Abstract
Introduction
The alarming rise in urinary tract infection (UTI) antimicrobial resistance has resulted from a combination of high prevalence, low specificity and the lack of a rapid, point-of-care (POC) antibiotic susceptibility test (AST), which has led to the overuse/inappropriate use of antibiotics.
Aim
This study aimed to evaluate the performance of a rapid POC phenotypic AST device in reporting susceptibility information within 2 h.
Methodology
Instrument calibration was performed with model bacteria and fluorescent microbeads to determine the dynamic range and limit of detection for quantifying concentrations of bacteria and demonstrate the ability to rapidly differentiate susceptible and resistant model bacteria. We then evaluated 30 presumptive UTI-positive patient urine samples in a clinical pilot study using a panel of 5 common UTI antibiotics plus a growth control and compared our results to the hospital standard of care AST.
Results
Our device was able to robustly detect and quantify bacteria concentrations from 50 to 105 colony-forming units (c.f.u.) ml−1. The high sensitivity of this measurement technique enabled the device to differentiate between susceptible and resistant model bacteria with 100 % specificity over a 2 h growth period. In the clinical pilot study, an overall categorical agreement (CA) of 90.7 % was observed (sensitivity=91.4 %, specificity=88.9 %, n=97) with performance for individual drugs ranging from 85 % CA (ceftazidime) to 100 % (nitrofurantoin).
Conclusions
By reducing the typical timeframe for susceptibility testing from 2–3 days to 2 h, our POC phenotypic AST can provide critical information to clinicians prior to the administration of antibiotic therapy.
Keywords: UTI, antimicrobial resistance, AST, cell counting, point of care, diagnostics
Introduction
The alarming rise in antimicrobial resistance (AMR) across the globe is a growing threat to food security and public health. An estimated 2 million illnesses and 23 000 deaths per year are due to antibiotic resistance in the USA and, under the executive order ‘combating antibiotic resistant bacteria’, it has become a national priority [1, 2]. Unlike infections due to antibiotic-susceptible micro-organisms, clinically significant forms of antibiotic resistance contribute to increased morbidity and mortality of infections [3]. Antibiotic-resistant infections are difficult and sometimes impossible to treat, require the use of costly/toxic alternatives to first-line antibiotics and require extended hospital stays or additional follow-up doctor visits. All of these factors contribute to poor clinical outcomes for individual patients and a growing financial burden on the healthcare system [4–6].
Other than novel therapeutics, the development of rapid and accurate antimicrobial susceptibility testing (AST) lies at the core of concerted efforts to uphold sound antibiotic stewardship practices and prevent the spread of drug resistance. Under the US National Action Plan for Combating Antibiotic-Resistant Bacteria, there is a call for action to ‘advance development and use of rapid and innovative diagnostic tests for identification and characterization of resistant bacteria’ [7]. Prior to AST, more accurate point-of-care (POC) screening tests have the potential to prevent overuse/inappropriate use of antibiotics (e.g. administration of antibiotics for inflammation or non-bacterial infections). Furthermore, rapid and accurate AST can help to prevent misuse of antibiotics (e.g. administration of first-line antibiotics for antibiotic-resistant infections). However, rapid, POC phenotypic AST devices with the ability to provide physicians with AST results within a timeframe that can help inform primary treatment decisions are severely lacking [8].
One of the most common forms of human bacterial infections are urinary tract infections (UTI), resulting in an estimated 7 million cases annually and associated costs of $1.6 billion in the USA alone [9, 10]. Patients with community-acquired UTIs or bacteriuria are among the top recipients of empirical outpatient antibiotic prescriptions [11]. UTIs have a high prevalence in females, affecting nearly 50 % of the female population at least once in their lifetime and necessitating antibiotic treatment for approximately one out of every three women before the age of 24 [12, 13]. For some of these outpatient cases of UTI, patient symptoms are not completely resolved within the first 2 weeks of empirical antibiotic treatment, leading to additional clinical visits and alternative antibiotic therapy. Furthermore, within less than 1 year of an initial infection, an estimated 30 % of women will experience a recurrent UTI, requiring additional visits to clinics and emergency rooms as well as antibiotic therapy [14]. These alarming trends, which include the rise of UTI antimicrobial resistance and the increase in morbidity and mortality of a once easily treatable infection, illustrate the critical clinical challenges for UTI diagnosis and treatment. For example, current rapid diagnostics such as the dipstick test for asymptomatic bacteriuria and UTI suffer from low specificity [15], resulting in high rates of misdiagnosis, which leads to the unnecessary use of antibiotics. In combination with high UTI prevalence and recurrence rates, the lack of technological advancement, especially in POC AST, has contributed to the misuse of antibiotics [16] and a rapid increase in antibiotic-resistant strains of uropathogens [17].
Although genotypic tests are reputably rapid when compared to the gold-standard culture test, many genotypic tests are still limited in detection sensitivity and/or coverage of resistance genes and may require pre-processing steps such as growth culture, which increases the overall sample-to-answer time. Additionally, these molecular tests do not always provide definitive results in instances where the absence of a resistance gene is not sufficient to conclude that the infectious agent is antibiotic-susceptible [18]. In the 24 h period (minimum turnaround time) that is required for the completion of gold-standard phenotypic AST, physicians must first administer antibiotic treatment to patients empirically, resulting in poor patient outcomes as described above. Accurate and sensitive phenotypic AST at the POC represents a new treatment paradigm, providing clinicians with vital information to improve initial antimicrobial therapy selection for patients with new or recurrent infections.
Here, we describe a rapid phenotypic AST for bacteriuria and UTI, with results on the antibiotic susceptibility/resistance of bacteria available in as little as 2 hours. The key advantage, when compared to standard phenotypic ASTs, lies in the instrument’s sensitivity, allowing for the detection of low concentrations of micro-organisms, eliminating the need for pre-culture/enrichment or extensive pre-processing of urine samples prior to AST. Regardless of the clinical presentation of the patient, the POC instrument is equally capable of screening urine from asymptomatic bacteriuria or UTI patients to give physicians information on sample bacterial load as well as conducting antimicrobial susceptibility testing of presumptive positive urine samples. The nature of UTI and progression of the disease throughout the urinary tract system is not expected to influence the performance of the rapid phenotypic AST; however, we foresee that the greatest clinical value of our technology will lie in the early testing of samples from patients with bacteriuria and acute cystitis in order to prevent the overuse and misuse of antibiotics, with an aim of improving antibiotic stewardship. The POC system that is under development is intended to provide rapid phenotypic susceptibility information to physicians within a timeframe that enables them to make more judicious choices when prescribing antibiotics for the majority of UTI cases. Since it operates with direct patient specimen, the user will only need to load the raw sample into the device, and the instrument will dispense the sample along with growth media to individual wells containing lyophilized drugs and reagents on a disposable cartridge.
In the current embodiment, aliquots of a patient urine sample are added to vials containing growth media with different antibiotics and incubated over a period of 2 hours. During this period, growth curve measurements are performed on each vial using a portable particle counting instrument and a fluorescent staining solution that specifically labels nucleotides within individual bacterial cells. The AST device allows for the sensitive detection of changes in the growth of antibiotic-treated and control samples by counting discrete, fluorescently labelled bacterial cells, analyses data in real time with integrated software, reports presumptive ‘susceptible’ or ‘resistant’ results and can test various panels of antibiotics typically used to treat UTI [19]. A panel of first-line antibiotics commonly used for UTI treatment was used for our AST panel, and since the majority of bacteriuria and uncomplicated UTIs are caused by members of the family Enterobacteriaceae , single drug concentrations based on European Committee on Antimicrobial Susceptibility Testing (EUCAST) minimum inhibitory concentration (MIC) breakpoints were used for the clinical evaluation [20]. In this study, we evaluated the technical sensitivity and accuracy of our device to enumerate bacteria across a wide dynamic range and demonstrate its utility for phenotypic antibiotic susceptibility testing with the use of laboratory models of susceptible and resistant bacteria and contrived samples. Finally, we investigated the clinical sensitivity and specificity of our prototype phenotypic AST POC device by running a clinical pilot evaluation on patient urine samples and compared our results with those of the standard hospital AST.
Methods
Bacteria rapid quantification instrument characteristics
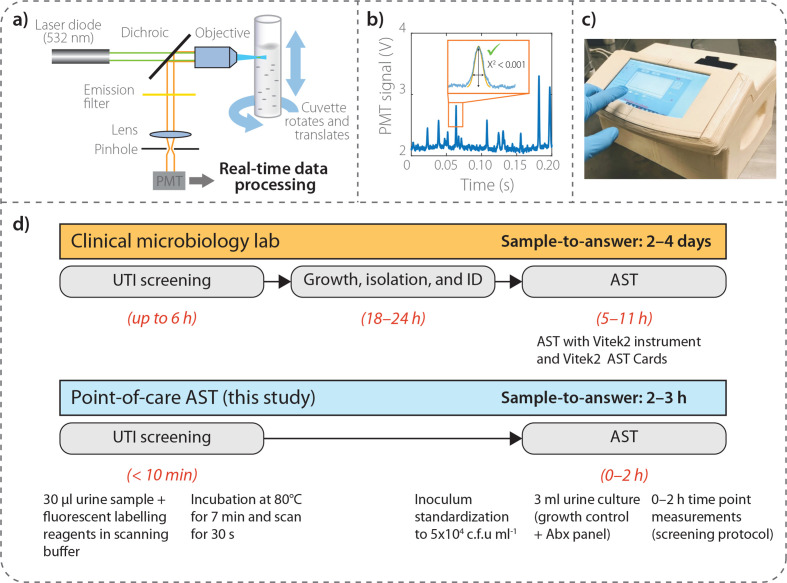
The portable, compact particle counter instrument is designed and manufactured by ASI Srl (Milan, Italy) for the quantification of fluorescent particles in liquid media, specifically fluorescently labelled microbes in this particular application [19]. The instrument consists primarily of a fluorescence confocal microscope oriented in a horizontal geometry (Fig. 1). Briefly, fast rotation (~5 rev s−1) and slower vertical inversion (~4 mm s−1) of the sample cuvette (1 cm in diameter), imparted by two synchronous motors (rotation: Johnson Electric UBR13NB1RN; vertical: Johnson Electric UFR10NB1NR+CEM reducer) results in a spiral trajectory that transports labelled bacteria through the optical analysis volume. Excitation light generated by a 532 nm laser diode (Apinex AGLM2-05) with optical power <3 mW is focused ~200 µm within the cuvette by a lens, thereby causing particles in the sample to fluoresce. The emitted fluorescence signal is collected by the same focusing lens and reflected by a dichroic mirror (ODL, SW 570) to the optical sensor through a lens that focuses the light on a 200 µm slit in front of a long-pass filter (HOYA, 600 nm). The photodetector [photomultiplier tube (PMT); Hamamatsu H10721-110] is in optical communication with the confocal microscope and receives a portion of the fluorescence from the observation volume, measuring its intensity as a function of time and generating a temporal profile of the fluorescence signal from the observation volume. The PMT output electric signal is amplified and digitized at 40 kHz and transmitted to a PC. In real time, the processor applies a pattern recognition algorithm to the temporal profile to determine the concentration of particles in the sample. The algorithm matches features in the temporal profile to predetermined patterns that correspond to the time-dependent fluorescence intensity of particles passing through the observation volume. The concentration of particles is determined by calculating the number of predetermined patterns matched to features in the temporal profile for a given sample scanning period compared to a calibration curve.
Fig. 1.
Instrument schematic and AST workflow. (a) Schematic of fluorescent particle counter instrument. By rotating and translating a cuvette containing the sample, fluorescently labeled bacteria pass in front of the observation volume of a horizontally oriented, miniature confocal microscope. (b) Real-time data processing of fluorescence time trace recorded by a photomultiplier tube (PMT) where a pre-defined shape is applied to correctly identify signatures originating from single bacteria. (c) Photograph of particle counter instrument with recent updated shell design incorporating touch screen user interface. (d) Comparison of clinical AST workflow with our AST device, whereby we are able to compress the typical timeframe from 2 to 4 days to 2–3 h for high-level antibiotic resistance status for a panel of antibiotics.
Fluorescent microbead dilution curve
To calibrate the particle counter instrument and assess its detection sensitivity across a wide dynamic range, stable, fluorescent beads used for flow cytometer instrument calibration were prepared and measured. Fluorescent microbeads [4 µm PS/DVB microspheres, Envy Green, Ex/Em 525/565 nm (Bangs Laboratories, Inc.)] were serially diluted in buffer solution (phosphate buffered saline (PBS), 0.02 % Tween 80) to prepare stock dilutions of 3×107 to 3×103 beads ml−1. Samples of 1×106 to 50 beads ml−1 were prepared by aliquoting 0.10 ml of stock dilutions into 2.90 ml of fresh buffer solution for a final sample volume of 3 ml. Negative samples (0 beads ml−1) were prepared by adding 0.10 ml of blank buffer to each negative sample cuvette. Technical replicates of each dilution were prepared in triplicate. Samples were scanned for 60 s and scanning was repeated three times for each sample. Hit counts from the three measurement replicates were averaged for each sample; standard deviations were reported for the three technical replicates. Logistic regression was used to fit the known bead concentrations to the hit rate (hits 60 s−1) for concentrations between 50 and 1×105 particles ml−1, representing the linear reportable range for the scanning instrument.
Isolation, proliferation and general preparation of model bacteria
Individual colonies of bacteria on plates were obtained by performing isolation streaks with cultures of antibiotic-susceptible wild-type Escherichia coli (WT E. coli ) [ATCC 295922 E. coli (Migula Castellani and Chalmers)] and antibiotic-resistant E. coli kan ( E. coli KR) on Lennox broth (LB) agar plates and LB agar plates under selective pressure with the addition of a working concentration of 50 µg ml−1 of kanamycin (Kan) (kanamycin sulfate, Sigma-Aldrich), respectively. For the proof-of-concept work with contrived samples performed in this study, both the antibiotic-susceptible and -resistant strains of E. coli were well-characterized, isogenic isolates. The working concentration of antibiotic was based on prior MIC and minimum bactericidal concentration (MBC) testing of antibiotic-susceptible and -resistant bacteria strains (data not shown). On three separate nights prior to the day of the experiment, individual colonies of bacteria were selected from isolation streak plates with a sterile loop and used for the inoculation of 5 ml overnight cultures of WT E. coli and E. coli KR in liquid growth medium. The following morning, single millilitre samples of overnight cultures grown to early stationary phase were harvested, washed and resuspended with fresh isotonic solution (0.9 % NaCl). Resuspended bacteria were diluted with isotonic solution in twofold intervals, and optical density measurements of undiluted to fourfold diluted culture were taken [absorbance was measured at λ=600 nm (OD600)] to determine the colony-forming units (c.f.u.) ml−1 concentrations.
E. coli dilution curve
To verify the results from the fluorescent microbead calibration exercise, solutions of bacteria were prepared and measured using our fluorescent staining protocol. Bacterial suspensions of WT E. coli were serially diluted to prepare stock concentrations from 3×107 to 3×103 c.f.u. ml−1. Glass cuvettes (which also serve as scanning vessels) with 2.90 ml of nucleic acid staining solution (0.5 µM SYTOX Orange in 0.02 % Tween 80, ddH20) were prepared for each bacteria concentration and 0.10 ml aliquots of stock concentrations were added directly to the staining solutions within the respective cuvettes for final concentrations from 1×106 to 1×102 c.f.u. ml−1. Negative samples (0 c.f.u. ml−1) were prepared by adding 0.10 ml of blank buffer to each negative sample cuvette. Staining reactions were heated for 7 min at 80 °C in a dry heat bath (Corning LSE Digital Dry Bath Heater) capped with sterile, disposable safety caps and scanned for 60 s each. Three separate biological replicates of each dilution were prepared and scanned three times each to determine the number of positive hits per minute, the variation among biological replicates (reported as standard deviation) and the linearity of the measurement response with respect to the calibration exercise performed with fluorescent microbeads.
AST growth curve
Prior to the launch of the clinical pilot study, contrived sensitive and resistant bacteria were tested according to the rapid AST measurement protocol. For each biological replicate, overnight cultures of WT E. coli and E. coli KR were prepared in liquid LB broth as described above, harvested during the mid-exponential phase, and resuspended in fresh isotonic buffer. The optical density (OD600) was measured for the resuspended bacterial stock and bacteria suspensions were serially diluted to a final concentration of 5×105 c.f.u. ml−1. Bacteria stock concentrations were verified by c.f.u. plate assays as described below. The contrived test sample concentration was based on the widely accepted clinical threshold of 1×105 c.f.u. ml−1 for a presumptive positive UTI screening test [11]. Initial concentrations for all growth cultures were kept constant at 5×104 c.f.u. ml−1 by spiking 2.70 ml of modified Lennox broth (M-LB) liquid media, with or without 50 µg ml−1 Kan, with 300 µl of our contrived test sample. Negative control cultures (with and without 50 µg ml−1 of Kan) were spiked with 300 µl of sterile isotonic solution. The complete set of AST growth culture samples included the following: WT E. coli – no antibiotic (antibiotic-susceptible positive control); WT E. coli +Kan (antibiotic-susceptible experimental sample); E. coli KR – no antibiotic (antibiotic-resistant positive control); E. coli KR+Kan (antibiotic-resistant experimental sample); sterile isotonic solution – no antibiotic and sterile isotonic solution+Kan (negative controls). Three biological replicates were prepared on separate days for each set of AST growth cultures. Sample measurements were taken for each time point, specifically at 0, 1, 2 and 3 hours (t 0–t 3). At each time point, glass cuvettes with 2.90 ml of staining solution were prepared for each sample, as described above. Specifically, 0.10 ml aliquots of each growth culture were added directly to staining solutions within respective cuvettes. Cuvettes were heated for 7 min at 80 °C and scanned for 60 s on the particle counter instrument. The average number of hits 60 s−1 was recorded for three measurement replicates; standard deviations were reported across the three independent biological replicates. Growth curves were plotted (Fig. 2b, c) and utilized in the determination of ‘susceptible’ and ‘resistant’ labels.
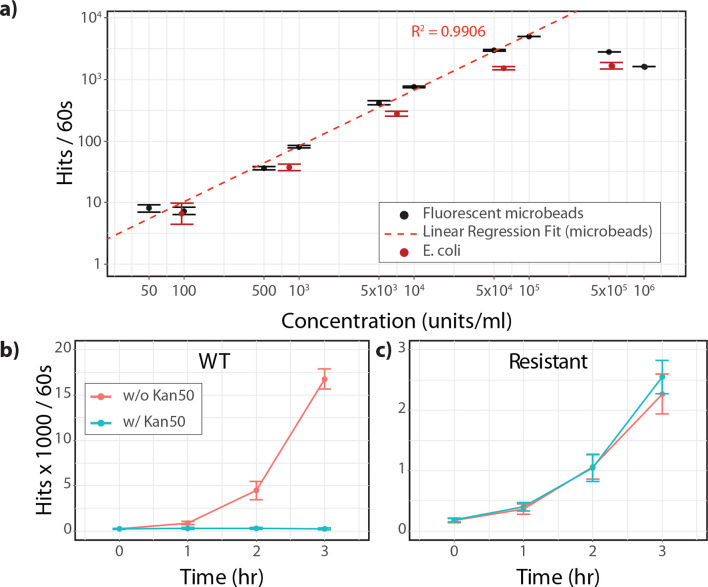
Fig. 2.
Performance demonstration of particle counter device for (a) bacterial quantification and (b, c) AST. (a) Linear response of particle counter instrument for quantifying bacterial concentrations [LoD=50 c.f.u. ml−1, linear range: 50–105 c.f.u. ml−1 (R 2=0.9906), based on fluorescent calibrator beads]. Average response, average of three measurement replicates; error bars, standard deviation across three independent biological replicates prepared on separate days. (b) Demonstration of AST growth curves for drug-treated (blue) and non-drug-treated (red) using model susceptible organism (WT), and (c) model resistant organisms. Average response, average of three measurement replicates; error bars, standard deviation across three independent biological replicates.
CFU verification assays
Bacteria concentrations for all biological replicates were verified by solid agar plate assays. LB agar plates were made in standard Petri dishes (9 cm diameter). Each sample was serially diluted to a final concentration of 103 c.f.u. ml−1 and spread (0.10 ml) onto three independent agar plates using a sterile loop and turntable. All plates were incubated overnight at 37 °C. Colonies were counted the following morning; the average c.f.u. count was used to determine the original c.f.u. ml−1 concentration of the stock dilution.
Clinical AST proof of concept
Patient enrolment
Prior to inclusion in the AST pilot evaluation, discarded urine samples collected as part of routine clinical evaluation were screened for ‘presumptive UTI-positive’ status using the particle counter system as described previously [19] (ASI, Milan, Italy). Samples were collected from both inpatients and outpatients from two clinical sites without any specific patient enrolment criteria. A total of 30 presumptive UTI-positive urine specimens were enrolled from Brotzu hospital (n=11) (Cagliari, Italy) and Monserrato University Hospital (n=19) (Cagliari, Italy) over a period of 13 days between 7 June 2018 and 16 July 2018 with informed consent from donors and approval from the University Hospital of Cagliari Institutional Review Board (PG/2018/5211). Due to personnel limitations, a maximum number of four presumptive positive samples per day were enrolled in the AST pilot evaluation (n=1–4 samples per day). Each sample was identified by a serial code with no patient information, such as sex or age. Urine specimens were stored at 4 °C without preservatives and tested with the rapid AST device within 3 h of sample collection.
Clinical antibiotic sensitivity testing using conventional systems
Clinical antibiotic sensitivity tests at each hospital were performed using a VITEK2 (BioMérieux). After primary isolation, presumptive and species identification of bacteria was performed using the LinearCount6 test and VITEK MS, and a standardized inoculum was prepared in 0.9 % saline solution at 0.5 McFarland turbidity with the use of VITEK2 DensiChek. Antibiotic sensitivity tests were performed using single-use AST-N376 cards for Gram-negative bacteria, AST-N659 for Staphylococcus spp. and AST-N658 for Enterococcus spp. Reading times varied from 5 (threshold value) to 11 h for slow-growing micro-organisms.
Clinical AST analysis with a rapid bacterial quantification system
A panel of five commonly used first-line UTI antibiotics [amoxicillin clavulanate (AX), ciprofloxacin (CP), ceftazidime (CZ), fosfomycin (FF) and nitrofurantoin (NF)] was selected with concentrations defined based on MIC thresholds outlined by the EUCAST guidelines [21]. Specifically, 4× maximum MIC concentrations were used for time-dependent drugs (AX, CZ, NF) and 10× maximum MIC concentrations were used for concentration-dependent drugs (CP, FF) [22–24].
For each patient sample, six tubes containing 3 ml of broth (LB, Sigma-Aldrich) were prepared, one for each of the antibiotics in the panel as well as a positive growth control without antibiotics. For this study, the non-selective, enriched growth medium was selected based on the highly referenced use of LB for growth of the majority of uropathogens, including E. coli (the primary infectious agent of UTI), as well as the relatively low autofluorescence of the medium. The hospital’s conventional AST employed standard, eugenic growth media approved for antimicrobial susceptibility testing. For each unique patient sample that tested positive for UTI screening, based on sample concentrations obtained during UTI screening, an aliquot of the raw urine sample was spiked into each of the six culture tubes for a final concentration of 5×104 c.f.u. ml−1. While this study used a common threshold for bacteriuria of 1×105 c.f.u. ml−1 and an inoculum of 5×104 c.f.u. ml−1 for the AST test, the sensitivity of the measurement technology is not limited to these values. In fact, the threshold for determining presumptive UTI-positive samples can be adjusted depending on the clinical setting, and the rapid AST test can utilize lower starting concentrations of bacteria (Figs 2a and S1, available in the online version of this article). At time points of 0, 1, 2 and 3 h, 30 µl of culture was added to cuvettes containing 3 ml of isotonic solution plus 0.5 µM SYTOX Orange (Thermo Fisher Scientific, Waltham, MA, USA) nucleic acid staining solution, heated for 7 min at 80 °C and scanned for 30 s on the particle counter unit to record bacteria concentrations. All culture tubes were incubated at 37 °C between subsequent time point measurements.
Data analysis
Bacterial growth in clinical samples was defined by a combination of different variables, including the growth rate of the antibiotic-treated sample [antibiotic growth rate (AGR), Fig. 3], the growth rate of the control sample, and a ratio of the relative difference between the two samples from the t 0 time point to the t 2 time point [antibiotic/control ratio (ACR), Fig. 3]. Due to the importance of comparing the growth of antibiotic-treated samples to that of their respective untreated control, samples in which the positive control failed to demonstrate growth within 3 h were omitted from the analysis (n=5 patients, defined as having an absolute value of less than 1000 hits 30 s−1 at the 3 h time point). While the screening method used to enroll samples has a very low false-negative rate of less than 1%, it carries a false-positive rate of approximately 22 % [19], which may explain the 17 % of samples that failed to grow in this patient cohort. Due to the rapid turnaround time of the POC AST, this test can also serve as an opportunity to detect presumptive negative UTI samples prior to the administration of antibiotics. Each of the remaining 25 patients had four time point measurements (0, 1, 2, 3 h) recorded for their control samples and 5 antibiotic-treated samples, resulting in a total of 600 measurements.
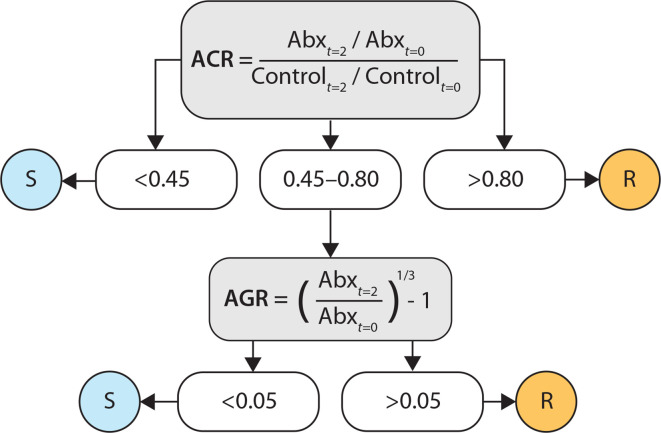
Fig. 3.
Labelling criteria for assigning ‘susceptible’ (S) and ‘resistant’ (R) categories for each sample based on antibiotic/control ratio (ACR) and antibiotic growth rate (AGR). Determination of S/R labels is dependent on relative growth rates of drug-treated sample (Abx) and growth control (control) between t=0 and t=2 h time points.
To evaluate the performance of the prototype AST device compared to the standard-of-care AST test adopted by the two clinical sites (VITEK2, BioMérieux) in terms of binary classification of susceptible (0) or resistant (1) labels, 2×2 confusion matrices were created using the ‘confusionMatrix’ function in the ‘caret’ package (version 6.0, https://topepo.github.io/caret/) in R (R Project, version 3.5.0) and used to quantify accuracy, sensitivity and specificity. A total of 97 VITEK results were reported from the clinical microbiology laboratories for the 125 sample–drug combinations. Final criteria were established for the determination of susceptible and resistant labels for the prototype AST test using a simple decision tree with user-defined breakpoints for antibiotic/control ratios and Antibiotic-growth rates (Fig. 3). Breakpoints for the ACR and AGR values were adjusted based on contrived sensitive and resistant model bacteria along with the first 5 sensitive and resistant sample–drug combinations, and then applied to the remaining 87 combinations.
Results
Rapid bacterial quantification
Conventional AST systems that rely on optical density-based measurements suffer from poor sensitivity and require massive proliferation of bacteria to induce a detectable change [8, 25]. Furthermore, these systems typically require starting concentrations of approximately 1×107 c.f.u. ml−1 (roughly equivalent to an OD600 of 0.01 for E. coli and similarly sized particles) to establish a reliable baseline [26, 27]. Since most common clinical thresholds for UTI bacterial load fall between 1×104 to 1×105 c.f.u. ml−1 [28], direct patient specimen is not suitable for most conventional AST systems without an initial enrichment step, further adding to the delay in AST information.
In comparison, the lower limit of detection for our instrument is <1×102 c.f.u. ml−1 [29, 30] (Fig. 2a), enabling starting concentrations that are at least 1000 to 10 000-fold lower than those for commercially available spectrophotometers, and it is sensitive enough to detect a change in concentration within a single doubling time (Fig. S1). Here, we demonstrate that we can reliably detect bacteria with clinically relevant stock concentrations of 1×104 c.f.u. ml−1 (with final concentrations of 1×103 c.f.u. ml−1 in scanned samples) (Fig. 2a). Since E. coli is the causative agent for approximately 75–95 % of uncomplicated UTIs, we used E. coli as a model bacterium for our proof-of-concept experiments and AST drug panel [20]. WT E. coli or fluorescent microbead suspensions were diluted into buffer or staining solution in triplicate and scanned as described in the Methods section. As seen in Fig. 2a, there was a linear correlation between expected particles or c.f.u. ml−1 and the average number of hits per scan with a dynamic range between 50 and 1×105 particles ml−1. Additionally, there was a linear correlation between the number of positive hits for respective concentrations of bacteria and standardized microbead samples, further verifying the accuracy and robustness of our prototype device. c.f.u. plate verification experiments also confirmed estimated quantities of bacteria in scanned samples (Table S1). With these data, we clearly show that our device is suitable for growth curve/AST studies with clinically relevant starting concentrations of bacteria without the need for enrichment or pre-culture.
AST method validation with model bacteria
Next, to demonstrate our ability to carry out AST directly on small aliquots of fresh urine without the need for a pre-culture or enrichment step, we ran experiments with contrived samples of laboratory models of antibiotic-resistant and antibiotic-susceptible E. coli [kanamycin-resistant E. coli ( E. coli KR) and WT E. coli , respectively] for three independent biological replicates. Contrived samples containing E. coli KR and WT E. coli were prepared with a starting concentration of 5×105 c.f.u. ml−1 to reflect the most widely accepted clinical threshold for UTI. Contrived sample aliquots of 0.30 ml were added to 2.70 ml of growth media, in the presence or absence of antibiotics, for a final starting concentration of 5×104 c.f.u. ml−1, as described in the Methods section. From these growth cultures, 0.10 ml aliquots were removed, fluorescently labelled and scanned from time zero (t 0) to 3 h (t 3) for a total of four time point measurements. As seen in Fig. 2b, c, we were able to detect dynamic changes in growth between doubling times for untreated WT E. coli , untreated drug-resistant E. coli and antibiotic-treated E. coli KR from as early as 60 min, while growth in antibiotic-treated WT E. coli was not detected. By as early as the 2 h time point, we were able to clearly differentiate growth patterns between antibiotic-resistant and -susceptible E. coli in the presence of antibiotics in growth media (Fig. 2b, c). Growth parameters including antibiotic/control ratios and antibiotic growth rates comparing 0 and 2 h time points demonstrated a marked difference between the antibiotic-susceptible model (ACR=0.07, AGR=0.12) and the antibiotic-resistant model (ACR=0.95, AGR=0.80). Aliquots of growth culture were plated in parallel for each time point as a standard reference and c.f.u. counts confirmed the results obtained with our prototype phenotypic AST device (Table S2).
Clinical AST performance
Using the objective criteria shown in Fig. 3, all sample–drug combinations were differentiated into susceptible or resistant cases based on quantitative growth measurements. Samples with antibiotic/control ratios at the lower end of the spectrum (ACR <0.45=susceptible) or at the higher end of the spectrum (ACR >0.80=resistant) made up 82.5 % of the total number of cases (n=80). By also comparing the antibiotic growth rates of the intermediate cases (AGR <0.05=susceptible, AGR >0.05=resistant), the remaining 17.5 % of cases were divided into susceptible and resistant classes.
Combined performance across all 5 antibiotics from the 25 patients from the Brotzu and Monserrato hospitals resulted in an average categorical agreement (accuracy) of 90.7 % (sensitivity=91.4 %, specificity=88.9 %, Tables 1 and S3) for a total of 64/70 susceptible cases and 24/27 resistant cases correctly identified with respect to reported clinical AST results (VITEK) for the same samples. Categorical agreement for specific drugs varied from 85 % (ceftazidime) to 100 % (nitrofuratonin), with an average of 19 cases for each specific drug based on the availability of conventional AST results reported from each hospital’s clinical microbiology laboratory.
Table 1.
Summary of results from patient data. ‘Minor errors’: rapid AST, R; standard AST, S. ‘Major errors’: rapid AST, S; standard AST, R
|
N |
Categorical agreement (%) |
Minor errors (%) |
Major errors (%) |
Sensitivity (%) |
Specificity (%) |
|
|---|---|---|---|---|---|---|
|
COMBO (AX, CP, CZ, FF, NF) |
97 |
90.7% |
6 (6.2 %) |
3 (3.1 %) |
91.4% |
88.9% |
|
AX |
19 |
89.5% |
2 (10.5 %) |
0 (0 %) |
83.3% |
100% |
|
CP |
22 |
90.9% |
1 (4.5 %) |
1 (4.5 %) |
92.9% |
87.5% |
|
CZ |
20 |
85.0% |
3 (15.0 %) |
0 (0 %) |
78.6% |
100% |
|
FF |
21 |
90.5% |
0 (0 %) |
2 (9.5 %) |
100% |
60.0% |
|
NF |
15 |
100% |
0 (0 %) |
0 (0 %) |
100% |
100% |
AX, amoxicillin clavulanate; CP, ciprofloxacin; CZ, ceftazidime; FF, fosfomycin; NF, nitrofurantoin.
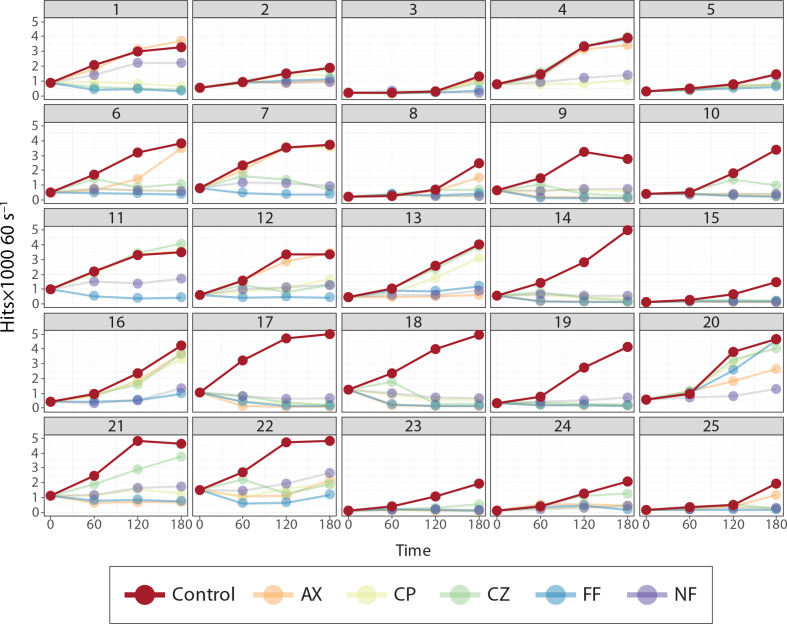
Although only initial and endpoint measurements at 0 and 2 h were used to identify resistant and susceptible bacteria from patient samples, additional measurements at 1 and 3 h were collected for increased granularity in the response of each sample to a panel of antibiotics with our rapid, highly sensitive bacterial quantification tool (Fig. 4).
Fig. 4.
AST panel measurements from 25 patient samples. Each panel (numbered 1–25 to signify patient ID) represents a single urine sample subjected to the five-drug AST panel plus growth control collected on the rapid POC prototype device.
Discussion
In this study, we describe the technical sensitivity and accuracy of our instrument for the detection and growth monitoring of bacterial models as well as the capability of our prototype POC device to perform rapid phenotypic UTI antibiotic susceptibility testing in a clinical pilot evaluation. Most importantly, we demonstrate the potential of our AST device to provide users with highly sensitive and specific phenotypic AST results in as little as 2 h. Cost considerations aside, while certain molecular AST products may also allow for rapid testing, the absence of a resistance gene is not always sufficient to draw conclusions about the antibiotic susceptibility of an infectious agent. This is especially true in the case of rare infectious micro-organisms or newly evolved sublineages, possibly leading to false negatives [31]. By leveraging our instrument’s detection sensitivity, we are able to detect bacterial growth with just a few time point measurements and can characterize changes in growth with 1000 to 10 000-fold lower starting concentrations of micro-organisms than are necessary for commercially available automated systems, eliminating the need for pre-culture/enrichment of urine samples prior to the AST [32]. Many currently available FDA-approved ASTs utilize established techniques such as microwell plating with periodic turbidimetric/colorimetric (BD Phoenix [33]) readings, photometric/fluorometric (Microscan Walkaway [34]) readings or custom AST cards (Vitek2, BioMérieux [35]). While the automation of laborious steps and turbidity/fluorometric readings have decreased the time associated with susceptibility testing, these tests still require hours of incubation prior to antibiotic susceptibility testing and are not suitable for POC applications. Thus, the AST technologies available today cannot provide physicians with antibiotic susceptibility results in time to inform early treatment decisions.
In addition to not requiring the enrichment of bacteria from urine specimens, our AST device does not require any pre-processing steps such as centrifugation or wash steps, simplifying the necessary instrumentation to be practical for POC applications. Furthermore, our instrument can directly detect micro-organisms at the single cell level, eliminating the need for c.f.u. plating, which has a time-to-answer of at least 24 h prior to obtaining visible bacterial growth. Unlike culture-based ASTs, our device provides objective, quantitative information to the end user and is not vulnerable to inaccurate or subjective interpretations due to variables such as user-dependent differences in sample preparations, plating techniques and zone diameter measurements [36].
While certain pathogenic bacteria, such as slow-growing species, may require incubation for longer than 2 h to provide accurate assessments of changes in growth in the presence or absence of antibiotics, E. coli , with a relatively short generation time, is one of the most common causative agents of UTI, accounting for the majority of uncomplicated UTI cases [20, 37, 38]. Regardless, difficulties with culturing certain species of bacteria or the necessity for prolonged growth periods are universal to any phenotypic AST and may explain some of our false-negative results in this early study (as time point growth measurements were not taken after 3 h) when compared to tests run in parallel for longer periods of time. However, it is certainly possible to continue taking measurements beyond the 2 h time point and our future system will have the built-in capacity to determine the need for extended time points by analysing growth rates in control samples in real time. Although we did not observe a correlation between mixed cultures and decreased accuracy of AST results, like all antimicrobial susceptibility tests, including the gold standard, mixed cultures affect the sensitivity and specificity of AST results, primarily due to contamination during the ‘clean catch’ urine collection process. More specifically, mixed cultures of susceptible and resistant bacteria variably result in sensitive, resistant or intermediate phenotypes during the testing process when compared to antibiotic susceptibility testing conducted with pure/uncontaminated cultures [39]. While our system is not currently immune to these same limitations, the ability to provide results rapidly allows for quickly re-evaluating fresh urine samples if gold-standard growth culture results indicate contamination.
As described in the Methods section, an LB formulation was used with the POC instrument instead of Mueller–Hinton (MH) medium due to its ability to support the growth of most common uropathogens and its relatively low fluorescence background. Although the hospital’s conventional AST used a eugenic growth media approved for antimicrobial susceptibility testing, we acknowledge the fact that not being able to verify the POC AST device results using MH broth poses as a technical limitation. Future studies will further evaluate the use of different growth media, including MH broth and/or derivations thereof with low fluorescence background.
For our clinical evaluation, EUCAST MIC breakpoints were used to finalize antibiotic concentrations [21]. Common first-line UTI antibiotics were chosen for the AST panel (as described and listed above) to assess the clinical performance of our device. That being said, different types of antibiotics do not pose a technical limitation for susceptibility testing with the POC device. The AST antibiotic panel can be customized depending on both the prevalence of AMR in the geographical region and patient history. The panel selected for this preliminary clinical evaluation was chosen based on the most commonly used antibiotics in the region where samples were obtained and consisted of three common drugs to treat uncomplicated UTIs [amoxicillin clavulanate (AX), fosfomycin (FF) and nitrofurantoin (NF)], and two commonly used to treat complicated UTIs or catheterized patients [ciprofloxacin (CP), ceftazidime (CZ)].
Since E. coli and other members of the family Enterobacteriaceae are the primary causative agents for the vast majority of uncomplicated UTI cases, the drug concentrations used in this study were based on the single EUCAST MIC breakpoints for the family Enterobacteriaceae . Except for the CP breakpoints, when common infectious agents are naturally susceptible to the antibiotics in our clinical AST panel, the MIC breakpoints are identical for Enterobacteriaceae as well as the less common causative agents of uncomplicated UTIs. Furthermore, the ciprofloxacin MIC breakpoints for Enterobacteriaceae are lower than those or Staphylococcus and Enterococcus spp. Although this factor has the potential to result in a resistant call (false positive) when an organism is susceptible, the use of Enterobacteriaceae breakpoints s not expected to result in false negatives for Staphylococcus and Enterococcus spp., as is evident with our clinical data. As expected, the vast majority of uropathogens from the clinical pilot study belonged to the family Enterobacteriaceae (88%, 22/25), as confirmed by culture. Although the remaining uropathogens only accounted for 12 % of the clinical samples, they were responsible for 4/6 of the reported false positives.
While we have demonstrated rapid antibiotic susceptibility testing in 2 h from direct patient specimens, interpretation of AST results routinely requires knowledge of bacteria identification, a requirement that typically involves lengthy culture and isolation steps. By leveraging the fact that the predominant bacteria that comprise up to 95 % of uropathogens share nearly identical MIC breakpoints [40], this study sought to evaluate a rapid AST intended to provide physicians with susceptibility information within a timeframe that enables them to make more judicious choices when prescribing antibiotics for the majority of UTI cases. Future development of this platform will seek to expand the number of concentrations tested as well as including high-level identification and Gram ID information within the 2 h test window to improve the interpretation of susceptibility results. When considering the relatively low prevalence of Staphylococcus and Enterococcus urinary tract infections, the use of single MIC breakpoints for UTI antibiotic susceptibility testing does not significantly affect the overall accuracy of the POC device. Changes in EUCAST/Clinical Laboratory Standards Institute (CLSI) breakpoints in the future will not pose a direct technical challenge to the POC AST device. However, as guidelines change, new breakpoints will be used to re-verify antibiotic concentrations with the use of clinical isolates and any necessary changes will be implemented in updated versions of antibiotic panels.
Fundamentally, the clinical value of our POC device lies in its unique ability to provide accurate AST results within a timeframe where it can help healthcare professionals make informed decisions about asymptomatic bacteriuria/UTI treatment, supporting antibiotic stewardship practices and playing a vital role in decreasing the spread of antimicrobial resistance. Our AST device can provide end users with rapid and accurate detection of changes in the growth of antibiotic-treated and control samples of urine and can be used to test susceptibility to various antibiotics typically used to treat UTIs. In order to increase sample capacity, we are working towards incorporating a more streamlined and automated workflow, including a faster fluorescence labelling step. Additionally, our technology will provide up-to-date epidemiological data as part of an integrated cloud reporting system to offer options for the use of different antibiotic panels (provided as individual product-specific kits) pertinent to the clinical setting or geographical location of the end user. By reducing the typical timeframe to receive susceptibility information from 2–3 days to 2 h, our POC AST has potential to enable same-day effective antibiotic selection compared to current empirical antibiotic treatment to combat the rise of antimicrobial resistance in UTIs.
Supplementary Data
Funding information
This study was supported, in part, by the National Institutes of Health (NIH)/ National Institute of Allergy and Infectious Diseases (NIAID) (1 R01 AI117061) and by Sardegna Ricerche (CUP:G46G17000340006).
Author contributions
Conceptualization: G. M., W. Z., E. N.; investigation: D. P., E. N., M. N. T., J. T. G.; resources: C. D., F. C., G. A.; data analysis: T. J. A., M. N. T; writing: M. N. T., J. T. G., E. N., T. J. A.; funding: G. M., B. S., W. Z.
Conflicts of interest
The University of Illinois is the owner of patent N: US 2009/0230324 A1. G. M. is the owner of the company ASI that has a license agreement with the University of Illinois. W. Z. is the founder of Velox Biosystems, Inc., which develops in vitro diagnostics. ASI and Velox Biosystems provided financial support, study design and data analysis for the present study. All other authors declare no conflicts of interest.
Ethical statement
This study was approved by the University Hospital of Cagliari Institutional Review Board (PG/2018/5211) and informed consent was obtained from donors.
Footnotes
Abbreviations: ACR, antibiotic/control ratio; AGR, antibiotic growth rate; AMR, antimicrobial resistance; AST, antibiotic susceptibility test; AX, amoxicillin clavulanate; CA, categorical agreement; CP, ciprofloxacin; CZ, ceftazidime; EUCAST, European Committee on Antimicrobial Susceptibility testing; FF, fosfomycin; Kan, Kanamycin; LB, Lennox-Broth; MBC, minimum bactericidal concentration; MIC, minimum inhibitory concentration; M-LB, modified Lennox-Broth; NF, nitrofurantoin; POC, point-of-care; UTI, urinary tract infection.
One supplementary figure and three supplementary tables are available with the online version of this article.
References
- 1.Roca I, Akova M, Baquero F, Carlet J, Cavaleri M, et al. The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect. 2015;6:22–29. doi: 10.1016/j.nmni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frieden T. Antibiotic resistance threats in the United States. Broch - US Cent Dis Control Prev. 2013 [Google Scholar]
- 3.Bartlett JG, Gilbert DN, Spellberg B. Seven ways to preserve the miracle of antibiotics. Clin Infect Dis. 2013;56:1445–1450. doi: 10.1093/cid/cit070. [DOI] [PubMed] [Google Scholar]
- 4.Shah NS, Wright A, Bai GH, Barrera L, Boulahbal F, et al. Worldwide emergence of extensively drug-resistant tuberculosis. Emerg Infect Dis. 2007 doi: 10.3201/eid1303.061400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright PM, Seiple IB, Myers AG. The evolving role of chemical synthesis in antibacterial drug discovery. Angew Chem Int Ed Engl. 2014;53:8840–8869. doi: 10.1002/anie.201310843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lushniak BD. Antibiotic resistance: a public health crisis. Public Health Rep. 2017 doi: 10.1177/003335491412900402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The White House Administration National action plan for combating antibiotic-resistant bacteria. Open Gov Natl Action Plans. 2015 [Google Scholar]
- 8.Doern CD. The slow March toward rapid phenotypic antimicrobial susceptibility testing: are we there yet? J Clin Microbiol. 2018;56:e01999-17. doi: 10.1128/JCM.01999-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicolle LE. Urinary tract infection: traditional pharmacologic therapies. Dis Mon. 2003;49:111–128. doi: 10.1067/mda.2003.11. [DOI] [PubMed] [Google Scholar]
- 10.Foxman B, Barlow R, D’Arcy H, Gillespie B, Sobel JD. Urinary tract infection: self-reported incidence and associated costs. Ann Epidemiol. 2000 doi: 10.1016/s1047-2797(00)00072-7. [DOI] [PubMed] [Google Scholar]
- 11.Grigoryan L, Trautner BW, Gupta K. Diagnosis and management of urinary tract infections in the outpatient setting: a review. JAMA - J Am Med Assoc. 2014 doi: 10.1001/jama.2014.12842. [DOI] [PubMed] [Google Scholar]
- 12.Suskind AM, Saigal CS, Hanley JM, Lai J, Setodji CM, et al. Incidence and management of uncomplicated recurrent urinary tract infections in a national sample of women in the United States. Urology. 2016;90:50–55. doi: 10.1016/j.urology.2015.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar S, Dave A, Wolf B, Lerma EV, Sampath Kumar MD. Urinary tract infections. Disease-a-Month. 2015;61:45–59. doi: 10.1016/j.disamonth.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Gupta K, Trautner BW. Diagnosis and management of recurrent urinary tract infections in non-pregnant women. BMJ. 2013;346:f3140. doi: 10.1136/bmj.f3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter LMJ, Devillé WL, Yzermans JC, Van Duijn NP, Bezemer D, et al. The urine dipstick test useful to rule out infections. A meta-analysis of the accuracy. BMC Urol. 2004 doi: 10.1186/1471-2490-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Belkum A, Dunne WM. Next-Generation antimicrobial susceptibility testing. J Clin Microbiol. 2013;51:2018–2024. doi: 10.1128/JCM.00313-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown PD, Freeman A, Foxman B. Prevalence and predictors of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli isolates in Michigan. Clin Infect Dis. 2002 doi: 10.1086/339491. [DOI] [PubMed] [Google Scholar]
- 18.Bard JD, Lee F. Why can’t we just use PCR? The role of genotypic versus phenotypic testing for antimicrobial resistance testing. Clin Microbiol Newsl. 2018 doi: 10.1016/j.clinmicnews.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolai E, Garau S, Favalli C, D’Agostini C, Gratton E, et al. Evaluation of BiesseBioscreen as a new methodology for bacteriuria screening. New Microbiol. 2014:495–501.:ISSN: 11217138. [PMC free article] [PubMed] [Google Scholar]
- 20.Masters BR. In: Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. 8th ed. Bennett JohnE, Dolin Raphael, Blaser MartinJ., editors. ISBN: 13-978-1-4557-4801-3, Elsevier Saunders. Graefe’s Arch Clin Exp Ophthalmol; 2015. [Google Scholar]
- 21.Rules E. The European Committee on antimicrobial susceptibility testing. Breakpoint tables for interpretation of MICs and zone diameters. 2019.
- 22.Quintiliani R. Pharmacodynamics of antimicrobial agents : time-Dependent vs concentration-dependent killing. Eur J Clin Microbiol Infect Desease. 2001 [Google Scholar]
- 23.Levison ME, Levison JH. Pharmacokinetics and pharmacodynamics of antibacterial agents. Infect Dis Clin North Am. 2009;23:791–815. doi: 10.1016/j.idc.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouton JW, Brown DFJ, Apfalter P, Cantón R, Giske CG, et al. The role of pharmacokinetics/pharmacodynamics in setting clinical MIC breakpoints: the EUCAST approach. Clin Microbiol Infect. 2012;18:E37–E45. doi: 10.1111/j.1469-0691.2011.03752.x. [DOI] [PubMed] [Google Scholar]
- 25.Avesar J, Rosenfeld D, Truman-Rosentsvit M, Ben-Arye T, Geffen Y, et al. Rapid phenotypic antimicrobial susceptibility testing using nanoliter arrays. Proc Natl Acad Sci USA. 2017;114:E5787–E5795. doi: 10.1073/pnas.1703736114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matlock BC, Beringer RW, Ash DL, Allen MW, Page AF. Analyzing differences in bacterial optical density measurements between Spectrophotometers. 2017.
- 27.Koch AL. Turbidity measurements of bacterial cultures in some available commercial instruments. Anal Biochem. 1970;38:252. doi: 10.1016/0003-2697(70)90174-0. [DOI] [PubMed] [Google Scholar]
- 28.Schmiemann G, Kniehl E, Gebhardt K, Matejczyk MM, Hummers-Pradier E. The diagnosis of urinary tract infection: a systematic review. Dtsch Arztebl Int. 2010;107:361–367. doi: 10.3238/arztebl.2010.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skinner JP, Swift KM, Ruan Q, Perfetto S, Gratton E, et al. Simplified confocal microscope for counting particles at low concentrations. Rev Sci Instrum. 2013;84:074301. doi: 10.1063/1.4812782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altamore I, Lanzano L, Gratton E. Dual channel detection of ultra low concentration of bacteria in real time by scanning fluorescence correlation spectroscopy. Meas Sci Technol. 2013 doi: 10.1088/0957-0233/24/6/065702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fastidious HDJ. And Furious: reporting antimicrobial susceptibility testing for fastidious or infrequently isolated bacteria. Clin Microbiol Newsl. 2017 [Google Scholar]
- 32.Hombach M, Maurer FP, Pfiffner T, Böttger EC, Furrer R. Standardization of Operator-Dependent variables affecting precision and accuracy of the disk diffusion method for antibiotic susceptibility testing. J Clin Microbiol. 2015 doi: 10.1128/JCM.02351-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carroll KC, Glanz BD, Borek AP, Burger C, Bhally HS, et al. Evaluation of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of Enterobacteriaceae. J Clin Microbiol. 2006 doi: 10.1128/JCM.00994-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGregor A, Schio F, Beaton S, Boulton V, Perman M, et al. The MicroScan WalkAway diagnostic microbiology system--an evaluation. Pathology. 1995;27:172–176. doi: 10.1080/00313029500169822. [DOI] [PubMed] [Google Scholar]
- 35.Joyanes P, del Carmen Conejo M, Martínez-Martínez L, Perea EJ. Evaluation of the Vitek 2 system for the identification and susceptibility testing of three species of nonfermenting gram-negative rods frequently isolated from clinical samples. J Clin Microbiol. 2001;39:3247. doi: 10.1128/JCM.39.9.3247-3253.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hombach M, Ochoa C, Maurer FP, Pfiffner T, Böttger EC, et al. Relative contribution of biological variation and technical variables to zone diameter variations of disc diffusion susceptibility testing. J Antimicrob Chemother. 2016;71:141–151. doi: 10.1093/jac/dkv309. [DOI] [PubMed] [Google Scholar]
- 37.Stamm WE. An epidemic of urinary tract infections? N Engl J Med. 2002 doi: 10.1056/NEJM200110043451409. [DOI] [PubMed] [Google Scholar]
- 38.Gupta K, Hooton TM, Stamm WE. Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann Intern Med. 2001 doi: 10.7326/0003-4819-135-1-200107030-00012. [DOI] [PubMed] [Google Scholar]
- 39.Shahidi A, Ellner PD. Effect of mixed cultures. Appl Microbiol. doi: 10.1128/am.18.5.766-770.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.George DB, Manges AR. A systematic review of outbreak and non-outbreak studies of extraintestinal pathogenic Escherichia coli causing community-acquired infections. Epidemiol Infect. 2010;138:1679–1690. doi: 10.1017/S0950268810001639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.