J Clin Pathol 2017;147:515–24. [DOI] [PubMed] [Google Scholar]
- [36].Gopisetty G, Xu J, Sampath D, et al. Epigenetic regulation of CD133/PROM1 expression in glioma stem cells by Sp1/myc and promoter methylation. Oncogene 2013;32:3119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Oliver JA, Ortiz R, Melguizo C, et al. Prognostic impact of MGMT promoter methylation and MGMT and CD133 expression in colorectal adenocarcinoma. BMC Cancer 2014;14:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lee CH, Wong TS, Chan JY, et al. Epigenetic regulation of the X-linked tumour suppressors BEX1 and LDOC1 in oral squamous cell carcinoma. J Pathol 2013;230:298–309. [DOI] [PubMed] [Google Scholar]
- [39].Ouadid-Ahidouch H, Rodat-Despoix L, Matifat F, et al. DNA methylation of channel-related genes in cancers. Biochim Biophys Acta 2015;1848(10 pt B):2621–8. [DOI] [PubMed] [Google Scholar]
- [40].Zhou C, Shen Z, Ye D, et al. The association and clinical significance of CDKN2A promoter methylation in head and neck squamous cell carcinoma: a meta-analysis. Cell Physiol Biochem 2018;50:868–82. [DOI] [PubMed] [Google Scholar]
- [41].Zhou C, Ye M, Ni S, et al. DNA methylation biomarkers for head and neck squamous cell carcinoma. Epigenetics 2018;13:398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nature Rev Cancer 2005;5:275–84. [DOI] [PubMed] [Google Scholar]
- [43].Diehn M, Clarke MF. Cancer stem cells and radiotherapy: new insights into tumor radioresistance. J Natl Cancer Inst 2006;98:1755–7. [DOI] [PubMed] [Google Scholar]
- [44].Tabu K, Sasai K, Kimura T, et al. Promoter hypomethylation regulates CD133 expression in human gliomas. Cell Res 2008;18:1037–46. [DOI] [PubMed] [Google Scholar]
- [45].Yi JM, Tsai HC, Glockner SC, et al. Abnormal DNA methylation of CD133 in colorectal and glioblastoma tumors. Cancer Res 2008;68:8094–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jethwa AR, Khariwala SS. Tobacco-related carcinogenesis in head and neck cancer. Cancer Metastasis Rev 2017;36:411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zeilinger S, Kuhnel B, Klopp N, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One 2013;8:e63812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Smith RA, Andrews KS, Brooks D, et al. Cancer screening in the United States, 2017: A review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin 2017;67:100–21. [DOI] [PubMed] [Google Scholar]
- [49].Choong N, Vokes E. Expanding role of the medical oncologist in the management of head and neck cancer. CA Cancer J Clin 2008;58:32–53. [DOI] [PubMed] [Google Scholar]
- [50].Vigneswaran N, Williams MD. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac Surg Clin North Am 2014;26:123–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Barak V, Meirovitz A, Leibovici V, et al. The diagnostic and prognostic value of tumor markers (CEA, SCC, CYFRA 21-1, TPS) in head and neck cancer patients. Anticancer Res 2015;35:5519–24. [PubMed] [Google Scholar]
- [52].Inal E, Lacin M, Asal K, et al. The significance of ferritin, lipid-associated sialic acid, CEA, squamous cell carcinoma (SCC) antigen, and CYFRA 21-1 levels in SCC of the head and neck. Kulak Burun Bogaz Ihtis Derg 2004;12:23–30. [PubMed] [Google Scholar]
- [53].Song B, Wang L, Zhang Y, et al. Combined detection of HER2, Ki67, and GSTP1 genes on the diagnosis and prognosis of breast cancer. Cancer Biother Radiopharm 2019;34:85–90. [DOI] [PubMed] [Google Scholar]
- [54].Yang Z, Qi W, Sun L, et al. DNA methylation analysis of selected genes for the detection of early-stage lung cancer using circulating cell-free DNA. Adv Clin Exp Med 2019;28:355–60. [DOI] [PubMed] [Google Scholar]
- [55].Bednarek K, Kostrzewska-Poczekaj M, Szaumkessel M, et al. Downregulation of CEACAM6 gene expression in laryngeal squamous cell carcinoma is an effect of DNA hypermethylation and correlates with disease progression. Am J Cancer Res 2018;8:1249–61. [PMC free article] [PubMed] [Google Scholar]


