Uropathogenic Escherichia coli (UPEC) is the leading cause of urinary tract infections. These bacteria undertake a multistage infection cycle involving invasion of and proliferation within urinary tract epithelial cells, leading to the rupture of the host cell and dispersal of the bacteria, some of which have a highly filamentous morphology. Here, we established a microfluidics-based model of UPEC infection of immortalized human bladder epithelial cells that recapitulates the main stages of bacterial morphological changes during the acute infection cycle in vivo and allows the development and fate of individual cells to be monitored in real time by fluorescence microscopy.
KEYWORDS: UPEC, infection model, microfluidics, morphological differentiation, stress response, urinary tract infection, urine
ABSTRACT
Uropathogenic Escherichia coli (UPEC) is the leading cause of urinary tract infections. These bacteria undertake a multistage infection cycle involving invasion of and proliferation within urinary tract epithelial cells, leading to the rupture of the host cell and dispersal of the bacteria, some of which have a highly filamentous morphology. Here, we established a microfluidics-based model of UPEC infection of immortalized human bladder epithelial cells that recapitulates the main stages of bacterial morphological changes during the acute infection cycle in vivo and allows the development and fate of individual cells to be monitored in real time by fluorescence microscopy. The UPEC-infected bladder cells remained alive and mobile in nonconfluent monolayers during the development of intracellular bacterial communities (IBCs). Switching from a flow of growth medium to human urine resulted in immobilization of both uninfected and infected bladder cells. Some IBCs continued to develop and then released many highly filamentous bacteria via an extrusion-like process, whereas other IBCs showed strong UPEC proliferation, and yet no filamentation was detected. The filamentation response was dependent on the weak acidity of human urine and required component(s) in a low molecular-mass (<3,000 Da) fraction from a mildly dehydrated donor. The developmental fate for bacteria therefore appears to be controlled by multiple factors that act at the level of the whole IBC, suggesting that variable local environments or stochastic differentiation pathways influence IBC developmental fates during infection.
INTRODUCTION
Uropathogenic Escherichia coli (UPEC) is responsible for more than 80% of urinary tract infections (UTIs), which are one of the most common infections that are worsening rapidly as a consequence of pandemic multiresistant strains spreading globally (1). These invasive infections proceed through a multistage intracellular cycle, beginning with the attachment of bacteria to the host urinary tract epithelial surfaces (2, 3). The bacteria are taken up by endocytosis, and some are released into the cytoplasm and grow to form tightly packed biofilm-like colonies called intracellular bacterial communities (IBCs) (4–8). Eventually the epithelial cell becomes overwhelmed by bacterial growth and ruptures, dispersing bacteria that may infect neighboring host cells (9). This cycle amplifies the infection and leads to widespread damage and shedding of layers of the epithelium, leading to the acute symptoms observed in UTI.
UPEC shows remarkable morphological variation during the infection cycle (10). Whereas UPEC strains appear as typical rod-shaped bacteria in laboratory cultures, during the infection cycle many of the intracellular bacteria exist as very short rods or almost-spherical smaller cells within well-established IBCs. However, while bacteria are released during IBC dispersal and host cell rupture, UPEC subpopulations appear as rods, motile rods, and viable highly filamentous bacteria (11). Filamentation occurs by bacterial cell growth (elongation) and ongoing chromosome replication without division and is a response to certain conditions or stresses (10). Filamentation is a characteristic response of UPEC during UTI and is thought to aid in dispersal and surface attachment after release from the host cell and might help prevent phagocytosis by immune cells (11–13). It has been demonstrated that UPEC filaments are capable of reverting back into rod-shaped cells, through numerous cell division events along the length of the filaments, that are then capable of infecting other host cells (14).
Many of the features of the UPEC infection cycle outlined above were identified with the mouse model of UTI, involving transurethral injection of UPEC into the bladder and analysis of collected urine/tissues or monitoring of infected bladder explants over time (4, 7, 9). IBCs and filamentous bacteria are also common in human cystitis (8). Cell culture models were more recently developed to allow further analyses of bacterial physiology during infection, based on UPEC infection of human bladder epithelial cell (BEC) surface culture and incubation with growth medium or human urine (14, 15). A static infection model was developed that could be performed in multiwell culture plates and was used to study features of the early stages of infection and quantify intracellular growth of UPEC within BECs (15). This was not suitable for analysis of the later stages of host cell rupture (bacterial dispersal and reinfection), and a flow chamber model was then developed, incorporating steps that allowed the analysis of later infection stages (14). This model involved UPEC infection of BECs in flow chambers that were maintained with a constant flow of medium during the early development of IBCs (day 1) and then switched to a flow of filtered human urine, which induced the dispersal/filamentation stage (day 2). Extensive bacterial filamentation was observed on the surface and dispersed into the effluent (14). Given that UPEC shows a complex pattern of morphological changes during the infection cycle and that the infection cycle in the population is likely to show a low degree of synchrony, the ability to use time-lapse microscopy to investigate the timing and transitions occurring at the single cell level is expected to be of significant benefit in understanding the sequence of morphological events occurring during the UPEC life cycle.
Here, we have established a microfluidics-based cell culture infection model for UPEC infection of immortalized human bladder epithelial cells, combined with a new robust green fluorescent protein (GFP)-based bacterial labeling for real-time fluorescence microscopy observation of infections at high resolution. The main advantages of using microfluidics are the use of greatly reduced quantities of reagents and laboratory animals, a high degree of control of conditions, and the ability to conduct multiple real-time infections experiments simultaneously while visualizing infection progression and the viability of individual cells. This approach has revealed details of how infected cultured cells behave during IBC development, that there are at least two distinct morphological fates of IBCs upon dispersal, and how filaments appear to extrude from infected bladder cells during dispersal. We also demonstrate that multiple conditions, including a urinary pH of ∼5, are necessary to elicit the UPEC filamentation response during the IBC dispersal and host cell rupture stage of infection.
RESULTS
Construction of fluorescent derivatives of uropathogenic E. coli UTI89.
We first aimed to establish a system for robust fluorescent labeling of live bacteria, to allow their visualization during the course of an intracellular infection. Lambda-red recombination (16) was initially used to replace the lacZ open reading frame on the chromosome of model UTI strain E. coli UTI89 with fluorescent proteins, GFP or mCherry, linked to a downstream Kmr marker; however, the fluorescence in the resulting bacteria was too faint to provide a useful label for bacteria in an infection (data not shown). Similar results were obtained with attempts involving transformation of UTI89 with the GFP expression plasmid pXG10 (17), whereas pEGFP-based plasmids reported previously for this purpose (14) gave stronger expression but required IPTG (isopropyl-β-d-thiogalactopyranoside) induction and showed a variegated fluorescence response between individual bacteria, particularly in the stationary phase of batch cultures (data not shown). A new low-copy-number plasmid was then constructed (pGI5) containing the pSC101 origin of replication and the PlacIQ1 strong constitutive promoter (18) driving expression of monomeric superfolder GFP (msfGFP) (19). This provided bright, robust, and uniform cytoplasmic fluorescent labeling of UTI89 (see Fig. S1 in the supplemental material). Equivalent plasmids based on the significantly weaker PlacI and PlacIQ promoters were also constructed (pGI3 and pGI4, respectively), although the fluorescence of UTI89 harboring these was considered too faint for use in most infections (Fig. S1).
Microfluidic infection model involving intracellular growth and dispersal of UPEC.
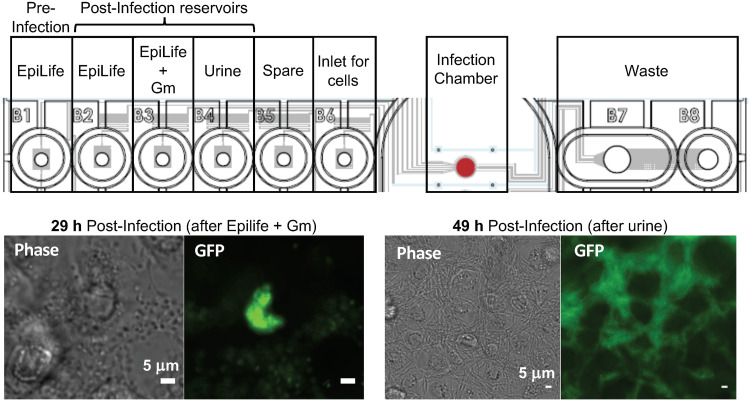
We sought to visualize the main infection stages of UPEC intracellular growth and release from BECs using a microfluidics-based culture system, based on the midscale flow-chamber model previously established (14). The microfluidics system used here (CellASIC Onix) has a thin glass base plate (170 μm), adhered to a cast set of microfluidic channels supplied by inlet wells filled with the different media needed for each stage of the infection (Fig. 1, upper panel). These wells have microfluidic channels connected to a main culture chamber, where the immortalized human BECs were initially grown. The flow of liquid from the wells through the culture chamber and into a waste reservoir is controlled by a manifold and air pressure controller wells through the culture chamber and into waste wells; each chamber can therefore be switched between the flow of different reservoir liquids (e.g., growth medium or urine). Four such chamber systems exist per plate (standard microtiter footprint), allowing multiple infections to be observed simultaneously in an incubated inverted microscope with automated stage and focusing.
FIG 1.
Microfluidic model for the intracellular UPEC and release stages of UTI. Diagrammatic representation of the CellASIC Onix microfluidics plate showing the contents of each well that were sequentially pumped through the main chamber during the defined stages of the infection. The lower panels show phase-contrast and fluorescence microscopy images of the infection chamber surface at the end of the intracellular growth stage (29 h postinfection), showing a well-developed IBC, and at the end of the dispersal phase (49 h postinfection), showing filamentous UTI89/pGI5 in among BECs. Scale bars, 5 μm.
A layer of PD07i BECs, initially grown for 2 days in the microfluidic culture chambers, was exposed to a capillary-driven flow of E. coli UTI89/pGI5 for 20 min to allow bacterial attachment, and then the flow was switched to the BEC growth medium (EpiLife) for 9 h to allow bacterial uptake and growth. The flow was then switched to medium containing the BEC-impermeable antibiotic gentamicin (Gm) for 20 h to kill and remove only the extracellular bacteria and allow IBC development and visualization. Finally, the flow was switched to human urine for a further 20 h to observe bacterial dispersal and host cell rupture. Microscope observation of the cells after the intracellular growth stage (i.e., at 29 h postinfection) showed densely packed discrete IBCs within BECs (Fig. 1, lower left) that often appeared near the edge of the nucleus or as multilobed colonies. The proportion of infected BECs was typically 2 to 10% of the total, which is consistent with previously observed rates of infection in vitro (14). At the 49-h stage—i.e., after a 20-h exposure to a flow of urine—many highly filamentous bacteria were evident among the BECs (Fig. 1, lower right).
UPEC-infected PD07i bladder epithelial cells retain surface migration capacity.
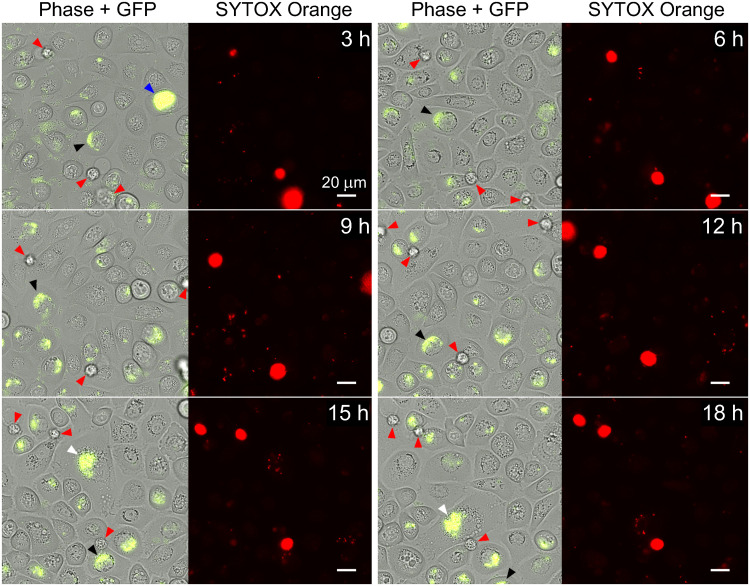
We then observed infections during the intracellular growth stage by recording time-lapse movies of the infections in the microfluidic culture chambers (i.e., during the flow of growth medium with Gm, 9 to 29 h postinfection). As shown in Fig. 2 (and in Movie S1 in the supplemental material), the bladder cells showed active migration on the surface of the culture chamber during this period. Infected BECs, as indicated by the presence of fluorescent IBCs, moved on the surface in a manner indistinguishable from the uninfected bladder cells in the same field of view, even when containing very large IBCs (Fig. 2, blue arrowhead). Some infected bladder cells were observed to move in or out of the field of view during the time lapse (Fig. 2, blue and white arrowheads; see also Movie S1). During these experiments, the live-cell-impermeant DNA stain SYTOX Orange was included in the medium (at 100 nM), with the aim of specifically staining and detecting permeable (dead) cells over the course of the infection (Fig. 2, red arrowheads), while simultaneously allowing the GFP-labeled bacteria to be located with green fluorescence. SYTOX Orange-stained BECs were occasionally seen that showed no evidence of infection, exhibited a rounded shape, and were partially or fully detached from the surface, which is typical of dead cells (Fig. 2, red arrowheads). In contrast, the surface-bound migratory bladder cells in the monolayer, both infected and uninfected, did not stain with SYTOX Orange (Fig. 2).
FIG 2.
PD07i bladder cells infected with UTI89/pGI5 are mobile. Time-lapse microscopy showing 3-h intervals during the 9- to 29-h stage of infection, with a flow of EpiLife+Gm, was performed. Phase-contrast and GFP channel overlays (left) with SYTOX Orange channel (right) are shown for the indicated time points after switching to Gm-containing medium. Infected bladder cells (black, white, and blue arrowheads, which track individual BECs) appeared at different positions at each time point. SYTOX Orange staining indicated that most infected and uninfected bladder cells were not permeable, whereas permeable, assigned-dead bladder cells are indicated by red arrowheads; these occasionally drifted detached from the surface during the movie. Scale bars, 20 μm (60× oil objective). Refer to Movie S1 in the supplemental material for the full set of time-lapse images.
PD07i cells cease surface migration and gain autofluorescence after urine exposure.
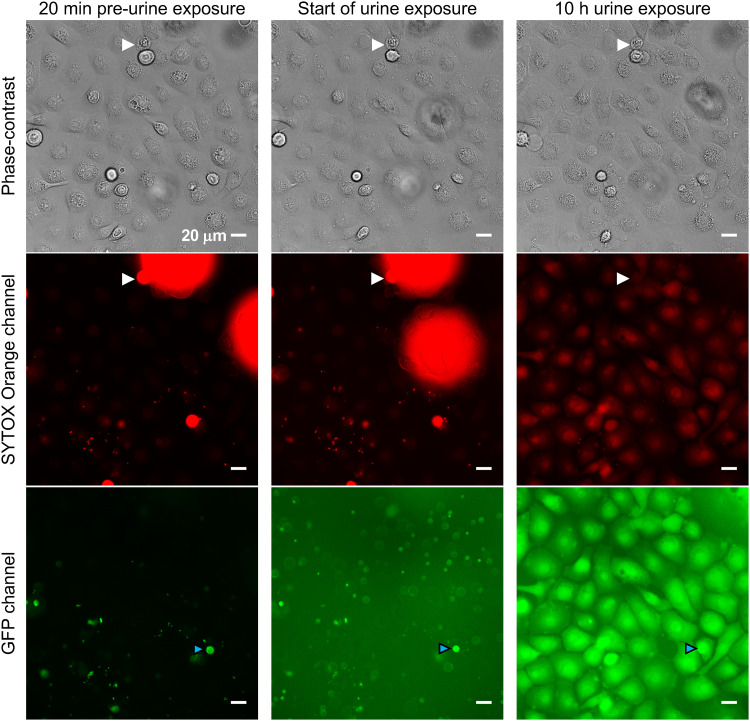
The viability and normal surface migration behavior of infected BECs (Fig. 2) clearly showed that IBCs did not globally incapacitate nor substantially affect the BECs during the early stages (first 29 h) of IBC establishment and growth. We then switched to a flow of filtered human urine, observing cells 20 min before the switch to urine, immediately after the switch to a flow of urine (at 29 h), and then 10 h into the urine exposure (Fig. 3). The lack of SYTOX Orange staining of BECs before the urine exposure indicated that most BECs were nonpermeable, apart from the occasional dead cell (e.g., Fig. 3; very bright drifting cells [out of focus], and some stationary cells indicated by white arrowheads). Upon commencement of urine exposure, the background fluorescence in the entire field of view increased significantly in the GFP channel. The brightly labeled intracellular UTI89/pGI5 bacteria were still detectable (e.g., Fig. 3, cyan arrowheads). After 10 h of urine exposure, the morphology of the BECs and their location in the field of view had remained unchanged, indicating that the urine treatment immobilized the BECs (Fig. 3, compare phase-contrast images). By 10 h, the BECs also showed much greater whole-cell autofluorescence in the red and especially in the green channels (Fig. 3).
FIG 3.
Bladder cells become immobile after exposure to urine. Microscopy images were taken 20 min before urine exposure, immediately after switching to urine (0 h), and 10 h into the urine exposure. Dead bladder cells have detached from the monolayer and taken up SYTOX Orange stain (e.g., white arrowheads), but the staining disappears soon after urine exposure (within 10 to 20 min). Scale bars, 20 μm (40× objective). All GFP exposures = 50 ms; all SYTOX exposures = 100 ms. Refer to Movie S2 for the full set of time-lapse images.
The above observations can be seen in greater temporal resolution in Movie S2, which shows time-lapse images recorded at 10-min intervals over the 20-h urine exposure (i.e., 29 to 49 h postinfection). Cells ceased surface migration immediately upon urine exposure. Then, during the first ∼6 h of urine flow, we observed the fluorescence of individual BECs (infected and uninfected) to become brighter at times that varied for individual cells, especially in the green channel. The transition for individual cells to the high autofluorescence state took ca. 10 to 40 min. This suggested a permeabilization or metabolic change in BECs in response to the urine. However, permeabilization was difficult to directly assess, since the SYTOX Orange (added to the urine) was not effectively reporting permeabilization in the urine conditions. We observed that the dead BECs that had been brightly stained in the preceding EpiLife medium plus gentamicin (EpiLife+Gm) stage had lost their bright red fluorescence during the first 10 to 20 min of urine exposure (Fig. 3, white arrowheads; Movie S2). This could be due to either a lack of dye fluorescence/DNA binding in urine or the DNA was lost (degraded) in dead cells upon urine exposure. In support of the latter, some bacteria retained both green and red fluorescence during the 20-h urine exposure, the red fluorescence of dead BECs took ∼20 min to completely disappear after switching to urine (Movie S2), and human urine contains DNases (20, 21).
UPEC emerges from infected PD07i bladder epithelial cells via at least two distinct IBC dispersal types.
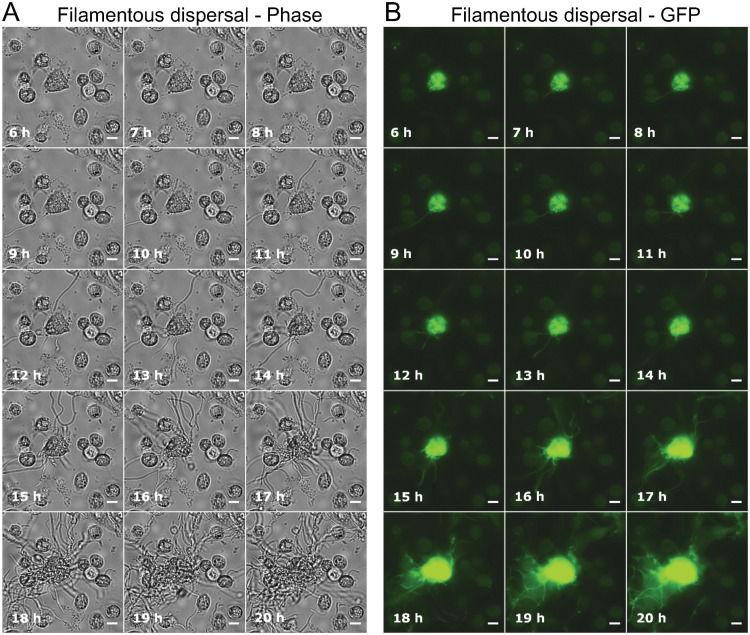
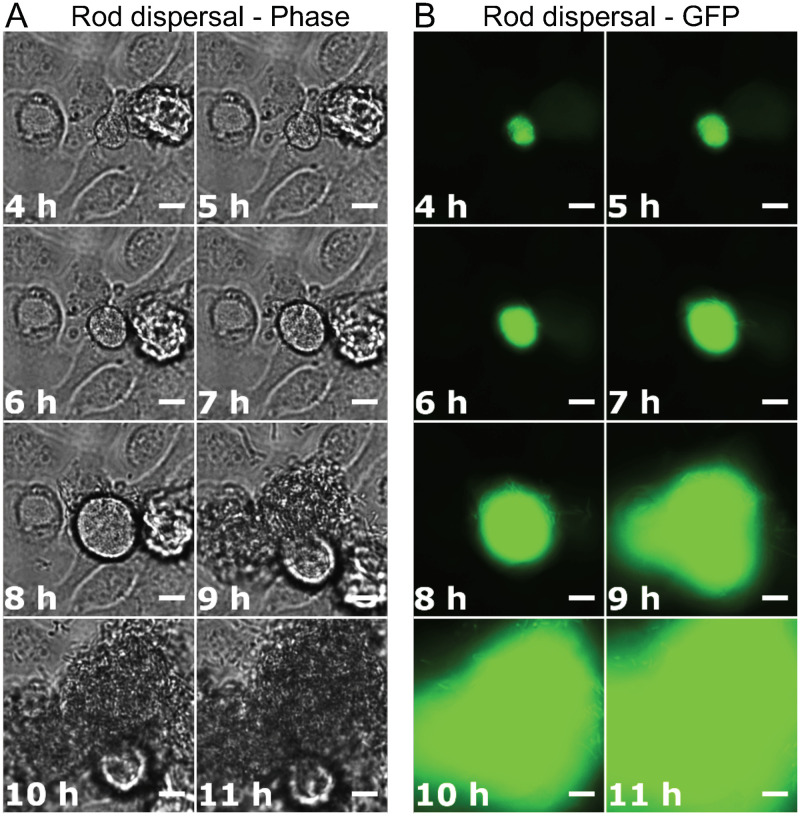
Bacteria within an IBC have been reported to grow from short rod-shaped cells into long filaments after exposure to high specific-gravity (SG) urine during the dispersal phase of infection (14). To directly visualize this stage of infection over time, phase-contrast and GFP fluorescence images were recorded at 10-min intervals during the 25- to 49-h postinfection period. This 24-h movie included the last 4 h of the growth medium flow conditions, followed by 20 h of urine flow conditions. Seven of the ten IBC dispersal events (individual BEC eruptions) that were monitored (in three flow separate infection channels with the same urine) resulted in the emergence of filamentous bacteria, and three resulted in emergence of rod-shaped bacteria. An example of an IBC that released filaments may be seen in Fig. 4 and Movie S3; infected BECs became overtaken by proliferation of an IBC, followed by the expulsion of filamentous bacteria via an extrusion-like process. The filamentous bacteria began to emerge 6 to 8 h after urine exposure in this cell, appearing initially as several filaments that grew out from the surface of the BEC, followed by a massive eruption of filamentous bacteria at ∼15 h after urine exposure (Fig. 4). Figure 5 shows another infected BEC with a growing IBC where no filaments were detected. The infected BEC became overwhelmed by the rapidly growing IBC, after a period of moderate development ca. 5 to 7 h after urine exposure (Fig. 5). Some bacteria then emerged at around 8 h after urine exposure—possibly motile bacteria—followed soon after by eruption of the IBC and release of a great number of rod-shaped bacteria (Fig. 5). After 13 h of urine exposure (Fig. 5), the bacteria from the ruptured BEC had occupied the entire field of view (Movie S4).
FIG 4.
Filamentous bacteria emerging from an infected bladder cell. (A and B) Phase-contrast (A) and GFP fluorescence (B) time-lapse microscopy images, showing an IBC developing from within an infected bladder cell during the 20-h urine exposure. The bacteria overwhelmed the bladder cell, causing it to rupture and release filamentous bacteria. Scale bars, 10 μm (40× objective). All GFP exposures = 50 ms. Refer to Movie S3 for the full set of time-lapse images.
FIG 5.
Short bacteria emerging from an infected bladder cell. (A and B) Phase-contrast (A) and GFP fluorescence (B) time-lapse microscopy showing an IBC developing from within an infected bladder cell during the 20-h urine exposure. The bacteria overwhelmed the bladder cell, causing it to rupture and release many short bacteria and no detected filaments. Scale bars, 10 μm, 40× objective. All GFP exposures = 50 ms. Refer to Movie S4 for the full set of time-lapse images.
Neutralized and acidified human urine do not induce UPEC filamentation.
Urine is thought to be required for inducing UPEC filamentation during development and dispersal of IBCs, the extent of which was positively correlated with the SG of the urine samples and was not detected in relatively dilute samples (14). Some samples with a similar SG also showed variation in the degree of filamentation detected, suggesting that other factors in urine might be involved (14). To identify additional conditions or factors triggering UPEC filamentation, we tested a potential role for urinary pH. For these experiments, we used a modified midscale flow chamber system, similar to that described previously (14), which follows the same time course of medium/urine flow described above, and allows collection of dispersed bacteria in the effluent from the flow chamber and quantification of the extent of filamentation by flow cytometry.
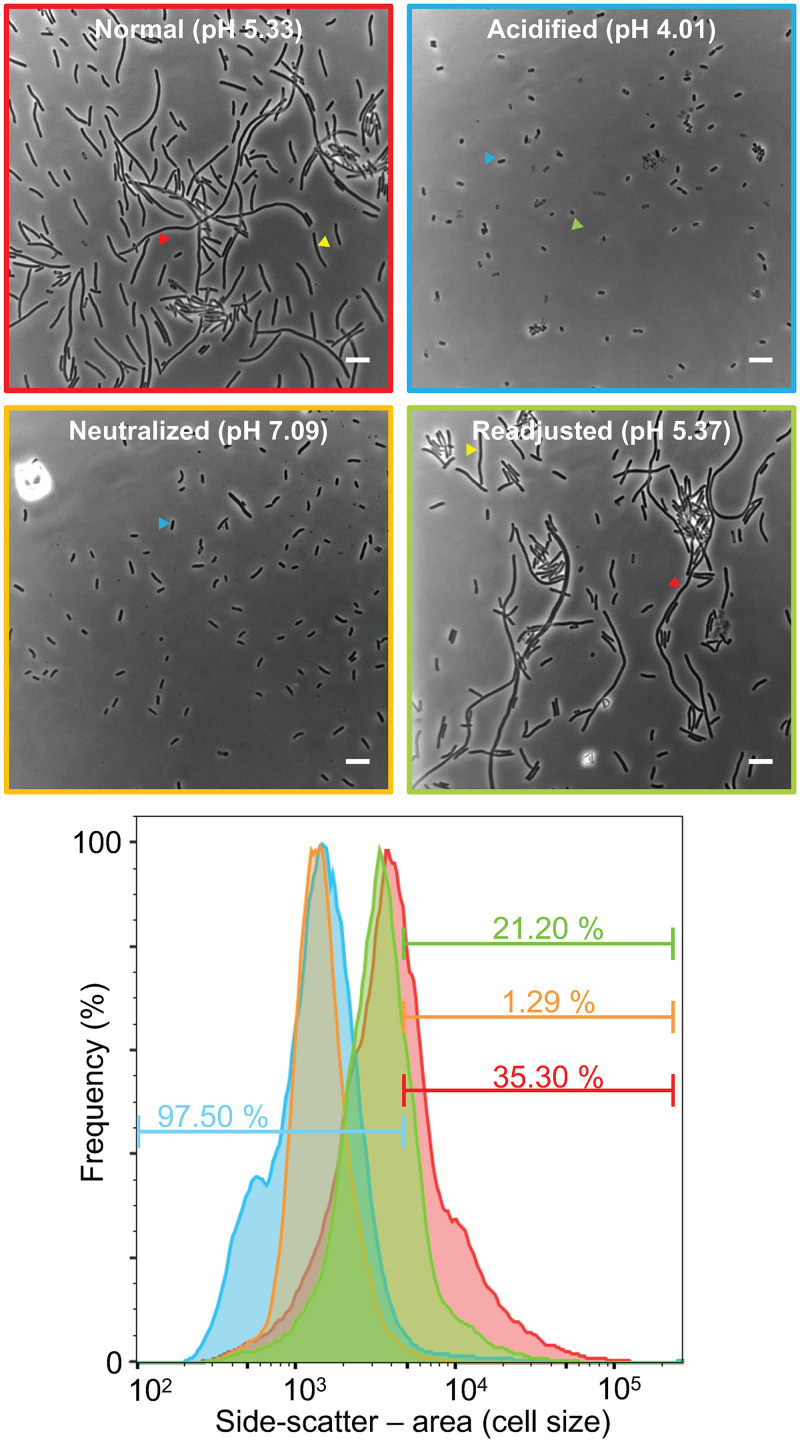
When human urine originally of pH 5.3 was adjusted to pH 4.0, the bacteria in the effluent after the dispersal stage (49 h) of infection appeared as mainly rod-shaped bacteria (Fig. 6). Some of the bacteria appeared smaller than typical E. coli—almost coccoid in shape and size. Flow cytometry confirmed that the bacterial cell size, as represented by the side-scatter (SSC-A) frequency distribution, in the acidified urine sample was significantly smaller than the control urine; the acidified urine gave 97.5% short bacteria (defined by a threshold, as described in Materials and Methods), compared to 64.7% short for the control urine (i.e., 35.3% filamentous; Fig. 6). Many bacteria within IBCs are known to be smaller coccoid-like cells compared to regular rods (9). We also observed that the bacterial yield from the flow chamber was much lower in the acidified urine experiments, e.g., achieving an optical density at 600 nm (OD600) of 0.038, compared to infection with the control urine (pH 5.3) OD600 of 0.571. These results suggested that bacteria released were killed or arrested at the IBC stage by the acidified urine. The apparent poor growth or viability of bacteria from the acidified urine experiment was investigated by inoculating Luria-Bertani (LB) medium with a sample of the effluent obtained from the flow chamber and incubating it for 3 h at 37°C. This sample showed no increase in absorbance during this time period, whereas bacteria obtained from a control infection at the same time point (urine pH 5.3) more than doubled in OD600 during this time, from 0.571 to 1.189. This trend was observed in both experimental duplicates.
FIG 6.
Urine pH controls UPEC filamentation. Phase-contrast microscopy and flow cytometry of UTI89/pGI5, harvested from the flow chamber infection model after day 2, with exposure to urine under different pH conditions, were performed on various samples: control (normal human urine, red, top left), acidified urine (blue, top right), neutralized urine (amber, bottom left), and pH-readjusted urine (green, bottom right). Scale bars, 5 μm. The lower panel shows flow cytometry frequency distributions, normalized to the sample mode, indicating the fraction of cells below or above a side-scatter area (SSC-A) cutoff value, defined as the SSC-A below which 99% of the cells are from a UTI89/pGI5 LB medium mid-log-phase culture. Sample colors correspond to the image frames above.
When a sample of the pH 5.3 urine was neutralized (pH 7.0) in preparation for an infection experiment, an obvious precipitate formed. This was cleared by centrifugation, and then half of the supernatant was readjusted back to pH 5.3; both urine samples were tested in infections for their effects on the growth and filamentation of dispersed bacteria to determine whether components in the precipitate or the pH change affected filamentation. Both urine samples caused substantial yields of bacteria in the effluent (at 49 h postinfection), but the pH 7.0 urine gave rise to no detected filamentation (Fig. 6). Similar results (high bacterial yields but no detected filamentation) were observed when just EpiLife medium was maintained for the 29- to 49-h period (not shown). Interestingly, the readjusted (pH 5.3) urine regained the capacity to induce robust filamentation of the bacteria to a similar degree as the original control urine (pH 5.3, unadjusted) (Fig. 6). Thus, the pH of human urine is a key factor in the induction of UPEC filamentation.
Synthetic human urine does not induce filamentation.
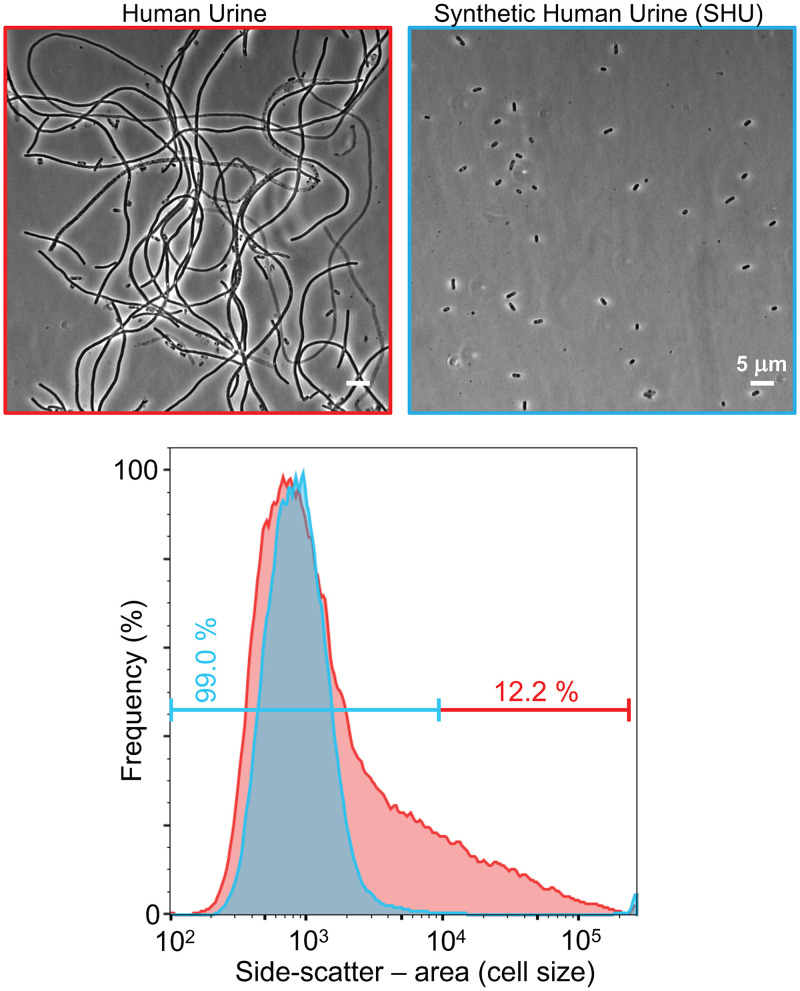
Since a relatively high urinary SG (14) and mildly acidic pH (above) are involved in triggering filamentation, we next investigated whether these factors were sufficient to trigger filamentation with a synthetic human urine (SHU) containing a mix of pure reagents representing the common solutes in human urine (22). The pH of the SHU was matched to that of a batch of human urine (pH 5.6) for comparison. The SGs of the real human urine and the SHU were 1.021 and 1.026 g/ml, respectively. Microscopy of infection effluent at 49 h postinfection showed that the human urine induced bacterial filamentation, as seen previously, but the SHU supported the normal high yields of bacteria in the effluent but did not induce any detectable filamentation (Fig. 6). The flow cytometry data from the human urine contained a peak consistent with normal rod-shaped bacteria and a large shoulder toward the right of the histogram indicating much longer bacteria (Fig. 7). Some variation in the bacterial size distributions obtained from infections with batches of human urine were commonly observed during this study, consistent with previous results (14). In contrast, the bacterial scatter distributions from infections with SHU consistently showed only one peak of small bacteria.
FIG 7.
Synthetic human urine does not induce UPEC filamentation. Phase-contrast microscopy and flow cytometry of UTI89/pGI5, harvested from the flow chamber infection model after day 2, with exposure to human urine (red) or synthetic human urine (SHU) (blue), were performed. Scale bars, 5 μm. Flow cytometry of infection effluent with human urine shows a left-hand peak with a large right-hand shoulder indicating a mixed population of short and filamentous bacteria (12.2% filamentous, according to the cutoff); the bacteria exposed to SHU are represented by a narrow left-hand peak indicating a population of short bacteria of a similar length (99% short).
A small molecular mass fraction of human urine can induce filamentation.
The above findings indicated that a high solute concentration (SG) and weak acid are necessary but insufficient to cause significant filamentation during infection, suggesting that filamentation also requires a urinary constituent(s) not present in SHU. To determine whether such a constituent is a small or macromolecule, urine was ultrafiltered to remove all molecules of ∼3,000 Da and higher. Microscopy of dispersed bacteria from infections (49 h) showed that bacteria exposed to both the whole urine and the ultrafiltered urine were both dispersed with high bacterial yields and were similarly filamentous, with only few short bacteria observed (Fig. 8). The fraction of the urine that had not passed through the filter, containing the large molecules of 3,000 Da and higher, was diluted using sterile water back to the starting volume, so the concentration of large molecules would be very similar to the whole urine, but the small molecules would be diluted. This enriched high-molecular-mass urine was also tested in the infection model to determine whether it too could induce a filamentous response in the bacteria. Microscopy of the sample of dispersed (49 h) bacteria showed that they remained short and rod-shaped (data not shown), indicating that this portion of the urine was not able to induce filamentation and supporting the finding that components with a molecular weight of <3,000 were required at a concentration found in high-SG human urine for triggering the filamentous response.
FIG 8.
The urine small-molecule fraction supports robust UPEC filamentation. Phase-contrast microscopy and flow cytometry of UTI89/pGI5, harvested from the flow chamber infection model after day 2, with exposure to whole human urine (red) or small molecule human urine (<3000-Da fraction, blue), were performed. Scale bars, 5 μm. Flow cytometry showed very similar curves for both urine types, a left-hand peak with a large right-hand shoulder indicating a mixed population of short and filamentous bacteria; 12.2% of bacteria for whole urine and 12.7% of small molecule urine were filamentous.
DISCUSSION
We have established a microfluidics-based infection model of UTI that enables real-time microscopic analysis of progression of UPEC development within infected bladder epithelial cells through to the IBC dispersal phase and release of rod-shaped and filamentous bacteria, thus reproducing the main morphological characteristics of UPEC IBC and filament development during infection. The mouse model of UTI showed similar IBCs and filamentous bacteria present (9), and urine samples collected during human UTI also commonly contain these characteristic features of infection (8). Our model is based on the midscale flow-chamber model developed previously (14), so the results obtained will be comparable, whereas the information obtainable from each is different. We have also incorporated continuous monitoring of cell viability and developed improved GFP labeling of UPEC that shows robust fluorescence in a range of growth conditions, including the main stages of the infection.
Our method creates a more automated and smaller-scale infection system suited for real-time microscopic visualization than previously available. Time-lapse microscopy was used to observe the maturation of IBCs in mouse bladder explants (9); however, to our knowledge the present study is the first to demonstrate a cell culture infection model that allows automated simultaneous visualization of multiple UPEC infection experiments in real time. Using the microfluidic model, we have further characterized the infection cycle, particularly in regard to the conditions required for filamentous IBC dispersal at a single-cell level, and we revealed some advantages and current limitations of cell culture infection models.
During IBC development within BECs over the first day of infection, real-time monitoring of cell impermeability by SYTOX Orange exclusion demonstrated that BECs maintain their integrity and surface migration while the bacteria grow internally, reflecting a likely bacterial survival mechanism in which IBCs do not cause substantial changes or damage to their host until the bacterial population has substantially increased in number. However, when exposed to urine for day 2 of the infection, both infected and uninfected BECs became almost immediately immobile (Fig. 2 and 3). Furthermore, within hours of urine exposure, individual BECs at different times showed a sudden increase in autofluorescence (see Movie S2), suggesting possible permeabilization or other rapid change in cell metabolism that could signify a stress response (23). If permeabilization occurs, the intracellular bacteria could thereby be directly exposed to the urine that triggers their filamentation and dispersal. If the increased autofluorescence does not reflect permeabilization, the bacteria must sense the BEC response to the urine conditions, leading to a subsequent bacterial dispersal response.
These results indicate that urine may not be able to support the growth or longer-term survival of these BECs in culture. In vivo, the surface of the bladder wall is a transitional epithelium where the uppermost layer of surface cells are eventually shed and replaced via differentiation of the lower layer cells as they come to the surface (21, 22). The uppermost layer is made from fully differentiated “umbrella” cells, with an apical surface that deals with urine exposure in a physically and chemically dynamic environment, and a basolateral surface that provides nutrition and other support for the cells. The current infection model, as with previous in vitro models, initially has the BECs growing in a monolayer of incompletely differentiated immortalized cells, likely resembling intermediate cells in the bladder transitional epithelium (15, 24); these cells are likely to be impacted differently by urine exposure compared to the fully differentiated surface umbrella cells. A previous infection study with the PD07i cell line noted that urine was well tolerated by the cultured BECs (14). By developing a real-time model, our results suggest that while the cell culture models are well suited to investigate bacterial pathophysiology, they might not ideally reflect how fully differentiated BECs behave late in the infection cycle in vivo, particularly in response to urine exposure. Nevertheless, we expect that IBCs experience very similar intracellular conditions in the culture models and in vivo, from the moment of internalization, through to establishment, development, and eventually dispersal into a flow of human urine.
While the microfluidics-based model developed here allows efficient real-time observation, it has limitations for preparative analysis of bacteria in the infection. It was found to be impractical to harvest useful quantities of the dispersed bacteria, particularly long filaments, unlike the midscale flow-chamber model where the yield of bacteria in the continuous effluent is sufficient for most modern molecular and cellular analytical techniques (14). On the other hand, the microfluidic infection model is more suitable for small-scale screening purposes involving direct observation of infected cells, allowing up to four simultaneous infections to be run with ∼100-fold less growth medium. It is more cost-effective and less cumbersome to perform. One possible application for the microfluidic UTI infection model could be to screen the effectiveness of newly developed drugs on different stages of the infection cycle, such as IBC development. This could aid the development of new treatments for UTI, since it has the advantage over non-infection-based growth assays of allowing assay of UPEC growth inside BECs and during the specific morphologies we have observed during dispersal. These are important stages of the infection cycle where potential treatments may be directed in future.
The results shown in Fig. 4 and 5 visualized IBCs growing and taking over bladder cells, causing rupture and dispersal of bacteria. This has allowed an approximate time frame to be placed on the progress of the infection. At ca. 6 to 8 h after the onset of urine exposure, some infected BECs began to rupture and release bacteria. However, this process initiated apparently randomly at later times for other IBCs too, suggesting that a certain stage of IBC development needs to be reached before dispersal of an individual IBC commences. It also underscores the low degree of infection synchrony in the later stages of this complex biological system.
We demonstrated that extensive bacterial filamentation frequently accompanies the dispersal and release from BECs (Fig. 4). Bacteria appear to encounter conditions that trigger filamentation (e.g., urine exposure) while inside the BEC, or its permeable carcass, prior to the development and eruption of the filamentous bacteria. This is consistent with in vivo data from non-real-time approaches that appeared to show filamentous bacteria emerging from within a bladder cell rather that forming after being released (7). Our observation that BECs may become permeabilized before filaments emerge is consistent with the notion that IBCs are directly exposed to urine through BEC permeabilization when they respond to these conditions by extensive bacterial filamentation and dispersal (Fig. 4). However, since the dye exclusion method cannot conclusively report on permeability during urine treatment (Movie S2), it is not yet certain how urine exposure may trigger the IBC response.
The data presented in Fig. 5 showed that some IBCs disperse from the residual BEC as a rapidly developing colony of rod-shaped bacteria, indicating that not all IBCs result in filamentous bacteria and suggesting that not all IBCs experience identical conditions, or do not respond to conditions in a consistent manner. The two distinct IBC developmental outcomes indicate that the developmental fate of bacteria during the dispersal stage of IBC development is decided at the level of whole IBC, not individual bacteria.
To examine the conditions that influence the switching between IBC developmental outcomes, a synthetic human urine (SHU) sample was tested and compared to donated human urine to determine the importance of pH, relative solute concentration (specific gravity), and urinary components (small and large molecules) in causing bacterial filamentation. A recently developed formulation of SHU was used (22). If this gave a similar response to human urine, we anticipated that the SHU could help improve the reproducibility of bacterial responses in the model, since different batches of human urine can introduce variability in the degree of filamentation observed (14). Our comparisons of SHU and human urine demonstrated that the filamentation response during IBC dispersal is strongly dependent on the slightly acidic pH values of the human urine. Use of neutralized or acidified human urine, or SHU of the same pH and SG as the human urine (approximately pH 5.3, SG >1.02), all failed to trigger UPEC filamentation in infection (Fig. 6). The previous flow chamber study noted no obvious correlation between pH and filamentation (14); however, the samples were unadjusted and are expected to show only a small pH range compared to the larger range of adjusted urine (pH 4 to 7) in the present study.
Our results suggested that either the factors in human urine that trigger filamentation are pH and concentration sensitive with regard to their interactions with UPEC or, alternatively, separate bacterial receptors sense pH and other urine components, and these all need to be activated in order for a filamentation response to be triggered. Another study suggested that UPEC does not filament in response to a simple artificial urine (25, 26), whereas the SHU used for the current research was a more recently developed artificial urine (22), indicating that neither synthetic urine contains the active components of human urine in triggering UPEC filamentation. Overall, our results strongly support the view that filamentation is not a response to the general solute concentration or pH, but it is a specific response triggered by a certain level of a component(s) in human urine and is also sensitive to pH. Our investigation of human urine separated into large- and small-molecular-weight fractions by ultrafiltration clearly demonstrated that the responsible factors are present in the <3,000-Da fraction. The extensive human urine metabolome includes thousands of urine metabolites, a large majority of which have a molecular weight of <3,000 Da (27). Identifying urinary factors that trigger filamentation or other developmental steps in the UPEC infection cycle would therefore benefit from metabolomic analyses or further “top-down” fractionation of urine to identify the responsible factors. Future studies on the influence and modulation of urinary pH on bacterial morphology and infection progression may be possible using the murine model of UTI, or larger human urine donor cohorts applying the in vitro model of infection described here, and potential clinical trials.
MATERIALS AND METHODS
Bacterial strains and plasmid construction.
E. coli UTI89 (28) was kindly provided by J. Møller-Jensen (University of Southern Denmark). Table 1 lists the oligonucleotides used in this study for plasmid construction. To construct plasmid pGI5, the BamHI+EcoRI fragment from pDG57 (29) containing the pSC101 origin of replication and aadA (for spectinomycin resistance) was ligated to a PCR product (with BamHI+EcoRI ends) containing the PlacIQ promoter from pNDM220 (30) to generate pGI1. The msfGFP region of pDG57 was then amplified in two parts and spliced by overlap extension PCR (to silently remove the internal NcoI site), followed by digestion with BglII and NcoI. This fragment was then ligated to pGI1, digested with BamHI and NcoI, to create pGI2. The PlacIQ promoter in pGI2 was then replaced by three minimal promoters with widely varying levels of expression—PlacI, PlacIQ, and PlacIQ1 (18)—by annealing primers containing the promoter sequences and ligating these (EcoRI+NcoI digested) to the large EcoRI+NcoI fragment of pGI2, to create the plasmids pGI3 (PlacI-msfGFP), pGI4 (PlacIQ-msfGFP), and pGI5 (PlacIQ1-msfGFP).
TABLE 1.
Oligonucleotides used in this studya
| Name | Sequence (5′–3′) | Description |
|---|---|---|
| PlacIq_F | GGGCCCGAATTCGCGGATTTGAACGTTGCG | Amplify the PlacIQ promoter from the plasmid pNDM220 |
| PlacIq_R | GGGCCCGGATCCATTAATTCCCATGGTCACCACCCTGAATTGACTC | |
| msfGFP_F | GGGCCCCCATGGGTAAAGGTGAAGAACTGTTCACC | Amplify first half of msfGFP from pDG57 |
| msfGFP_noNco_R | CCAGAGTCGGCCACGGAACCGGCAGTTTAC | |
| msfGFP_R | GGGCCCAGATCTTTAGGATCCTTTGTAGAGTTCATCCATGCC | Amplify second half of msfGFP from pDG57 |
| msfGFP_noNco_F | GTAAACTGCCGGTTCCGTGGCCGACTCTGG | |
| PlacI_F | AATTCGCGCAAAACCTTTCGCGGTATGGCATGATAGCGCCCGGTCTAGAGGAGGTACTAC | Annealed to create the promoter PlacI |
| PlacI_R | CATGGTAGTACCTCCTCTAGACCGGGCGCTATCATGCCATACCGCGAAAGGTTTTGCGCG | |
| PlacIQ_F | AATTCGTGCAAAACCTTTCGCGGTATGGCATGATAGCGCCCGGTCTAGAGGAGGTACTAC | Annealed to create the promoter PlacIQ |
| PlacIQ_R | CATGGTAGTACCTCCTCTAGACCGGGCGCTATCATGCCATACCGCGAAAGGTTTTGCACG | |
| PlacIQ1_F | AATTCTTGACACCACCTTTCGCGGTATGGCATGATAGCGCCCGGTCTAGAGGAGGTACTAC | Annealed to create the promoter PlacIQ1 |
| PlacIQ1_R | CATGGTAGTACCTCCTCTAGACCGGGCGCTATCATGCCATACCGCGAAAGGTGGTGTCAAG |
Relevant restriction sites are underlined. Complementary regions are indicated in boldface.
Bladder cell culture.
The human bladder epithelial cell line used in this project, PD07i (kindly provided by J. Møller-Jensen, University of Southern Denmark), was originally obtained from a human bladder and immortalized with human papillomavirus type 16 E6E7 (31). The cells were cultured in EpiLife medium with 60 mM calcium medium (Gibco), supplemented with 1% human keratinocyte growth supplement (HKGS; Gibco) and routinely included 1% penicillin-streptomycin (Gibco). The cells were seeded into a T-75 culture flask and left to incubate at 37°C with 5% CO2 until approximately 80% confluent, with changes of the medium every 2 days. The medium was removed from the flask, and the cells were washed once with phosphate-buffered saline (PBS). A solution of 0.5% trypsin-EDTA (Gibco) was then added to cover the base of the flask, followed by incubation incubated at 37°C with 5% CO2 for 10 min. The plate was agitated to aid release the cells from the surface; a 1× volume of defined trypsin inhibitor (Gibco) was then added, and the cell suspension was transferred to a centrifuge tube. The tube was centrifuged for 4 min at 1,000 × g, the supernatant removed, and the cell pellet was resuspended in PBS. A volume of 100 μl was taken to obtain a cell count per ml using an M4 Multisizer Coulter cytometer (Beckmann-Coulter) equipped with a 100-μm aperture tube. The cells were then centrifuged again as before, the supernatant was removed, and the pellet was then resuspended in EpiLife medium to a concentration of 3 × 106 cells/ml. The cells were dispensed into new T-75 culture flasks to continue the culture or into flow chambers to undergo an infection with bacteria.
Preparation of urine samples.
This study had human research ethics approval from the UTS Human Research Ethics Committee (HREC no. 2014000452). Urine was collected soon after waking in the morning, from a mildly dehydrated male donor, and stored at 4°C for 1 to 3 days. The samples were centrifuged at 4,000 × g for 10 min, and the supernatant was passed through a 0.2-μm membrane filter (Sartorius Minisart). The SG values for urine samples were determined by repeat measurements of the weight ratio compared to pure water, determined with a micropipette (1-ml volumes) and an analytical balance. Urine with an SG of 1.020 g/ml or higher was used for infections to promote reproducible filamentation. Filter-sterilized samples were stored frozen or at 4°C until required. To prepare urine samples with altered pH, a sample of urine with an SG of 1.025 g/ml and a pH of 5.33, which was known to cause filamentation, was divided into four samples. One of these was used unadjusted, and a second was acidified to pH 4.0 with HCl. The third sample was neutralized to pH 7.0 with NaOH, upon which a precipitate formed; this was removed by centrifugation. The fourth sample of urine was similarly neutralized, the precipitate was removed, and then the pH readjusted to the original pH of 5.3 with HCl. To obtain the low-molecular-weight urine fraction, filtered urine was fractionated using a Vivaspin 20 ultrafiltration spin column with a 3,000-Da membrane filter (GE Healthcare) until approximately 75% of the original volume had passed through the filter; the filtrate was used in infection studies. SHU was prepared as described previously (22); the pH was adjusted with HCl to match the donor human urine used for comparison (pH 5.0 to 5.5).
Small-scale microfluidics-based infection model.
This microfluidic infection model was established using a CellASIC Onix microfluidics system, with M04S-03 microfluidic plates for real-time visualization (Merck-Millipore). To prepare for an infection experiment, the storage PBS from the upper parts of wells 1 and 8 was removed, as was the PBS from wells 6 and 7, including the bottom holes. A 10-μl volume of EpiLife medium was pipetted into the bottom hole of well 6 to initiate capillary flow, followed by incubation at 37°C for 30 min. The EpiLife medium in the bottom of well 6 was then replaced with a 10-μl suspension of PD07i cells that had been trypsinized from an active culture and resuspended at a concentration of 3 × 106 cells/ml. Well 7 was aspirated, and the cell suspension was left for 10 min to fill the culture chamber. The progress was monitored under a light microscope to check qualitatively for a coverage of the chamber surface by PD07i cells. If more cells were needed, another 10 μl of suspended cells was added to the bottom of well 6, and then well 7 was aspirated again to reinitiate capillary flow. Once enough cells had filled the culture chamber, 350 μl of EpiLife medium was added to well 1, and 50 μl was added to well 7 to initiate gravity-driven perfusion. The plate was incubated at 37°C for 2 days to allow the bladder cells to grow and approach confluence.
Cultures of the required bacterial strains (e.g., UTI89/pGI5) in LB medium were inoculated the day before infection and incubated statically overnight at 37°C. The culture was centrifuged at 4,000 rpm for 10 min and resuspended in PBS to an OD600 of 0.2 in preparation for infection. Wells 6 and 7 of the flow system were aspirated, including the bottom hole. A volume of 10 μl of bacteria was pipetted into the bottom hole of well 6 to initiate capillary flow and draw the bacteria through the culture chamber for 20 min. Well 6 was then aspirated, and the bottom hole was filled with 10 μl of PBS. At this point, the well solutions were immediately added as described below, and we commenced recording the time postinfection.
The PBS solutions from wells 2 to 4 were immediately aspirated; 110 μl of EpiLife medium was added to well 2, 210 μl of EpiLife medium with 100 μg/ml Gm was added to well 3, and well 4 had 210 μl of human urine. Urine samples were obtained from a mildly dehydrated adult male donor (and first micturition of the day), stored for 1 to 3 days at 4°C, and centrifuged at 4,000 × g for 10 min, and then the supernatant was filter sterilized by passage through a 0.2-μm filter unit. Samples with a density of 1.020 g/ml or greater were used for infections. For some experiments observing cell permeability, 100 nM SYTOX Orange DNA stain (Thermo-Fisher) was included in the medium and urine. The upper part of wells 1 and 7 were aspirated. The plate was sealed to the microfluidic manifold and placed on the plate mount stage of a Nikon Ti inverted microscope at 37°C to record time-lapse images of the infections. The flow pressure from the microfluidic control unit was set to 1 lb/in2 for all wells. Well 2 was set to flow for 9 h, and wells 3 and 4 were each set to flow for 20 h in sequence (i.e., until 29 and 49 h postinfection, respectively). Multiple points within the chamber were selected for time-lapse microscopy. Each chamber was considered one independent replicate.
Midscale flow chamber infection model.
A modified version of the flow chamber infection model (14) was used to collect and analyze dispersed UPEC. Briefly, a sterile-ported medium bottle was connected with tubing via a bubble trap to the flow chamber (a channel slide) through a low-pulsation peristaltic pump (Ismatec) and into a waste container. To establish a PD07i monolayer, 100 μl of PD07i cells suspended in EpiLife medium (3 × 106 cells/ml) was added to the flow chamber (IBIDI μ-slide I [catalog no. 80176] instead of the custom apparatus used previously [14]), followed by incubation overnight at 37°C with 5% CO2. The flow system was then connected to the flow chamber, and a flow of EpiLife medium without antibiotics was applied overnight (50 μl/min); then, to prepare for infection, bacterial cultures (E. coli UTI89/pGI5) were inoculated, followed by incubation statically at 37°C overnight. The bacterial culture was centrifuged and gently resuspended in PBS to an OD600 of 0.2. The flow cell system was stopped, and the tubing before the flow chamber was disconnected. A length of sterile tubing was filled with the bacterial suspension, and one end was connected to the upstream port of the flow chamber, ensuring no bubbles could enter the system. The other end of the tubing was submerged in the bacterial suspension, avoiding bubbles, and the flow was resumed for 20 min. The pump was then stopped, the short length of tubing was disconnected, the medium line of tubing was reconnected, and the flow was then resumed for 9 h to allow the establishment of infection. The pump was then stopped, and all the tubing before the flow chamber was filled with EpiLife medium containing gentamicin (100 μg/ml). The flow was recommenced for 20 h to kill and remove extracellular bacteria and allow IBC growth and development. To harvest the bacteria at this stage, the flow chamber was disconnected and washed three times by applying 200 μl of PBS to the flow chamber in-port. BEC lysis buffer (0.5% trypsin-EDTA plus 0.1% [vol/vol] Triton X-100) was then added (200 μl) to the chamber, followed by incubation for 15 min at 37°C. The liquid was withdrawn with a pipette, and then another 200 μl of lysis buffer was applied. The two samples were pooled and centrifuged at 5,000 × g for 5 min, and then bacteria were fixed by resuspension in 2% (wt/vol) formaldehyde in PBS and stored at 4°C. To continue to the dispersal stage after the 20-h flow of EpiLife+Gm, the tubing before the flow chamber was disconnected, filled with prepared urine as described above, was then connected to the urine bottle and flow chamber, and the flow continued for another 20 h. To harvest the bacteria, the flow chamber was disconnected at both ends, and the liquid supernatant inside the chamber withdrawn and retained. PBS (200 μl) was then added to the chamber, pipetted back and forth, and then withdrawn and pooled with the supernatant. This sample was centrifuged at 5,000 × g for 5 min, and the pellet was gently resuspended in 2% formaldehyde and stored at 4°C.
Microscopy.
Samples of fixed bacteria (2 μl) were mounted on an ∼170-μm-thick 1% agarose pad in PBS prepared on a slide. The bacteria were viewed using a Nikon Ti epifluorescence microscope using a Plan Apo λ 1.4 NA phase-contrast oil objective. For fluorescence microscopy, Nikon filter sets for GFP or TRITC/Texas Red (for SYTOX Orange detection) were used. Images were recorded using a Nikon DS-Qi2 camera and visualized and analyzed using Fiji (32).
Flow cytometry.
Bacterial samples were analyzed with a BD LSRII flow cytometer to record forward scatter (FSC), side scatter (SSC, trigger), and fluorescence (GFP) signals for 100,000 events per sample, with the side scatter also set as the threshold. To further analyze and display the flow cytometry data, FlowJo (v10) was used. A histogram was generated, in which the frequency data were normalized to the mode of each sample, and then data were gated to distinguish between the short and filamentous bacteria by defining a “short population” based on a control (UTI89/pGI5 grown to mid-log phase in LB medium), which defined a gate (arbitrary cutoff) to contain 99% of events; this was then applied to each sample equally, to define the fraction of cells showing smaller (“short cells”) or larger (“‘filamentous”) values of the side-scatter (area) parameter (SSC-A) than the cutoff. The displayed frequency distributions are consistent and representative of at least two independent replicate experiments.
Supplementary Material
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
We thank Michael Johnson and the UTS Microbial Imaging Facility for technical support. I.G.D. acknowledges support from an Australian Research Council Future Fellowship (FT160100010).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Phan MD, Peters KM, Sarkar S, Lukowski SW, Allsopp LP, Gomes Moriel D, Achard ME, Totsika M, Marshall VM, Upton M, Beatson SA, Schembri MA. 2013. The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet 9:e1003834. doi: 10.1371/journal.pgen.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connell I, Agace W, Klemm P, Schembri M, Mărild S, Svanborg C. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci U S A 93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mulvey MA. 2002. Adhesion and entry of uropathogenic Escherichia coli. Cell Microbiol 4:257–271. doi: 10.1046/j.1462-5822.2002.00193.x. [DOI] [PubMed] [Google Scholar]
- 4.Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 5.Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. 2000. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J 19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 7.Mulvey MA, Schilling JD, Hultgren SJ. 2001. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun 69:4572–4579. doi: 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosen DA, Hooton TM, Stamm WE, Humphrey PA, Hultgren SJ. 2007. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med 4:e329. doi: 10.1371/journal.pmed.0040329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Justice SS, Hung C, Theriot JA, Fletcher DA, Anderson GG, Footer MJ, Hultgren SJ. 2004. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci U S A 101:1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Justice SS, Hunstad DA, Cegelski L, Hultgren SJ. 2008. Morphological plasticity as a bacterial survival strategy. Nat Rev Microbiol 6:162–168. doi: 10.1038/nrmicro1820. [DOI] [PubMed] [Google Scholar]
- 11.Justice SS, Hunstad DA, Seed PC, Hultgren SJ. 2006. Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc Natl Acad Sci U S A 103:19884–19889. doi: 10.1073/pnas.0606329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvath DJ Jr, Li B, Casper T, Partida-Sanchez S, Hunstad DA, Hultgren SJ, Justice SS. 2011. Morphological plasticity promotes resistance to phagocyte killing of uropathogenic Escherichia coli. Microbes Infect 13:426–437. doi: 10.1016/j.micinf.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moller J, Emge P, Vizcarra IA, Kollmannsberger P, Vogel V. 2013. Bacterial filamentation accelerates colonization of adhesive spots embedded in biopassive surfaces. New J Phys 15:125016. doi: 10.1088/1367-2630/15/12/125016. [DOI] [Google Scholar]
- 14.Andersen TE, Khandige S, Madelung M, Brewer J, Kolmos HJ, Moller-Jensen J. 2012. Escherichia coli uropathogenesis in vitro: invasion, cellular escape, and secondary infection analyzed in a human bladder cell infection model. Infect Immun 80:1858–1867. doi: 10.1128/IAI.06075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berry RE, Klumpp DJ, Schaeffer AJ. 2009. Urothelial cultures support intracellular bacterial community formation by uropathogenic Escherichia coli. Infect Immun 77:2762–2772. doi: 10.1128/IAI.00323-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urban JH, Vogel J. 2007. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res 35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glascock CB, Weickert MJ. 1998. Using chromosomal lacIQ1 to control expression of genes on high-copy-number plasmids in Escherichia coli. Gene 223:221–231. doi: 10.1016/s0378-1119(98)00240-6. [DOI] [PubMed] [Google Scholar]
- 19.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 20.Ito K, Minamiura N, Yamamoto T. 1984. Human urine DNase I: immunological identity with human pancreatic DNase I, and enzymic and proteochemical properties of the enzyme. J Biochem 95:1399–1406. doi: 10.1093/oxfordjournals.jbchem.a134747. [DOI] [PubMed] [Google Scholar]
- 21.Yasuda T, Nadano D, Awazu S, Kishi K. 1992. Human urine deoxyribonuclease II (DNase II) isoenzymes: a novel immunoaffinity purification, biochemical multiplicity, genetic heterogeneity and broad distribution among tissues and body fluids. Biochim Biophys Acta 1119:185–193. doi: 10.1016/0167-4838(92)90390-y. [DOI] [PubMed] [Google Scholar]
- 22.Ipe DS, Ulett GC. 2016. Evaluation of the in vitro growth of urinary tract infection-causing gram-negative and gram-positive bacteria in a proposed synthetic human urine (SHU) medium. J Microbiol Methods 127:164–171. doi: 10.1016/j.mimet.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Surre J, Saint-Ruf C, Collin V, Orenga S, Ramjeet M, Matic I. 2018. Strong increase in the autofluorescence of cells signals struggle for survival. Sci Rep 8:12088. doi: 10.1038/s41598-018-30623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apodaca G. 2004. The uroepithelium: not just a passive barrier. Traffic 5:117–128. doi: 10.1046/j.1600-0854.2003.00156.x. [DOI] [PubMed] [Google Scholar]
- 25.Brooks T, Keevil CW. 1997. A simple artificial urine for the growth of urinary pathogens. Lett Appl Microbiol 24:203–206. doi: 10.1046/j.1472-765x.1997.00378.x. [DOI] [PubMed] [Google Scholar]
- 26.Klein K, Palarasah Y, Kolmos HJ, Moller-Jensen J, Andersen TE. 2015. Quantification of filamentation by uropathogenic Escherichia coli during experimental bladder cell infection by using semi-automated image analysis. J Microbiol Methods 109:110–116. doi: 10.1016/j.mimet.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 27.Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, Dame ZT, Poelzer J, Huynh J, Yallou FS, Psychogios N, Dong E, Bogumil R, Roehring C, Wishart DS. 2013. The human urine metabolome. PLoS One 8:e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P, Armstrong JR, Fulton RS, Latreille JP, Spieth J, Hooton TM, Mardis ER, Hultgren SJ, Gordon JI. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A 103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosal D, Trambaiolo D, Amos LA, Lowe J. 2014. MinCD cell division proteins form alternating copolymeric cytomotive filaments. Nat Commun 5:5341. doi: 10.1038/ncomms6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gotfredsen M, Gerdes K. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol 29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- 31.Klumpp DJ, Weiser AC, Sengupta S, Forrestal SG, Batler RA, Schaeffer AJ. 2001. Uropathogenic Escherichia coli potentiates type 1 pilus-induced apoptosis by suppressing NF-κB. Infect Immun 69:6689–6695. doi: 10.1128/IAI.69.11.6689-6695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.