The gastrointestinal tracts of ruminants harbor a diverse microbial community that coevolved symbiotically with the host, influencing its nutrition, health, and performance. While the influence of environmental factors on rumen microbes is well documented, the process by which host genetics influences the establishment and colonization of the rumen microbiota still needs to be elucidated. This knowledge gap is due largely to our inability to easily sample the rumen microbiota. There are three common methods for rumen sampling but all of them present at least one disadvantage, including animal welfare, sample quality, labor, and scalability. The development and validation of noninvasive methods, such as buccal swabbing, for large-scale rumen sampling is needed to support studies that require large sample sizes to generate reliable results. The validation of buccal swabbing will also support the development of molecular tools for the early diagnosis of metabolic disorders associated with microbial changes in large herds.
KEYWORDS: bacteria, buccal swab, machine learning, oral community, random forest, rumen microbiota
ABSTRACT
Analysis of the cow microbiome, as well as host genetic influences on the establishment and colonization of the rumen microbiota, is critical for development of strategies to manipulate ruminal function toward more efficient and environmentally friendly milk production. To this end, the development and validation of noninvasive methods to sample the rumen microbiota at a large scale are required. In this study, we further optimized the analysis of buccal swab samples as a proxy for direct bacterial samples of the rumen of dairy cows. To identify an optimal time for sampling, we collected buccal swab and rumen samples at six different time points relative to animal feeding. We then evaluated several biases in these samples using a machine learning classifier (random forest) to select taxa that discriminate between buccal swab and rumen samples. Differences in the inverse Simpson’s diversity, Shannon’s evenness, and Bray-Curtis dissimilarities between methods were significantly less apparent when sampling was performed prior to morning feeding (P < 0.05), suggesting that this time point was optimal for representative sampling. In addition, the random forest classifier was able to accurately identify nonrumen taxa, including 10 oral and putative feed-associated taxa. Two highly prevalent (>60%) taxa in buccal and rumen samples had significant variance in relative abundances between sampling methods but could be qualitatively assessed via regular buccal swab sampling. This work not only provides new insights into the oral community of ruminants but also further validates and refines buccal swabbing as a method to assess the rumen bacterial in large herds.
IMPORTANCE The gastrointestinal tracts of ruminants harbor a diverse microbial community that coevolved symbiotically with the host, influencing its nutrition, health, and performance. While the influence of environmental factors on rumen microbes is well documented, the process by which host genetics influences the establishment and colonization of the rumen microbiota still needs to be elucidated. This knowledge gap is due largely to our inability to easily sample the rumen microbiota. There are three common methods for rumen sampling but all of them present at least one disadvantage, including animal welfare, sample quality, labor, and scalability. The development and validation of noninvasive methods, such as buccal swabbing, for large-scale rumen sampling is needed to support studies that require large sample sizes to generate reliable results. The validation of buccal swabbing will also support the development of molecular tools for the early diagnosis of metabolic disorders associated with microbial changes in large herds.
INTRODUCTION
The rumen is a specialized organ found in cattle that hosts a wide diversity of microorganisms from all three domains (for a review see references 1 and 2). Essential to the digestion of complex plant polymers by the host, the rumen microbiota consists of several species of specialized fibrolytic bacteria capable of degrading lignocellulose (3). Microbial changes following total rumen exchanges (4) and some preliminary genome-wide association data (5, 6) suggest that the microbial community composition is unique to each individual cow and that the genetics of the host animal may influence community development/maintenance in the rumen. The extent of host animal control over this phenomenon has not yet been validated with robust statistical analyses, as rumen samples are laborious to obtain.
Methods that directly sample the rumen contents of cattle are the rate-limiting step for generating a population-scale metric of the rumen microbiome. The gold standard method for assessing rumen microbial contents is via rumen cannulation; however, this requires invasive surgery and cannot be performed on hundreds of cows in a herd. Stomach tubing is another method of sampling that provides direct access to rumen contents, but this method is labor-intensive and is uncomfortable for the cow (7, 8). Given the requirements for surgery or labor-intensive sample collection, respectively, neither method is suitable for the development of a scalable industrial product. In light of the deficiencies of these methods, buccal swabbing has been proposed as a proxy for the rumen microbiota (9, 10). The ease of this method and its lower cost of implementation make it a tantalizing option for obtaining population-scale rumen microbial samples.
Buccal swabbing is a noninvasive method that takes advantage of cattle rumination, an innate behavioral process that characterizes the ruminant clade of mammals (11, 12). During this process, the cow regurgitates, masticates, moistens, and swallows a bolus from the rumen, which is a mixture of previously ingested plant material that is resistant to prolonged chemical degradation. This process exposes additional surface area of the digesting plant matter to continued microbial fermentation (12). However, rumen microbes are not effaced from the surface of the bolus prior to mastication, and microbial DNA in the oral cavity may constitute a representative proxy of the rumen microbiota.
Indeed, the oral cavity has its own resident microbiota that contains both transient facultative anaerobes and feed-associated microbes (13, 14) that can be concurrently sampled during buccal swabbing. The identification and exclusion of these contaminants constitute a prerequisite for the use of buccal swabs as a proxy for the rumen microbiota (9). In previous studies, the depletion of these contaminants was performed with mathematical filtering based on the comparison of the relative abundances of a given taxon between rumen and buccal swab samples (9, 10). However, these approaches noted the need for further statistical and qualitative validation for widespread adoption of the technique due to confounding factors that could impact predicted microbial taxon abundance (9). This is a necessary step toward the use of buccal swabbing as an independent method, as future surveys may not always have access to paired rumen samples for calibration.
Previous surveys have also not considered sampling time as a potential confounding factor for interrogating rumen microbial community profiles via buccal swabbing (15–18). Sampling time could be proximal to the time since last feeding as well as to the animal’s personal rumination pattern, which may affect the composition and abundance of the rumen microbial community present in the oral cavity. Previous studies have shown that rumen microbial species vary in terms of abundance at different intervals following feeding (19–21). Regardless of diet, Kamra et al. (21) observed a pronounced increase in the total protozoa within the first 2 h after feeding. However, total bacterial numbers appear to remain constant up to16 h after feeding (20). These changes are likely associated with factors such as (i) and increase of passage of digested materials from the rumen during at feeding, (ii) dilution of the ruminal contents with water and feed, (iii) microbial growth rate in response to feed intake and to incoming nutrients (19–22). The time since last feeding is also related to rumination behavior which normally peaks 4 h after feeding and results in an intense salivary production (12, 19). Therefore, salivary dilution coupled with contamination of epiphytic microbiota in fresh forages and microbiota in ensiled forages (for a review, see reference 23) can likely impact measured bacterial community composition and abundance. In this sense, it is possible that there is a specific window of time in which buccal swab samples best mirror the rumen contents of the sampled cow. Prior to its widespread adoption as a suitable proxy for rumen sampling, buccal swabbing data must be compared in a modeling experiment to identify the magnitude of these biases.
In this study, we applied statistical learning methods to buccal swab data obtained from 21 cannulated Holstein cows to identify bacterial taxa that are specific to the oral cavity. We hypothesize that the presence of nonrumen bacterial communities and the eventual salivary dilution of rumen microbial DNA impact the comparability of buccal swab samples with in situ rumen samples. We also tested if buccal swab operational taxonomic unit (OTU) abundances can be used in regression models to determine the approximate abundance of rumen bacterial OTUs in individual animals. Our analysis revealed an additional complexity in the diversity of microbes that colonize the ruminant gastrointestinal tract, and our findings support the future use of buccal swabs in population-scale surveys of the rumen bacterial community.
RESULTS
Amplicon sequencing and quality control.
To provide metrics for quality control and optimal parameter selection, we sampled buccal and rumen contents from several cohorts of cannulated cattle (Table 1). We sampled rumen strata (solids and liquids) from the anterior and ventral sections of the rumen lumen to further compare microbial compositions from different sections of the rumen. We hypothesized that buccal swab samples may have a closer resemblance to the anterior rumen samples than those taken from the ventral side of the cavity. Samples are here referred to by abbreviations that indicate the sample type (BS and R for buccal swab and rumen, respectively) and their location and content in the case of rumen samples (A, V, S, and L for anterior, ventral, solid, and liquid, respectively). For example, the abbreviation RAL refers to a rumen anterior liquid sample. All samples were sequenced using the same methods, and resulting data were processed using the same pipeline.
TABLE 1.
Samples and experimental design
| Sample set | Description | Sample count | Used in classification? | Used to train regression model? |
|---|---|---|---|---|
| Summer, time course, farm 1 (STC) | Six time points of sampling paired buccal and rumen contents | 8 animals | Yes | Yes |
| Spring sampling, farm 1 (SPS) | Paired rumen and buccal contents; taken 4 h after feeding | 5 animals | No | Yes |
| Summer sampling, farm 2 (SUS) | Paired rumen and buccal contents; taken 2 h prior to feeding | 8 animals | No | Yes |
After sequence quality filtering and normalization, a total of 1,392,036 reads (mean ± standard deviation [SD], 6,000.155 ± 132.615 per sample) and 8,147 unique OTUs (rounded mean and SD of redundant OTUs per sample, 846 ± 199) were obtained from 232 buccal, rumen solid, and rumen liquid samples. Good’s coverage estimation prior to normalization (0.969 ± 0.034 per sample) was deemed adequate and indicated that sequences sufficiently covered the diversity of the bacterial communities in our study. A full summary of sequencing statistics as well as rarefaction curves divided by sample type and time point is shown in Fig. S1 and Table S1 in the supplemental material.
Taxonomic composition analysis of the bacterial communities revealed a total of 2,031 OTUs (mean ± SD, 112.46 ± 32.91) present at relative abundances of ≥0.05% and representing 20 phyla, 116 families, and 279 genera. The average percentages of sequences unassigned to any phylum, family, or genus were 0.19 ± 0.15, 1.15 ± 0.45, and 10.49 ± 2.69, respectively. The most abundant OTUs, summarized at the phylum, family, and genus levels according to sampling time and sample type, are shown in Fig. S2.
Time course analysis and sampling method comparability.
We first sought to identify the effects of sampling method on the composition of observed bacterial communities in the rumen. For this analysis, we used paired rumen strata (solid and liquid) and buccal swab samples taken from the STC cohort (Table 1) in 2-h intervals, with the first time point (T1) taken 1 h prior to feeding. Rather than seeking a singular optimal time for sampling, we investigated the possibility that there are periods where the buccal bacterial community may be less representative in terms of species prevalence and relative abundance of the rumen community.
Sample type (i.e., buccal swabbing versus rumen cannula sampling) had the largest effect on observed bacterial content, as expected. Alpha diversity analysis revealed that the number of observed OTUs (Sobs) and inverse Simpson’s diversity index varied significantly with sampling time (P = 0.040; P = 0.044), with sample type (P < 0.001; P < 0.001) and with the interaction of these two factors (P < 0.001; P < 0.001). Meanwhile, variance in the Shannon’s evenness index was ascribed to sample type (P < 0.001) and the interaction terms (P < 0.001) but not to sampling time (P = 0.069).
In regard to interaction terms, the Sobs within buccal swabs displayed great variation but was significantly similar (Tukey’s honestly significant difference [HSD] test < 0.05) to all types of rumen samples at all sampling times (T1, T2, T4, and T6) with exception of time point T3, followed by T5, which displayed the lowest Sobs values. Shannon’s evenness of buccal swab samples was significantly similar to all types of rumen samples only at T1 and T4 (Tukey’s HSD test < 0.05). Likewise, inverse Simpson’s (Invsimpson’s) index of diversity of buccal swab samples was significantly higher than that for rumen samples and similar to those in all types of rumen samples taken at T1 and T4, respectively. In contrast, bacterial communities of buccal samples were less even and diverse than in all types of rumen samples taken at T3, followed by T2, T5, and T6 (Fig. S3A).
Regardless of sampling time, buccal swab samples displayed lower richness (i.e., Sobs), evenness (i.e., Shannon’s evenness,) and diversity (i.e., inverse Simpson’s diversity) than all types of rumen samples (Tukey’s HSD test < 0.05 [Fig. S3B]). For sample type, the Sobs was not significant different between time points (Tukey’s HSD test < 0.05 [Table S2 and Fig. S3B]) and bacterial communities sampled at T3 and T4 displayed the lowest and highest Invsimpson’s diversities, respectively (Tukey’s HSD test < 0.05 [Table S2 and Fig. S3C]).
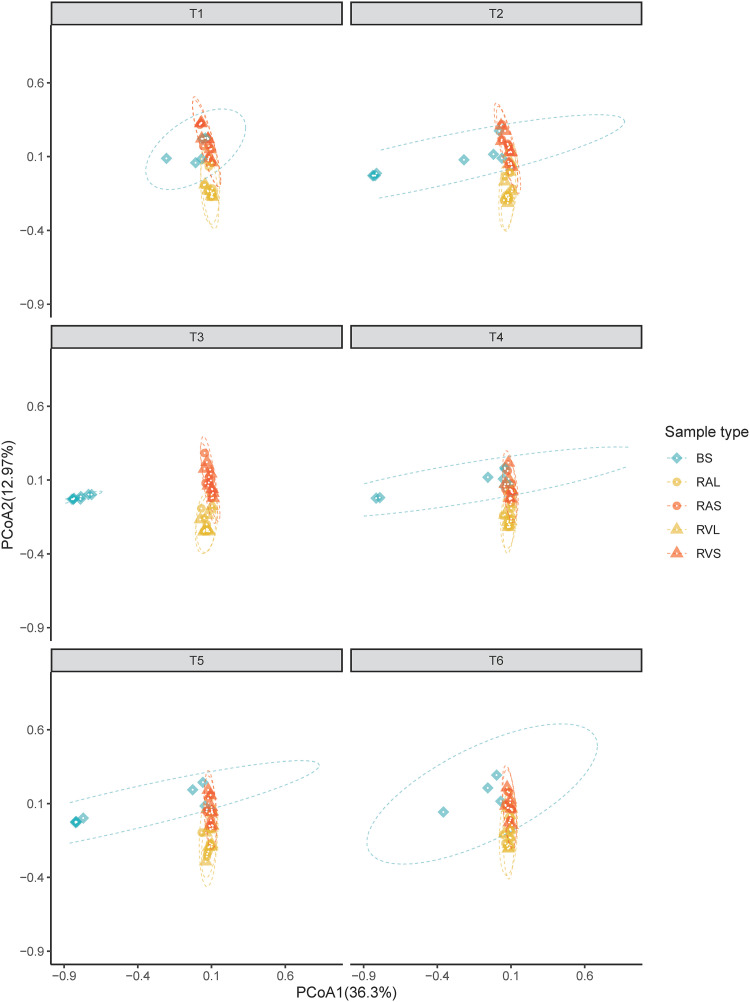
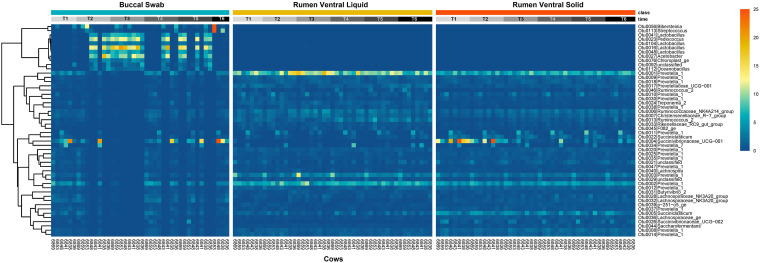
We used principal-coordinate analysis (PCoA) to visually inspect the similarity of buccal swab samples to contemporary rumen cannula samples. In general, rumen samples grouped by phase (i.e., L versus S) rather than location (i.e., A versus V). Additionally, we found that bacterial communities from buccal swab samples obtained just prior to morning feeding (T1) grouped most closely to rumen solid samples (RAS plus RVS) (Fig. 1). Moreover, ordination plots showed that T3 had the most pronounced differences between swab and rumen samples. The higher prevalence and abundance of putative silage-associated microbes belonging to the lactobacilli in T3 suggest that feed contamination was a major contributor to this discrepancy (Fig. 2 and Fig. S2).
FIG 1.
Principal-coordinate analysis (PCoA) showing Bray-Curtis dissimilarities in the composition of bacterial communities between sample types within each sampling time. Individual points in each plot represent a dairy cow, different colors and shapes represent a sample type (BS, buccal swab; RAL, rumen anterior liquid; RAS, rumen anterior solid; RVL, rumen ventral liquid; RVS, rumen ventral solid), and each facet represents a time point (T1 to T6). Percentages showed along the axes represent the proportion of dissimilarities captured by PCoA in two-dimensional (2D) coordinate space.
FIG 2.
Distribution of the most abundant bacterial taxa among individual dairy cows according to sample type and sampling time. The color key represents the relative abundance at gradient of color from dark blue (low abundance) to dark orange (high abundance). The hierarchical dendrogram was established using Pearson product-moment correlations as the distance measure and “complete” as a clustering method.
Permutational multivariate analysis of variance (PERMANOVA) showed that Bray-Curtis dissimilarities in the composition of bacterial communities were significantly driven by sampling time (R2 = 0.044; P < 0.001), by sample type (R2 = 0.284; P < 0.001), and by the interaction of these two factors (R2 = 0.106; P < 0.001). Pairwise comparisons between sample types showed that the composition of BS samples differs significantly from those of all types of rumen samples (P = 0.010). In addition, we found that bacterial composition at sampling time T1 was significantly different from those at T3 (P = 0.015) and T5 (P = 0.045). Lastly, comparisons between sample types within each sampling time indicates that the composition of bacterial communities in BS samples is similar to those observed in the RAS samples only at T1 (P = 0.054), confirming the clustering observed in the PCoA (Fig. 1, Fig. S4, and Table S3).
In addition to compositional dissimilarity, we assessed differences in the relative abundances of 277 bacterial OTUs (prevalence of at least 80% of all samples) in response to sampling time, sample type, and the interaction of these two factors (Fig. S5, S6, and S7 and Table S7). Overall, most of the variance in the relative abundance of bacterial communities in our study was ascribed to interaction terms given that 235 OTUs varied simultaneously with sampling time and sample type. Meanwhile, the differences ascribed to main effects were far less apparent, given that the relative abundances of only 42 and 22 OTUs varied independently in response to sample type and sampling time, respectively (Table S7).
Pairwise comparisons between oral and rumen samples within each sampling time showed that fewer OTUs had significantly different relative abundance between buccal and rumen samples taken at T1 followed by T4 and T6. In contrast, greater significant differences in the relative abundance of OTUs between buccal swabs and all rumen samples were observed at T3, followed by T5 and T2 (Fig. S5A and Table S7). We also observed that the magnitude of the differences in the relative abundance of bacterial OTUs was far less pronounced at T1 than at all others time points, especially at T3. At T1, the majority of significant differences in the relative abundances of bacterial OTUs was observed in pairwise comparisons between oral and rumen liquids, with buccal swabs showing lower relative abundance than RAL and RVL. However, we observed some exceptions for OTUs assigned to the Lachnospiraceae_NK3A20_group, Bifidobacterium, Ruminococcus_2, and Mogibacterium, whose relative abundances were significantly higher in BS than in rumen samples sampled at T1 (Tukey’s HSD test < 0.05 [Table S7]). We also observed that relative abundance of OTUs assigned to Prevotella_1, Succinivibrionaceae_UCG-001, Lachnospiraceae_NK3A20_group, Acetitomaculum, Lachnospiraceae_FE2018_group, and Lachnobacterium were significantly higher in BS than in rumen liquids (RVL and RAL, respectively; Tukey’s HSD test < 0.05) at T1, followed by T4 and T6 (Table S7).
In contrast, at all others time points, and especially at T3, the relative abundances of OTUs in oral samples were lower than those observed in all types of rumen samples (Fig. S5B and Table S7). In particular, the relative abundance of OTUs assigned to Bacteroidales_RF16_group_ge, Prevotella_1, Moryella, Lachnospira, Succiniclasticum, and Schwartzia were marked and significantly lower in buccal swabs than in all types of rumen samples (Tukey’s HSD test < 0.05 [Table S7]).
Pairwise comparisons between all types of rumen samples within each sampling time revealed no significant differences in the relative abundances of OTUs between RAS and RVS at T1, T2, T3, and T4. However, some OTUs varied in relative abundances between RAL and RVL at other sampling times, primarily at T3 (Fig. S6A and Table S7). Pronounced differences in the relative abundances of several OTUs between liquid and solid contents were observed at all time points. Specifically, the majority of the OTUs sampled at T1 and T2 displayed higher relative abundances in rumen liquids than in rumen solids (i.e., RAL versus RAS and RVL versus RVS), while the opposite was observed at other time points (Fig. S6B and Table S7).
In regard to the main effects, most of the significant differences in the relative abundances of bacterial OTUs between sampling points were observed in comparisons performed between T3 and T1, T4, T5, and T6. Regardless of sample type, the relative abundance of bacterial OTUs was significantly lower at T3 than at the other time points, particularly with T1, followed by T4 and T6 (Tukey’s HSD test ≤ 0.05 [Fig. S7A and B and Table S7]). Finally, comparisons performed between sample types showed that relative abundances of bacterial OTUs were significantly lower in buccal swabs than in all types of rumen samples (Tukey’s HSD test ≤ 0.05), regardless of sampling time. These differences were less apparent when buccal swab and rumen solids were compared (Fig. S7C and D and Table S7). However, a few exceptions were observed for OTUs assigned to Prevotellaceae_Ga6A1_group and Succinivibrionaceae_UCG-002, whose relative abundances were significantly higher in BS than in rumen liquids (RVL and RAL, respectively; Tukey’s HSD test < 0.05) (Table S7).
Random forest classifier analysis.
We next sought to identify key bacterial taxa present in the oral microbial community that contributed to discrepancies observed in our ordination plots. To distinguish statistically between taxa that had differences in relative abundance in each sample type, we trained a random forest classifier model using the STC cohort samples. Random forest is a supervised learning algorithm which uses an ensemble learning method (i.e., combine several trees base algorithms) to construct better predictive performance (for a review, see references 24 and 25) and has been widely and successfully employed for classification and regression purposes. In a classification problem, the algorithm returns a list of predictor variables (i.e., bacterial OTUs) that can be ranked according to their individual importance (i.e., variable importance [VIMP] score) in classifying the data.
Our preliminary analyses showed that the overall performance of the random forest classifier using five classification categories for sample type (BS, RAL, RAS, RVL, and RVS) was quite low (accuracy, 58.6%, and kappa, 48.2%), even after estimation and tuning of model hyperparameters (Table S4). This result supports the observation of high similarity between bacterial communities from rumen solid (RAS and RVS) and liquid (RAL and RVL) samples from different rumen lumen areas as observed in the PCoA (Fig. 1). We found improved classifier accuracy when rumen samples were merged based on rumen content strata (liquids and solids) into a single type in the training and testing sets (collectively referred to as RL and RS, respectively). This merger unbalanced our training set by providing a 2-fold increase in rumen categories (RL and RS = 95 samples each), and we thus implemented a resampling method for future model training to prevent misclassification of our minority class (BS = 42 samples). We tested three additional resampling methods (i.e., undersampling, oversampling, and synthetic minority oversampling technique [SMOTE]) to prevent classification bias toward the majority classes (26, 27). The results showed that random forest trained with additional resampling using the SMOTE had higher performance metrics than the other methods (Table S5).
Our final model was able to predict sample type-associated bacterial features with high accuracy (97.78% ± 3.7%) and Cohen’s kappa values (96.3% ± 5.4%). Cohen’s kappa is a frequently used statistic to assess the performance of machine learning models under a multiclass classification problem and or unbalanced data (28, 29). Other performance metrics, such as sensitivity, specificity, precision and recall, were also calculated for each sample type and are presented in Table S5. Additionally, our classifier returned the VIMP score, as a function of the mean decrease in Gini, of each bacterial OTU, which can be used to discriminate between oral and rumen samples (Table S6). Thus, higher values of VIMP score expressed as a percentage indicate higher feature importance (i.e., bacterial OTU) in discerning between classes and, in our case, between sample types.
OTU categorization based on variable importance estimates.
Bacterial OTUs with high VIMP scores (≥50% mean decrease in Gini) displayed patterns that allowed for manual categorization. Based on average taxon prevalence per sample type and sampling time, we categorized these OTUs into three categories: core, oral, and rumen (Table 2, Fig. 3, and Table S6). The remaining OTUs whose VIMP score was lower than 50% were also categorized for the sake of completeness but were not analyzed further (Table S6). The core category consisted of OTUs that displayed moderate to high prevalence (≥60% to 100%) in all sample types (both rumen and buccal) consistently across time points. The rumen category was defined as the community well represented (prevalence ≥ 75%) in rumen liquids and/or solids and was underrepresented in buccal swab samples (prevalence < 60%) at all time points (Fig. 3, Table 2, and Table S6). Finally, the oral group consisted of OTUs that were well represented in buccal swab samples (prevalence ≥ 60%) but were either absent or underrepresented in the rumen samples (<60% prevalence) across time points. The oral group was found to contain silage community microbes (i.e., lactobacilli) at time points when feed was provided to the animals (e.g., T3; see Fig. 4), further supporting our classification and the model’s accuracy.
TABLE 2.
Variable-importance analysis from the random forest classifier showing the most important bacterial OTUs (importance: scaled mean decrease in Gini ≥ 50%) that discriminate between buccal swab and rumen samples
| Taxa | Importance (%) | Samplea | T1 |
T2 |
T3 |
T4 |
T5 |
T6 |
Group | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Meanb | Prev.c | Mean | Prev. | Mean | Prev. | Mean | Prev. | Mean | Prev. | Mean | Prev. | ||||
| OTU0003-Prevotella_1d | 100 | BS | 1.38 | 100.0 | 0.73 | 75.0 | 0.08 | 62.5 | 1.07 | 100.0 | 0.40 | 87.5 | 0.68 | 100.0 | Core |
| RL | 3.17 | 100.0 | 3.03 | 100.0 | 4.09 | 100.0 | 3.49 | 100.0 | 3.63 | 100.0 | 2.95 | 100.0 | |||
| RS | 2.12 | 100.0 | 2.06 | 100.0 | 2.13 | 100.0 | 2.41 | 100.0 | 2.36 | 100.0 | 2.61 | 100.0 | |||
| Otu0405-p-1088-a5_gut_group | 96.8 | RS | 0.00 | 18.8 | 0.00 | 0.0 | 0.00 | 18.8 | 0.01 | 31.3 | 0.01 | 37.5 | 0.00 | 13.3 | Rumen |
| RL | 0.07 | 93.3 | 0.09 | 100.0 | 0.11 | 100.0 | 0.08 | 100.0 | 0.09 | 100.0 | 0.04 | 93.8 | |||
| BS | 0.01 | 33.3 | 0.00 | 12.5 | 0.00 | 12.5 | 0.01 | 37.5 | 0.00 | 0.0 | 0.00 | 25.0 | |||
| Otu0001-Prevotella_1d | 87.4 | BS | 3.13 | 100.0 | 1.16 | 87.5 | 0.17 | 62.5 | 2.78 | 100.0 | 0.90 | 100.0 | 2.21 | 100.0 | Core |
| RL | 8.08 | 100.0 | 9.27 | 100.0 | 12.13 | 100.0 | 9.15 | 100.0 | 9.83 | 100.0 | 7.97 | 100.0 | |||
| RS | 5.36 | 100.0 | 5.35 | 100.0 | 5.84 | 100.0 | 6.79 | 100.0 | 6.42 | 100.0 | 6.54 | 100.0 | |||
| Otu0241-Neisseriaceae | 86.5 | BS | 0.97 | 33.3 | 1.13 | 87.5 | 0.16 | 100.0 | 0.18 | 50.0 | 0.18 | 75.0 | 0.08 | 100.0 | Oral |
| RL | 0.00 | 20.0 | 0.00 | 6.3 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 12.5 | |||
| RS | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | |||
| Otu0113-Streptococcus | 86.1 | BS | 0.19 | 50.0 | 0.75 | 75.0 | 0.31 | 100.0 | 0.54 | 62.5 | 0.38 | 87.5 | 10.05 | 100.0 | Oral |
| RL | 0.00 | 6.7 | 0.00 | 6.3 | 0.00 | 6.3 | 0.00 | 0.0 | 0.00 | 6.3 | 0.00 | 6.3 | |||
| RS | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | |||
| Otu0401-Streptococcus | 84.9 | BS | 0.63 | 50.0 | 0.19 | 87.5 | 0.17 | 100.0 | 0.10 | 37.5 | 0.26 | 75.0 | 0.13 | 100.0 | Oral |
| RL | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | |||
| RS | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | |||
| Otu0434-Howardella | 83.3 | BS | 0.01 | 66.7 | 0.00 | 12.5 | 0.00 | 12.5 | 0.01 | 50.0 | 0.01 | 50.0 | 0.00 | 0.0 | Rumen |
| RL | 0.07 | 93.3 | 0.09 | 100.0 | 0.12 | 100.0 | 0.06 | 93.8 | 0.05 | 93.8 | 0.04 | 93.8 | |||
| RS | 0.00 | 0.0 | 0.00 | 18.8 | 0.00 | 12.5 | 0.01 | 31.3 | 0.00 | 25.0 | 0.00 | 13.3 | |||
| Otu0424-Ruminococcaceae_ge | 81.3 | BS | 0.01 | 50.0 | 0.00 | 12.5 | 0.00 | 0.0 | 0.01 | 37.5 | 0.00 | 12.5 | 0.00 | 0.0 | Rumen |
| RL | 0.06 | 93.3 | 0.06 | 87.5 | 0.14 | 93.8 | 0.06 | 81.3 | 0.08 | 100.0 | 0.07 | 93.8 | |||
| RS | 0.00 | 0.0 | 0.01 | 31.3 | 0.00 | 25.0 | 0.00 | 18.8 | 0.01 | 31.3 | 0.01 | 33.3 | |||
| Otu0838-Micrococcaceae | 79 | BS | 0.19 | 33.3 | 0.06 | 62.5 | 0.12 | 100.0 | 0.04 | 37.5 | 0.13 | 75.0 | 0.03 | 100.0 | Oral |
| RL | 0.00 | 6.7 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | |||
| RS | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | |||
| Otu0184-Pasteurellaceae | 76.3 | BS | 0.97 | 50.0 | 2.45 | 75.0 | 0.09 | 100.0 | 0.08 | 37.5 | 0.16 | 75.0 | 0.12 | 100.0 | Oral |
| RL | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | |||
| RS | 0.00 | 0.0 | 0.00 | 6.3 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 6.7 | |||
| Otu0720-Jeotgalicoccus | 75 | BS | 0.24 | 50.0 | 0.03 | 87.5 | 0.15 | 100.0 | 0.07 | 50.0 | 0.13 | 75.0 | 0.03 | 100.0 | Oral |
| RL | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 6.3 | |||
| RS | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | |||
| Otu0042-Ruminococcaceae_NK4A214_groupd | 70.7 | BS | 0.16 | 100.0 | 0.06 | 50.0 | 0.01 | 25.0 | 0.10 | 87.5 | 0.05 | 50.0 | 0.10 | 100.0 | Rumen |
| RL | 0.59 | 100.0 | 0.80 | 100.0 | 0.99 | 100.0 | 0.72 | 100.0 | 0.81 | 100.0 | 0.55 | 100.0 | |||
| RS | 0.09 | 100.0 | 0.15 | 100.0 | 0.18 | 100.0 | 0.15 | 100.0 | 0.18 | 100.0 | 0.15 | 100.0 | |||
| Otu0322-Streptococcus | 66.4 | BS | 0.19 | 50.0 | 0.05 | 62.5 | 0.60 | 75.0 | 0.03 | 62.5 | 0.83 | 75.0 | 1.67 | 75.0 | Oral |
| RL | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 6.3 | 0.00 | 0.0 | |||
| RS | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | |||
| Otu0115-Bacteroidales_RF16_group_ged | 63.8 | BS | 0.12 | 100.0 | 0.12 | 50.0 | 0.00 | 12.5 | 0.11 | 75.0 | 0.06 | 37.5 | 0.12 | 100.0 | Rumen |
| RL | 0.21 | 100.0 | 0.27 | 100.0 | 0.33 | 100.0 | 0.33 | 100.0 | 0.32 | 100.0 | 0.39 | 100.0 | |||
| RS | 0.02 | 81.3 | 0.02 | 87.5 | 0.02 | 75.0 | 0.03 | 81.3 | 0.01 | 68.8 | 0.02 | 66.7 | |||
| Otu1233-Planococcaceae | 62.3 | BS | 0.03 | 66.7 | 0.03 | 50.0 | 0.03 | 75.0 | 0.06 | 25.0 | 0.07 | 75.0 | 0.05 | 75.0 | Oral |
| RL | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | |||
| RS | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | |||
| Otu0780-Synergistes | 59.8 | BS | 0.00 | 16.7 | 0.00 | 12.5 | 0.00 | 0.0 | 0.01 | 37.5 | 0.00 | 12.5 | 0.00 | 0.0 | Rumen |
| RL | 0.02 | 80.0 | 0.02 | 87.5 | 0.06 | 93.8 | 0.03 | 75.0 | 0.03 | 87.5 | 0.03 | 75.0 | |||
| RS | 0.00 | 6.3 | 0.00 | 6.3 | 0.00 | 6.3 | 0.00 | 0.0 | 0.00 | 12.5 | 0.00 | 20.0 | |||
| Otu0239-Streptococcus | 58.3 | BS | 0.06 | 66.7 | 0.17 | 62.5 | 1.07 | 75.0 | 0.40 | 75.0 | 0.49 | 75.0 | 2.48 | 75.0 | Oral |
| RL | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 12.5 | 0.00 | 0.0 | 0.00 | 0.0 | |||
| RS | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | |||
| Otu0443-Prevotellaceae_UCG-001 | 55.4 | BS | 0.02 | 100.0 | 0.01 | 25.0 | 0.00 | 12.5 | 0.02 | 75.0 | 0.01 | 37.5 | 0.01 | 50.0 | Rumen |
| RL | 0.07 | 100.0 | 0.06 | 100.0 | 0.06 | 100.0 | 0.07 | 100.0 | 0.05 | 87.5 | 0.07 | 100.0 | |||
| RS | 0.01 | 62.5 | 0.01 | 56.3 | 0.01 | 37.5 | 0.01 | 62.5 | 0.00 | 25.0 | 0.01 | 60.0 | |||
| Otu0056-Bibersteinia | 53.3 | BS | 1.17 | 83.3 | 2.55 | 87.5 | 1.52 | 100.0 | 1.55 | 87.5 | 0.93 | 87.5 | 8.06 | 100.0 | Oral |
| RL | 0.00 | 6.7 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 12.5 | 0.00 | 18.8 | |||
| RS | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 6.3 | 0.00 | 6.7 | |||
| Otu0788-Rikenellaceae_RC9_gut_group | 52.7 | BS | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 25.0 | 0.00 | 12.5 | 0.00 | 25.0 | Rumen |
| RL | 0.03 | 86.7 | 0.03 | 87.5 | 0.04 | 87.5 | 0.03 | 87.5 | 0.03 | 87.5 | 0.03 | 62.5 | |||
| RS | 0.00 | 0.0 | 0.00 | 6.3 | 0.00 | 12.5 | 0.00 | 12.5 | 0.00 | 0.0 | 0.00 | 0.0 | |||
| Otu0356-Rikenellaceae_RC9_gut_group | 52.5 | BS | 0.01 | 33.3 | 0.00 | 12.5 | 0.00 | 12.5 | 0.01 | 37.5 | 0.01 | 25.0 | 0.01 | 50.0 | Rumen |
| RL | 0.09 | 100.0 | 0.09 | 100.0 | 0.13 | 100.0 | 0.11 | 100.0 | 0.09 | 100.0 | 0.07 | 87.5 | |||
| RS | 0.01 | 37.5 | 0.01 | 43.8 | 0.01 | 31.3 | 0.01 | 43.8 | 0.01 | 25.0 | 0.01 | 33.3 | |||
| Otu0120-Succiniclasticumd | 52.4 | BS | 0.07 | 100.0 | 0.02 | 50.0 | 0.00 | 0.0 | 0.05 | 75.0 | 0.02 | 37.5 | 0.03 | 50.0 | Rumen |
| RL | 0.18 | 100.0 | 0.20 | 100.0 | 0.22 | 100.0 | 0.19 | 100.0 | 0.17 | 100.0 | 0.26 | 100.0 | |||
| RS | 0.16 | 100.0 | 0.15 | 100.0 | 0.15 | 100.0 | 0.12 | 100.0 | 0.15 | 100.0 | 0.17 | 100.0 | |||
| Otu0096-Ruminococcus_1d | 50.2 | BS | 0.11 | 100.0 | 0.06 | 50.0 | 0.01 | 25.0 | 0.17 | 87.5 | 0.07 | 37.5 | 0.13 | 75.0 | Rumen |
| RL | 0.05 | 93.3 | 0.04 | 75.0 | 0.01 | 50.0 | 0.04 | 100.0 | 0.04 | 81.3 | 0.09 | 93.8 | |||
| RS | 0.40 | 100.0 | 0.44 | 100.0 | 0.36 | 100.0 | 0.24 | 100.0 | 0.32 | 100.0 | 0.38 | 100.0 | |||
| Otu0094-CPla-4_termite_group | 49.7 | BS | 0.02 | 66.7 | 0.01 | 25.0 | 0.00 | 12.5 | 0.03 | 87.5 | 0.01 | 25.0 | 0.00 | 0.0 | Rumen |
| RL | 0.25 | 100.0 | 0.31 | 100.0 | 0.56 | 100.0 | 0.37 | 100.0 | 0.45 | 100.0 | 0.26 | 100.0 | |||
| RS | 0.01 | 43.8 | 0.02 | 62.5 | 0.02 | 68.8 | 0.02 | 62.5 | 0.02 | 68.8 | 0.02 | 46.7 | |||
BS, buccal swab, rumen samples were merged based on rumen content strata; RL, rumen liquids (RAL plus RVL); RS, rumen solids (RAS plus RVS).
Average relative abundance.
Average prevalence (Prev.).
Varied with interaction of sampling time and sample type (Table S7); importance and prevalence are both expressed as percentages.
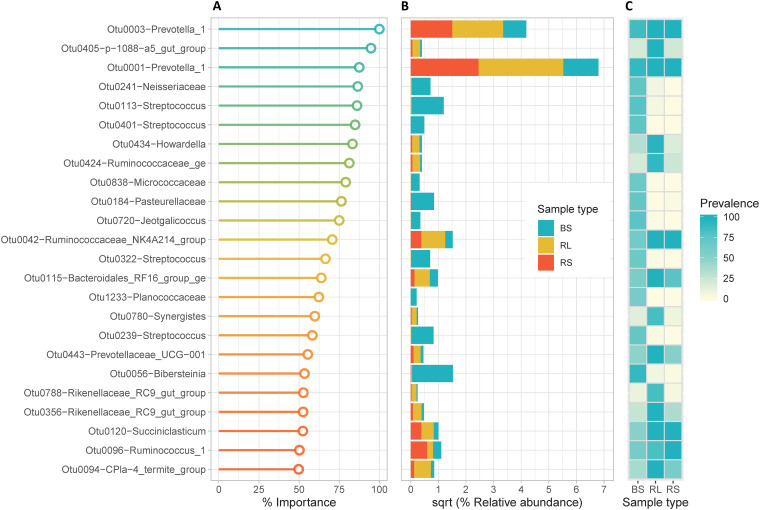
FIG 3.
Variable importance (VIMP) plot from the random forest classifier. (A) Lollipop chart showing the most important bacterial signatures that displayed importance (percent mean decrease in Gini ≥ 50) and that discriminate between buccal swab (BS), rumen liquid (RL), and rumen solid (RS) samples. (B) Bar plots of square root (sqrt) of the relative abundance of OTUs according to sample type. (C) Heat map of prevalence of OTUs in each sample type.
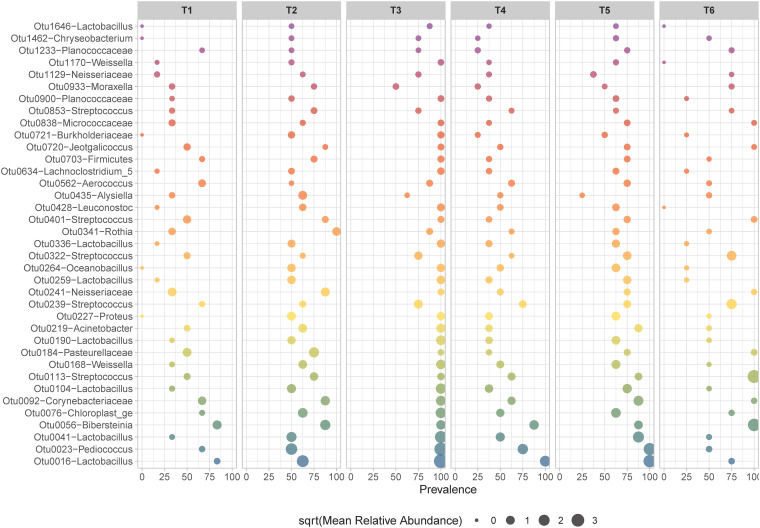
FIG 4.
Bubble chart showing the prevalence and relative abundance of the oral OTUs assigned to higher taxa (phylum, family, or genus level) according to sampling time.
In the core group, we identified two OTUs (VIMP > 80%) assigned to the genus Prevotella_1 (Fig. 3 and Table 2) that displayed high prevalence in both buccal swab and rumen (liquid and solid) samples. The relative abundances of these taxa were significantly lower (Tukey’s HSD test ≤ 0.05) in buccal swabs than in rumen samples (Tables S6 and S7). This suggests that these taxa can be reliably sampled via swabbing but that their relative abundances are greatly biased compared to the paired rumen samples.
We also identified taxa in the families Neisseriaceae, Pasteurellaceae, Micrococcaceae, and Planococcaceae, as well as in the genera Streptococcus, Jeotgalicoccus, and Bibersteinia, which displayed moderate to high VIMP scores (≥50%) and were assigned to the oral category. These taxa were overrepresented in terms of prevalence and abundance in buccal swab samples and displayed very low or zero abundance in rumen liquid and solid samples (Fig. 3, Table 2, and Table S6). In addition, we observed that several oral taxa (i.e., Oceanobacillus, Lactobacillus, Lachonoclostrium, Leuconostoc, Rothia, and Proteus) were underrepresented in terms of abundance and prevalence at specific time points, including T1, T4, and T6, relative to time points T2, T3, and T5 (Fig. 4 and Table S6).
Finally, the classifier also selected rumen stratum OTUs that have lower relative abundance in the buccal swab samples (rumen category). Several were specific to rumen liquids (0405-p-1088-a5_gut_group, Howardella, Ruminococcaceae_ge, Synergistes, Prevotellaceae_UCG-001, and Rikenellaceae_RC9_gut_group), and others were derived from the rumen solids (Ruminoccocus_1, Prevotellaceae_UCG-001 and Oribacterium) whose overall importance was ≥33% (Fig. 3, Table 2, and Table S6).
Random forest regression analysis.
We next sought to test whether the abundance of OTUs found in buccal swab samples could be used to predict the abundance of rumen OTUs. We tested the ability of four linear models (random forest regression, three log-linear models with either a Poisson distribution, zero inflated, or random generalized linear model [RGLM]) to characterize the relationship between bacterial OTUs of paired buccal swab and rumen liquid samples. In order to provide additional data for our training regression models, we incorporated data from 21 cows sampled in two other surveys (Table 1) processed with the same methods as used for the time course study. It is important to note that random forest regression was performed using sequence relative abundances, whereas log-linear models use the number of reads for each OTU, assuming a Poisson distribution of read counts. Our random forest and Poisson regression model converged, but they exhibited low accuracy in cross-validation studies, as shown by a low coefficient of determination (R2 = 0.39 ± 0.05) and high root mean square error (RMSE = 0.28 ± 0.09). We attempted to tune additional parameters in the random forest model but were unable to achieve an accuracy R2 above of 0.42 ± 0.07 on a per-OTU basis. Conversely, zero-inflated and RGLM trials failed to converge, despite several attempts to filter the OTU tables and tune model parameters. These results may be related to our use of a small data set as well as the nonlinear relationship between the buccal swab and rumen OTU abundance/counts on a per-sample basis.
DISCUSSION
In this study, we evaluated the ability of the buccal swabbing method to describe bacterial communities found in two types of rumen samples taken at six distinct sampling times over the course of 10 h. Buccal swab samples are an attractive alternative to more labor-intensive methods of sampling the rumen microbial community but may suffer from bias due to contamination by the surrounding oral community (9, 10). We first sought to identify the effect of sampling time on buccal swab community composition, as we hypothesized that animal rumination patterns and salivary flow may change the relative abundance of key members of the rumen community.
Our time course analysis suggested that there is a small, but statistically significant, effect of sampling time on the comparisons of several buccal swab bacterial taxa with contemporary rumen samples from the same animal. After dividing sampling times into 2-h intervals, we sampled buccal contents from each animal just prior to the start of morning feeding (T1), within regular intervals during and after feeding (T2, T3, T4, and T5), and prior to evening feeding (T6). We found that the only major outlier was at time point 3 (T3), where the greatest dissimilarities in the bacterial communities between buccal swabs and rumen samples were observed. It is possible that additional contamination by the silage microbial community and increased salivary flow induced by feeding changed the relative abundances of key rumen taxa in the oral samples of cows sampled at T3. This is evidenced by the presence of Lactobacillus from silage communities in the buccal swabs, but not in the rumen contents (Fig. 2 and 4). Our results support a hypothesis that there are brief windows of time in which buccal swab data best represent contemporary rumen bacterial data. This means that future surveys will need to record time of sampling relative to animal feeding in order to standardize results.
We also tested the possibility that buccal swab samples may be compositionally similar to rumen content fractions taken from different positions in the rumen (i.e., anterior versus ventral). Our comparisons of sampling time and sample types found no differences between the bacterial communities of the anterior and ventral rumen bacterial communities, which prevented us from finding such an association (Fig. 1). This result is likely associated with the constant mixing of rumen contents due to the contractions of the reticulorumen, which would result in indistinguishable variation in our observed rumen bacterial OTU abundance (12). This finding is supported by previously published work that did not identify noticeable differences in sample composition from five different locations of the rumen lumen via denaturing gradient gel electrophoresis (DGGE) surveys (30). However, we cannot rule out the possibility that our sampling and analysis methods could not identify the small effects that these locations have on the community.
We also found greater similarity between bacterial taxa present in buccal swabs and rumen solids than in rumen liquids (Fig. 1). We suspect that this reflects a key stage of the rumination process whereby, immediately after regurgitation, the liquid fraction of the bolus is swallowed (12). It is possible that the bacterial taxa that are predominant in the liquid phase of the rumen contents are evacuated from the oral cavity early in the process of rumination. During mastication of the bolus, bacteria from the solid phase of the rumen contents are more likely to adhere to oral mucosal surfaces and are more likely to be sampled during buccal swabbing.
In order to identify nonrumen taxa in buccal swab samples, we employed a machine learning classifier to assist in the filtering of oral and silage bacterial communities in buccal swab samples. As has been noted previously (9), the presence of the commensal oral microbial community in buccal swab samples prevents direct comparisons between rumen content samples and buccal swabs and must be filtered from buccal swab samples prior to analysis using manual and mathematical methods (9, 10). By using a random forest classifier, we were able to assign importance estimates to individual bacterial taxa based on their use as a feature in our classification models, as has been done previously (31, 32). The top OTUs, after variable importance analysis, consisted of microbes that were orally specific (oral, n = 10), those that were rumen biased (rumen, n = 12), and those with high prevalence regardless of sample type but that varied based on relative abundance (core, n = 2). These findings support our observations of the influence of sample type on OTU relative abundance and also identified members of the oral bacterial community that were prevalent only in buccal swab samples. In addition, the top OTUs identified by our VIMP analysis included two members of the Prevotella, which were found to vary substantially between buccal and rumen samples (Table S7). These two OTUs were prevalent in all samples and at all time points; however, their relative abundances in buccal swabs were lower than in the rumen samples. These differences were far less apparent at T1, which was just prior to feeding, than at any other sampling time. This observation of similarity at only one time point implies that sampling time had a large effect on the estimated relative abundance of this clade, as confirmed by our analysis of variance (ANOVA).
The OTUs present within the oral category represent taxa that are poorly represented in rumen samples. Indeed, we identified commensal oral microbes from the genus Rothia that were present only in the buccal swab samples (the oral category). These taxa can be safely removed from future buccal swab surveys. We also identified several oral taxa (i.e., Lactobacillus, Chryseobacterium, Burkholderiaceae, and Oceanobacillus) that were prevalent at some time points and underrepresented or even absent at others (Fig. 4), showing that sampling time is a critical factor to be considered in future studies. The higher prevalence of these taxa during (T2) and immediately after (T3) feeding suggests that these sampling times will result in buccal swab data that are least representative of the rumen contents of the animal.
Our use of random forest classifiers suggests that machine learning methods can be used to approximate the rumen bacterial community at the time of sampling. More accurate estimation of these communities will be beneficial to rumen microbial ecology experiments that suffer from low sample counts. However, we were unable to achieve an acceptable rate of error (measured via residual error of observed and predicted OTU counts) from our regression analysis. We found that multicollinearity of predictors and weak linear association between oral and rumen OTUs prevented accurate regression.
We suspect that other factors (i.e., sampling time, rumination frequency, herd, and diet) must be controlled in the modeling of these data, as evidenced by the significance of sampling time and interaction terms in our PERMANOVA and ANOVA. Of these, the time since last rumination is most likely to contribute to observed dissimilarities between buccal swabs and rumen contents, and future surveys should attempt to collect these data directly or through proxy estimations. Moreover, it is possible that the taxonomic affiliation of our OTU counts could be masking individual species-level abundances that provide far more variance than expected for the regression model. Similarly, our genus-level assignments could also contain inaccuracies due to strain abundance differences in the oral cavity versus the rumen contents. Finally, discrepancies in DNA yield variance for buccal swabs (Table S1) were identified that may indicate that cells from nonbacterial species or polysaccharides in the saliva may compete with bacteria for occupancy of the swab surface. Sampling shortly after rumination or accounting for the time since last rumination is likely to reduce this sampling discrepancy but will not overcome the compositional nature of the method.
Finally, we cannot rule out the possibility that several OTUs are metabolically active (i.e., facultative aerobes) in both locations and can proliferate in the oral cavity, thereby creating a nonlinear relationship between their abundance estimates in buccal swabs and rumen contents. Such a hypothesis could be tested through the use of RNA sequencing (RNA-seq) on buccal swab samples, which should preferentially target active microbial species in the oral cavity. However, we note that comparisons of RNA and DNA amplicon sequencing in the cattle rumen have revealed that such samples may not be directly comparable, potentially due to differences in sample preparation (33).
While this presents an impediment to the use of buccal swabs for classical microbial ecology experiments, we note that buccal swab data are still useful for other associative analysis. The ability to collect large numbers of samples from a diverse cohort of animals can present an opportunity for associations of microbial profiles with animal production and performance metrics, including milk production, health, and even fertility phenotypes. Such experiments would benefit from the removal of biases that we identified in this survey.
In summary, we have identified significant effects of sampling time and sample type on the composition of bacterial community abundance derived from buccal swabs and rumen samples. The buccal swab samples were prone to significant differences in bacterial profiles based on the time of sampling, with specific time points showing higher prevalence of the oral or feed-associated bacterial community than others. For future surveys using buccal swabs as a proxy for rumen bacterial abundance, we recommend buccal sampling at least 2 h prior to or 4 h after feeding. Our data also suggest that a portion of the rumen microbial community will remain inaccessible to buccal swab samples; however, this bias may not necessarily impede future association studies with host animal phenotypic traits.
MATERIALS AND METHODS
Animal care and use.
All animal procedures were conducted according to Research Animal Resource Center (RARC) protocol A005902-A02 approved 28 July 2017 by the University of Wisconsin—Madison College of Agriculture and Life Sciences Institutional Animal Care and Use Committee. This work was carried out at the U.S. Dairy Forage Research Center Farm, Prairie du Sac, WI, from November 2017 to June 2019 using a cohort of 21 cannulated lactating Holstein dairy cows (∼2.5 years old) fed a total mixed ration in a free-stall barn.
Sampling.
To identify the sampling time at which oral microbiota would best represent the rumen microbiota, paired oral (buccal swab [BS]) and ruminal (rumen anterior liquid [RAL], rumen anterior solid [RAS], rumen ventral liquid [RVL], and rumen ventral solid [RVS]) samples were collected from 8 cannulated Holstein cows every 2 h over the course of 10 h, starting 1 h prior to morning feeding (∼9 a.m.) and ending just prior to evening feeding (∼7 p.m.), totaling six time points (T1 to T6). This data set is referred to as the summer time course (STC) (Table 1).
Two other surveys of paired buccal swab and rumen content samplings were conducted on different animals in the same herd at two other time points separated by at least 3 months (Table 1). These data sets consist of a spring sampling (SPS; 5 cows) and a summer sampling (SUS; 8 cows) taken a year prior to the STC data set. Swabs and rumen contents were processed in the same manner as listed for the time course survey, but samples were collected from animals 4 h after feeding (all cows in SPS) or prior to feeding (all cows in SUS), representing equivalents to T4 and T1 from the time course trial, respectively. These samples were collected to provide additional power for training and testing regression models (see Table 1).
In all trials, two swabs (Puritan PurFlock ultrasterile flocked swab with an 80-mm breakpoint, Puritan Medical Products, Guilford, ME) were inserted in the buccal cavity of each cow and were gently scraped across the inner side of the right cheek for approximately 10 s. The buccal swabs were placed in a sterile conical tube (15 ml) containing 1 ml of sterile phosphate-buffered saline and stored on ice during sampling. Immediately after buccal swabbing, rumen contents were collected via the rumen cannula and squeezed through double layers of cheesecloth to obtain an aliquot of 40 ml of rumen liquids and 50 ml of a loosely packed rumen solid fraction. The solid fraction was squeezed once more to remove all liquids and the residual solid material was transferred to another container. All samples were stored and transported on wet ice and stored at −80°C until processing and DNA extraction.
DNA extraction and sequencing.
This study focused primarily on the detection of bacterial species and used primers specific for bacterial 16S rRNA amplification. Total genomic DNA was extracted from buccal swab, rumen liquid, and rumen solid samples as previously described (34, 35). The V4 hypervariable region of the bacterial 16S rRNA was amplified via PCR using universal bacterial primers (F-GTGCCAGCMGCCGCGGTAA and R-GGACTACHVGGGTWTCTAAT) described previously (36). These primers also included adapters suitable for sequencing using the Illumina technology (F-AATGATACGGCGACCACCGAGATCTACAC and R-CAAGCAGAAGACGGCATACGAGAT) and further included unique barcodes to facilitate multiplexing: the forward primers had 16 unique 8-bp barcodes, and the reverse primers had 24 unique 8-bp barcodes. PCR mixtures consisted of 2.5 to 25 ng of template DNA and 0.2 μM primer in a 25-μl reaction mixture with 2× KAPA HiFi HotStart ReadyMix (KAPA Biosystems, Wilmington, MA). The reactions were performed on a Bio-Rad S1000 thermocycler (Bio-Rad Laboratories, Hercules, CA) according to the following conditions: 95°C for 3 min, 25 cycles of 95° for 30 s, 55°C for 30 s, and 72°C for 30 s, and a final extension step at 72°C for 5 min. PCR products were visualized on a 1% (wt/vol) low-melt agarose gel using AquaPor low-melt agarose (National Diagnostics, Atlanta, GA) and SYBRSafe DNA gel stain (Invitrogen, Waltham, MA), and bands at ∼380 bp indicated successful amplification. These bands were excised, and then DNA was extracted and cleaned using a Zymoclean gel DNA recovery kit (Zymo Research, Irvine, CA). Gel-extracted DNA was then quantified using Qubit HS methodology (Invitrogen) and a 96-well plate spectrophotometer, and a library was created using a 4-nmol/liter equimolar pool of all PCR products. This library was then sequenced on an Illumina MiSeq (Illumina Inc., San Diego, CA) following standard Illumina sequencing protocols using a MiSeq v2 2 × 250 sequencing kit at 10 pmol/liter and with a 10% PhiX control.
Bioinformatics analysis.
DNA sequences were analyzed using mothur (v1.39.0) (37) as described in the pipeline available in the supplemental material. Briefly, paired-end reads were joined using default parameters in make.contigs, and sequences with a length shorter than 200 bp or longer than 500 bp containing ambiguous characters or exhibiting a homopolymer greater than 8 bp were removed. Sequences were aligned using the SILVA 16S rRNA gene reference database (release 132) (38), screened for sequences aligned to our region of interest (screen.seqs; start = 13,862 and end = 23,444), and then preclustered to remove sequencing errors. The Uchime algorithm was used to detect chimeric sequences (39), and sequences that did not align to the correct region or were chimeric were removed. Sequences were classified using the SILVA database, and those that were nonbacterial (i.e., Archaea, Eukaryota, cyanobacteria, and mitochondria) were removed. Those sequences that appeared only once in the data set were removed (split.abund) so as to minimize bias due to sequencing error, and the uncorrected pairwise distances between the sequences were calculated. The sequences were taxonomically assigned using the Wang method and Greengenes (August 2013 release) reference database (40) with a consensus confidence threshold of 80%. Finally, the sequences were grouped into operational taxonomic units (OTUs) by uncorrected pairwise distances clustered by the nearest-neighbor method with a similarity cutoff of 97%. Coverage was assessed by Good’s index (41), and samples that displayed coverage less than 93% were discarded prior to normalization. To address differences in sequencing depths, the OTU table was normalized by subsampling sequences to the sample with the smallest number of sequences and then normalizing across samples to produce equal sequence counts (3,000 sequences per sample). The normalized OTU table was used to calculate alpha diversity indices, including the number of observed OTUs (Sobs), Shannon’s index-based measure of evenness (Shannon’s evenness) (42), and the inverse Simpson’s (Invsimpson’s) index (43). Beta diversity, using the Bray-Curtis dissimilarity index (44), was also determined, as well as the relative abundance (reads/total reads in a sample × 100) of OTUs in each sample. Alpha diversity indices were obtained via mothur (v1.39.0) (21), whereas the Bray-Curtis dissimilarity index was calculated using function vegdist available in the vegan R package (v2.5-6) (45).
Statistical analysis.
All statistical analyses were performed in R (v3.6.1), and source code to reproduce these analyses is available in the supplemental material. Measurements of alpha diversity (Sobs, Shannon’s evenness, and inverse Simpson’s diversity) and relative abundance of OTUs were assessed for normality and were found to follow a nonnormal distribution. Only OTUs with relative abundances of ≥0.05% present in at least 80% of all samples were analyzed. Differences in the alpha diversity indices and OTU relative abundance values were analyzed, respectively, under gamma and Poisson distributions, using a repeated-measure generalized linear mixed model estimated via penalized quasilikelihood (46):
where is a vector of gamma- or Poisson-transformed of alpha diversity indices or OTU counts, Xi is a design matrix relating individual observations to levels of fixed effects, β is a vector of fixed effects (i.e., sampling time, sample type, and their interaction), Zi is the incidence matrix on random effects, b is the vector of random animal effects, and εi is a vector of random error terms. In order to identify differences in relative abundance, an offset variable was included in our Poisson mixed model. Given that the total number of sequence reads varies across samples, the offset term in our model refers to the log of the total number of reads in a given sample and was adjusted as a covariate in the model to ensure response to relative abundance rather than raw count data (47, 48). The resulting ANOVA P values were adjusted for false-discovery rate (FDR) using the Benjamini-Hochberg method, and values of ≤0.05 were considered significant. Pairwise comparisons among the least squares means (LSMEANS) were performed using Tukey’s honestly significant difference (HSD) method. In the presence of significant interaction effects, the LSMEANS of the sample types were compared within each sampling time. These analyses were performed using functions available in the R packages fitdistrplus (v1.0-14), MASS (v7.3-51.5), lsmeans (v2.30-0), and ggplot2 (v3.2.1) (49–52).
To visually explore the degree of dissimilarity between bacterial composition of oral and rumen samples collected at six distinct sampling times, principal-coordinate analysis (PCoA) was conducted on the Bray-Curtis distance matrix (44). In addition, permutational multivariate analysis of variance (PERMANOVA; nperm = 1,000) (53) with post hoc test using Benjamini-Hochberg correction was performed to assess differences in the composition of bacterial communities according to sample type, time points, and their interaction. These analyses were performed using functions available in the R packages ggplot2 (v3.2.1), vegan (v2.5-6), and EcolUtils (v0.1) (45, 54, 55).
To identify taxa that discriminate between oral and rumen samples, a random forest classifier was trained on a random selection of 70% (162 samples) of the database composed of 232 samples and 2,031 OTUs and validated using the remaining 30% (70 samples). Only OTUs with relative abundances of ≥0.05% present in at least one sample were included as input. The number of trees was set to 500, while the number of variables available for splitting at each tree node (mtry) was tuned and accuracy was used to select the optimal model using the largest value. In addition, to evaluate the capability of our model to predict on independent data set, we adopted a repeated k-fold cross validation method (10-fold repeated 3 times). Prediction performance metrics (i.e., accuracy, sensitivity, specificity, precision, and recall) and a confusion matrix were calculated and summarized by sample type. Finally, the mean decrease in Gini (i.e., Gini index) was used to calculate the variable importance score (VIMP) and select bacterial OTUs that were most predictive of sample types. To that end, we used the function varImp (56), which automatically scales the importance scores to be between 0 and 100. These results were plotted to show the most important sample type-associated bacterial OTUs with VIMP scores of ≥50%. These analyses were performed using the R packages randomForest (v4.6-14) and caret (v6.0-85) (24, 56).
In order to evaluate if abundance of oral microbiota can be used to predict the abundance of rumen microbiota, we tested distinct regression models (i.e., random forest, random generalized linear model, and GLMM zero-inflated quasi-Poisson). These analyses were performed using the R packages MASS (v7.3-51.5), caret (v6.0-85), randomForest (v4.6-14), and randomGLM (v1.02-1) (24, 56, 57).
Data availability.
The raw sequence reads from all samples analyzed in this study are available on the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/) under Bioproject accession number PRJNA623113.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by USDA Agricultural Research Service (Washington, DC) CRIS project 5090-31000-026-00-D supporting D.M.B., J.C.M., and J.Y., USDA ARS CRIS project 5090-31000-025-00D supporting K.F.K., USDA ARS CRIS project 8042-31000-001-00-D supporting D.M.B., and USDA ARS CRIS project 8042-31000-002-00-D supporting J.B.C. This work was also supported in part by USDA National Institute of Food and Agriculture (NIFA), Agricultural and Food Research Initiative (AFRI) Foundation grant no. 2019-05592 to G.S., D.M.B., and J.B.C. J.Y. was also partially supported by a USDA NIFA AFRI Foundation grant no. 2015-67015-22970. J.H.S. was supported by NIH National Research Service Award T32 GM07215.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. The USDA is an equal opportunity provider and employer.
We thank Paul J. Weimer for conversations related to the design of this study and for his careful reading of and suggestions for the manuscript.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Weimer PJ. 2015. Redundancy, resilience, and host specificity of the ruminal microbiota: implications for engineering improved ruminal fermentations. Front Microbiol 6:296. doi: 10.3389/fmicb.2015.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bickhart DM, Weimer PJ. 2018. Symposium review: host-rumen microbe interactions may be leveraged to improve the productivity of dairy cows. J Dairy Sci 101:7680–7689. doi: 10.3168/jds.2017-13328. [DOI] [PubMed] [Google Scholar]
- 3.Neumann AP, Suen G. 2018. The phylogenomic diversity of herbivore-associated Fibrobacter spp. is correlated to lignocellulose-degrading potential. mSphere 3:e00593-18. doi: 10.1128/mSphere.00593-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weimer PJ, Stevenson DM, Mantovani HC, Man S. 2010. Host specificity of the ruminal bacterial community in the dairy cow following near-total exchange of ruminal contents. J Dairy Sci 93:5902–5912. doi: 10.3168/jds.2010-3500. [DOI] [PubMed] [Google Scholar]
- 5.Li F, Li C, Chen Y, Liu J, Zhang C, Irving B, Fitzsimmons C, Plastow G, Guan LL. 2019. Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle. Microbiome 7:92. doi: 10.1186/s40168-019-0699-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace RJ, Rooke JA, McKain N, Duthie CA, Hyslop JJ, Ross DW, Waterhouse A, Watson M, Roehe R. 2015. The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics 16:1–14. doi: 10.1186/s12864-015-2032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henderson G, Cox F, Kittelmann S, Miri VH, Zethof M, Noel SJ, Waghorn GC, Janssen PH. 2013. Effect of DNA extraction methods and sampling techniques on the apparent structure of cow and sheep rumen microbial communities. PLoS One 8:e74787. doi: 10.1371/journal.pone.0074787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paz HA, Anderson CL, Muller MJ, Kononoff PJ, Fernando SC. 2016. Rumen bacterial community composition in Holstein and Jersey cows is different under same dietary condition and is not affected by sampling method. Front Microbiol 7:1206. doi: 10.3389/fmicb.2016.01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kittelmann S, Kirk MR, Jonker A, McCulloch A, Janssen PH. 2015. Buccal swabbing as a noninvasive method to determine bacterial, archaeal, and eukaryotic microbial community structures in the rumen. Appl Environ Microbiol 81:7470–7483. doi: 10.1128/AEM.02385-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tapio I, Shingfield KJ, McKain N, Bonin A, Fischer D, Bayat AR, Vilkki J, Taberlet P, Snelling TJ, Wallace RJ. 2016. Oral samples as non-invasive proxies for assessing the composition of the rumen microbial community. PLoS One 11:e0151220. doi: 10.1371/journal.pone.0151220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindström T, Redbo I. 2000. Effect of feeding duration and rumen fill on behaviour in dairy cows. Appl Anim Behav Sci 70:83–97. doi: 10.1016/S0168-1591(00)00148-9. [DOI] [PubMed] [Google Scholar]
- 12.Beauchemin KA. 2018. Invited review: current perspectives on eating and rumination activity in dairy cows. J Dairy Sci 101:4762–4784. doi: 10.3168/jds.2017-13706. [DOI] [PubMed] [Google Scholar]
- 13.Ramšak A, Peterka M, Tajima K, Martin JC, Wood J, Johnston MEA, Aminov RI, Flint HJ, Avguštin G. 2000. Unravelling the genetic diversity of ruminal bacteria belonging to the CFB phylum. FEMS Microbiol Ecol 33:69–79. doi: 10.1111/j.1574-6941.2000.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 14.Creevey CJ, Kelly WJ, Henderson G, Leahy SC. 2014. Determining the culturability of the rumen bacterial microbiome. Microb Biotechnol 7:467–479. doi: 10.1111/1751-7915.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Mulder T, Goossens K, Peiren N, Vandaele L, Haegeman A, de Tender C, Ruttink T, van de Wiele T, de Campeneere S. 2016. Exploring the methanogen and bacterial communities of rumen environments: solid adherent, fluid and epimural. FEMS Microbiol Ecol 93:fiw251. doi: 10.1093/femsec/fiw251. [DOI] [PubMed] [Google Scholar]
- 16.Jewell KA, McCormick CA, Odt CL, Weimer PJ, Suen G. 2015. Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Appl Environ Microbiol 81:4697–4710. doi: 10.1128/AEM.00720-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duffield T, Plaizier JC, Fairfield A, Bagg R, Vessie G, Dick P, Wilson J, Aramini J, McBride B. 2004. Comparison of techniques for measurement of rumen pH in lactating dairy cows. J Dairy Sci 87:59–66. doi: 10.3168/jds.S0022-0302(04)73142-2. [DOI] [PubMed] [Google Scholar]
- 18.Ji S, Zhang H, Yan H, Azarfar A, Shi H, Alugongo G, Li S, Cao Z, Wang Y. 2017. Comparison of rumen bacteria distribution in original rumen digesta, rumen liquid and solid fractions in lactating Holstein cows. J Anim Sci Biotechnol 8:16. doi: 10.1186/s40104-017-0142-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warner A. 1966. Diurnal changes in the concentrations of micro-organisms in the rumens of sheep fed limited diets once daily: with an appendix on the kinetics of rumen microbes and flow. J Gen Microbiol 45:213–235. doi: 10.1099/00221287-45-2-213. [DOI] [PubMed] [Google Scholar]
- 20.Leedle JA, Bryant MP, Hespell RB. 1982. Diurnal variations in bacterial numbers and fluid parameters in ruminal contents of animals fed low- or high-forage diets. Appl Environ Microbiol 44:402–412. doi: 10.1128/AEM.44.2.402-412.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamra DN, Sawal RK, Pathak NN, Kewalramani N, Agarwal N. 1991. Diurnal variation in ciliate protozoa in the rumen of black buck (Antilope cervicapra) fed green forage. Lett Appl Microbiol 13:165–167. doi: 10.1111/j.1472-765X.1991.tb00598.x. [DOI] [Google Scholar]
- 22.Schirmann K, Chapinal N, Weary DM, Heuwieser W, von Keyserlingk MA. 2012. Rumination and its relationship to feeding and lying behavior in Holstein dairy cows. J Dairy Sci 95:3212–3217. doi: 10.3168/jds.2011-4741. [DOI] [PubMed] [Google Scholar]
- 23.McAllister TA, Dunière L, Drouin P, Xu S, Wang Y, Munns K, Zaheer R. 2018. Silage review: using molecular approaches to define the microbial ecology of silage. J Dairy Sci 101:4060–4074. doi: 10.3168/jds.2017-13704. [DOI] [PubMed] [Google Scholar]
- 24.Breiman L. 2001. Random forests. Mach Learn 45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 25.Chen X, Ishwaran H. 2012. Random forests for genomic data analysis. Genomics 99:323–329. doi: 10.1016/j.ygeno.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. 2002. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357. doi: 10.1613/jair.953. [DOI] [Google Scholar]
- 27.Blagus R, Lusa L. 2013. SMOTE for high-dimensional class-imbalanced data. BMC Bioinformatics 14:106. doi: 10.1186/1471-2105-14-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landis JR, Koch GG. 1977. The measurement of observer agreement for categorical data. Biometrics 33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 29.Landis JR, Koch GG. 1977. An application of hierarchical kappa-type statistics in the assessment of majority agreement among multiple observers. Biometrics 33:363–374. doi: 10.2307/2529786. [DOI] [PubMed] [Google Scholar]
- 30.Li M, Penner GB, Hernandez-Sanabria E, Oba M, Guan LL. 2009. Effects of sampling location and time, and host animal on assessment of bacterial diversity and fermentation parameters in the bovine rumen. J Appl Microbiol 107:1924–1934. doi: 10.1111/j.1365-2672.2009.04376.x. [DOI] [PubMed] [Google Scholar]
- 31.Lv X, Chai J, Diao Q, Huang W, Zhuang Y, Zhang N. 2019. The signature microbiota drive rumen function shifts in goat kids introduced to solid diet regimes. Microorganisms 7:516. doi: 10.3390/microorganisms7110516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clemmons BA, Martino C, Powers JB, Campagna SR, Voy BH, Donohoe DR, Gaffney J, Embree MM, Myer PR. 2019. Rumen bacteria and serum metabolites predictive of feed efficiency phenotypes in beef cattle. Sci Rep 9:19265. doi: 10.1038/s41598-019-55978-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li F, Henderson G, Sun X, Cox F, Janssen PH, Guan LL. 2016. taxonomic assessment of rumen microbiota using total RNA and targeted amplicon sequencing approaches. Front Microbiol 7:987. doi: 10.3389/fmicb.2016.00987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevenson DM, Weimer PJ. 2007. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol 75:165–174. doi: 10.1007/s00253-006-0802-y. [DOI] [PubMed] [Google Scholar]
- 35.Skarlupka JH, Kamenetsky ME, Jewell KA, Suen G. 2019. The ruminal bacterial community in lactating dairy cows has limited variation on a day-to-day basis. J Anim Sci Biotechnol 10:66. doi: 10.1186/s40104-019-0375-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Good IJ. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264. doi: 10.2307/2333344. [DOI] [Google Scholar]
- 42.Shannon CE. 1948. A mathematical theory of communication. Bell Syst Tech J 27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 43.Simpson EH. 1949. Measurement of diversity. Nature 163:688–688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- 44.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of Southern Wisconsin. Ecol Monogr 27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 45.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2016. vegan: Community Ecology Package. https://CRAN.R-project.org/package=vegan.
- 46.Breslow NE, Clayton DG. 1993. Approximate inference in generalized linear mixed models. J Am Stat Assoc 88:9. doi: 10.2307/2290687. [DOI] [Google Scholar]
- 47.Zuur A, Ieno EN, Walker N, Saveliev AA, Smith GM. 2009. Mixed effects models and extensions in ecology with R. J Stat Softw 32:1–3. [Google Scholar]
- 48.Xia Y, Sun J, Chen D-G. 2018. Statistical analysis of microbiome data with R. Springer, Singapore. [Google Scholar]
- 49.Delignette-Muller ML, Dutang C. 2015. fitdistrplus: an R package for fitting distributions. J Stat Softw 64:1–34. doi: 10.18637/jss.v064.i04. [DOI] [Google Scholar]
- 50.Venables WN, Ripley BD. 2002. Modern applied statistics with S, 4th ed Springer, New York, NY. [Google Scholar]
- 51.Lenth RV. 2016. Least-squares means: the R package lsmeans. J Stat Softw 69:1–33. doi: 10.18637/jss.v069.i01. [DOI] [Google Scholar]
- 52.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York. [Google Scholar]
- 53.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 54.Salazar G. 2018. EcolUtils: utilities for community ecology analysis. R package version 0.1. https://github.com/GuillemSalazar/EcolUtils.
- 55.Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2:18–22. [Google Scholar]
- 56.Kuhn M. 2020. caret: classification and regression training. R package version 6.0-86. https://CRAN.R-project.org/package=caret.
- 57.Song L, Maintainer PL, Langfelder P. 2015. randomGLM: random general linear model prediction. R package version 1.02–1. https://CRAN.R-project.org/package=randomGLM.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence reads from all samples analyzed in this study are available on the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/) under Bioproject accession number PRJNA623113.