Abstract
We write to introduce our novel group formed to confront some of the issues raised by the COVID-19 pandemic. Information about the group, which we named “cure COVid for Ever and for All” (RxCOVEA), its dynamic membership (changing regularly), and some of its activities—described in more technical detail for expert perusal and commentary—are available upon request.
Keywords: COVID-19 research, bioinformatics, systems biology, immunology
1. WE ARE ORGANIZED DIFFERENTLY
The group is quite informal, rapidly evolving, easy to join and to leave. It now gathers about 100 members from more than 15 countries, representing a broad swath of disciplines relevant to the present crisis including, but not limited to, epidemiological modeling, artificial intelligence, immunology, game theory, drug development, diagnostic screening and testing, economics, and data management (Fig. 1). We are all volunteers, and our objectives are neither fame nor fortune; we simply hope to be helpful.
FIG. 1:
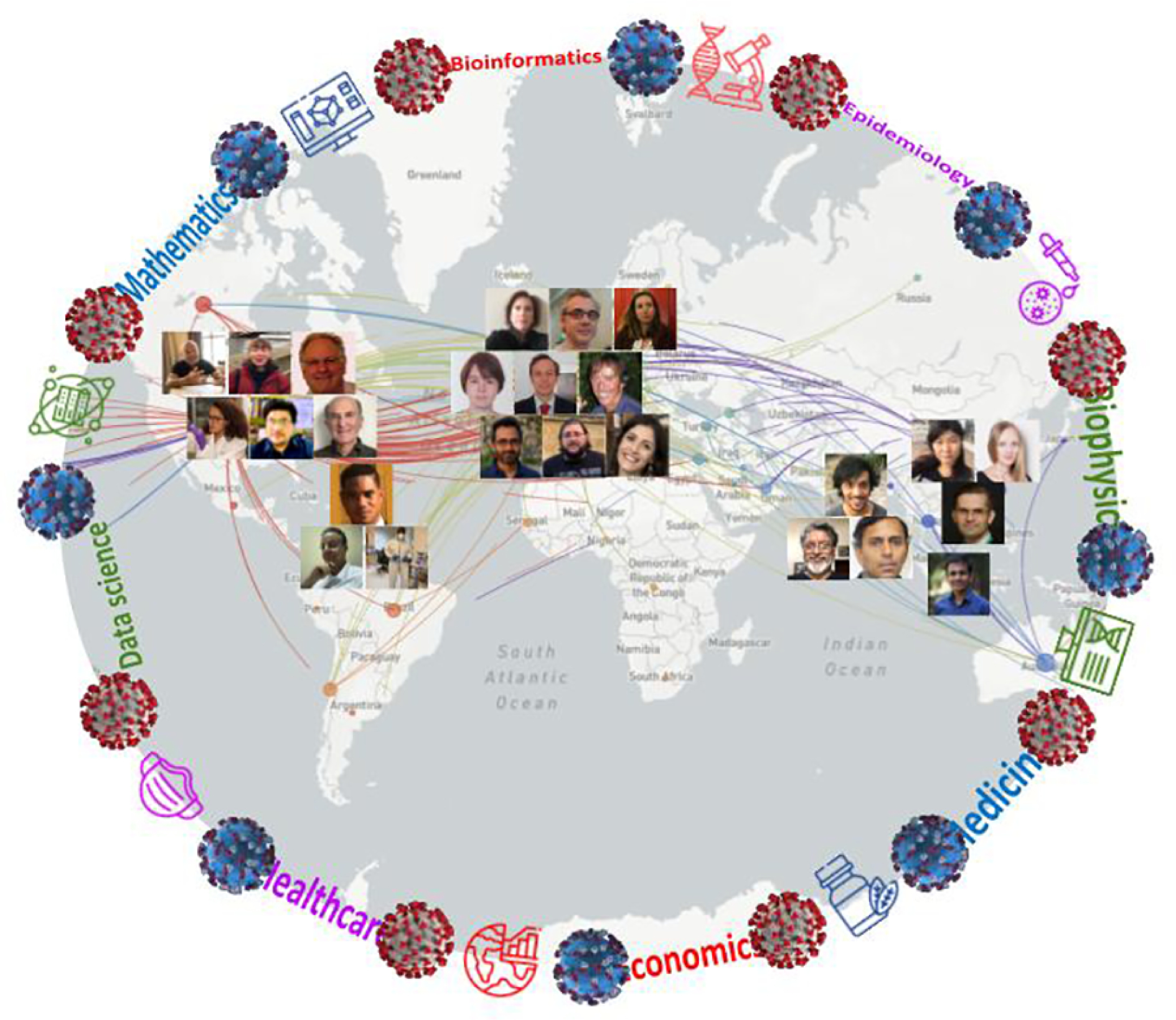
The “cure COVid for Ever and for All” (RxCOVEA) Framework: A Global Network. Pan-national members of RxCOVEA superimposed on the pandemic viral genomic tracing map of COVID-19 spread (Gisaid.org). The serious health and economic consequences of the interconnected world in spreading the infection are contrasted by the rigorous science and technology projects, self-assembling spontaneously and equally rapidly around the globe (representative images shown of RxCOVEA member locality).
2. YOUNG PEOPLE PLAY A MAJOR ROLE: BRIDGING THE GAP
Our premise is that existing methods to understand and manage this pandemic are inadequate, and that significant innovations are required to meet the current challenge, as well as to prepare to respond to any future pandemics effectively and efficiently. Innovations cannot be imposed, managed, or predicted, but the likelihood is greatly increased when disciplinary boundaries are bridged and young scientists and technologists can freely access mentors while strategizing against unprecedented challenges. Thus, from the outset, our members ranged widely in terms of ages and expertise, including more than 20 students. As technology has developed, so have our overall skills that reflect a particular generation. The gaps seen between the typical generations “Boomers” and “Millennials,” next to social and financial, include coding efficiency and tech fluency. Using the discipline, expertise, and mentorship of the older generations, in combination with the tech skills of the younger generations, we hope to not only bridge the gap, but use each individual’s talent(s) to come up with innovative strategies on how to solve the unique challenges of Coronavirus disease 2019 (COVID-19).
3. WE TRANSCEND ALL DISCIPLINARY BOUNDARIES
Our current activities are a mix of cross-discipline education to break down communication and other barriers that arise between subjects such as computer science and immunology, a journal club to keep abreast of related research, and core projects, e.g., building software for better epidemic modeling and related economic consequences, drug design and drug repurposing (combining artificial and human intelligence), and exploring non-mainstream tools for broad population screening and contact tracing, all taking into account the uniqueness of individual humans. The COVID-19 problem is too complex for any one group or discipline to handle. We encourage unconventional interactions and non-hierarchical organization within our group to generate ideas and tools, and then subject these to rigorous scrutiny and testing by proven experts. We are grappling with issues that mostly pertain to the difficult decisions involved in relaxing social distancing and confinement policies.
4. OUR CURRENT ACTIVITIES ARE CENTERED ON IMPROVED MODELING AND REASONING
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can apparently be easily transmitted by asymptomatic carriers (Gandhi et al., 2020; He et al., 2020). This introduces spatial and temporal instabilities into conventional epidemiological models (Jewell et al., 2020). More complex models are needed, but the question remains as to how one may estimate the necessary parameters needed for these models. Our models aim to capture the unique aspects of SARS-CoV-2, such as the host entry characteristics and spreading dynamics of infected persons. Using the combined efforts of data analysis and biological insight, the objective is to identify necessary parameters that reflect specific context and thus are able to provide suitable predictions about disease development.
It seems that massive and continual population testing will be required to spot inevitable pockets of COVID-19 recurrence before these spread into larger outbreaks. The costs, delays, and invasiveness of current methods are untenable. We are modeling measurements on large sample pools or other surrogates for individual testing, to determine if a quantum leap in testing efficiency can be achieved. Similarly, the costs and ethical risks of contact tracing on the scale needed for COVID-19 are staggering. We are modeling inexpensive ways to provide individuals with private estimates of their own risk and at the same time ways of making this information available for public health use, without a complete loss of individual privacy. To combat the pandemic, effective vaccines and therapeutics are indispensable—certainly for severe cases. Like many other entities, we are exploring why the disease course and progression in different individuals is so heterogeneous (De Clercq and Li, 2016). Progress is essential here for prioritizing treatment options, but also for more accurate and logical reasoning in the complex field of immunology and epidemiology; see, e.g., Du and Yuan (2020). Surely COVID-19 poses unique challenges to the innate and adaptive immune system and a successful vaccine may require unusual strategies. Development of effective antiviral drugs does not have a long history of success (De Clercq and Li, 2016). In the short term, any good candidates for drug repurposing need to be carefully explored (Zhavoronkov, 2020). In the long term, novel points of drug intervention in the viral life cycle need to be discovered and tested. Finally, the COVID-19 pandemic is playing out in very different ways in different nations and in different ethnic groups within nations. These differences need to be expeditiously and rigorously studied.
ACKNOWLEDGMENTS
T.S. is grateful for the support from the Division of Mathematical Science of NSF, RAPID Award 2030377, and from the National Institute of General Medical Sciences, NIH, Award R35GM122562.
APPENDIX
https://cs.nyu.edu/mishra/RXCovea.html
RxCOVEA Framework Members
Alexandra Acosta,1 Salvatore Alaimo,2 Ernest Battifarano,3 Vijay Chandru,4;5 Shirshendu Chatterjee,6 John Connolly,7 James Edmondson,8 Inavamsi Enaganti,9 Sheldon Gilbert,10 Ramesh Hariharan,5 Kose John,11 Judith Klein-Seetharaman,12;13;14 Ramón Luna de Maria,15 Jeffrey N. Maria,16; 17 Steven E. Massey,18 Kuldeep S. Meel,19 Alfredo Pulvirenti,2 Qianru (Cheryl) Qi,20 R. Andrea Ramirez,14 Valentina Rapicavoli,2 Javier Rionegro,14 Samir Saadi,20 Tariq Samad,21 Jaime Travesedo,14 Eva Xueyao Guo,22 Alex Zhavoronkov,23 Alessia Stasuzzo24
RxCOVEA Framework Affiliations
Pentaquark Consulting, Madrid, Spain
Department of Clinical and Experimental Medicine, Catania University, Italy
School of Professional Studies, New York University, New York, New York, USA
Indian Institute of Science, Bengaluru, India
Strand Life Sciences, Bengaluru, India
The City University of New York, City College and Graduate Center, New York, New York, USA
Institute of Molecular and Cell Biology, A*Star, Singapore
Galois, Portland Oregon, USA
Courant Institute, New York University, New York, New York, USA
Proclivity Systems, New York, New York, USA
Stern School, New York University, New York, New York, USA
Department of Chemistry, Colorado School of Mines, Golden, Colorado, USA
Biosciences and Bioengineering, Catalyst Health Tech Integrator, Denver, Colorado, USA
Initiative for AI in Bio and Health, Colorado School of Mines, Golden, Colorado, USA
Pentaquark Consulting, Madrid, Spain
ENT Clinic Curaçao, Willemstad, Curaçao
Curaçao Medical Center, Willemstad, Curaçao
Biology Department, University of Puerto Rico, Rio Piedras, San Juan, Puerto Rico
School of Computing, National University of Singapore, Singapore
Telfer School of Management, University of Ottawa, Ottawa, Ontario, Canada
Technological Leadership Institute, University of Minnesota, Minneapolis, Minnesota, USA
University of Rochester Computational Biology, Rochester, New York, USA
Insilico Medicine, Rockville, Maryland, USA
Department of Biomolecular Chemistry, Catania University, Italy
Footnotes
CONFLICT OF INTEREST
E.B. is an executive member of Women’s Brain Project, Switzerland.
REFERENCES
- De Clercq E and Li G, Approved Antiviral Drugs over the past 50 Years, Clin. Microbiol. Rev, vol. 29, no. 3, pp. 695–747, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du SQ and Yuan W, Mathematical Modeling of Interaction between Innate and Adaptive Immune Responses in COVID-19 and Implications for Viral Pathogenesis, J. Med. Virol, 2020. (ahead of print). DOI: 10.1002/jmv.25866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M, Yokoe DS, and Havlir DV, Asymptomatic Transmission, the Achilles’ Heel of Current Strategies to Control COVID-19, N. Engl. J. Med, 2020. (ahead of print). DOI: 10.1056/nejme2009758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Lau EH, Wu P, Deng X, Wang J, Hao X, Lau YC, Wong JY, Guan Y, Tan X, and Mo X, Temporal Dynamics in Viral Shedding and Transmissibility of COVID-19, Nat. Med, vol. 26, no. 5, pp. 672–675, 2020. [DOI] [PubMed] [Google Scholar]
- Jewell NP, Lewnard JA, and Jewell BL, Caution Warranted: Using the Institute for Health Metrics and Evaluation Model for Predicting the Course of the COVID-19 Pandemic, Ann. Intern. Med, 2020. (ahead of print). DOI: 10.7326/m20-1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhavoronkov A, Vanhaelen Q, and Oprea TI, Will Artificial Intelligence for Drug Discovery Impact Clinical Pharmacology?, Clin. Pharmacol. Ther, vol. 107, no. 4, pp. 780–785, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]


