Chlamydia trachomatis infection of the human fallopian tubes can lead to damaging inflammation and scarring, ultimately resulting in infertility. To study the human cellular responses to chlamydial infection, researchers have frequently used transformed cell lines that can have limited translational relevance. We developed a primary human fallopian tube epithelial cell model based on a method previously established for culture of primary human bronchial epithelial cells. After protease digestion and physical dissociation of excised fallopian tubes, epithelial cell precursors were expanded in growth factor-containing medium.
KEYWORDS: Chlamydia, fallopian tube, polarized epithelia, primary cells, sexually transmitted infection
ABSTRACT
Chlamydia trachomatis infection of the human fallopian tubes can lead to damaging inflammation and scarring, ultimately resulting in infertility. To study the human cellular responses to chlamydial infection, researchers have frequently used transformed cell lines that can have limited translational relevance. We developed a primary human fallopian tube epithelial cell model based on a method previously established for culture of primary human bronchial epithelial cells. After protease digestion and physical dissociation of excised fallopian tubes, epithelial cell precursors were expanded in growth factor-containing medium. Expanded cells were cryopreserved to generate a biobank of cells from multiple donors and cultured at an air-liquid interface. Culture conditions stimulated cellular differentiation into polarized mucin-secreting and multiciliated cells, recapitulating the architecture of human fallopian tube epithelium. The polarized and differentiated cells were infected with a clinical isolate of C. trachomatis, and inclusions containing chlamydial developmental forms were visualized by fluorescence and electron microscopy. Apical secretions from infected cells contained increased amounts of proteins associated with chlamydial growth and replication, including transferrin receptor protein 1, the amino acid transporters SLC3A2 and SLC1A5, and the T-cell chemoattractants CXCL10, CXCL11, and RANTES. Flow cytometry revealed that chlamydial infection induced cell surface expression of T-cell homing and activation proteins, including ICAM-1, VCAM-1, HLA class I and II, and interferon gamma receptor. This human fallopian tube epithelial cell culture model is an important tool with translational potential for studying cellular responses to Chlamydia and other sexually transmitted pathogens.
INTRODUCTION
Chlamydia trachomatis infects more than 1.7 million people in the United States annually, and cases have continued to rise since 2000 (1). Worldwide, an estimated 131 million new cases are reported each year (2). Infection is often asymptomatic. As a result, many women are undiagnosed and consequently untreated. Chlamydia trachomatis can ascend to the upper genital tract and infect the fallopian tubes. The resulting inflammation promotes long-term sequalae, such as tubal scarring, ectopic pregnancy, and infertility (3). Development of targeted therapies and vaccines to reduce and/or prevent such sequalae require improved understanding of the mechanisms that drive fallopian tube pathology during C. trachomatis infection.
Chlamydia trachomatis is an obligate intracellular Gram-negative pathogen with a unique biphasic developmental cycle involving an infectious, nonreplicative form called an elementary body (EB) and a noninfectious, replicative form called a reticulate body (RB). Infection is initiated by attachment and uptake of extracellular EBs to the apical surface of epithelial cells. Once internalized, EBs convert into RBs in an endosomal vacuole, which is modified to prevent fusion to lysosomes. The RBs replicate within this modified intracellular vacuole, called an inclusion, before converting to EBs later in the developmental cycle to propagate infection after release (4). In the female genital tract, infection is restricted to mucosal epithelial cells, which respond to infection by secreting proinflammatory cytokines and chemokines for recruitment and activation of immune cells to clear infection (5–8).
While the importance of cytokines and chemokines during genital chlamydial infection has been well described in mouse models and human transformed cell lines (5, 9–12), the response of primary human fallopian tube epithelium to chlamydial infection is less well characterized. Data related to chlamydial-epithelial interactions in humans have been gathered using the transformed cervical epithelial cell line HeLa. However, their increased metabolic rate, incidence of aneuploidy (13), and nonpolar secretion of cytokines/chemokines do not mirror the polarized columnar epithelia of the cervix and fallopian tubes (14).
Two ex vivo models to study epithelial responses to Chlamydia infection in primary cells have been established, namely fallopian tube explants (15) and polarized epithelial cells cultured directly from fallopian tube explants (16–18). An important barrier to investigation of epithelial cell responses to chlamydial infection using tissue from fallopian tube explants is the inability to distinguish epithelial-specific contributions from responses of resident immune cells. Similarly, polarized epithelial cell cultures generated directly from explants may still include immune cells. These cell populations can undermine establishment of pure primary epithelial cell cultures, while exposure to danger-associated molecular patterns (DAMPs) released from dead or dying cells during fallopian tissue processing could impact infectivity or host cellular responses to the pathogens being studied.
We have developed a primary human fallopian tube epithelial (FTE) cell culture model, which supports investigation of cellular responses to pathogens without risk of contaminating immune or stromal cells. We expanded isolated FTE cells in a nonproprietary epidermal growth factor-rich medium to generate purified FTE cell precursors. These cells were subsequently polarized on porous support membranes using an approach adapted from a method used to generate polarized bronchial epithelial cells at an air-liquid interface (ALI) (19). Transition to the ALI format resulted in columnar epithelium containing mucin-secreting goblet and multiciliated epithelial cells that recapitulate the morphology of cells previously imaged in human fallopian tube specimens by histology or scanning electron microscopy (20, 21). We also infected the FTE cells with a clinical isolate of C. trachomatis and observed inclusion development, EB to RB transition, RB division, and generation of new EBs. Proteins important for chlamydial growth and replication, and neutrophil and T cell chemoattractants, were detected in apical washes of infected FTE cell cultures. Furthermore, infected cells increased surface expression of proteins involved in cell-cell adhesion and antigen presentation. Together, our data indicate that the primary human FTE cell culture model provides a useful tool to examine pathogen-driven responses specific to FTE cells in a controlled and reproducible fashion.
RESULTS
Primary human FTE cells polarize, secrete mucus, have beating cilia, and form tight junctions.
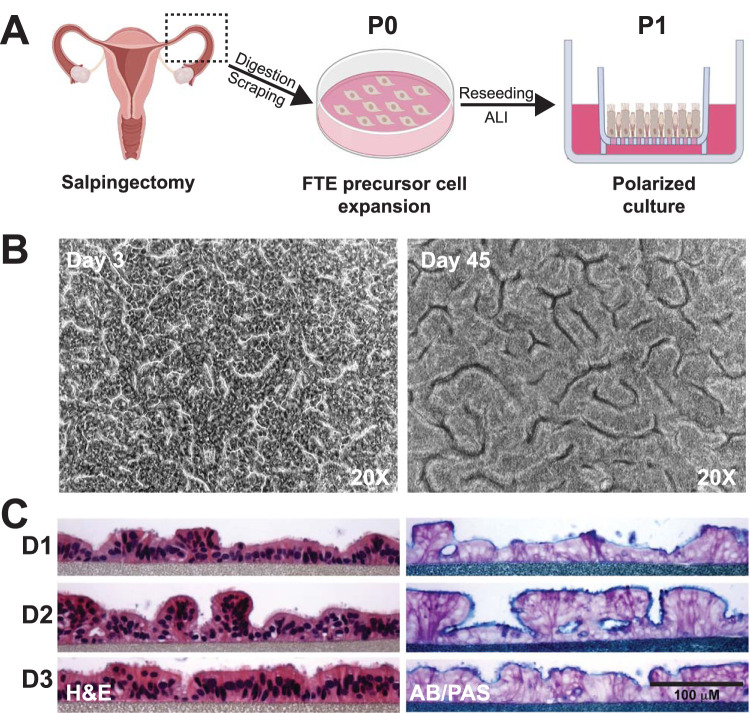
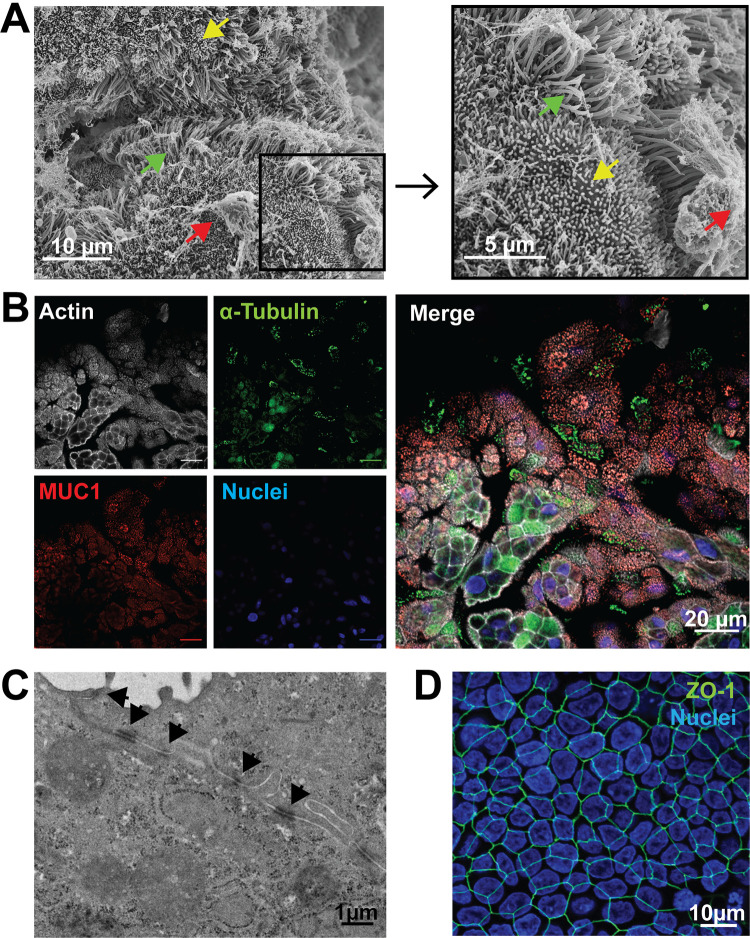
To establish a primary human FTE cell culture that recapitulates native morphology and function, we isolated and expanded cells from fallopian tube explants (N = 12 donors) to obtain undifferentiated epithelial cell populations. Purified epithelial precursor cells ranged from 15 to 200 million cells/donor, with 4 donors yielding >140 million cells. These cells were either cryopreserved for future use or cultured immediately on microporous membranes. When confluent (3 to 5 days), overlying medium was removed from the apical surface to establish an ALI (Fig. 1A). Multicellular, cobblestone-like structures were observed by light microscopy as early as 3 days after transition to ALI (post-ALI). These structures were maintained through 45 days post-ALI (Fig. 1B) without medium leakage from the basolateral to apical surface, indicating continuous barrier integrity. Additionally, we observed cilia movement as early as 7 days post-ALI (see Video S1 in the supplemental material), and through 45 days (see Video S2 in the supplemental material). Hematoxylin and eosin (H&E) staining of histological sections at 21 days post-ALI revealed ciliated and nonciliated polarized columnar epithelial cells, while alcian blue and periodic acid-Schiff (AB/PAS) staining indicated mucin production at the apical surface (Fig. 1C). Polarized FTE cells produced copious secretions which were removed by washing every 2 to 3 days to preserve viability. Scanning electron microscopy (SEM) of cells at 24 days post-ALI revealed structures consistent with cilia and microvilli at high density (Fig. 2A). Using antibodies directed against α-tubulin, a structural protein in cilia, and MUC1, a major protein in mucoid secretions (22), we confirmed their expression by FTE cells at 12 days post-ALI (Fig. 2B). Junctional complexes were observed with transmission electron microscopy (TEM) between adjacent cells 24 days post-ALI (Fig. 2C). Tight junctions between the polarized columnar FTE cells were detected as early as 5 days post-ALI after immunostaining for ZO-1, a protein found in epithelial cell tight junctions (23, 24) (Fig. 2D). Transepithelial electrical resistance (TEER) was transiently decreased after removal of the apical medium. However, FTE cells subsequently maintained consistent TEER (>157 ± 9.7 Ω · cm2), starting 7 days post-ALI (11 days postseeding) (see Fig. S2 in the supplemental material). Thus, donor-derived epithelial precursors established polarized ciliated and mucin-producing cells with tight junctions, mimicking the epithelial morphology of human fallopian tube explants (15, 16) after transition to ALI.
FIG 1.
Primary human fallopian tube epithelial (FTE) cells cultured in an air-liquid interface (ALI) polarize and secrete mucins. (A) Simplified model of the procedure to generate primary human FTE cell cultures. Fallopian tubes were obtained from women undergoing elective salpingectomies. Fallopian tubes were opened, diced into pieces, and enzymatically digested, and epithelial cells were obtained by gentle scraping of the lumen. Primary passage (P0) cells were then expanded on collagen-coated tissue culture dishes in a growth factor-rich medium, followed by dissociation and seeding of P1 cells on inserts or cryopreservation for later use. Creation of an ALI was induced on inserts by removing medium from the apical surface, which promoted epithelial polarization and cell differentiation. Images created with BioRender. (B) Cobblestone-like structures in FTE cell cultures were observed by phase-contrast light microscopy 3 days post-ALI, which was maintained through 45 days post-ALI. (C) Columnar polarized multiciliated and secretory cells were observed by hematoxylin and eosin (H&E) staining, and mucopolysaccharides were observed by alcian blue and periodic acid-Schiff (AB/PAS) staining of histological sections of FTE cell cultures from 3 donors 23 days post-ALI.
FIG 2.
Primary human FTE cells cultured in an ALI form cilia, produce mucin-rich secretions, and form tight junctions. (A) Mucin, cilia, and microvilli were observed by scanning electron microscopy (SEM) on polarized FTE cells 24 days post-ALI. Arrows indicate mucin (red), microvilli (yellow), and cilia (green). (B) Mucin and structural protein of cilia were immunostained and observed by immunofluorescence in a single plane with individual channels (left) and a merged image (right) at 12 days post-ALI. Phalloidin for filamentous actin (white), α-tubulin for cilia (green), MUC1 for mucin (red), and DAPI (4′,6-diamidino-2-phenylindole; blue). Bars, 20 μm. (C) Cellular junctions, including tight junctions, were observed by transmission electron microscopy (TEM) of a cross section of an FTE cell junction 24 days post-ALI. Black arrows indicate cellular junctions. (D) Presence of the structural tight junction protein ZO-1 was observed by immunofluorescence at 5 days post-ALI (anti-ZO-1; green). Image contains DAPI staining by an extended focus view of the three-dimensional (3D) FTE cells.
Chlamydia trachomatis infects and replicates in FTE cells.
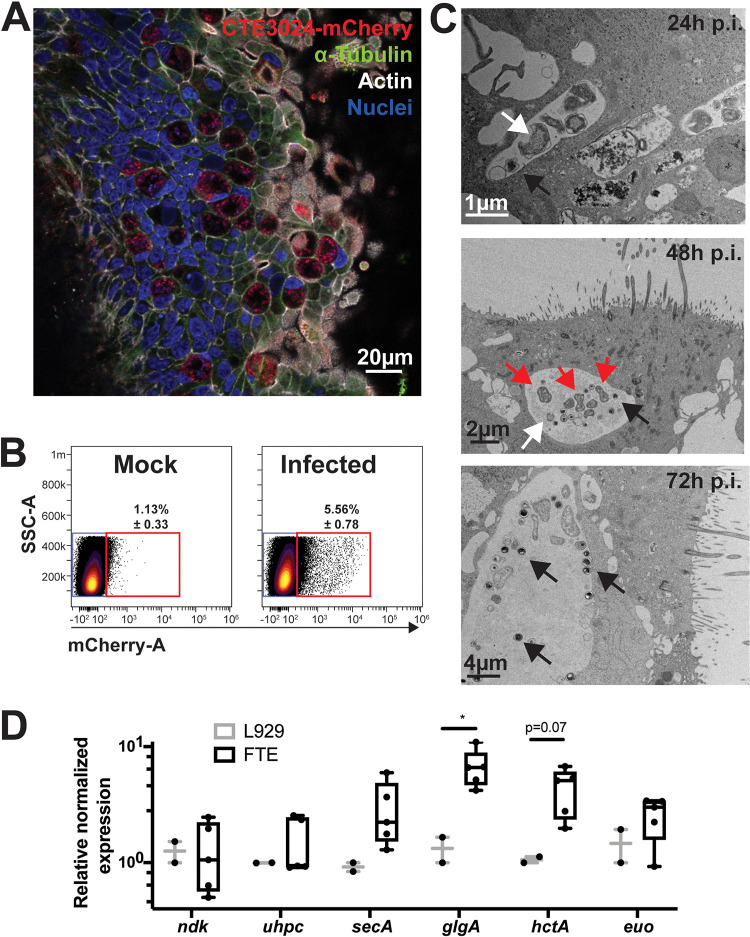
Polarized FTE cells at 8 to 10 days post-ALI were inoculated with a low-passage, endometrial isolate of C. trachomatis strain CTE3024 (serovar E) transformed with a chimeric plasmid encoding the fluorescent protein mCherry. Chlamydial inclusions were visualized by confocal microscopy 48 h postinfection by immunofluorescent staining using a monoclonal antibody directed against chlamydial lipopolysaccharide (LPS) (Fig. 3A). Inclusions were unevenly distributed throughout the culture, and this “patchiness” was not remediated with an increased multiplicity of infection (MOI) of 10 and centrifugation. Photoimaging of infected cultures for quantitation of infection rate by microscopy was compromised by uneven infection of the FTE, while the distribution of chlamydial inclusions to multiple planes within cells made inclusion counts of infected regions uninterpretable. Therefore, we used flow cytometry to quantitate infection rates. Flow cytometry of FTE cells at 48 h postinfection revealed that 4.43% ± 0.26% of the cells were mCherry positive (Fig. 3B), suggesting a low infection rate. To compare infection rates quantitated by flow cytometry to microscopy, nonpolarized FTE cells were infected and analyzed by flow cytometry and immunofluorescent microscopy 36 h postinfection. We observed low infectivity rates by both flow cytometry and immunofluorescent microscopy with no significant differences in the percentages of infected cells (see Fig. S3 in the supplemental material). Inoculations using highly characterized strains frequently used for cell culture studies, such as C. trachomatis L2/434/Bu or D/UW-3/Cx, did not improve the infection rate in polarized FTE cells (data not shown). Treatment of polarized FTE cells with progesterone and estrogen or washing the cells with the mucolytic agent dithiothreitol also did not improve infection rates (data not shown). Interestingly, infection with CTE3024-mCherry did not destroy the structural integrity of polarized FTE cells when cells remained in culture for up to 32 days (data not shown). This was reflected in TEER measurements that were unchanged in infected cells compared to mock-infected cells (see Fig. S2 in the supplemental material).
FIG 3.
Chlamydia trachomatis infects polarized FTE cells and completes its developmental cycle. Primary human polarized FTE cells were infected with CTE3024-mCherry (MOI = 10). (A) Immunofluorescent microscopy of a representative culture stained with DAPI (blue), phalloidin (white), antichlamydial lipopolysaccharide (LPS; red), and anti-α-tubulin (green) 48 h postinfection. (B) Percentage of infected cells was determined by flow cytometry. Concatenated flow cytometry dot plots of mock-infected (left) or infected (right) cultures measuring mCherry expression 48 h postinfection. N = 3 donors. Percentages in gate are ± standard deviation (SD). (C) Developmental cycles were observed by TEM at 24, 48, or 72 h postinfection. Arrows indicate elementary bodies (black), reticulate bodies (white), or dividing reticulate bodies (red). (D) Polarized FTE (N = 5 donors) or L929 monolayers (N = 2) were infected with CTE3024-mCherry. Total RNA was harvested 24 h after infection, and cDNAs were generated, then assayed by quantitative PCR (qPCR). Data were normalized to omcA, and gene expression is shown relative to that of L929 cells. Statistical analysis was performed using ANOVA and Sidak’s multiple comparisons. *, P < 0.05.
Transmission electron microscopy of infected polarized FTE cells revealed inclusions containing small, electron-dense EBs and larger, less dense RBs, indicating conversion of EB to RB by 24 h postinfection. Dividing RBs were observed inside inclusions at 48 h postinfection. We observed increased numbers of EBs compared to RBs in the inclusions 72 h postinfection, suggesting conversion of RBs to new EB progeny (Fig. 3C). Consistent with these observations, transcription of chlamydial genes was detected by quantitative reverse transcription-PCR (RT-PCR) at 24 h postinfection at levels comparable to those detected in infected fibroblast monolayers. Specifically, no difference was detected between high-level (ndk) or low-level constitutively expressed (uhpC and secA) genes in either cell type. Transcription of “late” developmental genes, hctA and glgA, appeared mildly elevated, but expression of the developmental regulator euo was unaltered (Fig. 3D). Overall, detection of chlamydial forms and gene expression indicated that chlamydial development was supported by FTE, although the infection rate was low.
FTE cells respond to C. trachomatis infection by releasing amino acid and iron transporters and chemokines and by upregulating cell adhesion molecules.
Apical culture washes were collected 24 h postinfection from mock-infected and infected cultures of four donors to characterize their innate response to chlamydial infection by mass spectrometry-based proteomics. Host-derived proteins whose abundance in the secretions significantly differed (P < 0.05) between infected and mock-infected cultures are presented in Table 1, and a complete list of apically detected proteins is provided in Table S2 in the supplemental material. The mucin proteins MUC1, MUC4, MUC5AC, MUC5B, and MUC16 were abundant in both mock-infected and infected cultures, and their levels were unaltered by infection (Table S2). In apical washes from Chlamydia-infected cells, we observed increases in the amino acid transporter SLC3A2 and the iron transporter TFRC, neutrophil chemokines (CXCL1 and CXCL8), proteins involved in cell cycle regulation (TSG101), metabolism (CPS1), and the multifunctional transmembrane glycoprotein basigin (Table 1). In contrast, proteins that were significantly downregulated in secretions from infected cells included those involved in host protein synthesis and posttranslational modification (EEF1A2, RPS25, RPL35, DARS, and GALNT1), intracellular trafficking (SNX2), fatty acid metabolism (DECR1), retinoic acid transport (CRABP2), antiviral response (PARP4), and Toll-like receptor (TLR) signaling (TMED7-TICAM2).
TABLE 1.
Differential levels of proteins in apical washes of infected cells at 24 h postinfection, detected by mass spectrometrya
| Protein | Gene name | Fold change (×) | P valueb |
|---|---|---|---|
| Carbamoyl-phosphate synthase, mitochondrial | CPS1 | INF | 0.011 |
| Growth-regulated alpha protein | CXCL1 | 42 | 0.050 |
| 4F2 cell-surface antigen heavy chain SLC3A2 | SLC3A2 | 25 | 0.005 |
| Transferrin receptor protein 1 | TFRC | 8.4 | 0.040 |
| Basigin | BSG | 4.6 | 0.024 |
| Interleukin-8 | CXCL8 | 4.3 | 0.005 |
| Tumor susceptibility gene 101 | TSG101 | 2.9 | 0.033 |
| Elongation factor 1-alpha 2 | EEF1A2 | 0.4 | 0.013 |
| 2,4-dienoyl-CoA reductase, mitochondrialc | DECR1 | 0.4 | 0.025 |
| 40S ribosomal protein S25 | RPS25 | 0.4 | 0.048 |
| Polypeptide N-acetylgalactosaminyl transferase | GALNT1 | 0.3 | 0.009 |
| Poly [ADP-ribose] polymerase 4 | PARP4 | 0.3 | 0.018 |
| TMED7-TICAM2 | TMED7-TICAM2 | 0.3 | 0.035 |
| Cellular retinoic acid-binding protein 2 | CRABP2 | 0.3 | 0.045 |
| Sorting nexin-2 | SNX2 | 0.2 | 0.038 |
| 60S ribosomal protein L35 | RPL35 | 0.2 | 0.040 |
| Aspartate-tRNA ligase, cytoplasmic | DARS | 0.2 | 0.044 |
Shaded and nonshaded cells indicate proteins that were increased and decreased, respectively, in infected cells compared to those in mock-infected FTE cells. Data shown here include proteins with differential of about 3-fold with P < 0.05. For a complete list, see Table S2 in the supplemental material. INF, infinity.
P value is from a paired t test with mock-infected and infected FTE cells from 4 donors.
CoA, coenzyme A.
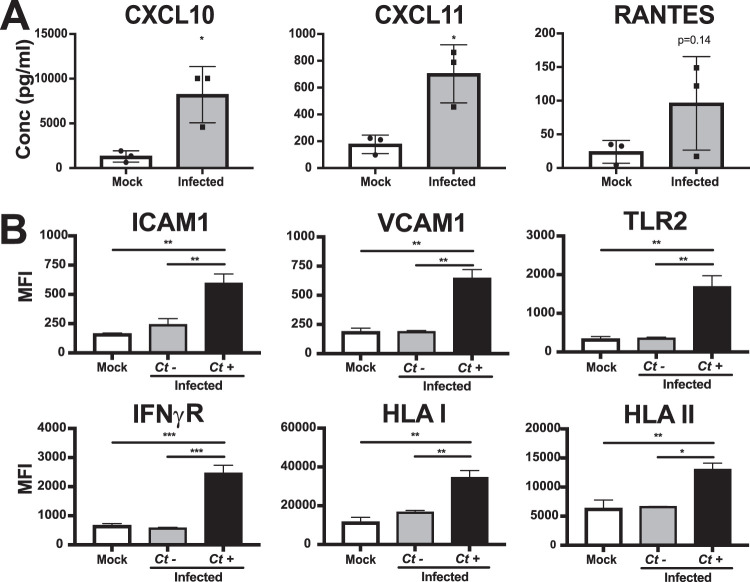
To investigate the innate inflammatory responses of FTE cells to C. trachomatis infection, apical washes and basolateral medium were tested for the presence of cytokines and chemokines by Luminex bead array at 48 h postinfection with CTE3024-mCherry. Washes collected in parallel from mock-infected cells were used as controls. Increased levels of CXCL10, CXCL11, and RANTES were detected in the apical washes of infected FTE cells compared to those in mock-infected cells (Fig. 4A), with no significant change in basolateral cytokines or chemokines (data not shown). Many of the proinflammatory and/or signaling proteins probed for were below the level of detection, possibly a result of the overall low level of infection and of dilution in washing solution or medium. To examine responses specific to cells containing a chlamydial inclusion, we infected FTE cells with CTE3024-mCherry, then removed the cells at 48 h postinfection from their underlying membrane before staining them with antibodies to various cell surface molecules. Flow cytometry analysis of mCherry-positive cells revealed significantly increased expression of ICAM-1, VCAM-1, TLR2, interferon gamma receptor (IFN-γR), and human leukocyte antigen (HLA) class I and II molecules compared to mCherry negative-cells of the same culture (Fig. 4B). We did not detect significant differences in surface expression of these proteins between mock-infected and mCherry-negative cells.
FIG 4.
FTE cells respond to C. trachomatis infection by increasing secretion of cytokines and expression of surface proteins. Primary human polarized FTE cells were infected with CTE3024-mCherry (MOI = 10). (A) Luminex assay of apical secretions from mock-infected and infected cultures 48 h postinfection. Individual dots represent each donor (N = 3). Statistical analysis performed using a paired t test. *, P < 0.05. (B) Mean fluorescent intensities (MFI) of surface protein expression in mock-infected and infected cultures measured by flow cytometry 48 h postinfection. The infected cultures were then gated for mCherry-negative (Ct−) and mCherry-positive (Ct+) cells. N = 3 donors with 2 replicates per donor. Statistical analysis was performed using Tukey’s multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.005.
Overall, our findings demonstrate that FTE cells respond to chlamydial infection by increasing expression of host proteins that support chlamydial growth, cellular adhesion, microbial detection, and immune responsiveness, as well as increasing apical secretion of several chemokines. Interestingly, we also observed reduced levels of several host proteins directly involved in cellular protein synthesis, metabolism, and immune signaling.
DISCUSSION
Genital epithelial cell responses to Chlamydia infection have been studied using transformed cell lines and mouse models of infection. Data from these models have greatly improved our understanding of chlamydial pathogenesis. However, limitations of these approaches include the potential for decreased responses in primary transformed cell lines to chlamydial infection (25–27). Furthermore, responses resulting from mismatched host/Chlamydia species, particularly with respect to p65 and p53 degradation and to gamma interferon (IFN-γ)-mediated inhibition of chlamydiae (28–30), need further validation in a relevant human cell type. Development of therapies and/or vaccines that limit or prevent fallopian tube pathology during genital chlamydial infection requires an improved understanding of pathogen-host responses that occur during species-specific infection of this vulnerable tissue. To advance study in all of these areas, we have generated a primary human FTE cell culture model that recapitulates native cell morphology with polarized multiciliated and mucin-producing cells. Furthermore, C. trachomatis infection of FTE cells induces expression of proteins associated with chlamydial growth, secretion of chemokines, and expression of cell adhesion and immune response molecules, indicating the translational potential for this model to study chlamydial pathogenesis.
Our method has distinct advantages over previously developed approaches to polarize primary human FTE cells (16–18), in which harvested FTE cells were directly cultured in microporous supports immediately after extraction/dispersion from fallopian tube tissue. First, we expand FTE cell precursors over several days. This removes dead and terminally differentiated cells collected after enzymatic digestion and luminal scraping, limiting exposure to DAMPs released from dead cells that could dysregulate cellular responses during insert seeding and polarization. Second, expansion of FTE cell precursors enables the cryopreservation of relatively large numbers of viable cells and the generation of a biobank with multiple donors. Following cryopreservation and subsequent recovery, these cells polarize into mucin-secreting goblet and multiciliated cells and maintain tight junctions after transition to the ALI. The proteomic analysis of apical washes revealed peptides corresponding to mucin proteins, including MUC5AC, MUC5B, MUC4, MUC16, and MUC1 (see Table S1 in the supplemental material). A previous study did not detect mRNA for MUC5AC, MUC5B, or MUC4 in human fallopian tube explants (22). It is possible that we detect these proteins in our cell culture system due to enrichment of mucins or lack of cell-extrinsic regulators of secretion during in vitro culture. However, consistent with the findings of Gipson et al. (31), we did not detect peptides derived from MUC2, MUC3, MUC6, or MUC7.
Previous studies have demonstrated C. trachomatis infection and reported responses to infection in polarized endometrial and cervical cells (25, 32, 33). We now extend these investigations by profiling the responses of the human fallopian tube epithelium, where infection can lead to irreversible sequelae. However, we observed a low percentage of infected FTE cells despite multiple vigorous washings with phosphate-buffered saline (PBS) with and without mucolytics such as dithiothreitol and hormonal treatment. Furthermore, we were only able to establish infection by use of a high MOI and centrifugation, as previously reported for primary cervical and endometrial cells (25, 33) (Fig. 3B). One explanation could be the host cell cycle state. Previous studies using other mucosal pathogens indicate that surface proteins utilized for entry into the host cell are differentially expressed during specific phases of the cell cycle (34–36), thereby indicating that host cells are more susceptible to infection during particular phases of the cell cycle. Interestingly, we observed similar low infection rates in nonpolarized FTE cells (see Fig. S3A to C in the supplemental material) compared with those in polarized FTE cells (<10%) (Fig. 3B), suggesting that morphological changes may have a minimal role in C. trachomatis infectivity. Our data corroborate the low infection rates seen in fallopian tube explants (37). Thus, it appears that FTE cells are intrinsically resistant to C. trachomatis infection compared to transformed cell lines. The low infection rates observed for FTE cells may contribute to the low incidence of long-term tubal complications in women (38–40) despite the high prevalence of chlamydial infection (1). We acknowledge that our model lacks immune cells or cytokines present in vivo, which can disrupt the ordered epithelium and promote spread of infection to neighboring cells, a topic for future investigation.
Once internalized, the chlamydial developmental cycle appeared normal with respect to inclusion formation, differentiation to RB, gene transcription, and generation of new EB within polarized FTE. The mildly elevated expression of the late genes glgA and hctA by chlamydiae in FTE cells may reflect chlamydial sensing of the cellular environment; highly abundant glgA transcripts have been detected in a clinical cervical specimen (41), and anti-chlamydial glycogen synthase antibody was prevalent in a large human cohort (42). Alternatively, these increases may be artifacts of RNA carryover from the high MOI needed to achieve infection. These preliminary observations appear to be consistent across donors and nevertheless highlight the potential that polarized FTE can be used to study chlamydial gene expression effectively ex vivo.
We observed mature inclusions that contained both chlamydial developmental forms in both multiciliated and nonciliated cells (Fig. 3C). Our results are consistent with prior observations of chlamydial inclusions in secretory (43) and ciliated cells (37, 44) by electron microscopy. Whether mucin-producing cells or ciliated cells are more susceptible to infection, and whether these cells respond differently to infection, remains to be determined. Although chlamydial inclusions were observed in only a fraction of cells, infection led to significant changes in the content of proteins in apical washes. Consistent with a previous study (45), increased levels of amino acid transporters SLC3A2 and SLC15A (P = 0.06) (Table 1; see also Table S2 in the supplemental material) were observed in washes from cultures of infected cells. SLC3A2 is a transporter for large neutral amino acids, such as tryptophan, while SLC1A5 is a transporter for glutamine. Both are important for chlamydial replication (45–47). We also observed increased levels of TFRC, which is responsible for iron uptake. Iron is also essential for chlamydial growth and replication (48, 49). These data suggest that the increases of SLC3A2, SLC1A5, and TFRC could be actively modulated by Chlamydia. The detection of neutrophil chemoattractants CXCL8 (interleukin-8 [IL-8]) and CXCL1 in apical washes from infected cells was expected given many reports of the release of these proteins from Chlamydia-infected cells in vitro (5) and early on during in vivo infection (6). Our failure to detect these proteins in the basolateral medium from infected FTE cells was likely due to dilution effects. Proteins that were downregulated in apical secretions of infected cells are involved in host protein synthesis and posttranslational modification, intracellular trafficking, fatty acid metabolism, retinoic acid transport, the antiviral response, and TLR signaling. However, their impact during Chlamydia infection remains unknown. Intracellular pathogens are known to dysregulate elements of host protein synthesis (50), upregulating factors that benefit bacterial growth and replication (45) and downregulating factors that may be detrimental to survival (51).
We detected increased levels of the chemokines CXCL10, CXCL11, and RANTES in the apical washes of infected FTE cell cultures. CXCL10, CXCL11 and RANTES are chemoattractants for multiple immune cell types, including T cells (52–55). We have previously detected these proteins in cervical secretions from C. trachomatis-infected women (6) and CXCL10 mRNA in endometrial biopsy specimens of women with endometrial infection (56). Multiple studies have established that an adaptive CD4 Th1 T-cell response is essential to combat chlamydial infection (57–62). Thus, it is likely that secretion of these chemokines serves a protective role. We did not detect a significant increase in many cytokines frequently associated with chlamydial infection in epithelial cells, such as IL-6, CXCL8 (IL-8), CXCL1 (GROα), GM-CSF, or tumor necrosis factor alpha (TNF-α) (5, 25), likely a result of the low levels of infection. However, we did detect peptides for CXCL8 and CXCL1 by mass spectrometry (Table 1). Similarly, infection did not influence the abundance of any apically secreted mucins.
We observed increased cell surface expression of the adhesion molecules ICAM-1 and VCAM-1 in Chlamydia-infected FTE cells; these were also upregulated in human and mouse genital tracts during chlamydial infection (63–65). Both HLA class I and II proteins were increased on the surface of infected FTE cells at 48 h postinfection. Prior studies suggested that C. trachomatis decreases HLA class 1 and II expression (66–68), a process possibly mediated by the chlamydial secreted protease CPAF (69). However, a subsequent study failed to detect altered HLA expression in response to Chlamydia infection (70), and the investigators suggested that CPAF might instead degrade chlamydial peptides to impair pathogen recognition by the adaptive immune system. Collectively, increased expression of ICAM-1, VCAM-1, IFN-γR, and HLA class I and II proteins indicate that FTE cell cultures may be useful for the study of T cell-FTE cell interactions during Chlamydia infection, using donor- or HLA-matched FTE cells and T cells.
In conclusion, the findings of this study indicate that polarized FTE cell cultures will serve as a valuable tool to study cellular responses to C. trachomatis infection, infection mechanisms, and immune cell-epithelial interactions. Furthermore, this approach provides a mechanism to biobank samples of FTE cells that will allow researchers to study human genetic variation that may modulate responses to Chlamydia infection. The FTE cell culture model also enables future studies examining their response to other sexually transmitted pathogens, alone or in combination with Chlamydia.
MATERIALS AND METHODS
Isolation, expansion, and polarization of primary human FTE cells.
Fallopian tubes were obtained from healthy premenopausal women after elective salpingectomies performed at the University of North Carolina Hospital’s Hillsborough Campus. The Office of Human Research Ethics at UNC determined that the use of these tissues in this research study did not require institutional review board (IRB) approval (IRB no. 15-2805). About 6 to 10 cm of deidentified tissue was provided. Tissues were placed in phenol red-free Ham’s F12 medium (catalog no. 21700075; Gibco) immediately after surgery and refrigerated. Tissues were then transported to the lab on ice and processed on the same day.
(i) Isolation. The following procedures for isolation of FTE cells from the tissue reflect modifications to a protocol for isolation and culture of human airway epithelial cells from human lungs (19). Fallopian tubes were opened with sterile scissors to expose the luminal space, cut into ∼1-cm sections, and placed in 30 ml Joklik minimal essential medium (MEM) (catalog no. M8028; Millipore Sigma) containing 2 ml of 10× 1.0% protease-0.01% DNase stock (catalog no. P5147 and DN-25; Sigma), 35 μl of gentamicin (50 μg/ml, catalog no. 400-108; Gemini Bio-Products), and 175 μl of amphotericin B (250 μg/ml, catalog no. 400-104; Gemini Bio-Products) in 50-ml conical tubes overnight at 4°C while gently rocking. Tissues were removed, and the epithelial cells were gently scraped free using a no. 10 scalpel. The protease-DNase was neutralized with 10% fetal bovine serum (FBS), and cells were centrifuged at 600 × g at 4°C for 5 min. Cells were resuspended in Accutase (catalog no. AT104; Innovative Cell Technologies, Inc.) with 0.5 mM EDTA (as supplied) for 30 min at 37°C to declump. Red blood cell (RBC) lysis was performed with ACK lysis buffer (catalog no. A1049201; Gibco) as needed per tissue. All isolated cells (including precursors) were described as passage 0 (P0), and were counted and resuspended in bronchial epithelial cell growth medium (BEGM) (19) with supplemental antibiotics (ceftazidime, 100 μg/ml; vancomycin, 100 μg/ml; tobramycin, 80 μg/ml) and antifungals (fluconazole, 2.5 μg/ml; mycamine, 20 μg/ml; amphotericin B, 1 μg/ml).
(ii) Expansion. To expand epithelial precursors, cells were cultured in BEGM, an epidermal growth factor-rich medium (71), on 100-mm tissue culture plates previously coated with 1.5% PureCol collagen type I (catalog no. 5005; Advanced BioMatrix) at 4 × 106 to 6 × 106 cells per plate. The adherent cells were washed with PBS at 24 h to remove dead or unbound cells and debris before replacing the BEGM medium with fresh antibiotics and antifungals. After 3 days in culture, fallopian tube cells were washed, and fresh BEGM without supplemental antibiotics or antifungals was applied. Upon reaching ∼80% to 90% confluence (4 to 7 days), precursor cells were removed using Accutase for 30 min at 37°C, resuspended in Ham’s F-12 medium, and counted. At this stage, cells were described as P1 and were cryopreserved at 2 × 106 cells/ml in freezing medium (78% Ham’s F-12, 10% dimethyl sulfoxide [DMSO], 10% FBS, and 2% 1.5 M HEPES) for future use or immediately transferred to collagen-coated porous inserts to develop into polarized cultures.
(iii) Polarization. To develop polarized, differentiated FTE cell cultures, cells were seeded at 2.5 × 105 cells on 0.4-μm polytetrafluoroethylene 12-mm inserts (catalog no. PICM01250; Millipore Sigma) in air-liquid interface (ALI) differentiation medium. Inserts were previously coated with 150 μl of 50 μg/ml human placenta collagen type IV (catalog no. C-7521; Sigma) (71). Upon reaching 100% confluence (3 to 5 days), apical medium was removed to establish an ALI and to promote multiciliated and goblet cell differentiation. Mucoid secretions on the apical surface were removed by washing with 100 μl of 37°C PBS twice carefully every 48 to 72 h, making sure that the tip of the pipette did not touch the cells. Medium on the basolateral side was replaced at the same time. Transepithelial electrical resistance measurements (TEER) were obtained using an EVOM2 instrument (World Precision Instruments, Sarasota, FL, USA) with STX2 electrodes, and Ohm resistance was normalized to a cell-free blank and calculated by culture area.
To quantify infection rate (see Fig. S3 in the supplemental material), nonpolarized FTE cells were seeded at 2 × 105 cells/well on a 24-well plate or 5 × 105 cells/well in a 12-well plate in ALI differentiation medium. HeLa cells were seeded at the same density and used in parallel as controls. HeLa cells were cultured in Dulbecco's Modified Eagle medium plus 10% FBS. After infection, cycloheximide (500 ng/ml) was added to the HeLa medium to limit overgrowth.
Chlamydia trachomatis infection of FTE cells.
Chlamydia trachomatis CTE3024 (serovar E) is a low-passage clinical isolate cultured from an endometrial biopsy specimen using procedures previously described (72) and propagated in McCoy cells in our laboratory. CTE3024 was transformed with either p2TK2-SW2mCherry (73) or p2TK2mCherry CmR (see Fig. S1 in the supplemental material), generating CTE3024 expressing mCherry (passage 5). CTE3024-mCherry was also propagated in McCoy cells. CTE3024-mCherry harvested from infected cells was treated with 233 μg/ml DNase I (catalog no. D0876; Sigma) and 2.1 mg/ml RNase A (catalog no. R5503; Sigma) for 60 min at 37°C and then centrifuged at 39,200 × g over a 32% Renografin density gradient to remove cellular debris and prepare concentrated purified EB/RB suspensions. Titers of chlamydial stocks were determined by infection of L929 monolayers and determination of inclusion-forming units (IFUs) as previously described (74, 75). Primary FTE cell cultures, 12- to 15-day-old cultures (7 to 10 days post-ALI) were prepared for infection by removing mucus by washing the apical surface with 37°C PBS 4 times for 20 min each. Inserts were placed in a 24-well plate containing 1 ml of ALI medium, and cells were infected with 50 μl CTE3024-mCherry containing ALI medium at an MOI of 10 for 1 h by centrifuging the plate at 1,800 × g at 37°C. Apical medium was aspirated after centrifugation. Additional efforts to increase infection rate included washing the apical side of FTE cells with 10 mM dithiothreitol for 20 min prior to infection or treatment of cells with 10 nM β-estradiol (catalog no. 2824; Tocris) and/or 400 nM progesterone (catalog no. 2835; Tocris) for 24 to 72 h prior to infection.
Immunostaining and confocal microscopy.
Cells were fixed with methanol-free 4% formaldehyde, prepared from 20% paraformaldehyde, for 15 min at room temperature (RT). Fixed cells were washed with PBS three times and permeabilized with 0.2% Triton X-100 (catalog no. T8787; Sigma) for 30 min. Permeabilized cells were washed 3 times with PBS and blocked overnight with a solution comprised of 1% fish gelatin (catalog no. G7765; Sigma), 0.1% bovine serum albumin (BSA) (catalog no. A4503; Sigma), and 0.1% Triton X-100 (catalog no. 9002-93-1; Sigma) at 4°C in PBS. Immunostaining with primary antibodies was carried out in blocking solution overnight at 4°C. Cilia were stained with rat anti-human α-tubulin (catalog no. MAB1864; Sigma) at 5 μg/ml for cilia, mouse anti-human MUC1 (catalog no. MA5-13168; Thermo Fisher) at 3 μg/ml for mucus, rabbit anti-human ZO-1 (catalog no. 40-2200; Thermo Fisher) at 2 μg/ml for tight junctions, and/or with mouse anti-chlamydial LPS (catalog no. MCA2718; Bio-Rad) at 3 μg/ml for chlamydial inclusions. HeLa cells and nonpolarized FTE cells were stained with rabbit anti-red fluorescent protein (RFP) (catalog no. 600-401-379; Rockland) at 1:1,000 dilution to enhance mCherry fluorescence. Cells were washed 3 times with 20% blocking solution in PBS. Secondary antibodies used were goat anti-rat 488 (catalog no. AB150165; abcam) at 2 μg/ml, goat anti-rabbit 488 (catalog no. A11034; Invitrogen) at 4 μg/ml, goat anti-rabbit 594 (catalog no. A11037; Invitrogen) at 1 μg/ml, and/or donkey anti-mouse AF546 (catalog no. A10036; Invitrogen) at 4 μg/ml. Secondary antibodies were applied for 2 h at room temperature, followed by 3 washes with PBS. Cultures were counterstained for actin with phalloidin AF647 (catalog no. A22287; Invitrogen) at 1:50 dilution and DNA with Hoechst 33342 (catalog no. H1399; Invitrogen) at 1:200 dilution for 30 min at RT. Membranes were carefully excised from inserts with a no. 12 scalpel and mounted with ProLong Gold antifade mount (catalog no. P10144; Invitrogen). Fluorescently labeled cells were visualized on a Zeiss 800 upright confocal microscope and analyzed using Zen software.
Transmission electron microscopy.
CTE3024-mCherry-infected FTE cell cultures grown on inserts were fixed in 2% formaldehyde/2.5% glutaraldehyde/0.15 M sodium phosphate buffer (pH 7.4) for 1 h at room temperature and stored in fixative at 4°C overnight up to several days. Following three rinses with 150 mM sodium phosphate buffer (pH 7.4), the cells were postfixed with 1% osmium tetroxide-150 mM sodium phosphate buffer for 1 h. After washes in deionized water, cells were dehydrated using increasing concentrations of ethanol (30%, 50%, 75%, 100%, and 100%; 10 min each) and embedded in Polybed 812 epoxy resin (Polysciences, Inc., Warrington, PA). Cross sections of the filter/cell layer were ultrathin sectioned at an 80-nm thickness using a diamond knife and a Leica Ultracut UCT ultramicrotome (Leica Microsystems, Inc., Buffalo Grove, IL). Ultrathin sections were collected on 200 mesh copper grids and stained with 4% aqueous uranyl acetate for 12 min, followed by Reynolds’s lead citrate for 8 min (76). Samples were observed with a JEM-1230 transmission electron microscope operating at 80 kV (JEOL USA, Peabody, MA), and digital images were acquired using a Gatan Orius SC1000 charge-coupled device (CCD) camera and Microscopy Suite 3.0 software (Gatan, Inc., Pleasanton, CA).
Scanning electron microscopy.
For SEM, the cells were fixed and dehydrated as above and transferred in 100% ethanol to a Samdri-795 critical point dryer (Tousimis Research Corporation, Rockville, MD). Samples were critical point dried with liquid carbon dioxide as the transitional solvent. The filter membrane was removed from the well insert support and mounted cell side up onto 13-mm diameter aluminum stubs with carbon adhesive tabs. The samples were coated with 10 nm of gold/palladium alloy (60 Au:40 Pd) using a Hummer X sputter coater (Anatech USA, Union City, CA). Images were taken using a Zeiss Supra 25 field emission scanning electron microscope operating at 5 kV with the an in-lens secondary electron detector, a 30-μm aperture, and an approximate working distance of 5 mm (Carl Zeiss Microscopy, LLC, Peabody, MA).
Gene expression and Luminex assay.
Polarized FTE cells were processed for RNA using miRCURY isolation kits (Exiqon). RNA (500 ng) was processed for reverse transcription and quantitative PCR using SsoAdvanced SYBR mix (Bio-Rad) and a CFX iCycler as previously described (74). Primer sequences for chlamydial gene expression were selected based on genes annotated in the C. trachomatis D/UW-3/Cx genome (77) (see Table S1 in the supplemental material). Gene expression was normalized using omcA rather than 16S and 23S rRNA because these transcripts were carried over in the high MOI needed to infect the FTE. Gene expression in FTE cells relative to that in L929 cells was determined by the cycle threshold (ΔΔCT) method using Bio-Rad Maestro software. Luminex assays were performed on apical washes and basolateral conditioned medium using a multiplex panel containing 48 cytokines/chemokines as described previously (56). Apical washes were collected 48 h postinfection by applying 100 μl 37°C PBS for 5 min twice and aspirating from mock-infected and infected FTE cell cultures. Basolateral medium (1 ml) was also collected. Luminex data were not adjusted by false discovery rate (FDR) due to small sample size.
Mass spectrometry.
Mass spectrometry-based proteomic analysis was performed on apical washes of 4 mock-infected and 4 CTE3024-mCherry-infected FTE cell cultures 24 h postinfection. Apical FTE cell culture washes (200 μl) were prepared for mass spectrometry analysis utilizing filter-aided sample preparation (78). Proteins were digested overnight using trypsin (20 ng/μl) in 50 mM ammonium bicarbonate at 37°C. The resulting peptide digests were eluted using Amicon Ultra 4 10-kDa spin filters. Peptides were vacuum freeze-dried and dissolved in 25 μl of 1% acetonitrile and 0.1% trifluoroacetic acid. Solubilized peptide material (5 μl) was injected for proteomic analysis in a Q Exactive (Thermo Scientific) mass spectrometer coupled to an UltiMate 3000 (Thermo Scientific) nano-high-performance liquid chromatography (HPLC) system, and data acquisition was performed as described previously (79). The acquired raw data were processed using Proteome Discoverer 1.4 (Thermo Scientific) software and searched against the UniProt protein database (Homo sapiens; August 2015) using the SEQUEST search engine with parameters set as follows: 10 ppm mass accuracy for parent ions and 0.02-Da accuracy for fragment ions, with 2 missed cleavages allowed. Carbamidomethyl of cysteine was specified as a fixed modification, and oxidation of methionine was specified in SEQUEST as a variable modification. Scaffold 4.7.5 (Proteome Software, Inc.) was used to validate tandem mass spectrometry (MS/MS)-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the scaffold local FDR algorithm. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the ProteinProphet algorithm (80). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Relative protein quantification was performed by summarizing the intensities of identified precursor ions for each protein as total precursor intensities. Individual protein intensities were normalized to the total intensity of all identified proteins in each sample. Analysis of statistical significance between the Chlamydia-infected and mock-infected control group was determined on pairs for each donor using paired t test analysis.
Flow cytometry.
Mock-infected and CTE3024-mCherry-infected HeLa and FTE cells were removed from cultures 36 or 48 h postinfection using Accutase for up to 1 h at 37°C. Cells were washed and resuspended in cell-staining buffer (catalog no. 420201; BioLegend). Cells were either analyzed for infection rates or stained for surface markers by incubating with manufacturer recommended concentrations of either mouse IgG anti-ICAM1-PacBlue (catalog no. 322715; BioLegend), anti-VCAM1-fluorescein isothiocyanate (FITC) (catalog no. 551146; BD), anti-TLR2-allophycocyanin (APC) (catalog no. 392304; BioLegend), anti-IFN-γR-phycoerythrin (PE) (catalog no. 308606; BioLegend), anti-HLA A, B, C-AF700 (catalog no. 311438; BioLegend), or anti-HLA DR, DP, DQ-PerCP/Cy5.5 (catalog no. 361710;BioLegend) for 30 min at room temperature in the dark. Cells were then washed three times with cell stain buffer and fixed with 2% formaldehyde. Cells were analyzed on an Attune NxT cytometer at the UNC Flow Cytometry Core Facility. Data analysis was performed using Cytobank (81). The percentage of infected cells was determined by drawing gates on ∼1% of mock-infected cells to include infected cells whose inclusions may be smaller than others and have a decreased mean fluorescent intensity (MFI). The percentage of infected cells was then normalized to the mock-infected percentage.
Supplementary Material
ACKNOWLEDGMENTS
The study was primarily funded through a Translational Team Science Award (UNC School of Medicine and NC TRaCS) and also funded by NIAID NIH R01 AI067678 to U.M.N., NIAID NIH R01 AI119164 to T.D., and NIH grant DK065988 and Cystic Fibrosis Foundation grant BOUCHE15R0 to S.H.R. B.E.M. was supported by STI T32 grant AI007001. The Microscopy Services Laboratory, Department of Pathology and Laboratory Medicine, is supported in part by Cancer Center core support grant P30 CA016086 to the UNC Lineberger Comprehensive Cancer Center. The UNC Flow Cytometry Core Facility is supported in part by Cancer Center core support grant P30 CA016086 to the UNC Lineberger Comprehensive Cancer Center. The flow cytometry research reported in this publication was supported in part by North Carolina Biotech Center Institutional support grant 2017-IDG-1025 and by National Institutes of Health grant 1UM2AI30836-01.
We thank the University of North Carolina Hospital, Department of OB/GYN, Hillsborough Campus, for fallopian tube tissues. We specifically thank Mathew Zerden, M.D., and the OB/GYN pathology core staff for providing the tissues in a timely manner.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We declare no conflicts of interest.
The work was conceived by a team comprising S.H.R., T.D., U.M.N., C.M.O., P.W., M.K., and B.E.M. Culture conditions for FTE cells were developed by S.H.R. and M.L.F., and carried out by M.L., E.P., K.P., and B.E.M. Isolation of CTE3024 from endometrium tissue was carried out by R.J.S., and C.M.O. generated the mCherry-expressing derivative CTE3024-mCherry and propagated the culture to prepare stocks. Optimization of infection, confocal microscopy, and imaging were done by U.M.N., A. Kiatthanapaiboon, and B.E.M. Flow cytometry experiments were designed and conducted by B.E.M. and A. Kollipara. V.M. performed sample preparation for electron microscopy and V.M., P.W., and B.E.M. performed imaging. B.R. and M.K. carried out the mass spectrometry analysis. B.E.M. and U.M.N. directed the laboratory work related to Chlamydia infection. B.E.M. wrote the manuscript with T.D. and U.M.N. All senior authors participated in reviewing and editing the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2018. Sexually transmitted disease surveillance 2018. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 2.Newman L, Rowley J, Vander Hoorn S, Wijesooriya NS, Unemo M, Low N, Stevens G, Gottlieb S, Kiarie J, Temmerman M. 2015. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One 10:e0143304. doi: 10.1371/journal.pone.0143304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Connell CM, Ferone ME. 2016. Chlamydia trachomatis genital infections. Microb Cell 3:390–403. doi: 10.15698/mic2016.09.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elwell C, Mirrashidi K, Engel J. 2016. Chlamydia cell biology and pathogenesis. Nat Rev Microbiol 14:385–400. doi: 10.1038/nrmicro.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasmussen SJ, Eckmann L, Quayle AJ, Shen L, Zhang YX, Anderson DJ, Fierer J, Stephens RS, Kagnoff MF. 1997. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J Clin Invest 99:77–87. doi: 10.1172/JCI119136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rank RG, Lacy HM, Goodwin A, Sikes J, Whittimore J, Wyrick PB, Nagarajan UM. 2010. Host chemokine and cytokine response in the endocervix within the first developmental cycle of Chlamydia muridarum. Infect Immun 78:536–544. doi: 10.1128/IAI.00772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quayle AJ. 2002. The innate and early immune response to pathogen challenge in the female genital tract and the pivotal role of epithelial cells. J Reprod Immunol 57:61–79. doi: 10.1016/s0165-0378(02)00019-0. [DOI] [PubMed] [Google Scholar]
- 8.Johnson RM. 2004. Murine oviduct epithelial cell cytokine responses to Chlamydia muridarum infection include interleukin-12-p70 secretion. Infect Immun 72:3951–3960. doi: 10.1128/IAI.72.7.3951-3960.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasilevsky S, Greub G, Nardelli-Haefliger D, Baud D. 2014. Genital Chlamydia trachomatis: understanding the roles of innate and adaptive immunity in vaccine research. Clin Microbiol Rev 27:346–370. doi: 10.1128/CMR.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehr S, Vier J, Hacker G, Kirschnek S. 2018. Activation of neutrophils by Chlamydia trachomatis-infected epithelial cells is modulated by the chlamydial plasmid. Microbes Infect 20:284–292. doi: 10.1016/j.micinf.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Mukura LR, Hickey DK, Rodriguez-Garcia M, Fahey JV, Wira CR. 2017. Chlamydia trachomatis regulates innate immune barrier integrity and mediates cytokine and antimicrobial responses in human uterine ECC-1 epithelial cells. Am J Reprod Immunol 78:e12764. doi: 10.1111/aji.12764. [DOI] [PubMed] [Google Scholar]
- 12.Maxion HK, Kelly KA. 2002. Chemokine expression patterns differ within anatomically distinct regions of the genital tract during Chlamydia trachomatis infection. Infect Immun 70:1538–1546. doi: 10.1128/iai.70.3.1538-1546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frattini A, Fabbri M, Valli R, De Paoli E, Montalbano G, Gribaldo L, Pasquali F, Maserati E. 2015. High variability of genomic instability and gene expression profiling in different HeLa clones. Sci Rep 5:15377. doi: 10.1038/srep15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahey JV, Schaefer TM, Channon JY, Wira CR. 2005. Secretion of cytokines and chemokines by polarized human epithelial cells from the female reproductive tract. Hum Reprod 20:1439–1446. doi: 10.1093/humrep/deh806. [DOI] [PubMed] [Google Scholar]
- 15.Hvid M, Baczynska A, Deleuran B, Fedder J, Knudsen HJ, Christiansen G, Birkelund S. 2007. Interleukin-1 is the initiator of fallopian tube destruction during Chlamydia trachomatis infection. Cell Microbiol 9:2795–2803. doi: 10.1111/j.1462-5822.2007.00996.x. [DOI] [PubMed] [Google Scholar]
- 16.Levanon K, Ng V, Piao HY, Zhang Y, Chang MC, Roh MH, Kindelberger DW, Hirsch MS, Crum CP, Marto JA, Drapkin R. 2010. Primary ex vivo cultures of human fallopian tube epithelium as a model for serous ovarian carcinogenesis. Oncogene 29:1103–1113. doi: 10.1038/onc.2009.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickens CJ, Comer MT, Southgate J, Leese HJ. 1996. Human fallopian tubal epithelial cells in vitro: establishment of polarity and potential role of intracellular calcium and extracellular ATP in fluid secretion. Hum Reprod 11:212–217. doi: 10.1093/oxfordjournals.humrep.a019021. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh M, Schaefer TM, Fahey JV, Wright JA, Wira CR. 2008. Antiviral responses of human fallopian tube epithelial cells to Toll-like receptor 3 agonist poly(I:C). Fertil Steril 89:1497–1506. doi: 10.1016/j.fertnstert.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fulcher ML, Randell SH. 2013. Human nasal and tracheo-bronchial respiratory epithelial cell culture. Methods Mol Biol 945:109–121. doi: 10.1007/978-1-62703-125-7_8. [DOI] [PubMed] [Google Scholar]
- 20.Correr S, Makabe S, Heyn R, Relucenti M, Naguro T, Familiari G. 2006. Microplicae-like structures of the fallopian tube in postmenopausal women as shown by electron microscopy. Histol Histopathol 21:219–226. doi: 10.14670/HH-21.219. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Chen X, Zhou J. 1996. Ultrastructural study on the epithelium of ligated fallopian tubes in women of reproductive age. Ann Anat 178:317–320. doi: 10.1016/S0940-9602(96)80082-3. [DOI] [PubMed] [Google Scholar]
- 22.Gipson IK, Ho SB, Spurr-Michaud SJ, Tisdale AS, Zhan Q, Torlakovic E, Pudney J, Anderson DJ, Toribara NW, Hill JA 3rd. 1997. Mucin genes expressed by human female reproductive tract epithelia. Biol Reprod 56:999–1011. doi: 10.1095/biolreprod56.4.999. [DOI] [PubMed] [Google Scholar]
- 23.Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. 1986. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol 103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zihni C, Mills C, Matter K, Balda MS. 2016. Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol 17:564–580. doi: 10.1038/nrm.2016.80. [DOI] [PubMed] [Google Scholar]
- 25.Buckner LR, Lewis ME, Greene SJ, Foster TP, Quayle AJ. 2013. Chlamydia trachomatis infection results in a modest pro-inflammatory cytokine response and a decrease in T cell chemokine secretion in human polarized endocervical epithelial cells. Cytokine 63:151–165. doi: 10.1016/j.cyto.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boucher A, Mourad W, Mailloux J, Lemay A, Akoum A. 2000. Ovarian hormones modulate monocyte chemotactic protein-1 expression in endometrial cells of women with endometriosis. Mol Hum Reprod 6:618–626. doi: 10.1093/molehr/6.7.618. [DOI] [PubMed] [Google Scholar]
- 27.Cunningham K, Stansfield SH, Patel P, Menon S, Kienzle V, Allan JA, Huston WM. 2013. The IL-6 response to Chlamydia from primary reproductive epithelial cells is highly variable and may be involved in differential susceptibility to the immunopathological consequences of chlamydial infection. BMC Immunol 14:50. doi: 10.1186/1471-2172-14-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lad SP, Li J, da Silva Correia J, Pan Q, Gadwal S, Ulevitch RJ, Li E. 2007. Cleavage of p65/RelA of the NF-kappaB pathway by Chlamydia. Proc Natl Acad Sci U S A 104:2933–2938. doi: 10.1073/pnas.0608393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haldar AK, Piro AS, Finethy R, Espenschied ST, Brown HE, Giebel AM, Frickel EM, Nelson DE, Coers J. 2016. Chlamydia trachomatis Is resistant to inclusion ubiquitination and associated host defense in gamma interferon-primed human epithelial cells. mBio 7:e01417-16. doi: 10.1128/mBio.01417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegl C, Prusty BK, Karunakaran K, Wischhusen J, Rudel T. 2014. Tumor suppressor p53 alters host cell metabolism to limit Chlamydia trachomatis infection. Cell Rep 9:918–929. doi: 10.1016/j.celrep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Gipson IK. 2001. Mucins of the human endocervix. Front Biosci 6:D1245–D1255. doi: 10.2741/gipson. [DOI] [PubMed] [Google Scholar]
- 32.Hall JV, Schell M, Dessus-Babus S, Moore CG, Whittimore JD, Sal M, Dill BD, Wyrick PB. 2011. The multifaceted role of oestrogen in enhancing Chlamydia trachomatis infection in polarized human endometrial epithelial cells. Cell Microbiol 13:1183–1199. doi: 10.1111/j.1462-5822.2011.01608.x. [DOI] [PubMed] [Google Scholar]
- 33.Wyrick PB, Choong J, Davis CH, Knight ST, Royal MO, Maslow AS, Bagnell CR. 1989. Entry of genital Chlamydia trachomatis into polarized human epithelial cells. Infect Immun 57:2378–2389. doi: 10.1128/IAI.57.8.2378-2389.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al-Taweel FB, Douglas CWI, Whawell SA. 2016. The periodontal pathogen Porphyromonas gingivalis preferentially interacts with oral epithelial cells in S phase of the cell cycle. Infect Immun 84:1966–1974. doi: 10.1128/IAI.00111-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santos AJ, Meinecke M, Fessler MB, Holden DW, Boucrot E. 2013. Preferential invasion of mitotic cells by Salmonella reveals that cell surface cholesterol is maximal during metaphase. J Cell Sci 126:2990–2996. doi: 10.1242/jcs.115253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueda R, Sugiura T, Kume S, Ichikawa A, Larsen S, Miyoshi H, Hiramatsu H, Nagatsuka Y, Arai F, Suzuki Y, Hirabayashi Y, Fukuda T, Honda A. 2013. A novel single virus infection system reveals that influenza virus preferentially infects cells in G1 phase. PLoS One 8:e67011. doi: 10.1371/journal.pone.0067011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooper MD, Rapp J, Jeffery-Wiseman C, Barnes RC, Stephens DS. 1990. Chlamydia trachomatis infection of human fallopian tube organ cultures. J Gen Microbiol 136:1109–1115. doi: 10.1099/00221287-136-6-1109. [DOI] [PubMed] [Google Scholar]
- 38.Hoenderboom BM, van Benthem BHB, van Bergen J, Dukers-Muijrers N, Gotz HM, Hoebe C, Hogewoning AA, Land JA, van der Sande M, Morre SA, van den Broek I. 2019. Relation between Chlamydia trachomatis infection and pelvic inflammatory disease, ectopic pregnancy and tubal factor infertility in a Dutch cohort of women previously tested for chlamydia in a chlamydia screening trial. Sex Transm Infect 95:300–306. doi: 10.1136/sextrans-2018-053778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herzog SA, Althaus CL, Heijne JC, Oakeshott P, Kerry S, Hay P, Low N. 2012. Timing of progression from Chlamydia trachomatis infection to pelvic inflammatory disease: a mathematical modelling study. BMC Infect Dis 12:187. doi: 10.1186/1471-2334-12-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oakeshott P, Kerry S, Aghaizu A, Atherton H, Hay S, Taylor-Robinson D, Simms I, Hay P. 2010. Randomised controlled trial of screening for Chlamydia trachomatis to prevent pelvic inflammatory disease: the POPI (prevention of pelvic infection) trial. BMJ 340:c1642. doi: 10.1136/bmj.c1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Connell CM, Brochu H, Girardi J, Harrell E, Jones A, Darville T, Sena AC, Peng X. 2019. Simultaneous profiling of sexually transmitted bacterial pathogens, microbiome, and concordant host response in cervical samples using whole transcriptome sequencing analysis. Microb Cell 6:177–183. doi: 10.15698/mic2019.03.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hufnagel K, Lueong S, Willhauck-Fleckenstein M, Hotz-Wagenblatt A, Miao B, Bauer A, Michel A, Butt J, Pawlita M, Hoheisel JD, Waterboer T. 2018. Immunoprofiling of Chlamydia trachomatis using whole-proteome microarrays generated by on-chip in situ expression. Sci Rep 8:7503. doi: 10.1038/s41598-018-25918-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patton DL, Halbert SA, Kuo CC, Wang SP, Holmes KK. 1983. Host response to primary Chlamydia trachomatis infection of the fallopian tube in pig-tailed monkeys. Fertil Steril 40:829–840. doi: 10.1016/S0015-0282(16)47489-3. [DOI] [PubMed] [Google Scholar]
- 44.Phillips DM, Swenson CE, Schachter J. 1984. Ultrastructure of Chlamydia trachomatis infection of the mouse oviduct. J Ultrastruct Res 88:244–256. doi: 10.1016/s0022-5320(84)90122-9. [DOI] [PubMed] [Google Scholar]
- 45.Rajeeve K, Vollmuth N, Janaki-Raman S, Wulff T, Schmalhofer M, Schmitz W, Baluapuri A, Huber C, Fink J, Dejure FR, Wolf E, Eisenreich W, Schulze A, Seibel J, Rudel T. 2019. A central role of glutamine in Chlamydia infection. bioRxiv 10.1101/742817. [DOI]
- 46.Singla M. 2007. Role of tryptophan supplementation in the treatment of Chlamydia. Med Hypotheses 68:278–280. doi: 10.1016/j.mehy.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 47.Leonhardt RM, Lee SJ, Kavathas PB, Cresswell P. 2007. Severe tryptophan starvation blocks onset of conventional persistence and reduces reactivation of Chlamydia trachomatis. Infect Immun 75:5105–5117. doi: 10.1128/IAI.00668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raulston JE. 1997. Response of Chlamydia trachomatis serovar E to iron restriction in vitro and evidence for iron-regulated chlamydial proteins. Infect Immun 65:4539–4547. doi: 10.1128/IAI.65.11.4539-4547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al-Younes HM, Rudel T, Brinkmann V, Szczepek AJ, Meyer TF. 2001. Low iron availability modulates the course of Chlamydia pneumoniae infection. Cell Microbiol 3:427–437. doi: 10.1046/j.1462-5822.2001.00125.x. [DOI] [PubMed] [Google Scholar]
- 50.Kumar Y, Valdivia RH. 2009. Leading a sheltered life: intracellular pathogens and maintenance of vacuolar compartments. Cell Host Microbe 5:593–601. doi: 10.1016/j.chom.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rother M, Gonzalez E, Teixeira da Costa AR, Wask L, Gravenstein I, Pardo M, Pietzke M, Gurumurthy RK, Angermann J, Laudeley R, Glage S, Meyer M, Chumduri C, Kempa S, Dinkel K, Unger A, Klebl B, Klos A, Meyer TF. 2018. Combined human genome-wide RNAi and metabolite analyses identify IMPDH as a host-directed target against Chlamydia infection. Cell Host Microbe 23:661–671.e8. doi: 10.1016/j.chom.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 52.Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S, McSkane M, Baba H, Lenz HJ. 2018. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation—a target for novel cancer therapy. Cancer Treat Rev 63:40–47. doi: 10.1016/j.ctrv.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sokol CL, Luster AD. 2015. The chemokine system in innate immunity. Cold Spring Harb Perspect Biol 7:a016303. doi: 10.1101/cshperspect.a016303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hussen J, Frank C, Duvel A, Koy M, Schuberth HJ. 2014. The chemokine CCL5 induces selective migration of bovine classical monocytes and drives their differentiation into LPS-hyporesponsive macrophages in vitro. Dev Comp Immunol 47:169–177. doi: 10.1016/j.dci.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 55.Aswad M, Assi S, Schif-Zuck S, Ariel A. 2017. CCL5 promotes resolution-phase macrophage reprogramming in concert with the atypical chemokine receptor D6 and apoptotic polymorphonuclear cells. J Immunol 199:1393–1404. doi: 10.4049/jimmunol.1502542. [DOI] [PubMed] [Google Scholar]
- 56.Poston TB, Lee DE, Darville T, Zhong W, Dong L, O’Connell CM, Wiesenfeld HC, Hillier SL, Sempowski GD, Zheng X. 2019. Cervical cytokines associated with Chlamydia trachomatis susceptibility and protection. J Infect Dis 220:330–339. doi: 10.1093/infdis/jiz087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramsey KH, Rank RG. 1991. Resolution of chlamydial genital infection with antigen-specific T-lymphocyte lines. Infect Immun 59:925–931. doi: 10.1128/IAI.59.3.925-931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roan NR, Starnbach MN. 2006. Antigen-specific CD8+ T cells respond to Chlamydia trachomatis in the genital mucosa. J Immunol 177:7974–7979. doi: 10.4049/jimmunol.177.11.7974. [DOI] [PubMed] [Google Scholar]
- 59.Bakshi RK, Gupta K, Jordan SJ, Chi X, Lensing SY, Press CG, Geisler WM. 2018. An adaptive Chlamydia trachomatis-specific IFN-gamma-producing CD4+ T cell response is associated with protection against Chlamydia reinfection in women. Front Immunol 9:1981. doi: 10.3389/fimmu.2018.01981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morrison RP, Feilzer K, Tumas DB. 1995. Gene knockout mice establish a primary protective role for major histocompatibility complex class II-restricted responses in Chlamydia trachomatis genital tract infection. Infect Immun 63:4661–4668. doi: 10.1128/IAI.63.12.4661-4668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morrison SG, Su H, Caldwell HD, Morrison RP. 2000. Immunity to murine Chlamydia trachomatis genital tract reinfection involves B cells and CD4+ T cells but not CD8+ T cells. Infect Immun 68:6979–6987. doi: 10.1128/iai.68.12.6979-6987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gondek DC, Olive AJ, Stary G, Starnbach MN. 2012. CD4+ T cells are necessary and sufficient to confer protection against Chlamydia trachomatis infection in the murine upper genital tract. J Immunol 189:2441–2449. doi: 10.4049/jimmunol.1103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kelly KA, Rank RG. 1997. Identification of homing receptors that mediate the recruitment of CD4 T cells to the genital tract following intravaginal infection with Chlamydia trachomatis. Infect Immun 65:5198–5208. doi: 10.1128/IAI.65.12.5198-5208.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perry LL, Feilzer K, Portis JL, Caldwell HD. 1998. Distinct homing pathways direct T lymphocytes to the genital and intestinal mucosae in Chlamydia-infected mice. J Immunol 160:2905–2914. [PubMed] [Google Scholar]
- 65.Kelly KA, Natarajan S, Ruther P, Wisse A, Chang MH, Ault KA. 2001. Chlamydia trachomatis infection induces mucosal addressin cell adhesion molecule-1 and vascular cell adhesion molecule-1, providing an immunologic link between the fallopian tube and other mucosal tissues. J Infect Dis 184:885–891. doi: 10.1086/323341. [DOI] [PubMed] [Google Scholar]
- 66.Ibana JA, Schust DJ, Sugimoto J, Nagamatsu T, Greene SJ, Quayle AJ. 2011. Chlamydia trachomatis immune evasion via downregulation of MHC class I surface expression involves direct and indirect mechanisms. Infect Dis Obstet Gynecol 2011:420905. doi: 10.1155/2011/420905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhong G, Fan T, Liu L. 1999. Chlamydia inhibits interferon gamma-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J Exp Med 189:1931–1938. doi: 10.1084/jem.189.12.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhong G, Liu L, Fan T, Fan P, Ji H. 2000. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in chlamydia-infected cells. J Exp Med 191:1525–1534. doi: 10.1084/jem.191.9.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhong G, Fan P, Ji H, Dong F, Huang Y. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J Exp Med 193:935–942. doi: 10.1084/jem.193.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cram ED, Simmons RS, Palmer AL, Hildebrand WH, Rockey DD, Dolan BP. 2016. Enhanced direct major histocompatibility complex class I self-antigen presentation induced by Chlamydia infection. Infect Immun 84:480–490. doi: 10.1128/IAI.01254-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. 2005. Well-differentiated human airway epithelial cell cultures. Methods Mol Med 107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 72.Suchland RJ, Stamm WE. 1991. Simplified microtiter cell culture method for rapid immunotyping of Chlamydia trachomatis. J Clin Microbiol 29:1333–1338. doi: 10.1128/JCM.29.7.1333-1338.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Agaisse H, Derre I. 2013. A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PLoS One 8:e57090. doi: 10.1371/journal.pone.0057090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prantner D, Darville T, Nagarajan UM. 2010. Stimulator of IFN gene is critical for induction of IFN-beta during Chlamydia muridarum infection. J Immunol 184:2551–2560. doi: 10.4049/jimmunol.0903704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dessus-Babus S, Darville TL, Cuozzo FP, Ferguson K, Wyrick PB. 2002. Differences in innate immune responses (in vitro) to HeLa cells infected with nondisseminating serovar E and disseminating serovar L2 of Chlamydia trachomatis. Infect Immun 70:3234–3248. doi: 10.1128/iai.70.6.3234-3248.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reynolds ES. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stephens R, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R, Zhao Q, Koonin E, Davis R. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 78.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 79.Kesimer M, Cullen J, Cao R, Radicioni G, Mathews KG, Seiler G, Gookin JL. 2015. Excess secretion of gel-forming mucins and associated innate defense proteins with defective mucin un-packaging underpin gallbladder mucocele formation in dogs. PLoS One 10:e0138988. doi: 10.1371/journal.pone.0138988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 81.Kotecha N, Krutzik PO, Irish JM. 2010. Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom 53: 10.17.1–10.17.24. doi: 10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.