Abstract
PURPOSE
Using nonenrichment-based, potentially more sensitive Epic Sciences circulating tumor cell (CTC) platform, we sought to detect and characterize CTCs in untreated, high-risk localized prostate cancer and to evaluate their clinical implication.
METHODS
Between 2012 and 2015, blood samples were prospectively collected from patients with National Comprehensive Cancer Network high-risk localized prostate cancer undergoing either radiotherapy (XRT) plus androgen deprivation therapy or radical prostatectomy (RP) with curative intent. Samples were analyzed with the Epic Sciences platform with 4′,6-diamidino-2-phenylindole, CD45, cytokeratin (CK), and androgen receptor (AR) N-terminal staining. CTC counts were correlated with biochemical recurrence (BCR).
RESULTS
A diversity of CTC subtypes, including CK-positive, CK-negative, AR-positive, and CTC clusters, were observed in 73.3% (33 of 45) of patients with evaluable data. The median follow-up was 14.2 months (range, 0.5 to 43.7 months). BCR occurred more frequently in the RP group than XRT (15 of 26 v one of 19), with most patients in the XRT group continuing to receive androgen deprivation therapy. A higher proportion of metastatic events were observed in the RP group (five of 26 v one of 19). In the RP group, BCR and development of metastases were associated with a higher total number of CTCs, AR-positive CTCs, and CTC phenotypic heterogeneity. One patient who developed BCR and metastases quickly after RP had diverse phenotypical CTC subtypes, and single-cell genomic analyses of all detectable CTCs confirmed common prostate cancer copy number alterations and PTEN loss.
CONCLUSION
CTCs can be identified in most patients with high-risk localized prostate cancer before definitive therapy using the Epic Sciences platform. If confirmed in a larger cohort with longer follow-up, phenotypic and genomic characterization of CTCs pretherapy may provide an additional means of risk stratifying patients with newly diagnosed high-risk disease and potentially help identify patients who could require multimodal therapy.
INTRODUCTION
The existence of disseminated tumor cells detectable in the blood or bone marrow of men with clinically localized prostate cancer has long been known.1-4 Yet, molecular characterization of these cells in the localized setting has remained elusive because of the extremely low ratio of tumor to normal cells in circulation. With recent advances in molecular imaging, we now know that many men with clinically localized disease have positron emission tomography–visible metastatic disease,5-9 yet these modalities are expensive and may only detect lesions consisting of at least several million tumor cells. Additional methods of detecting otherwise occult, disseminated disease are still needed, because these may inform primary treatment decisions such as the use of systemic therapy.
Circulating tumor cells (CTCs) may provide an approach for identifying men with micrometastatic disease at the time of diagnosis. Men with high-risk, clinically localized disease are often undertreated, contributing to the high risk of prostate cancer–specific death in this patient population. Many of these men would benefit from multimodal therapy. Ongoing efforts to improve risk stratification include investigation of novel imaging techniques (eg, A Prospective Phase 2/3 Multicenter Study of 18F-DCFPyL PET/CT Imaging in Patients With Prostate Cancer: Examination of Diagnostic Accuracy [OSPREY] trial; ClinicalTrials.gov identifier: NCT02981368), tissue-based gene expression classifiers, and liquid biopsy approaches, such as characterization of CTCs and circulating tumor DNA.
CONTEXT
Key Objective
To determine if circulating tumor cells (CTCs) can predict treatment response in high-risk localized prostate cancer.
Knowledge Generated
Phenotypic and genomic features of CTCs may predict biochemical recurrence in high-risk localized prostate cancer. Individual CTCs can be sequenced in this setting, confirming their tumor origin.
Relevance
Detection and characterization of CTCs may facilitate identification of patients with high-risk prostate cancer who could benefit from multimodal therapy.
In the advanced prostate cancer setting, the identification and characterization of CTCs is now poised to help guide appropriate treatment selection, because detection of the AR-V7 splice variant in CTCs predicts resistance to second-generation antiandrogens in men with metastatic castration-resistant prostate cancer (mCRPC).10-12 Previous efforts to reliably detect and analyze CTCs in localized prostate cancer, however, have not led to clinically useful assays because of the low clinical sensitivity and challenges associated with characterizing the individual detected CTCs.13-23 In men with mCRPC, a high-throughput digital imaging approach has been shown to increase the sensitivity of CTC detection while also providing additional molecular and morphologic granularity at the single-cell level.24-26 This same approach may offer one avenue to improving the sensitivity and clinical implications of CTC detection in the localized disease setting. To date, these investigations have not yet been performed in patients with localized disease.
In the current study, we sought to use a digital imaging–based platform to determine the molecular characteristics and clinical relevance of CTCs in men undergoing primary therapy for high-risk localized prostate cancer.
METHODS
Cohort Description
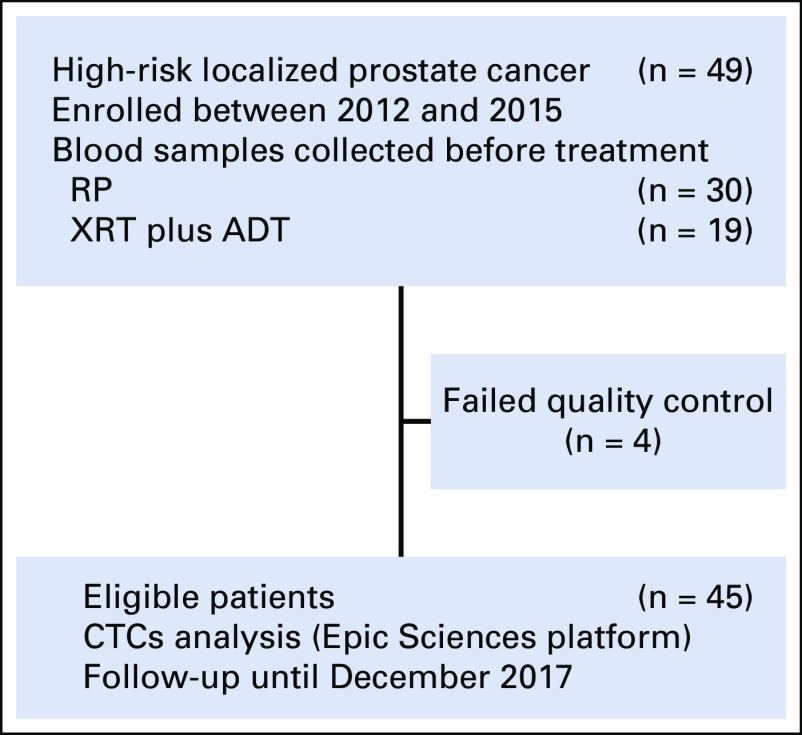
After institutional review board approval, we conducted a prospective study of 49 patients (Fig 1) with untreated National Comprehensive Cancer Network (NCCN)–defined high-risk prostate cancer.27 These men had at least one of the following: grade group 4 or 5 disease, prostate-specific antigen (PSA) greater than 20, and/or clinical stage T2c or greater. All patients completed bone scans and computerized tomography (CT) scans of the abdomen and pelvis demonstrating no evidence of metastatic disease before undergoing local therapy (either radiotherapy [XRT] plus androgen deprivation therapy [ADT] or radical prostatectomy [RP]) with curative intent. Biochemical recurrence (BCR) was defined as PSA of 0.2 ng/mL or greater and included patients who did not experience a nadir PSA post radical prostatectomy less than 0.1 ng/mL.
FIG 1.
Study cohort. ADT, androgen deprivation therapy; CTC, circulating tumor cell; RP, radical prostatectomy; XRT, radiation therapy.
CTC Sample Processing and Identification
Blood samples were prospectively collected from each patient before therapy between 2012 and 2015. The samples were analyzed with the Epic Sciences CTC platform (Epic Sciences, San Diego, CA), which uses an enrichment-free approach to CTC detection, with all nucleated cells analyzed on a glass slide using digital pathology. The details of CTC collection, sample processing, androgen receptor (AR) staining and CTC classification have been described previously.24-26 Briefly, a 10-mL blood sample was collected from each patient using Cell Free DNA BCT tubes (Streck, Omaha, NE) and shipped to Epic Sciences at ambient temperature within 72 hours. On receipt, sample processing consisted of RBC lysis, nucleated blood cell deposition onto 10 to 12 glass slides, storage at −80°C, and AR testing on two slides from each sample by immunofluorescence staining of AR N-terminus, cytokeratins (CKs), CD45, and 4′,6-diamidino-2-phenylindole counterstain. Approximately 3 million cells were deposited onto each slide, imaged, and then analyzed by software that characterizes each cell according to multiple predefined parameters. CTCs were characterized for traditional CTC markers (CK-positive, intact nuclei) as well as other CTC subtypes inclusive of CK-negative CTCs, CTC clusters, and apoptotic CTCs (fragmented nuclei).28 Additional parameters included cell size, shape, nuclear area, and presence of macronucleoli, along with AR expression, uniformity, and cellular localization. CTC candidates were identified in an interactive report and reviewed by trained technicians. CTC phenotypic heterogeneity was quantified using the Shannon index, as previously described.29 Samples from four patients failed quality control testing, leaving a final analyzable cohort size of 45 patients (Fig 1).
Single-Cell Whole-Genome Sequencing and Copy Number Variation Analysis
Single CTC copy number variation (CNV) analysis based on low-pass whole-genome sequencing was performed as previously described.30,31 Briefly, individual CTCs were isolated from the slides, dropped into Eppendorf tubes, and whole-genome amplified using SeqPlex enhanced DNA amplification kit from Sigma (Sigma Aldrich, St Louis, MO). Amplified DNA was used for library construction using NEBNext Ultra DNA Library Prep Kit for Illumina (New England BioLabs, Ipswich, MA) and 2 × 150 paired-end sequencing was performed using Illumina NextSEquation 500 sequencer. DNA CNVs were determined using an existing, standardized single-cell CNV analysis pipeline.30
Data Collection and Statistical Analyses
We tracked relevant demographic, clinical, and pathologic data for each patient and entered all data into a secure electronic Health Insurance Portability and Accountability Act–compliant database. Fisher’s exact test was used to compare categorical data, and population means were compared using a Wilcoxon rank sum test. Statistical analyses were performed with R Foundation for Statistical Computing (http://www.R-project.org), and two-sided P values < 0.05 were considered statistically significant.
RESULTS
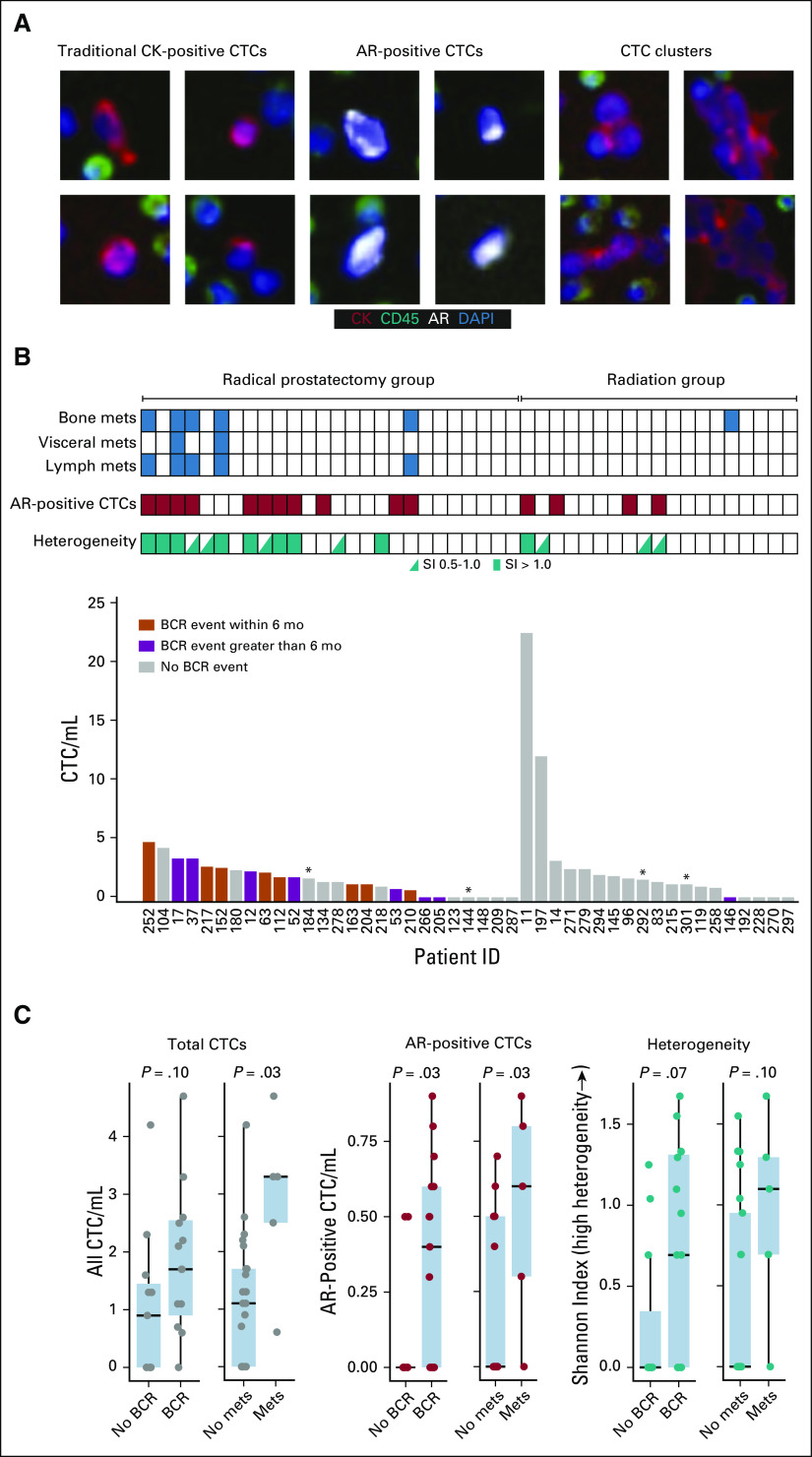
The 45-patient cohort consisted of 26 men (58%) who subsequently underwent RP and 19 (42%) who subsequently underwent XRT with or without ADT (Fig 1). The demographic and clinical characteristics of the study cohort are presented in Table 1. The median post-treatment follow-up was 14.2 months (range, 0.5 to 43.7 months). A diversity of CTC subtypes was detected, including AR-positive cells, CK-positive cells, CK-negative CTCs,28 and clusters of CTCs (Fig 2A). At least one CTC was present in 73.3% (33 of 45) of patients and more than 3 CTCs/mL in seven of 45 (15.6%) patients (Table 2). CTCs with detectable AR expression were observed in 24.4% (11 of 45) of the cohort.
TABLE 1.
Demographic and Clinical Characteristics of the Study Cohort (N = 45)
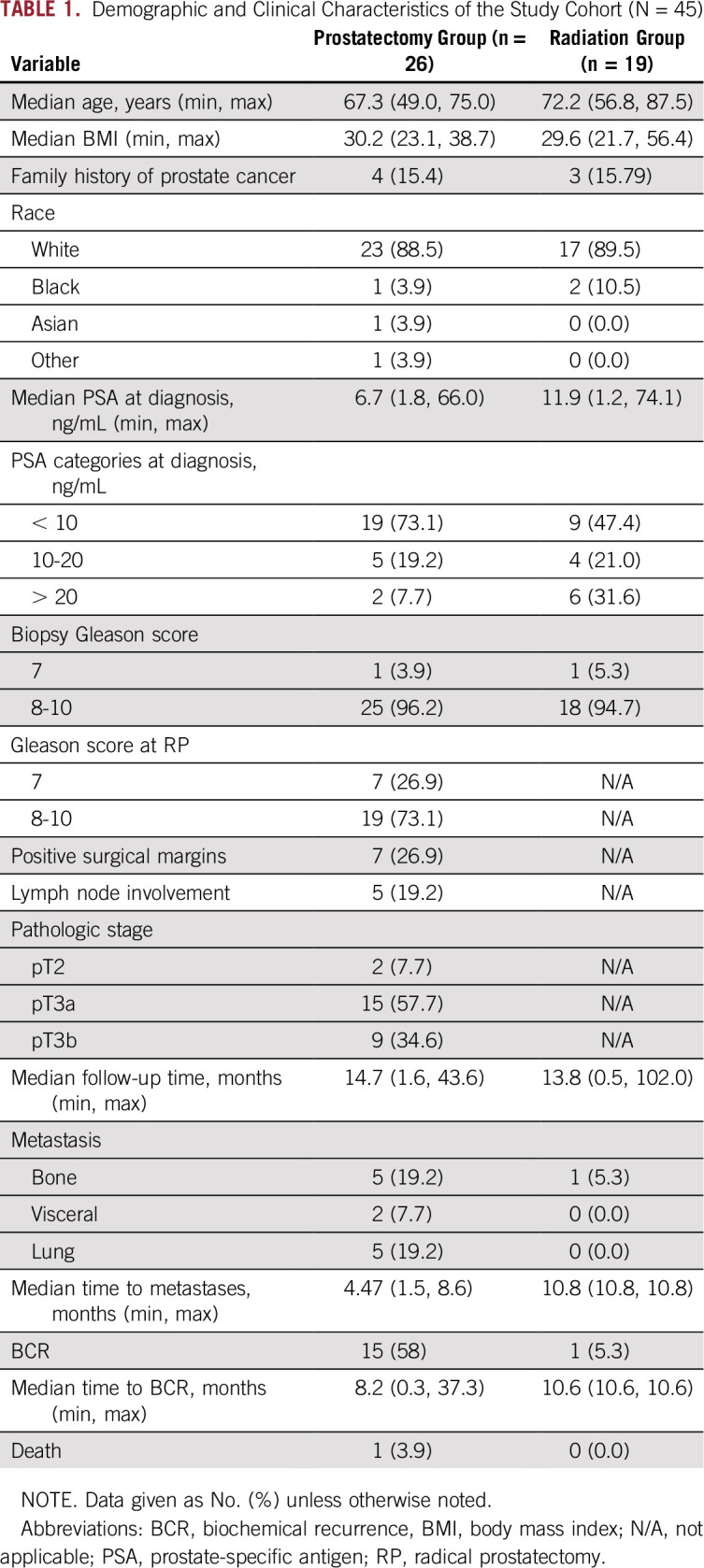
FIG 2.
Circulating tumor cell (CTC) images and relationship of CTCs to biochemical recurrence (BCR) and development of metastases. (A) Representative CTC images. (B) Comparison of total CTC/mL of blood, presence of androgen receptor (AR)–positive CTCs, CTC phenotypic heterogeneity or Shannon index (SI), metastases, and BCR for each patient. A high SI is indicative of a heterogeneous population of detected CTCs. Each box represents an individual patient. (C) Association of BCR and development of metastases after radical prostatectomy to total CTC/mL, AR-positive CTC/mL, and CTC phenotypic heterogeneity. Wilcoxon rank sum test was used to compare means and calculate P values. (*) Follow-up time less than 6 months. CK, cytokeratin; DAPI, 4′,6-diamidino-2-phenylindole; mets, metastases.
TABLE 2.
Types of CTCs Detected in the Study Cohort
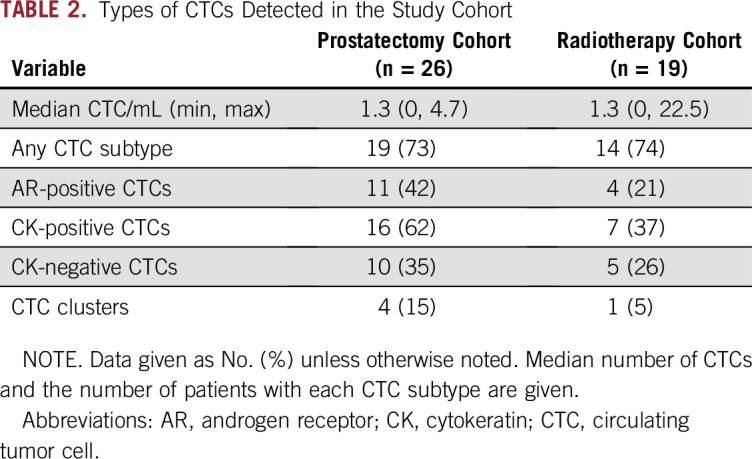
Compared with patients who underwent XRT, those who underwent RP had a higher rate of BCR (15 of 26 v one of 19; P < .001), although most patients in the XRT group continued receiving ADT. Rates of metastasis also were higher, although not statistically significant (five of 26 v one of 19; P = .22). In the RP cohort, the median time to BCR was 3.6 months (range, 0.3 to 37.3 months). BCR occurred post-XRT in one patient at 11 months after treatment. In exploratory analyses, patients who had RP who experienced BCR had a slightly higher median pretreatment serum PSA than those without BCR (7.5 v 4.4 ng/mL; P = .012). There were no differences in age, Gleason scores, pathologic T-stage, and margin status at the time of RP between patients with and without BCR (all P > .05). Five (33%) of the patients with BCR had lymph node metastasis at the time of RP, whereas no patients in the group without BCR had lymph node involvement (P = .052). All of the patients who underwent RP and XRT who experienced metastasis (to bone, regional lymph nodes, and abdominal viscera) during follow-up were preceded by BCR. Recurrence and metastasis were associated with significant differences in baseline CTC detection. Patients experiencing BCR had significantly greater numbers of AR-positive CTCs, and patients developing metastases had significantly more total CTCs and AR-positive CTCs (Figs 2B and 2C).
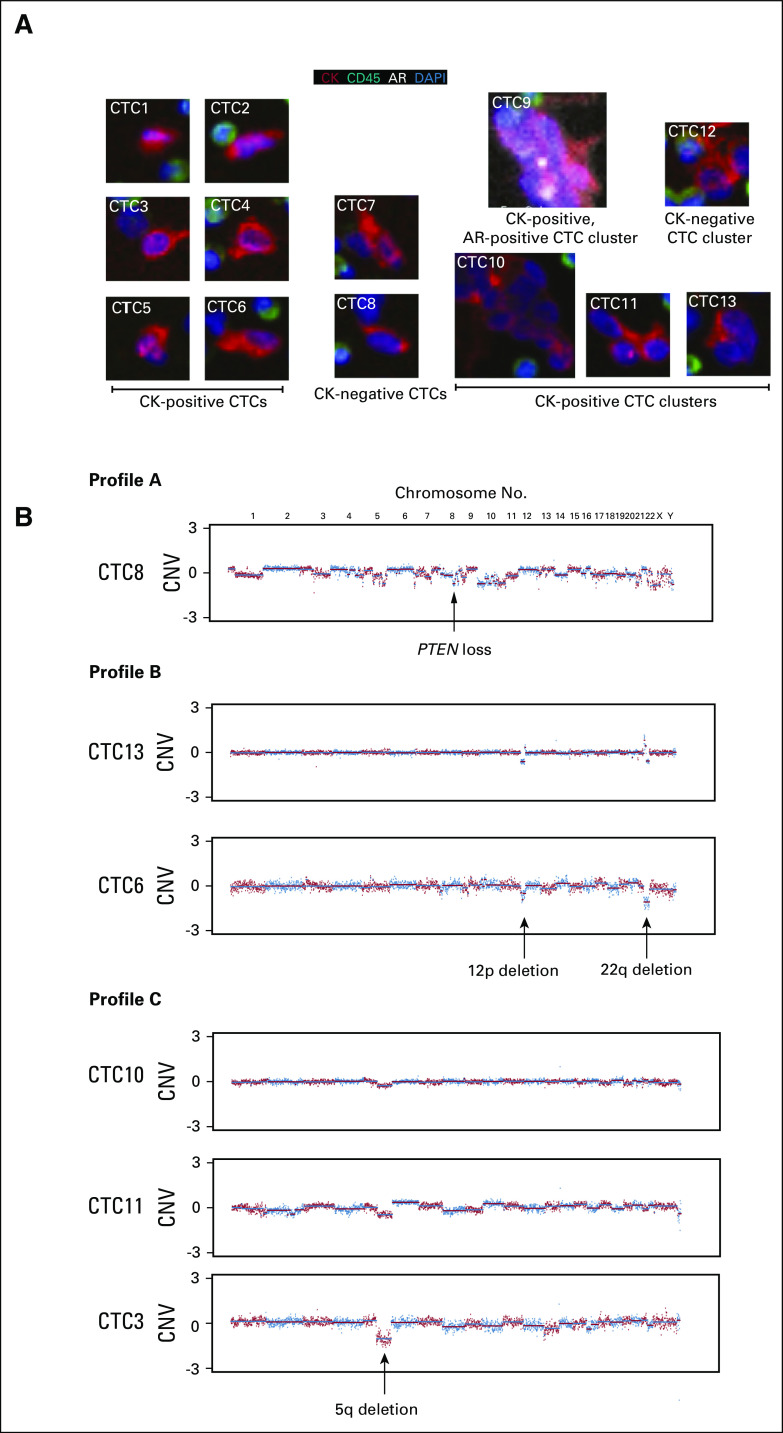
Single-cell sequencing and morphology analysis of all detected CTCs was performed on patient 252, who had the highest CTC count in the RP group (4.7 CTCs/mL of blood). This patient had negative preoperative imaging and was found to have lymph node metastasis at the time of surgery, and the initial postoperative PSA was 7.5 ng/mL. We detected a heterogeneous population (Shannon index > 1) of CTCs in this patient, including AR-positive, CK-positive, and CK-negative CTCs, along with CTC clusters (Fig 3A). Single-cell sequencing results identified genomic aberrations consistent with malignant origin in six (46%) of the 13 CTCs sequenced (Fig 3B). Moreover, heterogeneous genomic profiles were observed in these six CTCs. One CTC had high genomic instability with dozens of break points observed across almost all chromosomes (Profile A). PTEN loss was detected in this CTC akin to those CTCs found in patients with mCRPC previously analyzed with similar methodology.24,29 Two CTCs had 12p loss and 22q loss (Profile B); both of these alterations have been reported for prostate cancer.32,33 Three cells shared the same 5q loss (Profile C).
FIG 3.
Phenotypic and single-cell genomic analyses of patient 252 who had lymph node metastases at the time of radical prostatectomy. (A) Images of all 13 circulating tumor cells (CTCs) sequenced. (B) Genomic copy number variation (CNV) profiles across chromosomes from the six CTCs that had genomic alterations consistent with malignant origin. AR, androgen receptor; CK, cytokeratin; DAPI, 4′,6-diamidino-2-phenylindole.
DISCUSSION
In this study, we detected CTCs in more than two-thirds of patients, demonstrating the feasibility of detecting and characterizing CTC morphology and genomics in men with localized prostate cancer. In addition, we found the presence of CTCs with high AR protein expression to be associated with BCR and metastatic progression in patients undergoing radical prostatectomy. Oncologic end points were not evaluable in this time frame for patients who underwent radiotherapy, because most of these patients continued to receive ADT.
Various liquid biopsy approaches have been interrogated in the past for risk stratifying patients with apparent clinically localized prostate cancer with varying success. For example, reverse transcriptase polymerase chain reaction assay for PSA,34-36 an indirect approach to detect PSA-expressing cells in circulation, demonstrated no significant advantage in preoperative staging of prostate cancer. Similarly, recent approaches to identifying prostate cancer CTCs have widely varied in their capacity to detect CTCs in men with localized disease.13-23,37,38 Thus, unlike in metastatic disease, where CTC number is associated with survival, the clinical or prognostic relevance of CTC detection in localized prostate cancer remains unclear. This stands in contrast to other tumor types, where the detection of CTCs in localized disease is considered prognostic.39,40
There are a number of existing, tissue-based molecular markers for risk stratification in prostate cancer; therefore, a key question is whether the molecular information obtained through analysis of CTCs could have any clinical relevance beyond tissue-based approaches.41-44 Yet, prostate cancer is characterized by tumor multifocality as well as intra- and interfocal genomic and transcriptomic heterogeneity, which may affect the clinical performance of tests assessing a single tumor focus.45-49 Similarly, in metastatic prostate cancer, CTCs are diverse in their phenotypic appearance and genomic makeup.29 A liquid biopsy approach may provide one avenue for addressing tumor heterogeneity and biopsy under-sampling by allowing a broader sample of potentially clinically significant tumor cells.
In line with this hypothesis, we observed significant phenotypic diversity within the same patient and from patient to patient in our cohort of men with untreated high-risk localized prostate cancer. In the patient who underwent single-cell sequencing, we observed distinguishable copy number alterations, which in primary tumors has previously been linked to BCR and metastasis.50 Furthermore, one of these individual CTCs had confirmed PTEN loss, which is a common event in localized prostate cancer and is associated with a poor prognosis.51 Collectively, the results suggest that tumor cell heterogeneity is measurable in CTCs, providing a potential source of prognostic information beyond simple enumeration. This molecular information could allow for clinical utility related to risk stratification and prediction of treatment response.
The role for molecular diagnostics in localized prostate cancer continues to evolve and now also includes advanced molecular imaging techniques.5-9 The ultimate goal is the prediction and/or identification of early metastatic disease to guide improved treatment and long-term oncologic outcomes. Compared with men who underwent radical prostatectomy, we observed a relatively lower rate of BCR in men who received radiation therapy, likely due to the effect of ADT in this subgroup. These data suggest that the presence of CTCs in a patient with no visible extraprostatic or distant disease on imaging may be an indicator of micrometastatic disease spread. Furthermore, the phenotypic and genomic make-up of CTCs may provide additional prognostic and predictive information.
To our knowledge, the current study is the first to characterize CTCs at the combined protein and whole-genome single-cell level in localized prostate cancer. The finding that CTCs with high AR protein expression are associated with metastatic progression is consistent with previous reports in metastatic prostate cancer. For example, AR-V7 expression on CTCs strongly predicts response to second-generation antiandrogen therapy in the metastatic setting.10-12
This study has several limitations. First, our study was performed in a small cohort of patients, and we were therefore not powered to make between-group comparisons. Second, the follow-up duration was short. Before routine clinical implementation, larger studies with longer follow-up are needed to validate our findings and further investigate the association between CTC profiles and the likelihood of subsequent metastasis. Third, we did not measure CTCs intra- or postoperatively to account for the impact of surgery on CTC count or evaluate the clearance of CTCs after surgery.
In this study, we demonstrate that CTCs are not only detectable but also can be further profiled in men with localized high-risk prostate cancer. The association of CTC subtypes, including those with high AR protein expression, with BCR and metastases suggests that characterization of CTCs may provide useful information for risk stratification and the need for multimodal therapy. Larger studies with longer follow-up are needed to fully elucidate the potential for CTC profiling to predict long-term outcomes and treatment response in men with localized disease.
Footnotes
Supported by the Prostate Cancer Foundation (S.S.S. and T.M.M), Department of Defense Grants No. W81XWH-14-1-0287 (T.M.M.) and W81XWH-18-1-0219 (S.S.S.), and the A. Alfred Taubman Medical Research Institute (S.A.T. and T.M.M.).
Presented at the 17th Society of Urologic Oncology annual meeting, San Antonio, TX, November 30-December 2, 2016.
AUTHOR CONTRIBUTIONS
Conception and design: Simpa S. Salami, Udit Singhal, Ganesh S. Palapattu, Scott A. Tomlins, Ryan Dittamore, Felix Y. Feng, Todd M. Morgan
Financial support: Ganesh S. Palapattu, Felix Y. Feng
Administrative support: Daniel E. Spratt, Ganesh S. Palapattu, Lyndsey Dugan
Provision of study material or patients: Daniel E. Spratt, Brent K. Hollenbeck, Felix Y. Feng, Todd M. Morgan
Collection and assembly of data: Simpa S. Salami, Udit Singhal, Daniel E. Spratt, Brent K. Hollenbeck, Joseph D. Schonhoft, Ryon Graf, Jessica Louw, Lyndsey Dugan, Yipeng Wang, Felix Y. Feng, Todd M. Morgan
Data analysis and interpretation: Simpa S. Salami, Udit Singhal, Daniel E. Spratt, Joseph D. Schonhoft, Ryon Graf, Adam Jendrisak, Yipeng Wang, Ryan Dittamore, Felix Y. Feng, Todd M. Morgan
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Ganesh S. Palapattu
Stock and Other Ownership Interests: NantKwest
Consulting or Advisory Role: Janssen Scientific Affairs
Research Funding: Minomic
Patents, Royalties, Other Intellectual Property: implantable nanotechnology for long-term testosterone delivery functionalized fiducial marker for drug delivery
Brent K. Hollenbeck
Other Relationship: Elsevier
Joseph D. Schonhoft
Employment: Epic Sciences
Stock and Other Ownership Interests: Epic Sciences
Ryon Graf
Employment: Epic Sciences
Research Funding: Epic Sciences
Travel, Accommodations, Expenses: Epic Sciences
Jessica Louw
Employment: Epic Sciences, Epic Sciences (I), NAVICAN Genomics
Stock and Other Ownership Interests: NAVICAN Genomics
Travel, Accommodations, Expenses: NAVICAN Genomics
Adam Jendrisak
Employment: Epic Sciences
Stock and Other Ownership Interests: Epic Sciences
Research Funding: Epic Sciences
Lyndsey Dugan
Employment: NAVICAN Genomics, Human Longevity
Stock and Other Ownership Interests: NAVICAN Genomics
Consulting or Advisory Role: Human Longevity (Inst), ChromaCode
Yipeng Wang
Employment: Epic Sciences
Stock and Other Ownership Interests: Epic Sciences
Scott A. Tomlins
Employment: Strata Oncology
Leadership: Strata Oncology
Stock and Other Ownership Interests: Strata Oncology
Consulting or Advisory Role: AbbVie, Janssen, Astellas Medivation, Strata Oncology, Sanofi, Almac Diagnostics
Research Funding: Astellas Medivation (Inst), GenomeDx (Inst)
Patents, Royalties, Other Intellectual Property: I am a coauthor on a patent issued to the University of Michigan on ETS gene fusions in prostate cancer. The diagnostic field of use has been licensed to Hologic/Gen-Probe, who has sublicensed some rights to Ventana Medical Systems/Roche.
Travel, Accommodations, Expenses: Strata Oncology
Ryan Dittamore
Employment: Epic Sciences
Leadership: Epic Sciences
Stock and Other Ownership Interests: Epic Sciences
Patents, Royalties, Other Intellectual Property: Patents pending
Felix Y. Feng
Leadership: PFS Genomics
Stock and Other Ownership Interests: PFS Genomics
Consulting or Advisory Role: Dendreon, EMD Serono, Janssen Oncology, Ferring Pharmaceuticals, Sanofi, Bayer, Blue Earth Diagnostics, Celgene, Medivation/Astellas
Research Funding: Zenith Epigenetics
Patents, Royalties, Other Intellectual Property: I helped develop a molecular signature to predict radiation resistance in breast cancer, and this signature was patented by the University of Michigan, my employer. It is in the process of being licensed to PFS Genomics, a company that I helped found. (Inst)
Todd M. Morgan
Consulting or Advisory Role: Myriad Genetics, TerumoBCT
Research Funding: Myriad Genetics (Inst), MDxHealth (Inst), GenomeDx (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1.Chéry L, Lam HM, Coleman I, et al. Characterization of single disseminated prostate cancer cells reveals tumor cell heterogeneity and identifies dormancy associated pathways. Oncotarget. 2014;5:9939–9951. doi: 10.18632/oncotarget.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Todenhöfer T, Hennenlotter J, Faber F, et al. Significance of apoptotic and non-apoptotic disseminated tumor cells in the bone marrow of patients with clinically localized prostate cancer. Prostate. 2015;75:637–645. doi: 10.1002/pros.22947. [DOI] [PubMed] [Google Scholar]
- 3.Lilleby W, Stensvold A, Mills IG, et al. Disseminated tumor cells and their prognostic significance in nonmetastatic prostate cancer patients. Int J Cancer. 2013;133:149–155. doi: 10.1002/ijc.28002. [DOI] [PubMed] [Google Scholar]
- 4.Morgan TM, Lange PH, Porter MP, et al. Disseminated tumor cells in prostate cancer patients after radical prostatectomy and without evidence of disease predicts biochemical recurrence. Clin Cancer Res. 2009;15:677–683. doi: 10.1158/1078-0432.CCR-08-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herlemann A, Kretschmer A, Buchner A, et al. Salvage lymph node dissection after 68Ga-PSMA or 18F-FEC PET/CT for nodal recurrence in prostate cancer patients. Oncotarget. 2017;8:84180–84192. doi: 10.18632/oncotarget.21118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selnæs KM, Krüger-Stokke B, Elschot M, et al. 18F-Fluciclovine PET/MRI for preoperative lymph node staging in high-risk prostate cancer patients. Eur Radiol. 2018;28:3151–3159. doi: 10.1007/s00330-017-5213-1. [DOI] [PubMed] [Google Scholar]
- 7. Bauman G, Martin P, Thiessen JD, et al: [18F]-DCFPyL positron emission tomography/magnetic resonance imaging for localization of dominant intraprostatic foci: First experience. Eur Urol Focus 4:702-706, 2016. [DOI] [PubMed]
- 8.Schmuck S, von Klot CA, Henkenberens C, et al. Initial experience with volumetric 68Ga-PSMA I&T PET/CT for assessment of whole-body tumor burden as a quantitative imaging biomarker in patients with prostate cancer. J Nucl Med. 2017;58:1962–1968. doi: 10.2967/jnumed.117.193581. [DOI] [PubMed] [Google Scholar]
- 9.Budäus L, Leyh-Bannurah SR, Salomon G, et al. Initial experience of (68)Ga-PSMA PET/CT imaging in high-risk prostate cancer patients prior to radical prostatectomy. Eur Urol. 2016;69:393–396. doi: 10.1016/j.eururo.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 10.Scher HI, Graf RP, Schreiber NA, et al. Nuclear-specific AR-V7 protein localization is necessary to guide treatment selection in metastatic castration-resistant prostate cancer. Eur Urol. 2017;71:874–882. doi: 10.1016/j.eururo.2016.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scher HI, Graf RP, Schreiber NA, et al. Assessment of the validity of nuclear-localized androgen receptor splice variant 7 in circulating tumor cells as a predictive biomarker for castration-resistant prostate cancer. JAMA Oncol. 2018;4:1179–1186. doi: 10.1001/jamaoncol.2018.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scher HI, Lu D, Schreiber NA, et al. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol. 2016;2:1441–1449. doi: 10.1001/jamaoncol.2016.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meyer CP, Pantel K, Tennstedt P, et al: Limited prognostic value of preoperative circulating tumor cells for early biochemical recurrence in patients with localized prostate cancer. Urol Oncol 34:235:e11-e16, 2016. [DOI] [PubMed]
- 14.Pal SK, He M, Wilson T, et al. Detection and phenotyping of circulating tumor cells in high-risk localized prostate cancer. Clin Genitourin Cancer. 2015;13:130–136. doi: 10.1016/j.clgc.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aragon-Ching JB, Siegel RS, Frazier H, II, et al. Circulating tumor cells in biochemical recurrence of prostate cancer. Clin Genitourin Cancer. 2015;13:e341–e345. doi: 10.1016/j.clgc.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Shao C, Liao CP, Hu P, et al. Detection of live circulating tumor cells by a class of near-infrared heptamethine carbocyanine dyes in patients with localized and metastatic prostate cancer. PLoS One. 2014;9:e88967. doi: 10.1371/journal.pone.0088967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thalgott M, Rack B, Maurer T, et al. Detection of circulating tumor cells in different stages of prostate cancer. J Cancer Res Clin Oncol. 2013;139:755–763. doi: 10.1007/s00432-013-1377-5. [DOI] [PubMed] [Google Scholar]
- 18.Khurana KK, Grane R, Borden EC, et al. Prevalence of circulating tumor cells in localized prostate cancer. Curr Urol. 2013;7:65–69. doi: 10.1159/000356251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stott SL, Lee RJ, Nagrath S, et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010;2:25ra23. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maestro LM, Sastre J, Rafael SB, et al. Circulating tumor cells in solid tumor in metastatic and localized stages. Anticancer Res. 2009;29:4839–4843. [PubMed] [Google Scholar]
- 21.Davis JW, Nakanishi H, Kumar VS, et al. Circulating tumor cells in peripheral blood samples from patients with increased serum prostate specific antigen: Initial results in early prostate cancer. J Urol. 2008;179:2187–2191. doi: 10.1016/j.juro.2008.01.102. [DOI] [PubMed] [Google Scholar]
- 22.Eschwège P, Moutereau S, Droupy S, et al. Prognostic value of prostate circulating cells detection in prostate cancer patients: A prospective study. Br J Cancer. 2009;100:608–610. doi: 10.1038/sj.bjc.6604912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kauffman EC, Lee MJ, Alarcon SV, et al. Lack of impact of robotic assisted laparoscopic radical prostatectomy on intraoperative levels of prostate cancer circulating tumor cells. J Urol. 2016;195:1136–1142. doi: 10.1016/j.juro.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Punnoose EA, Ferraldeschi R, Szafer-Glusman E, et al. PTEN loss in circulating tumour cells correlates with PTEN loss in fresh tumour tissue from castration-resistant prostate cancer patients. Br J Cancer. 2015;113:1225–1233. doi: 10.1038/bjc.2015.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Werner SL, Graf RP, Landers M, et al. Analytical validation and capabilities of the epic CTC platform: Enrichment-free circulating tumour cell detection and characterization. J Circ Biomark. 2015;4:3. doi: 10.5772/60725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marrinucci D, Bethel K, Kolatkar A, et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol. 2012;9:016003. doi: 10.1088/1478-3975/9/1/016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohler JL, Kantoff PW, Armstrong AJ, et al. Prostate cancer, version 2.2014. J Natl Compr Canc Netw. 2014;12:686–718. doi: 10.6004/jnccn.2014.0072. [DOI] [PubMed] [Google Scholar]
- 28.McDaniel AS, Ferraldeschi R, Krupa R, et al. Phenotypic diversity of circulating tumour cells in patients with metastatic castration-resistant prostate cancer. BJU Int. 2017;120:E30–E44. doi: 10.1111/bju.13631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scher HI, Graf RP, Schreiber NA, et al. Phenotypic heterogeneity of circulating tumor cells informs clinical decisions between AR signaling inhibitors and taxanes in metastatic prostate cancer. Cancer Res. 2017;77:5687–5698. doi: 10.1158/0008-5472.CAN-17-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greene SB, Dago AE, Leitz LJ, et al. Chromosomal instability estimation based on next generation sequencing and single cell genome wide copy number variation analysis. PLoS One. 2016;11:e0165089. doi: 10.1371/journal.pone.0165089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anantharaman A, Friedlander T, Lu D, et al. Programmed death-ligand 1 (PD-L1) characterization of circulating tumor cells (CTCs) in muscle invasive and metastatic bladder cancer patients. BMC Cancer. 2016;16:744. doi: 10.1186/s12885-016-2758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camp NJ, Farnham JM, Cannon-Albright LA. Localization of a prostate cancer predisposition gene to an 880-kb region on chromosome 22q12.3 in Utah high-risk pedigrees. Cancer Res. 2006;66:10205–10212. doi: 10.1158/0008-5472.CAN-06-1233. [DOI] [PubMed] [Google Scholar]
- 33. doi: 10.18632/oncotarget.14557. Kluth M, Ahrary R, Hube-Magg C, et al: Genomic deletion of chromosome 12p is an independent prognostic marker in prostate cancer. Oncotarget 6:17966-27979, 2015 [Erratum: Oncotarget 8:3761, 2017] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katz AE, Olsson CA, Raffo AJ, et al. Molecular staging of prostate cancer with the use of an enhanced reverse transcriptase-PCR assay. Urology. 1994;43:765–775. doi: 10.1016/0090-4295(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 35.Ignatoff JM, Oefelein MG, Watkin W, et al. Prostate specific antigen reverse transcriptase-polymerase chain reaction assay in preoperative staging of prostate cancer. J Urol. 1997;158:1870–1874. doi: 10.1016/s0022-5347(01)64150-8. [DOI] [PubMed] [Google Scholar]
- 36.Shariat SF, Gottenger E, Nguyen C, et al. Preoperative blood reverse transcriptase-PCR assays for prostate-specific antigen and human glandular kallikrein for prediction of prostate cancer progression after radical prostatectomy. Cancer Res. 2002;62:5974–5979. [PubMed] [Google Scholar]
- 37.Helo P, Cronin AM, Danila DC, et al. Circulating prostate tumor cells detected by reverse transcription-PCR in men with localized or castration-refractory prostate cancer: Concordance with CellSearch assay and association with bone metastases and with survival. Clin Chem. 2009;55:765–773. doi: 10.1373/clinchem.2008.117952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreno JG, Miller MC, Gross S, et al. Circulating tumor cells predict survival in patients with metastatic prostate cancer. Urology. 2005;65:713–718. doi: 10.1016/j.urology.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 39. doi: 10.1093/jnci/dju066. Rack B, Schindlbeck C, Jückstock J, et al: Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J Natl Cancer Inst 106:dju066, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch M, Kienle P, Kastrati D, et al. Prognostic impact of hematogenous tumor cell dissemination in patients with stage II colorectal cancer. Int J Cancer. 2006;118:3072–3077. doi: 10.1002/ijc.21784. [DOI] [PubMed] [Google Scholar]
- 41.Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8:e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: A retrospective study. Lancet Oncol. 2011;12:245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuzick J, Berney DM, Fisher G, et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer. 2012;106:1095–1099. doi: 10.1038/bjc.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein EA, Cooperberg MR, Magi-Galluzzi C, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol. 2014;66:550–560. doi: 10.1016/j.eururo.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Kumar A, Coleman I, Morrissey C, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22:369–378. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. doi: 10.1016/j.cell.2015.10.025. Cancer Genome Atlas Research Network: The molecular taxonomy of primary prostate cancer. Cell 163:1011-1025, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Taylor BS, Schultz N, Hieronymus H, et al: Integrative genomic profiling of human prostate cancer. Cancer Cell 18:11-22, 2010. [DOI] [PMC free article] [PubMed]
- 48.Armenia J, Wankowicz SAM, Liu D, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet. 2018;50:645–651. doi: 10.1038/s41588-018-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. doi: 10.1172/jci.insight.123468. Salami SS, Hovelson DH, Kaplan JB, et al: Transcriptomic heterogeneity in multifocal prostate cancer. JCI Insight 3:123468, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hieronymus H, Schultz N, Gopalan A, et al. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci USA. 2014;111:11139–11144. doi: 10.1073/pnas.1411446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarker D, Reid AH, Yap TA, et al. Targeting the PI3K/AKT pathway for the treatment of prostate cancer. Clin Cancer Res. 2009;15:4799–4805. doi: 10.1158/1078-0432.CCR-08-0125. [DOI] [PubMed] [Google Scholar]