Gut microbial communities affect their animal hosts in numerous ways, motivating investigations of the factors that shape the gut microbiota and the consequences of gut microbiota variation for host traits. In this study, we tested the effects of increases in environmental temperatures on the gut microbiota of fence lizards, a vertebrate ectotherm threatened by warming climates. By monitoring lizards and their gut microbes during an experimental temperature treatment, we showed that the warming altered and destabilized the lizard gut microbiota. Moreover, measuring thermal performance of lizard hosts at the end of the experiment indicated that the composition of the gut microbiota was associated with host thermal tolerance. These results indicate that warming temperatures can alter the gut microbiota of vertebrate ectotherms and suggest relationships between variation in the gut microbiota and the thermal physiology of natural host populations.
KEYWORDS: amplicon sequence variant, global warming, metagenome, plasticity, thermal physiology
ABSTRACT
Vertebrates harbor trillions of microorganisms in the gut, collectively termed the gut microbiota, which affect a wide range of host functions. Recent experiments in lab-reared vertebrates have shown that changes in environmental temperature can induce shifts in the gut microbiota, and in some cases these shifts have been shown to affect host thermal physiology. However, there is a lack of information about the effects of temperature on the gut microbiota of wild-caught vertebrates. Moreover, in ectotherms, which are particularly vulnerable to changing temperature regimens, the extent to which microbiota composition is shaped by temperature and associated with host thermal tolerance has not been investigated. To address these issues, we monitored the gut microbiota composition of wild-caught western fence lizards (Sceloporus occidentalis) experimentally exposed to a cool-to-warm temperature transition. Comparing experimentally exposed and control lizards indicated that warm temperatures altered and destabilized the composition of the S. occidentalis gut microbiota. Warming drove a significant reduction in the relative abundances of a clade of Firmicutes, a significant increase in the rate of compositional turnover in the gut microbiota within individual lizards, and increases in the abundances of bacteria from predicted pathogenic clades. In addition, the composition of the microbiota was significantly associated with the thermal tolerance of lizards measured at the end of the experiment. These results suggest that temperature can alter the lizard gut microbiota, with potential implications for the physiological performance and fitness of natural populations.
IMPORTANCE Gut microbial communities affect their animal hosts in numerous ways, motivating investigations of the factors that shape the gut microbiota and the consequences of gut microbiota variation for host traits. In this study, we tested the effects of increases in environmental temperatures on the gut microbiota of fence lizards, a vertebrate ectotherm threatened by warming climates. By monitoring lizards and their gut microbes during an experimental temperature treatment, we showed that the warming altered and destabilized the lizard gut microbiota. Moreover, measuring thermal performance of lizard hosts at the end of the experiment indicated that the composition of the gut microbiota was associated with host thermal tolerance. These results indicate that warming temperatures can alter the gut microbiota of vertebrate ectotherms and suggest relationships between variation in the gut microbiota and the thermal physiology of natural host populations.
INTRODUCTION
The gastrointestinal tracts of animals are colonized by diverse assemblages of microorganisms that can have profound effects on host phenotypes (1), demanding investigation of the causes and consequences of variation in the animal gut microbiota. There is mounting evidence that environmental temperature shapes the composition of gut microbial communities in many animal taxa, including invertebrates and vertebrates (2–6). Moreover, recent studies have provided evidence that temperature-induced changes in the microbiota can have cascading consequences for host phenotypes relevant to thermal tolerance (7–10). For example, transplantation of the gut microbiota from mice acclimated to cool conditions into germfree mice caused remodeling of gut morphology in a manner consistent with greater cold tolerance in recipient hosts (10).
Despite the progress that has been made, understanding of how temperature affects the animal gut microbiota remains limited. This is particularly true for reptiles, despite the fact that rising global temperatures are expected to have extreme negative effects on biodiversity in this vertebrate clade (11). Prior studies of lizards have shown that temperature increases can affect the composition of the gut microbiota (12); however, these studies relied on hosts descended from captive-bred populations, and hosts were sampled at only one or two time points. The responses of the gut microbiotas of wild-caught reptiles to temperature variation remain unclear, and the temporal dynamics of gut microbiota responses to temperature in reptiles have not been investigated. Moreover, the contribution of gut microbiota variation to host thermal tolerance in reptiles is unknown.
To address these issues, we conducted an experiment to test the effects of environmental temperature on the gut microbiotas of wild-caught western fence lizards (Sceloporus occidentalis). We acclimated animals to two different temperatures and measured the progression of changes in gut microbiota. Combining gut microbiota data with physiological measurements of host heat tolerances allowed us to test for effects of temperature on gut microbiota composition in S. occidentalis and for associations between aspects of gut community composition and individual resistance to heat. Results suggest that changing thermal conditions can significantly impact lizard gut microbiota and are consistent with a role of the gut microbiota in shaping lizard thermal physiology.
RESULTS
We tracked the gut microbiota composition of 25 wild-caught lizards in two experimental groups: a control group that experienced 25°C for 16 days and a treatment group that experienced 25°C for 7 days and 35°C for 10 days (see Table S1 in the supplemental material). To this end, we generated 5,841,604 16S rRNA gene reads across 89 fecal samples collected throughout the experiment, yielding 2,069,546 high-quality reads after quality filtering, trimming, and deblur filtering (interquartile range of reads per sample, 15,242 to 28,790). The final data set included 9,037 amplicon sequence variants (ASVs) in total. To include all samples in downstream analyses, rarefaction was performed, yielding an even depth of 7,500 reads per sample. Rarefaction curves supported the idea that this sequencing depth was sufficient to enumerate the majority of ASVs present at appreciable abundances in the fecal samples (Fig. S1). Taxonomy assignments of the ASVs are presented in Table S2. A diagram displaying the experimental design, sampling strategy, and relative abundances of bacterial phyla within each sample is presented in Fig. S2.
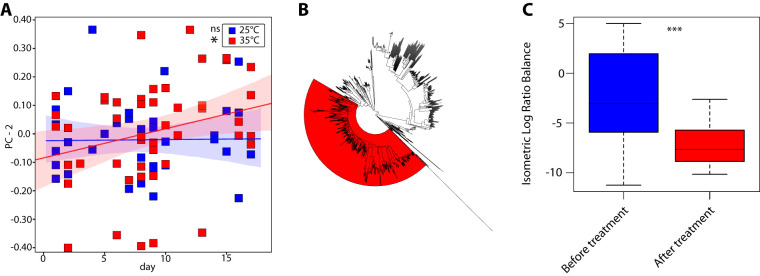
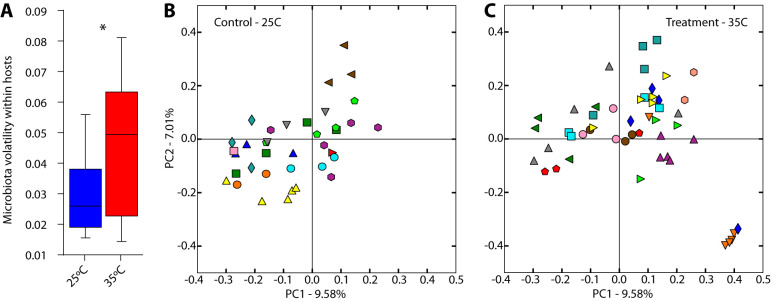
Visualizing the dissimilarities among all samples through principal-coordinate analyses indicated that lizard hosts in the treatment group displayed a compositional shift in their gut microbiotas following movement from 25°C to 35°C, a shift not observed in control lizards kept at 25°C (Fig. 1A and B). To test the significance of the compositional shift, we compared treated and control lizards over time on the first four principal coordinates. These analyses show that the gut microbiotas of treatment lizards displayed a positive association between the second principal component (PC2) and time (P value for nonzero slope = 0.029, df = 23, R2 = 0.0759), whereas the gut microbiotas of control lizards displayed no temporal relationship with PC2 (P value for nonzero slope = 0.69, df = 23, R2 = 0.0002) (Fig. 2A). Linear mixed-effects modeling and likelihood ratio tests further supported this finding, indicating that PC2 values for each sample were better explained by a model containing “treatment group,” “host individual,” “day,” and “sex” than a model containing only “host individual,” “day,” and “sex” (χ2 = 8.1582, P = 0.01692) (see the supplemental material). In contrast, these two nested models did not significantly differ in their abilities to explain PC1, PC3, or PC4 values (see the supplemental material).
FIG 1.
Exposure to 35°C induces compositional shifts in the gut microbiota not seen at 25°C. Plots display the first two principal coordinates of Bray-Curtis dissimilarities among samples collected from control lizards (A) and treated lizards (B). Each point represents the gut microbiota of a lizard measured in a single fecal sample; colors indicate collection before (blue) or after (red) treatment. Blue and red polygons denote regions containing samples collected before or after the treatment day, respectively. Note that the samples from heat-treated lizards occupy a region of compositional space not occupied by lizards before heat treatment (B) and that this effect is not seen in the control group (A).
FIG 2.
A clade within the Firmicutes displayed reduced relative abundance after warming. (A) Regression of PC2 of Bray-Curtis dissimilarities against time for treated (red) and control (blue) lizards. Shaded regions indicate 95% confidence bands. ns, P > 0.05; *, P < 0.05. (B) Phylogeny of all bacterial ASVs detected across all lizard samples analyzed. The region highlighted in red corresponds to a clade within the Firmicutes identified by Phylofactor analysis that displayed significantly reduced mean relative abundance within individual lizards following warming relative to the same lizards sampled before warming. Labeled phylogeny in Newick format is presented in Data File S2. (C) Isometric log-ratio balance of the clade shown in panel B within treated individuals sampled before warming (left) and after warming (right). ***, P < 0.001.
An effect of treatment at 35°C on gut microbiota composition was further supported by analysis of variance using distance matrices (i.e., adonis2), which indicated a significant effect of an interaction between “treatment” and “day of experiment” on beta diversity among microbiota profiles recovered from fecal samples (F = 1.477, df = 1, P = 0.028; see the supplemental material). Variance partitioning analyses further indicated that “treatment” explained the second-highest proportion of variation in beta diversity of all metadata variables, with “host individual” explaining the most variation (see the supplemental material). Phylofactor analyses of isometric log-ratio balances comparing the gut microbiotas of treatment lizards before and after treatment at 35°C showed that the shift in gut microbiota composition was underlain by a reduction in the relative abundances of a clade of Firmicutes (Fig. 2B and C) (F = 8.568, P = 0.00083). The phylogeny underlying Phylofactor analyses is presented in Data File S2 in the supplemental material.
The microbiotas of treated lizards after transfer to 35°C did not display significantly different alpha diversity, in terms of observed ASVs, relative to the microbiotas of control lizards after the treatment began (t test; T = 0.74, df = 23, P = 0.43). In addition, mixed-effects modeling controlling for sex and individual detected no differences in observed ASVs between control and treatment groups (see the supplemental material). Corresponding analyses of Shannon diversity also revealed no differences between control and treatment groups (see the supplemental material). Similarly, ANCOM (analysis of composition of microbiomes) revealed no individual ASVs with significantly different relative abundances between treatment and control lizards after the initiation of exposure to 35°C (Fig. S3).
To test whether microbiotas of treatment and control groups differed in terms of bacterial phenotypes, we assigned functional trait categories to bacterial species in our data set through the BugBase pipeline (13). Analyses of these categories indicated that 35°C drove increases in the relative abundances of bacterial lineages related to obligate and opportunistic pathogens compared to 25°C (Fig. 3). Taxonomic assignments of bacterial lineages classified by BugBase as obligate and opportunistic pathogens are presented in Table S3. Mann-Whitney–Wilcoxon tests indicated a significant difference between groups (P = 0.039). Analyses of other functional axes revealed no significant differences between groups, including aerobicity (P = 0.13), anaerobicity (P = 0.55), oxygen stress tolerance (P = 0.24), formation of biofilms (P = 0.61), Gram-negative status (P = 0.62), and Gram-positive status (P = 0.62).
FIG 3.

Increased abundances of putative pathogens at 35°C. Plots show the relative abundances of putatively pathogenic bacterial species within the gut microbiota of lizards after treatment at 35°C compared to control lizards sampled at the same time period. Putative pathogenic species were inferred with BugBase (13). *, P < 0.05 (Mann-Whitney–Wilcoxon test).
Lizards housed at 35°C displayed an elevated rate of compositional turnover in their gut microbiotas relative to lizards housed at 25°C. Volatility analyses indicated that the compositional divergence between consecutive samples weighted by the number of days separating the samples was significantly higher within treatment individuals after treatment than within control individuals (Fig. 4A) (t test; T = 2.59, df = 7, P = 0.035). This difference in the within-individual variation in the microbiota between treatment and control groups is evident when samples are plotted on the first two principal coordinates of microbiota beta diversity (Fig. 4B and C). In addition, microbiome beta diversity among samples collected from the same individual was significantly higher on average in the treatment group than in the control group (nonparametric permutation test with 1,000 permutations; P = 0.001). Moreover, microbiome beta diversity among samples within the treatment group was significantly higher than in the control group (nonparametric permutation test with 1,000 permutations; P = 0.001) (Fig. S4). Feature volatility analysis revealed that an ASV belonging to the Firmicutes genus Coprobacillus displayed the highest volatility in relative abundance in the treatment lizards, with a mean decrease in relative abundance within individuals of 1.7% per day. A complete list of ASVs identified by feature volatility analyses and the statistics resulting from these analyses is presented in Table S4.
FIG 4.
Elevated compositional turnover in the gut microbiota at 35°C. (A) Mean gut microbiota turnover per day between consecutive samples collected from the same individuals after the initiation of heat exposure on day 9 in control lizards (left) and warmed lizards (right). Microbiota turnover between consecutive samples was calculated based on Bray-Curtis dissimilarities. *, P < 0.05 (t test). Plots display the first two principal coordinates of Bray-Curtis dissimilarities among samples collected from control lizards (B) and treated lizards (C). Each color represents an individual lizard. On average, samples in the control group (B) displayed higher compositional similarity than did samples in the treatment group (C) (Fig. S5).
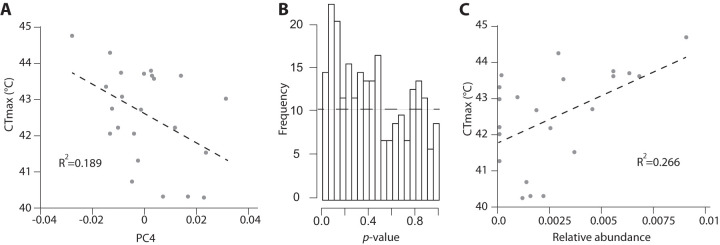
We also tested whether the critical thermal maximum (CTmax) of lizards was associated with the relative abundances of ASVs. CTmax did not differ at the end of the experiment between control and treated lizards (df = 23, T = 0.86, P = 0.40) (Fig. S5; Table S5), but CTmax was significantly associated with aspects of gut microbiota composition. Regressing CTmax against principal coordinates of Bray-Curtis dissimilarities among all samples revealed that PC4 was significantly associated with CTmax (df = 23, T = −2.31, P = 0.029) (Fig. 5A; also, see the supplemental material). The significant association between CTmax and PC4 were further supported by likelihood ratio tests of nested linear models (see the supplemental material). Regression was also employed to test for associations of CTmax with log-transformed relative abundances of individual ASVs. These analyses revealed no individual ASVs significantly associated with CTmax after false-discovery correction (Table S5; Table S6). However, Fisher’s combined P value test across all ASVs revealed that the distribution of P values across ASVs differed from the null expectation (P = 0.043), although this result should be interpreted with caution, given that relative abundances of ASVs are not independent due to compositionality. A plot of the distribution of P values for regressions of individual ASVs against the time to loss of righting response is presented in Fig. 5B, and a list of these P values is presented in Table S3. The ASV that displayed the strongest association with CTmax belonged to the genus Anaerotignum (Fig. 5C). This ASV was also identified as associated with CTmax by selbal analyses (14), which identified balances (i.e., ratios of ASV relative abundances) that explain variation in the response variable of interest. Additional methods and results from selbal results are presented in the supplemental material (Fig. S6 and S7; Table S7).
FIG 5.
Microbiota composition was associated with host thermal tolerance. (A) The regression line (dashed line) shows the relationship between lizard thermal tolerance, measured as loss-of-righting-response temperature (CTmax), and PC4 of Bray-Curtis microbiota dissimilarities among samples. A complete list of regression coefficients between ASVs and CTmax is presented in Table S3. (B) Histogram of P values of regression significance for each ASV with CTmax. The horizontal line indicates null distribution. (C) The regression line (dashed line) shows the relationship between CTmax and an ASV belonging to the genus Anaerotignum.
DISCUSSION
Our experiments indicate that temperature has a significant effect on the composition and dynamics of the gut microbiota of wild-caught lizards. Exposure to 35°C led to an expansion of variation in microbiota composition among lizard hosts (Fig. 1A and B) and a decrease in the relative abundances of a large clade of bacterial lineages within the Firmicutes (Fig. 2B and C). These results mirror previous studies of a diversity of vertebrate taxa that found effects of temperature on the relative abundances of Firmicutes in the gut microbiota (15). For example, previous studies in amphibians observed that several Firmicutes decreased in relative abundances within the gut microbiotas of adult salamanders (2) and frog tadpoles (6) reared at high relative to low temperatures. Similarly, a study of hens found that the gut microbiota displayed reduced relative abundances of Firmicutes during summer months relative to spring months (16). These parallels suggest that some responses of the tetrapod gut microbiota to temperature may be conserved across host taxa. However, we observed no decrease in alpha diversity (i.e., species richness) when lizards were moved to higher temperatures, contrasting recent experiments with the lizard Zootoca vivipara (12).
We also observed an elevated rate of turnover in microbiota composition in treatment lizards after they were moved to 35°C compared to control lizards maintained at 25°C throughout the experiment (Fig. 4A). Wild-caught control lizards at 25°C maintained individual-specific gut microbiotas throughout the duration of the experiment, but treatment lizards in some cases lost their compositional distinctiveness after exposure to 35°C (Fig. 4B and C). These results mirror previous observations that microbiotas associated with corals display increased variability in response to elevated temperatures (17), suggesting the possibility that temperature may have some common effects on the temporal dynamics of host-associated microbial communities. The effects that this destabilization of the microbiota may have on host fitness are unclear. For example, 35°C is in the range of preferred body temperatures (i.e., the body temperature they seek out in a thermally variable environment) of S. occidentalis (18). Preferred body temperatures correlate with the body temperatures at which physiological performance is maximized (19), and indeed, locomotor performance in S. occidentalis is at or near maximum at 35°C (20). Nevertheless, the observations that 35°C destabilizes the gut microbiota (Fig. 4) raises questions about the potential costs of long-term exposure of hosts to these elevated temperatures.
Functional categorization of bacterial species in our data set through BugBase (13) further corroborated the possibility that the compositional shifts in the lizard gut microbiota that occurred at 35°C may have deleterious effects on some hosts. We observed overrepresentation of potentially pathogenic bacterial lineages within the gut microbiota of lizard hosts at 35°C compared to 25°C (Fig. 3). However, the classification of bacterial taxa as pathogens primarily reflects data from humans (13), and the degree to which these taxa display pathogenic qualities in fence lizards remains an area for further investigation.
In addition, the mechanisms underlying our observations that temperature altered the fence lizard gut microbiota remain unclear. In endotherms, the shifts in composition of the gut microbiota in response to changes in ambient temperature that have been observed (e.g., see reference 10) are likely due to changes in host physiology caused by changes in temperature, rather than changes in temperature directly. However, in ectotherms, whose internal temperatures can vary widely, temperature may exert a more direct effect on microbiota composition. Future experiments to study the performance of bacterial lineages under different thermal and host physiological conditions will be required to assess the relative contributions of these mechanisms to the effects of temperature that we observed.
Another important and unresolved question is whether microbiota composition can affect host thermal physiology and tolerance. For example, it remains unclear based on our experiments whether the decreased relative abundances of Firmicutes that we observed could have functional effects on host physiology that might impact thermal performance. The ratio of Firmicutes and Bacteroidetes within the guts of individual mammals has been associated with altered capacities of the microbiota for energy harvest and functional effects on host metabolism (21). In addition, previous experiments in germfree mice have shown that cold-associated microbiota compositions are marked by increased Firmicutes relative abundances and can confer phenotypes associated with cold tolerance in hosts (e.g., white-fat browning) (10). However, the effects of temperature-driven shifts in the gut microbiota on host metabolism and physiology have not been tested by experiments in nonmammalian tetrapods. We identified components of the S. occidentalis gut microbiota that were significantly associated with CTmax at the end of the experiment. Specifically, CTmax was associated with PC4 of microbiota dissimilarities (Fig. 5A), and the distribution of ASV regression P values differed significantly from the null expectation, suggesting a scenario in which multiple ASVs were weakly associated with CTmax (Fig. 5B). We did not observe differences in CTmax between groups (Fig. S2), and the components of the microbiota associated with CTmax were distinct from those associated with treatment temperature. These results highlight the need for microbiota transplant experiments designed to test the physiological consequences of standing variation in the lizard gut microbiota and of the changes in the microbiota that occur in response to temperature.
Cumulatively, our results indicate that temperature significantly affects the gut microbiota of lizards and that the composition of the gut microbiota is associated with host thermal performance. These observations raise the question of whether temperature-induced changes in the lizard gut microbiota contribute to host thermal performance in natural populations, which should prompt experimental manipulations of the lizard gut microbiota designed to test the functional effects of temperature-driven microbiota variation on host phenotypes and fitness. Research into the effects of microbiome composition on wildlife is in its infancy. This study contributes to an emerging view that microbiomes are strongly affected by environmental perturbations such as temperature change (22) and that microbiome changes could have significant consequences for hosts (23, 24). Understanding these host-symbiont relationships is likely to prove critical for our ability to predict and mitigate the effects of global change on wildlife (25).
MATERIALS AND METHODS
Animal husbandry and fecal collection.
Sceloporus occidentalis (n = 25; 4 females and 21 males) were collected with hand-held lassos on the campus of the California Polytechnic State University (Cal Poly), San Luis Obispo, California (35.302077°N, −120.659446°W) in February 2018. In San Luis Obispo in February, mean high air temperatures are 18°C and average low air temperatures are 7°C. However, lizards emerge to bask on sunny days and can increase their body temperatures well above air temperature through absorption of solar radiation and conduction from warm rocks (26), allowing opportunities for capturing individuals. Lizards were brought back to the laboratory, placed in mesh wire cages (7.8 cm wide, 7.3 cm tall, and 19.5 cm long), and acclimated to the laboratory at room temperature for 3 days prior to treatment. During this time, fecal pellets were collected in the morning (0800) and the evening (2000) using forceps that were heat sterilized between collections. Samples were placed in sterile 1.5-ml collection tube, labeled, and placed in a −80°C freezer. During the entirety of this study, lizards were fed 1 or 2 crickets/day to recapitulate their insectivorous diet and had access to water ad libitum.
Lizards were randomly assigned to control (n = 12; 3 females and 9 males) or treatment (n = 13; 1 female and 12 males) groups. Lizards were transferred to two environmental chambers (treatment and control chambers, both set to a constant temperature of 25°C with a 12:12 photoperiod). Lizard cages in each treatment were separated from one another by cardboard so the lizards could not see one another, to avoid potential stress due to interaction. We continued to collect fecal pellets at 0800 and 2000 every day from each lizard, as available.
After 7 days, lizards in the treatment group were switched to a constant temperature of 35°C, and control lizards were kept at a constant 25°C. All fecal samples collected in the morning on the day of transfer were labeled as 25°C and samples collected at night were then labeled according to the treatment they were in. Feeding and collection were continued according to the methods described above.
Upper thermal tolerance assays.
Lizard upper thermal tolerance was recorded as the temperature at which lizards lost their righting response (ability to right themselves when flipped over). This is a measure of the critical thermal maximum (CTmax), the temperature at which an organism loses its locomotor ability and at which continued heating would result in death (27–29). We measured CTmax for each lizard at the end of the experiment with a Cal Poly-engineered device, the gas analysis temperature oxygen regulation system (GATORS). For the GATORS, lizards were fitted with cloacal resistance temperature detectors to measure body temperature, equilibrated to 30°C, and placed in individual cylindrical chambers (18-cm length, 4-cm diameter) encased in larger transparent chambers (25-cm length, 10-cm diameter), with ambient air inside the inner chambers heated at exactly 1°C per minute. As lizards began gaping and panting at high temperatures, the chambers were turned so that lizards were flipped onto their backs and attempted to right themselves; the body temperature at which they could no longer do so was considered their CTmax. Lizards were then rapidly removed from the chambers and cooled to room temperature; no lizards perished during this process.
Microbiome sequencing.
Total genomic DNA from fecal samples (n = 89) was extracted using the MoBio Powerlyzer PowerSoil bead-beating kit. PCR amplification of the V4-V5 region (515F + 926R) of 16S rRNA genes was performed in triplicate as previously described (30). All samples and four negative-control wells were sequenced on a single lane of an Illumina MiSeq sequencer using 2 × 300 bp PE v3 chemistry. Samples were sequenced to a minimum depth of 7,500 reads per sample.
16S data processing and functional inference.
Reads were initially processed in QIIME 2.0 (31) implemented on the Qiita webserver. Reads were filtered for quality through Split libraries FASTQ and trimmed through Trimmed Demultiplexed. Trimmed reads were further processed and grouped into amplicon sequence variants (ASVs) with the deblur pipeline. ASVs detected in negative-control wells were removed from all downstream analyses. Rarefaction curves were constructed, and all samples were rarefied to an even depth of 7,500 reads per sample in order to retain all samples for downstream analyses. Taxonomy was assigned using the q2-fragment-insertion method in QIIME 2 against the Silva 128 SEPP reference database. Relative abundances of bacterial functional groups and phenotypes were inferred through the BugBase web server (13). To enable phenotypic profiling through the BugBase web server, taxonomic identities of ASVs for this analysis were assigned against the Greengenes 97% OTU (operational taxonomic unit) database gg_13_5.
Statistical analyses.
Pairwise beta diversity dissimilarities were calculated from ASV tables. Bray-Curtis (BC) dissimilarities, which consider relative abundances of ASVs, and binary Sorensen-Dice (BSD) dissimilarities, which consider only presence and absence of ASVs, were calculated for every pair of samples. These measures of beta diversity are agnostic to the phylogenetic relationships among ASVs and enable detection of differences in beta diversity underlain by the distributions of closely related ASVs. The ASV table underlying these analyses is presented in Data File S1.
We used adonis2 in the vegan package in R (32) to test for independent significant effects of sex, individual, treatment, time (measured as day number of the experiment), and the interaction between treatment and time on BC dissimilarities using the “margin” option. Principal-coordinate analyses (PCoA) were calculated from BC and BSD dissimilarities and plotted in QIIME 2.0. Statistical support for differences in bacterial functional groups between treatment and control lizards was assessed by Mann-Whitney–Wilcoxon tests through the BugBase web server.
Phylofactor (33) was employed using the variance method to test for clades in the bacterial phylogeny that displayed significantly different relative abundances between treatment and control individuals after the warming treatment. This method allowed us to test for differences in relative isometric log ratio balances between groups at all phylogenetic levels in our data set (33). The phylogenetic tree underlying Phylofactor analyses is presented in Data File S2. In addition, we employed ANCOM (34), which compares compositional log ratios of ASVs between groups of samples, to test for ASVs that displayed significantly different relative abundances between treatment and control individuals after the warming treatment. Phylofactor and ANCOM analyses were based on mean gut microbiota compositions in each individual after the treatment. Mean gut microbiota compositions were calculated for each individual host by averaging relative abundances of each ASV across samples collected from the host.
The script compare_trajectories.py was employed in QIIME 1.9 to test whether host individuals in the treatment group displayed elevated rates of compositional turnover in their gut microbiotas relative to individuals in the control group. This analysis employed the “trajectory” algorithm and was based on BC dissimilarities weighted by the distance in days between consecutive samples. In addition, time series analyses were conducted with q2-longitudinal (35). Feature volatility analyses was performed using default settings to identify specific ASVs that displayed the most rapid fluctuations in relative abundances within the treatment group.
Regression analyses of the time to loss of righting response and microbiome features were conducted in R with host treatment group included as an effect in the model. False discovery rate-corrected P values were calculated for each ASV’s regression against the time to loss of righting response. Fisher’s combined probability test was employed on uncorrected P values using the sumlog command in the metap package in R. Balances (i.e., ratios of ASV abundances) most associated with loss-of-righting-response temperature (lrr) were determined using the selbal R package (14) (see the supplemental material).
Data availability.
All 16S rRNA gene sequences produced for this study are available in the European Nucleotide Archive under accession no. PRJEB39005.
Supplementary Material
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fontaine SS, Novarro AJ, Kohl KD. 2018. Environmental temperature alters the digestive performance and gut microbiota of a terrestrial amphibian. J Exp Biol 221:jeb187559. doi: 10.1242/jeb.187559. [DOI] [PubMed] [Google Scholar]
- 3.Moghadam NN, Thorshauge PM, Kristensen TN, de Jonge N, Bahrndorff S, Kjeldal H, Nielsen JL. 2018. Strong responses of Drosophila melanogaster microbiota to developmental temperature. Fly (Austin) 12:1–12. doi: 10.1080/19336934.2017.1394558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferguson LV, Dhakal P, Lebenzon JE, Heinrichs DE, Bucking C, Sinclair BJ. 2018. Seasonal shifts in the insect gut microbiome are concurrent with changes in cold tolerance and immunity. Funct Ecol 32:2357–2368. doi: 10.1111/1365-2435.13153. [DOI] [Google Scholar]
- 5.Li YF, Yang N, Liang X, Yoshida A, Osatomi K, Power D, Batista FM, Yang JL. 2018. Elevated seawater temperatures decrease microbial diversity in the gut of Mytilus coruscus. Front Physiol 9:839. doi: 10.3389/fphys.2018.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohl KD, Yahn J. 2016. Effects of environmental temperature on the gut microbial communities of tadpoles. Environ Microbiol 18:1561–1565. doi: 10.1111/1462-2920.13255. [DOI] [PubMed] [Google Scholar]
- 7.Montllor CB, Maxmen A, Purcell AH. 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol 27:189–195. doi: 10.1046/j.1365-2311.2002.00393.x. [DOI] [Google Scholar]
- 8.Dunbar HE, Wilson AC, Ferguson NR, Moran NA. 2007. Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol 5:e96. doi: 10.1371/journal.pbio.0050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doremus MR, Smith AH, Kim KL, Holder AJ, Russell JA, Oliver KM. 2018. Breakdown of a defensive symbiosis, but not endogenous defences, at elevated temperatures. Mol Ecol 27:2138–2151. doi: 10.1111/mec.14399. [DOI] [PubMed] [Google Scholar]
- 10.Chevalier C, Stojanović O, Colin DJ, Suarez-Zamorano N, Tarallo V, Veyrat-Durebex C, Rigo D, Fabbiano S, Stevanović A, Hagemann S, Montet X, Seimbille Y, Zamboni N, Hapfelmeier S, Trajkovski M. 2015. Gut microbiota orchestrates energy homeostasis during cold. Cell 163:1360–1374. doi: 10.1016/j.cell.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Urban MC. 2015. Accelerating extinction risk from climate change. Science 348:571–573. doi: 10.1126/science.aaa4984. [DOI] [PubMed] [Google Scholar]
- 12.Bestion E, Jacob S, Zinger L, Di Gesu L, Richard M, White J, Cote J. 2017. Climate warming reduces gut microbiota diversity in a vertebrate ectotherm. Nat Ecol Evol 1:0161. doi: 10.1038/s41559-017-0161. [DOI] [PubMed] [Google Scholar]
- 13.Ward T, Larson J, Meulemans J, Hillmann B, Lynch J, Sidiropoulos D, Spear J, Caporaso G, Blekhman R, Knight R, Fink R. 2017. BugBase predicts organism level microbiome phenotypes. bioRxiv doi: 10.1101/133462. [DOI]
- 14.Rivera-Pinto J, Egozcue JJ, Pawlowsky-Glahn V, Paredes R, Noguera-Julian M, Calle ML. 2018. Balances: a new perspective for microbiome analysis. mSystems 3:e00053-18. doi: 10.1128/mSystems.00053-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sepulveda J, Moeller AH. 2020. The effects of temperature on animal gut microbiomes. Front Microbiol 11:384. doi: 10.3389/fmicb.2020.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu L, Liao R, Wu N, Zhu G, Yang C. 2019. Heat stress mediates changes in fecal microbiome and functional pathways of laying hens. Appl Microbiol Biotechnol 103:461–472. doi: 10.1007/s00253-018-9465-8. [DOI] [PubMed] [Google Scholar]
- 17.McDevitt-Irwin JM, Baum JK, Garren M, Vega Thurber RL. 2017. Responses of coral-associated bacterial communities to local and global stressors. Front Mar Sci 4:262. doi: 10.3389/fmars.2017.00262. [DOI] [Google Scholar]
- 18.McGinnis SM. 1966. Sceloporus occidentalis: preferred body temperature of the western fence lizard. Science 152:1090–1091. doi: 10.1126/science.152.3725.1090. [DOI] [PubMed] [Google Scholar]
- 19.Martin TL, Huey RB. 2008. Why “suboptimal” is optimal: Jensen’s inequality and ectotherm thermal preferences. Am Nat 171:E102–E118. doi: 10.1086/527502. [DOI] [PubMed] [Google Scholar]
- 20.Marsh RL, Bennett AF. 1986. Thermal dependence of sprint performance of the lizard Sceloporus occidentalis. J Exp Biol 126:79–87. [DOI] [PubMed] [Google Scholar]
- 21.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Oppen MJ, Blackall LL. 2019. Coral microbiome dynamics, functions and design in a changing world. Nat Rev Microbiol 17:557–567. doi: 10.1038/s41579-019-0223-4. [DOI] [PubMed] [Google Scholar]
- 23.Zhang B, Leonard SP, Li Y, Moran NA. 2019. Obligate bacterial endosymbionts limit thermal tolerance of insect host species. Proc Natl Acad Sci U S A 116:24712–24718. doi: 10.1073/pnas.1915307116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.West AG, Waite DW, Deines P, Bourne DG, Digby A, McKenzie VJ, Taylor MW. 2019. The microbiome in threatened species conservation. Biological Conservation 229:85–98. doi: 10.1016/j.biocon.2018.11.016. [DOI] [Google Scholar]
- 25.Trevelline BK, Fontaine SS, Hartup BK, Kohl KD. 2019. Conservation biology needs a microbial renaissance: a call for the consideration of host-associated microbiota in wildlife management practices. Proc R Soc B Biol Sci 286:20182448. doi: 10.1098/rspb.2018.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adolph SC. 1990. Influence of behavioral thermoregulation on microhabitat use by two Sceloporus lizards. Ecology 71:315–327. doi: 10.2307/1940271. [DOI] [Google Scholar]
- 27.Larson MW. 1961. The critical thermal maximum of the lizard Sceloporus occidentalis occidentalis Baird and Girard. Herpetologica 17:113–122. [Google Scholar]
- 28.Prieto AA Jr, Whitford WG. 1971. Physiological responses to temperature in the horned lizards, Phrynosoma cornutum and Phrynosoma douglassii. Copeia 1971:498–504. doi: 10.2307/1442447. [DOI] [Google Scholar]
- 29.Shea TK, DuBois PM, Claunch NM, Murphey NE, Rucker KA, Brewster RA, Taylor EN. 2016. Oxygen concentration affects upper thermal tolerance in a terrestrial vertebrate. Comp Biochem Physiol A Mol Integr Physiol 199:87–94. doi: 10.1016/j.cbpa.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 30.Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA, Apprill A, Knight R. 2016. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi: 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 33.Washburne AD, Silverman JD, Leff JW, Bennett DJ, Darcy JL, Mukherjee S, Fierer N, David LA. 2017. Phylogenetic factorization of compositional data yields lineage-level associations in microbiome datasets. PeerJ 5:e2969. doi: 10.7717/peerj.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandal S, Van Treuren W, White RA, Eggesbø M, Knight R, Peddada SD. 2015. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis 26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bokulich NA, Dillon MR, Zhang Y, Rideout JR, Bolyen E, Li H, Albert PS, Caporaso JG. 2018. q2-longitudinal: longitudinal and paired-sample analyses of microbiome data. mSystems 3:e00219-18. doi: 10.1128/mSystems.00219-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All 16S rRNA gene sequences produced for this study are available in the European Nucleotide Archive under accession no. PRJEB39005.