Dissimilatory nitrate/nitrite reduction to ammonium (DNRA) is an anaerobic microbial pathway that competes with denitrification for common substrates NO3− and NO2−. Unlike denitrification, which leads to nitrogen loss and N2O emission, DNRA reduces NO3− and NO2− to NH4+, a reactive nitrogen compound with a higher tendency to be retained in the soil matrix. Therefore, stimulation of DNRA has often been proposed as a strategy to improve fertilizer efficiency and reduce greenhouse gas emissions. Such attempts have been hampered by lack of insights into soil DNRA bacterial ecophysiology. Here, we have developed a new screening method for isolating DNRA-catalyzing organisms from agricultural soils without apparent DNRA activity. Physiological characteristics of six DNRA isolates were closely examined, disclosing a previously overlooked link between NO3− repression of NO2−-to-NH4+ reduction and the C-to-N ratio regulation of DNRA activity, which may be a key to understanding why DNRA activity is rarely observed at substantial levels in nitrogen-rich agricultural soils.
KEYWORDS: agricultural soil, DNRA, denitrification, nitrogen cycle, nitrous oxide
ABSTRACT
Dissimilatory nitrate/nitrite reduction to ammonium (DNRA) has recently regained attention as a nitrogen retention pathway that may potentially be harnessed to alleviate nitrogen loss resulting from denitrification. Until recently, the ecophysiology of DNRA bacteria inhabiting agricultural soils has remained largely unexplored, due to the difficulty in targeted enrichment and isolation of DNRA microorganisms. In this study, >100 DNRA bacteria were isolated from NO3−-reducing anoxic enrichment cultures established with rice paddy soils using a newly developed colorimetric screening method. Six of these isolates, each assigned to a different genus, were characterized to improve the understanding of DNRA physiology. All the isolates carried nrfA and/or nirB, and the Bacillus sp. strain possessed a clade II nosZ gene conferring the capacity for N2O reduction. A common prominent physiological feature observed in the isolates was NO2− accumulation before NH4+ production, which was further examined with Citrobacter sp. strain DNRA3 (possessing nrfA and nirB) and Enterobacter sp. strain DNRA5 (possessing only nirB). Both isolates showed inhibition of NO2−-to-NH4+ reduction at submillimolar NO3− concentrations and downregulation of nrfA or nirB transcription when NO3− was being reduced to NO2−. In batch and chemostat experiments, both isolates produced NH4+ from NO3− reduction when incubated with excess organic electron donors, while incubation with excess NO3− resulted in NO2− buildup but no substantial NH4+ production, presumably due to inhibitory NO3− concentrations. This previously overlooked link between NO3− repression of NO2−-to-NH4+ reduction and the C-to-N ratio regulation of DNRA activity may be a key mechanism underpinning denitrification-versus-DNRA competition in soil.
IMPORTANCE Dissimilatory nitrate/nitrite reduction to ammonium (DNRA) is an anaerobic microbial pathway that competes with denitrification for common substrates NO3− and NO2−. Unlike denitrification, which leads to nitrogen loss and N2O emission, DNRA reduces NO3− and NO2− to NH4+, a reactive nitrogen compound with a higher tendency to be retained in the soil matrix. Therefore, stimulation of DNRA has often been proposed as a strategy to improve fertilizer efficiency and reduce greenhouse gas emissions. Such attempts have been hampered by lack of insights into soil DNRA bacterial ecophysiology. Here, we have developed a new screening method for isolating DNRA-catalyzing organisms from agricultural soils without apparent DNRA activity. Physiological characteristics of six DNRA isolates were closely examined, disclosing a previously overlooked link between NO3− repression of NO2−-to-NH4+ reduction and the C-to-N ratio regulation of DNRA activity, which may be a key to understanding why DNRA activity is rarely observed at substantial levels in nitrogen-rich agricultural soils.
INTRODUCTION
Nitrogen is an essential element for plant growth. Today, the Haber-Bosch process, used primarily for the production of nitrogen fertilizers, is singled out as the largest energy-consuming industrial process, with global energy consumption summing up to 2.5% of the total energy consumed across the globe, and naturally, nitrogen is one of the largest sources of greenhouse gases (1, 2). The increased nitrogen flux in the soil and aquatic environments as a consequence of fertilizer application to agricultural soils has also led to aggravation of various nitrogen-related environmental problems, e.g., enrichment of NO3− in groundwater and harmful algal blooms as a symptom of eutrophication in surface water (3). Thus, mitigation of the “nitrogen dilemma” has been regarded as one of the most pressing issues for environmental sustainability (4).
Despite the environmental consequences, nitrogen is not used efficiently in agroecosystems. Nitrogen fertilizer efficiency, i.e., the proportion of applied fertilizer nitrogen that eventually ends up in crop biomass, rarely exceeds 40% (5). One of the primary nitrogen loss pathways is nitrification followed by denitrification. Both nitrification and denitrification are also the major culprits of N2O emissions. Several different strategies have been devised to limit nitrogen loss and N2O emissions from soil systems, including the use of nitrification inhibitors and slow-release fertilizers (6, 7). Another possible strategy recently proposed for improved soil nitrogen management is to outcompete the denitrification pathway with the nitrogen-retaining process of dissimilatory nitrate/nitrite reduction to ammonium (DNRA) (8–11). The reduction of NO3− to NH4+ via DNRA also increases the tendency of N to be retained in the soil matrix, thereby reducing NO3− leaching, another substantial nitrogen loss avenue in agricultural soils (12). Dissimilatory nitrate/nitrite reduction to ammonium is catalyzed by the microorganisms carrying cytochrome c552 nitrite reductases (encoded by nrfA genes) or NADH-dependent nitrite reductases (encoded by nirB genes), often incorrectly generalized as assimilatory nitrite reductases (13). According to the current limited knowledge, NO2−-to-NH4+ reduction may serve as the electron acceptor reaction for respiration (respiratory DNRA) or the electron dump for NADH regeneration in fermentation of complex organics (fermentative DNRA) (14–16).
Denitrification and DNRA pathways compete for common substrates, NO3−/NO2−, and thus, stimulating one would repress the other (17, 18). Previous investigations suggested that DNRA is favored in environments with high organic carbon (C) content and a limiting supply of nitrogenous electron acceptors (NO3−/NO2−) (19–21). This hypothesis was further corroborated by recent laboratory experiments with microbial enrichments and axenic microbial cultures harboring both denitrification and DNRA pathways; however, conflicting observations (e.g., in experiments with Intrasporangium calvum and Deltaproteobacteria-dominated wastewater enrichments) suggest the possibility that the observed correlation between DNRA activity and the C-to-N ratio (the ratio of C in bioavailable organic compounds to N in NO3−/NO2− in this context) may be circumstantial (18, 22–24).
The potential significance of DNRA as a key reaction determining the fate of reactive nitrogen in the environment has been considered for decades, albeit lacking sufficient evidence (25–27). Observation of dominance of DNRA over denitrification, i.e., higher NH4+ production than N2O-plus-N2 production from NO3−/NO2− reduction, has been limited to several specific highly reduced marine environments (28, 29). Nevertheless, recovery of 15NH4+ from 15NO3− reduction in both in situ column studies and ex situ soil incubation experiments supported the presence of DNRA activity in soil environments (8, 17). Furthermore, the abundance of nrfA genes in several sequenced soil metagenomes suggested that microbes capable of DNRA activity may be abundant in soil communities (30, 31). Few attempts have been made to isolate and examine DNRA organisms from soils, however, presumably due to the relative insignificance of the contribution of DNRA in nitrogen-rich soils, e.g., fertilized agricultural soils, where the fate of nitrogen is most relevant to the global biogeochemical cycle (8, 17, 32).
In this era dominated by molecular microbial ecology and meta-omics, the importance of culture-based studies is often overlooked; however, meta-omics data can be effectively interpreted along ecological and biogeochemical contexts only with sound understanding of microbial physiology and metabolism (33). In this aspect, that DNRA deserves further culture-based investigation for improved understanding of soil nitrogen fate in agroecosystems is beyond doubt, even with broad availability of molecular tools and bioinformatics pipelines for culture-independent analyses of DNRA-related functional genes, e.g., nrfA and nirB (30, 34). The major bottleneck in investigation of soil DNRA ecophysiology, however, has been the difficulty in efficiently securing diverse DNRA isolates from soils, where the contribution of DNRA to anoxic NO3−/NO2− turnover is, in most cases, minor (9). Due to this difficulty, investigations of DNRA ecophysiology have relied on extrapolation of findings from experiments with limited numbers of isolates, mostly acquired from nonsoil environments (16, 18, 35–37). Furthermore, many of these isolates had been aerobically isolated and cultured for decades in laboratory settings before they were recognized as being capable of DNRA (16, 18, 37–40). Thus, the use of these isolates as representatives of soil DNRA bacteria has received criticism as lacking ecological relevance to the fate of NO3− in anoxic agricultural soils.
To address this issue of ecological relevance in examining soil DNRA ecophysiology, a less onerous and time-consuming method for the isolation of DNRA bacteria in denitrification-dominant agricultural soils was needed. Here, a rapid, inexpensive, high-throughput screening method was developed utilizing the well-established salicylate method for NH4+ detection and quantification (41). Reductive transformation of NO3− was examined with six DNRA organisms isolated from rice paddy soils using this novel screening method. The isolates were taxonomically assigned as Bacillus (belonging to the Firmicutes phylum) and Aeromonas, Citrobacter, Enterobacter, Klebsiella, and Shewanella (belonging to the Proteobacteria phylum) at the genus level. Their most obvious common physiological feature was NO3− inhibition of NO2−-to-NH4+ reduction, which had also been observed previously with nrfA- and-nirB-harboring organisms Escherichia coli and Bacillus vireti (35, 42). With a series of batch and continuous culture experiments, we identified the NO3− repression of DNRA activity as one of the mechanisms underpinning the widely acknowledged but controversial C-to-N ratio regulation of DNRA-versus-denitrification competition.
RESULTS
Isolation of DNRA bacteria from denitrification-dominant agricultural soil.
Out of 192 colonies each from lactate- and glucose-amended rice paddy soil enrichments, both with negligible NH4+ production from NO3− reduction, 126 and 12 colonies tested DNRA positive, respectively (Fig. S1 in the supplemental material). Sequencing of the 16S rRNA gene amplicons of the positive colonies (30 randomly selected colonies from the lactate-amended enrichment and all 12 colonies from the glucose-amended enrichment) identified six bacterial genera: Aeromonas, Bacillus, and Shewanella (lactate-amended enrichment), Enterobacter and Klebsiella (glucose-amended enrichment), and Citrobacter (both enrichments) (Fig. S2). The DNRA activities in six of these isolates, each randomly selected from the isolates belonging to a unique genus, were further examined.
Identification of functional genes relevant to dissimilatory nitrogen reduction.
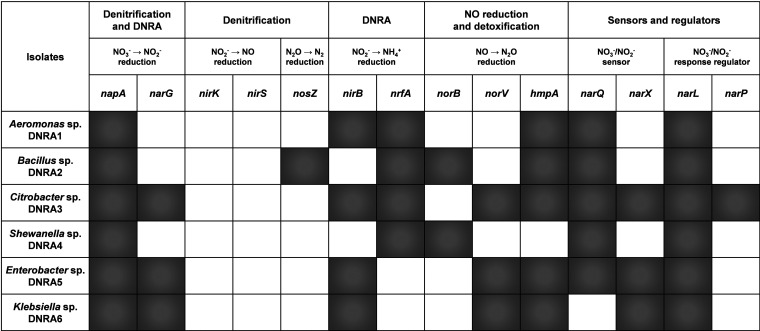
The draft genomes of the six DNRA isolates were constructed from HiSeq sequencing reads (sequencing statistics are presented in Table S2). The functional genes potentially relevant to turnover of reactive nitrogen species or regulation of nitrogen metabolism were then analyzed in these draft genomes (Fig. 1, Table S3). The isolates that originated from lactate-enriched cultures all possessed nrfA genes, encoding NH4+-forming cytochrome c552 nitrite reductases. The two isolates from glucose-enriched cultures lacked nrfA genes but possessed nirB genes, suggesting that NirB-type nitrite reductase was responsible for dissimilatory reduction of NO2− to NH4+ in these organisms. Aeromonas sp. strain DNRA1 and Citrobacter sp. strain DNRA3 possessed both nrfA and nirB. All six isolates had napA in their genomes, and Citrobacter sp. DNRA3, Enterobacter sp. strain DNRA5, and Klebsiella sp. strain DNRA6 carried narG, indicating the genomic potential of these organisms to reduce NO3− to NO2−. Neither nirK nor nirS (both of which encode NO-forming nitrite reductases) was present in any of the isolates; however, a clade II nosZ gene was identified in the draft genome of Bacillus sp. strain DNRA2, suggesting N2O-reducing capability. nosD and nosL genes, encoding a maturation protein and a copper chaperone, respectively, were identified in the draft genome of Shewanella sp. strain DNRA4, possibly as vestiges of a functional nos operon. Bacillus sp. DRNA2 and Shewanella sp. DNRA4 possessed norB genes encoding quinol-dependent nitric oxide reductases, and norV and/or hmpA genes that encode enzymes involved in detoxification of NO were recovered in all of the sequenced draft genomes.
FIG 1.
Functional genes identified in the draft genomes of the six DNRA bacteria that are potentially relevant to turnover of reactive nitrogen species or regulation of dissimilatory nitrogen metabolism. The genes that were recovered in the draft genome are represented as shaded boxes. Detailed information that includes the lists of the accessory genes and their closest BLAST hits is provided in Table S3 in the supplemental material.
NO3− reduction by the DNRA isolates.
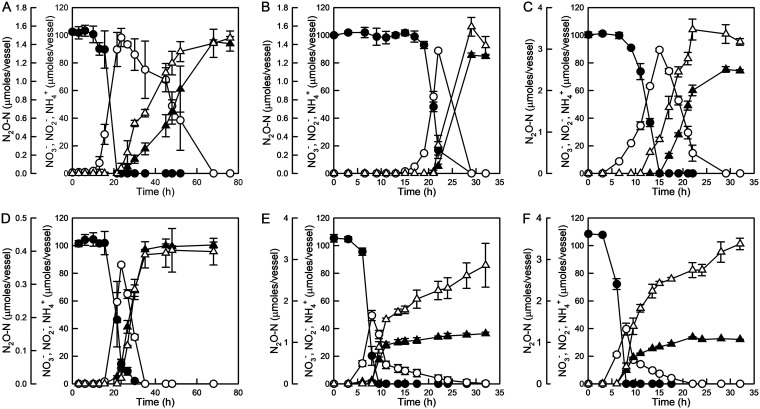
Reductive transformation of NO3− was observed with the axenic cultures of the six DNRA isolates with or without 10% C2H2 in the headspace (Fig. 2, Fig. S3 and S4). The six isolates completely reduced the initially supplemented NO3− to NH4+ via NO2− with lactate or glucose as the source of electrons. Lactate-coupled NO3− reduction in Aeromonas sp. DNRA1, Bacillus sp. DNRA2, Citrobacter sp. DNRA3, and Shewanella sp. DNRA4 resulted in near-stoichiometric production of NH4+ from NO3−. Reduction of NO3− to NH4+ was also observed in Enterobacter sp. DNRA5 and Klebsiella sp. DNRA6 grown on glucose; however, the NH4+ produced only amounted to 36.3 ± 1.1 (mean ± standard deviation [SD]) and 32.1 ± 0.2 μmol, respectively, which were less than half of the added NO3−. As the cell densities of the glucose-fed Enterobacter sp. DNRA5 and Klebsiella sp. DNRA6 reached at least 2.5-fold higher than those of the lactate-consuming isolates, the missing nitrogen was likely due to assimilation. Despite the absence of nirK or nirS genes, N2O production was observed in all of the isolates during NO3− reduction when incubated with C2H2. The amounts of N2O produced varied across the isolates, ranging from 0.40 ± 0.06 μmol N2O-N (0.4% of the added NO3−) for Shewanella sp. DNRA4 to 3.5 ± 0.3 μmol N2O-N (3.5% of the added NO3−) for Citrobacter sp. DNRA3. In all six isolates examined, the start of N2O production corresponded with the start of NH4+ production, suggesting that N2O was a by-product of NO2−-to-NH4+ reduction, not NO3−-to-NO2− reduction. Of the six isolates, only Bacillus sp. DNRA2 showed a substantially different N2O-N time series profile when incubated without C2H2 (Fig. S4). The absence of N2O accumulation suggested that N2O consumption occurred simultaneously with DNRA in this clade II nosZ-harboring organism.
FIG 2.
NO3− reduction monitored in 100-ml batch cultures (prepared in sealed 160-ml serum bottles with headspace consisting of 90% N2 and 10% C2H2). (A) Aeromonas sp. DNRA1; (B) Bacillus sp. DNRA2; (C) Citrobacter sp. DNRA3; (D) Shewanella sp. DNRA4; (E) Enterobacter sp. DNRA5; (F) Klebsiella sp. DNRA6. The average values from biological replicates (n = 3) are presented, with error bars representing the standard deviations (●, NO3−; ○, NO2−; ▲, NH4+; ▵, N2O-N).
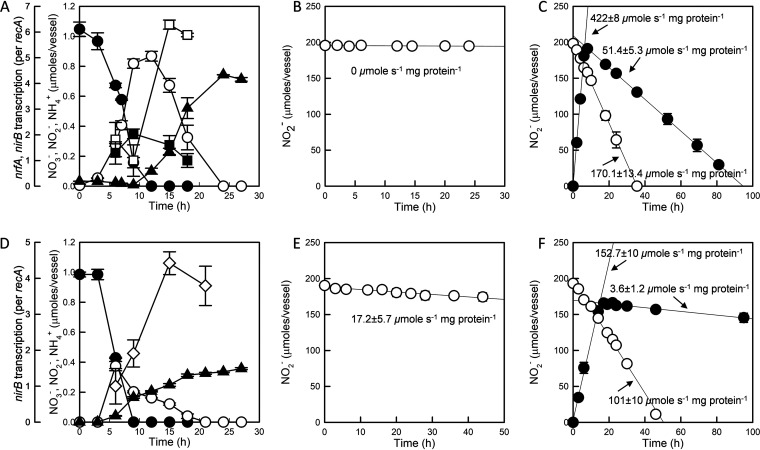
Accumulation of NO2− before reduction to NH4+ was consistently observed in all six isolates. Reduction of NO2− to NH4+ did not commence until >80% of NO3− was consumed, suggesting that NrfA- or NirB-catalyzed NO2−-to-NH4+ reduction was affected by changing NO2− or NO3− concentrations. The DNRA activities of Citrobacter sp. DNRA3 and Enterobacter sp. DNRA5 were further investigated to identify whether possible causality exists between the NO2− or NO3− concentration and DNRA activity (Fig. 3). The transcription levels of nrfA in Citrobacter sp. DNRA3 and nirB in Enterobacter sp. DNRA5 were significantly higher (P < 0.05) after NO3− was depleted than before. Transcription of nrfA in Citrobacter sp. DNRA3 increased significantly (P < 0.05), from an nrfA/recA transcript ratio of 1.0 ± 0.6 at t = 9 h (0.16 ± 0.03 mM NO3− and 0.82 ± 0.03 mM NO2− remaining) to an nrfA/recA transcript ratio of 6.3 ± 0.2 at t = 15 h (0.33 ± 0.08 mM NO2− remaining). No significant change was observed with nirB transcription (from an nirB/recA transcript ratio of 2.04 ± 0.19 at t = 9 h to an nirB/recA transcript ratio of 1.61 ± 0.37 at t = 15 h), suggesting that NirB-type nitrite reductase was irrelevant to respiratory DNRA. Transcription of nirB in Enterobacter sp. DNRA5 followed a trend similar to that of nrfA in Citrobacter sp. DNRA3, increasing significantly from an nirB/recA transcript ratio of 1.0 ± 0.5 at t = 6 h (0.43 ± 0.01 mM NO3− and 0.37 ± 0.03 mM NO2− remaining) to an nirB/recA transcript ratio of 4.4 ± 0.3 at t = 15 h (0.31 ± 0.008 mM NO2− remaining) upon NO3− depletion (P < 0.05). Substrate (NO2−) regulation of transcription was unlikely for either nrfA in Citrobacter sp. DNRA3 or nirB in Enterobacter sp. DNRA5, as the transcription of these genes appeared unresponsive to elevated NO2− concentrations as long as NO3− was present in the medium at >0.15 mM. Thus, the NO3− concentration was the most probable environmental factor that affected the transcription of the genes encoding these DNRA-catalyzing nitrite reductases. The significant differences in the rates of NO2− reduction measured with Citrobacter sp. DNRA3 or Enterobacter sp. DNRA5 cells harvested before and after the NO3− depletion and treated with chloramphenicol also supported the idea that expression of the NH4+-forming nitrite reductases was downregulated by the presence of NO3− (Fig. 3B, C, E, and F). Citrobacter sp. DNRA3 cells extracted before NO3− depletion did not exhibit significant NO2− reduction activity, while the cells extracted after NO3− depletion readily reduced NO2−, at a rate of 170 ± 13 μmol s−1 mg protein−1. NO2− reduction by Enterobacter sp. DNRA5 cells was also ∼6 times higher with the cells harvested after NO3− depletion (101 ± 10 μmol s−1 mg protein−1) than with the cells harvested before NO3− depletion (17.2 ± 5.7 μmol s−1 mg protein−1) (P < 0.05).
FIG 3.
(A and D) Transcription of nrfA (□) and nirB (■) in Citrobacter sp. DNRA3 (A) and nirB (◇) in Enterobacter sp. DNRA5 (D) cells as 1 mM NO3− (●) was reduced to NH4+ (▲) via NO2− (○). (B, C, E, and F) Changes to the amounts of NO2− were monitored in the chloramphenicol-treated resting cultures of Citrobacter sp. DNRA3 (B and C) and Enterobacter sp. DNRA5 (E and F) harvested before (B and E) and after (C and F) NO3− depletion and resuspended in fresh medium containing 2 mM NO2− (○) or 2 mM NO3− (●). All experiments were performed in biological replicates (n = 3), and error bars represent the standard deviations.
In the resting-cell experiments with 2 mM NO3− added to chloramphenicol-treated Citrobacter sp. DNRA3 cells harvested after NO3− depletion, NO2− accumulated up to 1.93 ± 0.04 mM at a rate of 422 ± 8 μmol s−1 mg protein−1 before it was consumed at a rate of 51.4 ± 5.3 μmol s−1 mg protein−1 (Fig. 3C). The negligible NO2− reduction activity before NO3− depletion suggested an additional NO3−-mediated inhibitory mechanism in NrfA-type nitrite reductase activity apart from transcriptional regulation of the nrfA gene. Such repression of NO2− reduction activity by the presence of NO3− was not observed in the parallel experiment performed with Enterobacter sp. DNRA5 (lacking nrfA) and presumably utilizing NirB-type nitrite reductase (Fig. 3F).
DNRA reaction at various C-to-N ratios in batch and continuous cultivation.
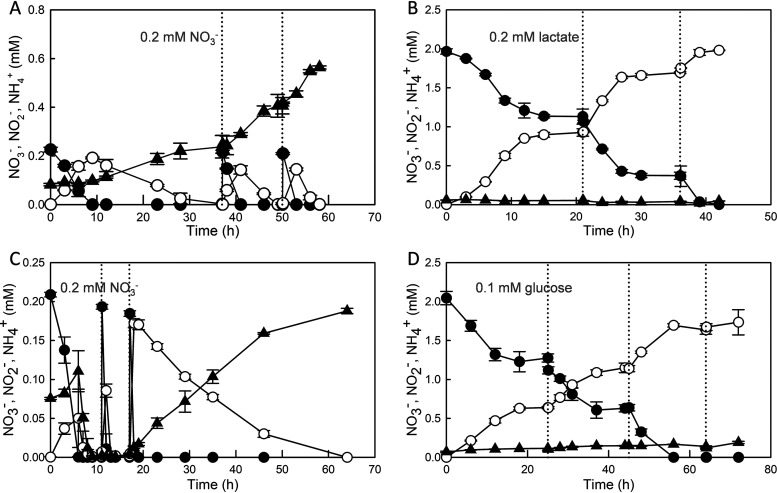
Citrobacter sp. DNRA3 and Enterobacter sp. DNRA5 were grown in batch and continuous cultures, each with two different C-to-N ratios, and NO3− reduction was monitored to investigate whether the generally perceived positive correlation between C-to-N ratio and DNRA activity may be related to the NO3− repression of NO2−-to-NH4+ reduction (Fig. 4 and 5). When grown at the initial C-to-N ratio of 75 in batch cultures, Citrobacter sp. DNRA3 produced NH4+ from NO3− reduction, and each addition of 20 μmol NO3− resulted in near-stoichiometric turnover to NH4+. In contrast, growth of Citrobacter sp. DNRA3 at the initial C-to-N ratio of 0.3 did not result in a significant increase in NH4+ concentration but did lead to stoichiometric NO2− accumulation, as NO3− reduction had produced 0.87 ± 0.06 mM NO2− when the initial reaction stopped at t = 16 h due to depletion of lactate. Reduction of NO3− to NO2− resumed after replenishment with 0.2 mM lactate at t = 21 h. Under this low C-to-N ratio incubation condition, NO3− was present in the culture medium throughout incubation, and the presence of NO3− was likely the reason for the absence of sensu stricto DNRA activity.
FIG 4.
NO3− reduction observed with batch cultures of Citrobacter sp. DNRA3 (A and B) and Enterobacter sp. DNRA5 (C and D) prepared with two different initial C-to-N ratios. The high C-to-N conditions were prepared with 0.2 mM NO3− and 5 mM lactate (A) or 2.5 mM glucose (C), and NO3− was replenished to 0.2 mM upon NO3−/NO2− depletion. The low C-to-N conditions were prepared with 2.0 mM NO3− and 0.2 mM lactate (B) or 0.1 mM glucose (D), and the carbon sources were replenished when NO3−/NO2− reduction stopped. The dotted lines denote the time points where the limiting nutrients were replenished. The average values from biological replicates (n = 3) are presented, with error bars representing the standard deviations (●, NO3−; ○, NO2−; ▲, NH4+).
FIG 5.
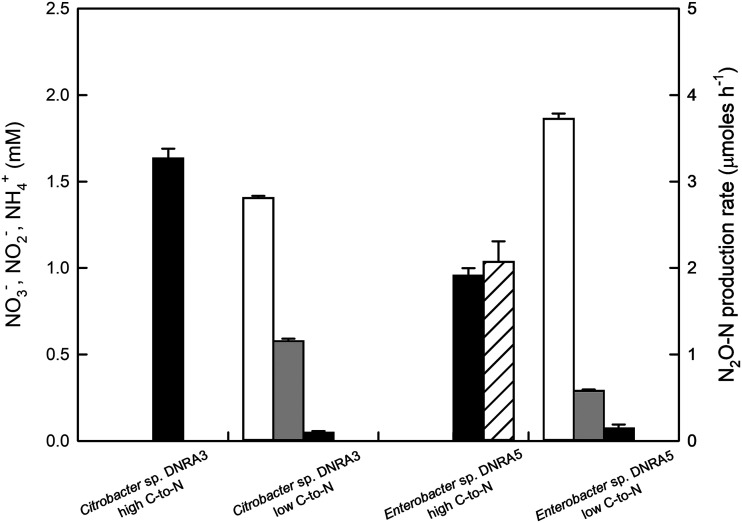
Steady-state concentrations of NO3− (white bars), NO2− (gray bars), and NH4+ (black bars) in electron acceptor limiting (high C-to-N ratio) and electron donor limiting (low C-to-N ratio) chemostat cultures of Citrobacter sp. DNRA3 and Enterobacter sp. DNRA5. The N2O-N production rate (hatched bars) is also presented for Enterobacter sp. DNRA5 cultivated under the electron acceptor limiting condition, which was the only reactor culture with observed N2O production. The average values of the three measurements taken at 6-h intervals are presented, with error bars representing the standard deviations.
Enterobacter sp. DNRA5 incubated on glucose at a C-to-N ratio of 75 produced a significant amount of NH4+ only after the initially added glucose (2.19 ± 0.8 mM) was fully consumed, suggesting that substantial portions of NO3− and its reduction products, NO2− and NH4+, were assimilated. Upon the third addition of 0.2 mM NO3−, with no glucose remaining in the medium, the sequential NO3−-to-NO2−-to-NH4+ reduction was stoichiometric, suggesting that NirB-catalyzed NO2−-to-NH4+ reduction was coupled to oxidation of the fermentation products. At the low C-to-N ratio, where the culture medium was replenished with 0.1 mM glucose upon a halt in NO3− reduction, the time series profiles of the N species concentrations were indistinguishable from those of Citrobacter sp. DNRA3, save for the imperfect stoichiometry between consumed NO3− and produced NO2− and the modest, albeit significant, increase in NH4+ concentration from 0.07 ± 0.01 mM at t = 0 h to 0.19 ± 0.01 mM at t = 72 h. The modest production of NH4+ was in line with the reduced but still significant NO2− reduction rate observed in the resting-cell cultures of Enterobacter sp. DNRA5 extracted before NO3− depletion.
A continuous culture of Citrobacter sp. DNRA3 that was fed with medium carrying 10 mM lactate and 2 mM NO3− (C-to-N ratio of 15), after attaining steady state, contained 1.68 ± 0.09 mM NH4+ as the only dissolved inorganic nitrogen, indicating that NO3− and NO2− were readily reduced in the chemostat. When the reactor was fed with medium carrying 0.2 mM lactate and 2 mM NO3− (C-to-N ratio of 0.3), 1.41 ± 0.04 mM NO3− remained in the medium at steady state, due to carbon limitation. That NO2− was the major product of NO3− reduction (0.58 ± 0.04 mM at steady state) and the NH4+ concentration did not differ significantly from the concentration in the fresh medium (P > 0.05) indicated that DNRA did not proceed further beyond NO2−. Similarly, with Enterobacter sp. DNRA5, significant NH4+ formation was observed only in a continuous culture operated under the electron-acceptor-limiting condition, i.e., at a C-to-N ratio of 15. The steady-state NH4+ concentration was 0.95 ± 0.04 mM in this chemostat. In the continuous culture operated under the electron-donor-limiting condition, NO2− was the only dissolved nitrogen species with a significantly higher concentration than in the influent medium. Production of N2O (2.07 ± 0.24 μmol h−1) was observed only in the high C-to-N chemostat of Enterobacter sp. DNRA5. The absence of significant NO2−-to-NH4+ reduction in the low C-to-N ratio batch and chemostat cultures, regardless of whether mediated by NrfA-type or NirB-type nitrite reductase, could be best explained as the inhibitory effect of NO3−.
DISCUSSION
The soil DNRA isolates newly acquired with the screening method developed in this study were assigned to six genera according to their 16S rRNA gene sequences. Several of these genera have been previously confirmed to include strains capable of carrying out DNRA (Bacillus, Citrobacter, Enterobacter, and Klebsiella). The list also included a genus generally perceived as a marine organism (Shewanella) and a genus without physiologically confirmed DNRA activity (Aeromonas) (36, 43–45). The genomic analyses of six DNRA isolates, one from each of these genera, confirmed that the possession of nrfA or nirB is necessary for a DNRA phenotype (35, 37). All four isolates utilizing lactate as the electron donor were of the nrfA genotype. Enterobacter sp. DNRA5 and Klebsiella sp. DNRA6 that lacked nrfA failed to grow on lactate under a NO2−-reducing condition, suggesting that NrfA-type nitrite reductase is needed for respiratory NO2− reduction to NH4+. Thus, physiological functions of NirB-catalyzed NO2−-to-NH4+ reduction in these organisms may be NAD+ regeneration for fermentation, detoxification of NO2−, and/or assimilatory reduction, as previously suggested (15, 37). Neither nirS nor nirK was found in any of the sequenced draft genomes; however, an nosZ gene was recovered in the genome of the nrfA-possessing Bacillus sp. DNRA2. Observation of NO3− reduction with and without C2H2 confirmed N2O reduction activity in this isolate amid active DNRA, which was probably catalyzed by the NosZ encoded by this gene.
The most prominent common phenotype of the DNRA isolates was NO2− accumulation before NH4+ production, suggesting NO3− repression of NO2−-to-NH4+ reduction. This phenotype has been consistently observed in previously studied DNRA bacteria (35, 42, 46). These previous studies attributed the NO3−-induced repression to transcriptional regulation involving NO3− sensor proteins NarQ and NarX and the transcript abundances of nrfA in Escherichia coli, and transcripts of both nrfA and nirB in B. vireti were significantly lower when the culture was supplied with higher NO3− concentrations, supporting their claims. In agreement with these previous studies, nrfA in Citrobacter sp. DNRA3 and nirB in Enterobacter sp. DNRA5 exhibited at least 4.4-fold higher transcription after NO3− depletion than before (P < 0.05). Furthermore, the results from the resting-cell experiments with these isolates showed clear indications that the presence of NO3− at submillimolar concentrations was sufficient to inhibit activities of expressed NrfA-type nitrite reductase. However, whether the apparent inhibition was due to the redirection of electron flow analogous to what was observed with NosZ-catalyzed N2O reduction in the presence of O2 or to inhibition of the NrfA enzyme itself cannot be determined and is outside the scope of the current study (47). In denitrifiers, such NO3−-mediated repression of dissimilatory NO2− reduction, either via transcription regulation or enzyme inhibition, has not yet been reported, and near-stoichiometric NO2− accumulation during NO3− reduction has been observed only as isolated cases (48–50). Therefore, as long as NO3− is present in soil matrices harboring diverse denitrifiers and DNRA-catalyzing organisms, NO2− produced from NO3− would be reduced mostly to N2O and N2 via denitrification, with the DNRA phenotype remaining silent.
The environmental physicochemical parameter that has been most frequently associated with DNRA activity is the C-to-N ratio. Multiple experimental evidences from culture-based experiments and field measurements have supported that DNRA is favored at high C-to-N ratios, i.e., electron acceptor limiting conditions, while denitrification is favored at low C-to-N ratios, i.e., electron donor limiting conditions (18, 20, 23). The observations from the incubation of the two isolates at the two different C-to-N ratios indicated that the NO3− repression of NO2−-to-NH4+ reduction activity may actually be directly linked to this C-to-N ratio regulation of DNRA activity in the environment. The C-to-N ratios of soils or sediments are often inversely related to the NO3− contents (51). In soils with low C-to-N ratios, the NO2−-to-NH4+ reduction may thus be deactivated in the DNRA-catalyzing organisms due to the high NO3− contents while NO2−-to-N2O/N2 reduction activity remains intact in denitrifiers cohabiting the ecological niches. Even with an abundant DNRA-catalyzing population, the NO3− fate would still be determined by denitrification in such soils. Thus, what was previously regarded as the effect of the C-to-N ratio on the denitrification-versus-DNRA competition may be, at least in part, explained as the consequence of NO3− inhibition of sensu stricto DNRA (32).
Production of N2O has consistently been observed in nondenitrifying organisms with DNRA phenotypes, with recovery of up to ∼50% of NO3−-N as N2O-N (36, 37, 46, 52). Likewise, all DNRA isolates examined in this study produced N2O during the course of NO3− reduction to NH4+ despite the absence of nirS or nirK genes in their genomes. The previously hypothesized mechanisms of N2O production from DNRA invariably have implicated involvement of NO (37, 42, 46). Considering that norB, norV, and/or hmp genes were identified in the genomes of DNRA isolates, it is plausible to regard NO as the precursor of N2O in these organisms; however, the mechanism leading to NO production from NO3− or NO2− remains unclear. The absence of N2O production before NO3− depletion in all six isolates suggested that direct N2O production from NapA- or NarG-type nitrate reductases was unlikely. The more plausible source of NO would be NO2−-to-NH4+ reduction by NrfA- and NirB-type nitrite reductases, although a reaction mechanism leading to formation of NO as a by-product has not been elucidated for either enzyme (46, 53). The results from the chemostat experiments with Enterobacter sp. DNRA5 further support this hypothesis, as detectable N2O production was observed only under the high C-to-N ratio operating condition where active NO2−-to-NH4+ reduction occurred.
Another noteworthy observation in this study was the absence of detectable N2O production in the NH4+-producing Citrobacter sp. DNRA3 chemostat (under the high C-to-N ratio condition), which was contradictory to the result from batch incubation of the same DNRA isolate. This absence of N2O production from the NO3−-limiting chemostat was in line with the previous observations from chemostat studies of Shewanella loihica strain PV-4 with NO3−/NO2− as the limiting substrate, in that no detectable N2O production was observed from the DNRA-dominant chemostat cultures (18, 54). Furthermore, in a previous study with nrfA-utilizing DNRA isolates of the Citrobacter and Bacillus genera, larger proportions of NO3− were released as N2O when the initial C-to-N ratios were lower, i.e., the NO3− concentrations were higher (36). Together, these observations suggest a positive correlation between NO3−/NO2− concentrations in the surrounding environment and N2O formation by nrfA-utilizing organisms, such that N2O produced from NrfA-mediated DNRA may be negligible in environments with limiting influx of NO3−/NO2−. Also, considering that nosZ-possessing DNRA organisms are often found to be capable of simultaneous N2O consumption with NO2−-to-NH4+ reduction, as observed with Bacillus sp. DNRA2 in this study, the possibility exists that nrfA-type DNRA organisms may function as sinks, rather than sources, of N2O in anoxic environments with consistent but limiting NO3−/NO2− influx.
In summary, close examination of the physiology of the DNRA organisms isolated and screened using the newly developed targeted isolation method substantially enhanced the understanding of DNRA ecophysiology. The analyses of the genomic and physiological features of lactate-oxidizing DNRA phenotypes and glucose-oxidizing DNRA phenotypes evidenced clear distinctions between NrfA-mediated respiratory DNRA and NirB-mediated fermentative DNRA. The NO3− inhibition of NO2−-to-NH4+ reduction observed in both nrfA-type and nirB-type DNRA organisms suggested a plausible mechanistic explanation for the oft-observed C-to-N ratio effects on DNRA-versus-denitrification competition. Significant production of N2O as a by-product of NO2−-to-NH4+ reduction was also confirmed in batch cultures of all six closely examined nrfA- and nirB-type isolates; however, the observations from the chemostat incubation experiments suggested dependence of N2O production associated with NrfA-mediated DNRA, but not NirB-mediated DNRA, on the extracellular NO3−/NO2− concentrations. The number and diversity of DNRA organisms isolated with the new isolation and screening method may appear limited to skeptics; however, the method is easily expandable in volume and is open to modifications incorporating various selective cultivation techniques that may enable the isolation of more phylogenetically and metabolically diverse DNRA organisms, which would include difficult-to-culture microorganisms. Understanding the physiology of more diverse DNRA isolates would be a sensible starting point for developing soil management techniques for enhancing the nitrogen-retaining DNRA pathway.
MATERIALS AND METHODS
Soil sampling and initial characterization.
The agricultural soil used in this study was sampled from an experimental rice paddy located at the Chungnam National University (CNU) agricultural research site in Daejeon, South Korea (36°22′01.6″N, 127°21′14.3″E) in October 2018. Harvesting had been completed and there was no standing water at the time of sampling. Cover soil and plant materials were carefully removed before sampling, and approximately 1 kg of soil at 5 to 30 cm depth from the surface was collected with a stainless steel tubular soil sampler with an inner diameter of 2 cm. The sampled soils were transported to the laboratory in coolers filled with ice and stored at 4°C until use. The physicochemical characteristics of this soil, including the pH, textures, and total carbon and nitrogen contents, were analyzed using standardized protocols (11).
Culture medium and growth conditions.
The minimal salts medium (MSM) for enrichment, isolation, and cultivation of soil DNRA bacteria was prepared by adding, per liter of deionized distilled water, 10 mmol NaCl, 3.24 mmol Na2HPO4, 1.76 mmol KH2PO4, 0.1 mmol NH4Cl, and 1 ml 1,000× trace element stock solution (55). For enrichment and isolation, dehydrated R2A broth (Kisanbio, Seoul, South Korea) was added to the medium as a growth supplement. To minimize the interference of NH4+ derived from mineralization of organic nitrogen in the ensuing DNRA-screening process, the R2A broth concentration in the medium was limited to 6.2 mg liter−1. The pH of the medium was adjusted to 7.0. Pure-culture incubation and experiments were performed with 100 ml medium dispensed to 160-ml serum vials. The serum bottles were flushed with N2 gas (≥99.999%; Special Gas, Inc., Daejeon, South Korea) for 10 min, sealed with black butyl-rubber stoppers (Geo-Microbial Technologies, Inc., Ochelata, OK), and crimped with aluminum crimp seals before autoclaving. The degassed, filter-sterilized, 200× vitamin stock solution was then added to the medium (55). Immediately before inoculation, sodium lactate or glucose was added as the electron donor and organic carbon source and KNO3 as the electron acceptor. Lactate and glucose were chosen as the nonfermentable and fermentable electron donors, respectively, as both substrates had been previously reported to support DNRA reactions (18, 56, 57). Enrichment with acetate as the electron donor was also attempted but failed to yield any DNRA-positive isolates. Thus, acetate was not further considered as a potential electron donor in this study. The culture bottles were incubated with shaking at 150 rpm in the dark at room temperature (25°C). Agar plates were prepared by adding 15 g liter−1 Bacto agar (Becton, Dickinson, Franklin Lakes, NJ) to the liquid medium prepared with elevated concentrations of KNO3 (12 mM) and sodium lactate or glucose (120 mM as C). The 96-well plates for DNRA screening were prepared by distributing 200-μl aliquots of the prepared culture medium to the wells in UV-sterilized 96-well clear flat-bottom microplates (Corning, Inc., Corning, NY). Agar plates and 96-well plates were prepared and incubated in an anaerobic chamber (Coy Laboratory Products, Inc., Grass Lake, MI) with atmosphere consisting of 96% N2 and 4% H2.
High-throughput DNRA phenotype screening.
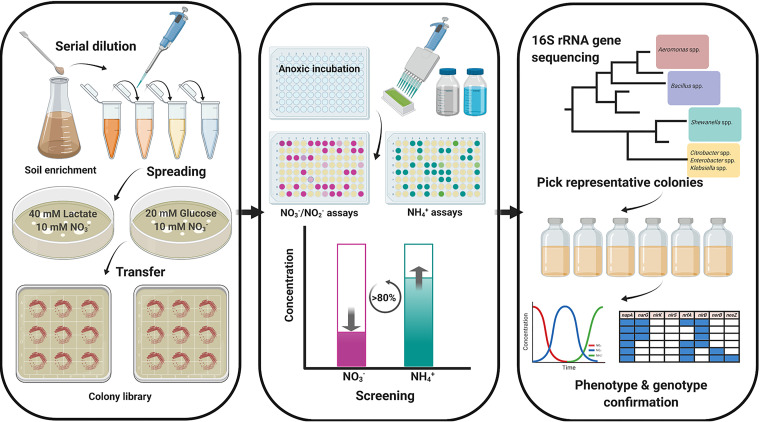
A simple, novel high-throughput screening method was developed in this study for isolating DNRA-catalyzing organisms from agricultural soil enrichments without apparent DNRA activity, i.e., significant NH4+ production from NO3−/NO2− (Fig. 6). Anoxic soil enrichments were prepared in 250-ml Erlenmeyer flasks (Duran Group, Wertheim, Germany) in an anaerobic chamber. Rice paddy soil samples were suspended at a 1:10 (wt/vol) soil-to-medium ratio in 200 ml MSM amended with 2 mM KNO3 and 6.67 mM sodium lactate or 3.33 mM glucose (20 mM total C concentration) and incubated for 2 weeks in the dark without shaking. The aqueous NO3−-N, NO2−-N, and NH4+-N concentrations were measured to confirm depletion of the electron acceptors and to check the extent of DNRA reaction in the enrichments. Serial dilutions of the soil enrichment cultures were spread onto agar plates, and after incubation in the anaerobic chamber, single colonies were picked to inoculate the 96-well plates loaded with fresh medium containing 1 mM NO3− and 3.34 mM lactate or 1.67 mM glucose. The inoculated 96-well plates were covered with an optical adhesive cover (Applied Biosystems, Foster City, CA) to prevent contamination and evaporation of the culture medium and incubated for a week.
FIG 6.
Schematic overview of the high-throughput screening method developed for isolation of DNRA-catalyzing organisms from agricultural soil.
After incubation, 100 μl of the 200-μl culture in each well was transferred to its corresponding position on a new 96-well plate. The absorbances at 600 nm, 660 nm, and 540 nm were determined using a Sunrise microplate reader (Tecan, Männedorf, Switzerland) with one of the duplicated plates to measure the possible interference of cell turbidity in the colorimetric determination of inorganic nitrogen concentrations. This plate was used to screen the wells with increased NH4+ concentrations indicative of DNRA activity, using salicylate-nitroprusside chemistry. To each well, 80 μl of the color reagent (containing 0.2 M sodium hydroxide, 1 M sodium salicylate, and 5.88 mM sodium nitroprusside dihydrate) and 20 μl of 5.1 mM sodium dichloroisocyanurate solution were added sequentially. The absorbance at 660 nm was measured after 30 min of incubation at 25°C, and the wells with optical density values at 660 nm (OD660) that were higher than 1.6 (equivalent to 0.8 mM NH4+) after subtracting the OD660 resulting from cell turbidity were considered positive. The duplicated 96-well plate was used for sequential measurements of NO2−-N and NO3−-N (58). The Griess reagent was added to each well to a total volume of 200 μl, and the absorbance at 540 nm was measured after 30 min of incubation at 25°C for determination of the NO2−-N concentration. The NO3−-N concentration was determined after the initial Griess assay. After reducing NO3− to NO2− by adding, per well, 20 μl of 1% wt/vol vanadium(III) chloride (VCl3; Sigma-Aldrich) prepared in 1 M HCl aqueous solution, the absorbance at 540 nm was measured to obtain the NO3−-plus-NO2− concentration, from which the NO2− concentration was subtracted. The colonies corresponding to the wells with NH4+ concentrations higher than 0.8 mM and with NO2− and NO3− absent were transferred to gridded fresh agar plates and stored at 4°C until use.
Characterization of DNRA isolates.
The partial 16S rRNA genes of these candidate DNRA isolates were amplified with the 27F/1492R primer set and sequenced to identify their phylogenetic affiliations. Based on these initial 16S rRNA sequencing data, one isolate per genus was randomly selected and subjected to further analyses. The DNRA activity of each isolate was confirmed by incubating the isolate with NO3− as the sole electron acceptor in 100 ml MSM in 160-ml serum bottle with and without 10% vol/vol C2H2 in the anoxic N2 headspace. C2H2 inhibits N2O reduction by NosZ, thus enabling observation of N2O production from NO3− and/or NO2− reduction in an NosZ-harboring organism (59). The changes to the dissolved concentrations of NO3−-N, NO2−-N, and NH4+-N, headspace concentrations of N2O, and microbial growth (OD600) were monitored until no further change was observed.
After incubation, a culture sample was collected from each of these six DNRA-positive isolates and the genomic DNA was extracted using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Genome sequencing was performed using the HiSeq 4000 platform (Illumina, San Diego, CA) at Macrogen, Inc. (Seoul, South Korea). Quality trimming and removal of adapter sequences from raw reads was performed using Cutadapt version 2.9, and de novo assembly was done using SPAdes (version 3.14.0) with the minimum contig length set to 200 bp (60, 61). The quality of the draft genomes was assessed using CheckM software version 1.0.18 (62). The NCBI Prokaryotic Genome Annotation Pipeline (PGAP) was used for genome annotation (63). The presence or absence of the nitrogen dissimilation functional genes was double-checked by running the hmmsearch command of HMMER software package version 3.1b1 with hidden Markov models (HMM), downloaded from the FunGene database (http://fungene.cme.msu.edu/), accessed on 14 October 2019 (64). This process ensured that the missing genes were not due to incompleteness of the draft genomes. The genes encoding the regulatory proteins putatively involved in nitrogen dissimilation were also searched in the annotated genome. The draft genome sequences of the six isolates were deposited to NCBI’s GenBank database.
NO3− inhibition of NO2−-to-NH4+ reduction.
Citrobacter sp. DNRA3 carrying single copies of nrfA and nirB genes and Enterobacter sp. DNRA5 carrying nirB genes were subjected to further physiological characterization to examine whether and how NO3− affected NO2−-to-NH4+ reduction. The resting-cell NO3− and NO2− reduction activities were examined with the cells harvested from the two distinct phases of the DNRA reaction, i.e., NO3−-to-NO2− and NO2−-to-NH4+ reduction. The DNRA-catalyzing isolates were grown with 5 mM NO3− as the electron acceptor and 40 mM lactate or 10 mM glucose as the electron donor and carbon source. The cells were harvested before and after NO3− depletion. Cell pellets were collected by centrifuging 200 ml culture at 10,000 × g for 20 min at 4°C and resuspended in 10 ml MSM. One milliliter of the cell suspension was added to a 160-ml stopper-sealed serum vial containing 100 ml of fresh MSM with N2 headspace. Chloramphenicol (water soluble; Sigma-Aldrich) was added to a final concentration of 25 μg ml−1 to arrest de novo protein synthesis (65). These cell suspensions were then amended with 2 mM NO3− or NO2− and 6.67 mM lactate (Citrobacter sp. DNRA3) or 3.34 mM glucose (Enterobacter sp. DNRA5). The rates of change in the amounts of NO2− were measured and normalized to the protein mass of the resting-cell cultures.
To observe the effects of changing NO3− and NO2− concentrations on transcriptional expression of the nitrite reductase genes directly relevant to DNRA, the transcript abundances of nrfA and nirB genes in Citrobacter sp. DNRA3 and the nirB gene in Enterobacter sp. DNRA5 were monitored as the cells were grown with 1 mM NO3− and 3.34 mM lactate or 1.67 mM glucose. Collection and treatment of the samples, including extraction, purification, and reverse transcription processes, were performed using established protocols (18). Quantitative PCR (qPCR) was performed with a QuantStudio 3 real-time PCR instrument (Thermo Fisher Scientific, Waltham, MA) using SYBR green detection chemistry, targeting nrfA and nirB in Citrobacter sp. DNRA3 and nirB in Enterobacter sp. DNRA5 using the primer sets listed in Table 1, as described in detail in the supplemental material.
TABLE 1.
Primer sets used for PCR and qPCR
| Primer | Purpose | Target gene (locus tag) | Sequence (5ʹ to 3ʹ) | Amplicon size (bp) | Slope | y intercept | Amplification efficiency (%) | R2 | Reference or source |
|---|---|---|---|---|---|---|---|---|---|
| 27F | PCR | Bacterial 16S rRNA | AGAGTTTGATCMTGGCTCAG | 69 | |||||
| 1492R | TACGGYTACCTTGTTACGACTT | ||||||||
| Cit_nrfA_F | qPCR | nrfA (HG548_21145) | ACATGCCGAAAGTGCAAAACGC | 154 | −3.146 | 34.312 | 102.9 | 0.991 | This study |
| Cit_nrfA_R | TGAATGGCCTGTTTACGCTCGG | ||||||||
| Cit_nirB_F | qPCR | nirB (HG548_14725) | ACACCAACGACAACTTCCTGGC | 166 | −3.287 | 36.225 | 101.5 | 0.998 | This study |
| Cit_nirB_R | AAGCCGATACGTTGAGAACCGG | ||||||||
| Cit_recA_F | qPCR | recA (HG548_20545) | GGTAAAACAACGCTGACCCTGC | 186 | −3.381 | 34.183 | 97.6 | 0.999 | This study |
| Cit_recA_R | CAGCGCATCACAGATTTCCAGC | ||||||||
| Ente_nirB_F | qPCR | nirB (HG551_19495) | TGAAAGCGGAAACCAAAGCCG | 178 | −3.336 | 33.848 | 99.4 | 0.994 | This study |
| Ente_nirB_R | AAGGACTTAATGCCCTCCACGC | ||||||||
| Ente_recA_F | qPCR | recA (HG551_15920) | TGGTGTGATGTTCGGTAACCCG | 151 | −3.255 | 33.021 | 102.8 | 0.996 | This study |
| Ente_recA_R | GTTCTTCACAACCTTCACGCGG |
Batch and chemostat incubation of the DNRA-catalyzing isolates with various C-to-N ratios.
Batch cultures of Citrobacter sp. DNRA3 and Enterobacter sp. DNRA5 were prepared with two different C-to-N (carbon in the organic electron donor/nitrogen in NO3−) ratios, and the DNRA reaction in these vessels was observed. For high C-to-N ratio incubation, the culture medium was initially prepared with 0.2 mM NO3− and 5 mM lactate or 2.5 mM glucose, and after each NO3−/NO2− depletion event, the culture vessels were amended with an additional batch of 0.2 mM NO3−. For low C-to-N ratio incubation, the initial medium contained 2 mM NO3− and 0.2 mM lactate or 0.1 mM glucose, and the organic electron donors were replenished upon depletion, indicated by discontinued NO3− reduction. The concentrations of NO3−, NO2−, and NH4+ were monitored throughout the incubation periods.
The chemostat cultures of the DNRA isolates were set up with a 300-ml culture in a continuously stirred 620-ml glass reactor fed fresh medium at a dilution rate of 0.05 h−1 (Fig. S5). The medium bottle and the reactor vessel were consistently purged with N2 gas to maintain anoxic culture conditions during incubation. The reactor was operated with high (10 mM lactate or 5 mM glucose and 2 mM NO3− in the feed) and low (0.2 mM lactate or 0.1 mM glucose and 2 mM NO3−) C-to-N ratios. The concentrations of NO2−, NO3−, and NH4+ in the effluent were monitored until the reactor reached steady state, as indicated by three statistically similar NO2−, NO3−, and NH4+ concentrations measured at 6-h intervals. The N2O production rate was measured after steady state was established by closing the gas inlet and outlet of the reactor and monitoring linear N2O production.
Analytical methods.
The concentrations of NH4+, NO2−, and NO3− were determined calorimetrically. At each sampling event, a 1-ml sample was extracted with a disposable syringe and the cell-free supernatant was subjected to spectrophotometric assays. The NH4+-N concentration was measured using the salicylate method and the NO2−-N and/or NO3−-N concentrations were determined using the Griess method (41, 66). Headspace N2O concentrations were determined using an HP 6890 series gas chromatograph equipped with an HP-PLOT Q column and a 63Ni electron capture detector (Agilent Technologies, Santa Clara, CA) (67). Helium (≥99.999%; Special Gas, Inc., Daejeon, South Korea) and 5% CH4–95% Ar mixed gas were used as the carrier gas and the make-up gas, respectively. The injector, oven, and detector temperatures were set to 200, 85, and 250°C, respectively. Assuming equilibrium between the aqueous and gas phases, the total amounts of N2O-N in reaction vessels were calculated using the dimensionless Henry’s law constant of 1.68 at 25°C (68). The concentrations of glucose, lactate, and acetate were measured using a Prominence high-performance liquid chromatograph (Shimadzu, Kyoto, Japan) equipped with an Aminex HPX-87H column (Bio-Rad Laboratories, Inc., Hercules, CA). Protein concentrations were determined with the Quick Start Bradford protein assay kit (Bio-Rad Laboratories, Hercules, CA) using a concentration series of bovine serum albumin solution as the standard.
Statistical analysis.
With the exception of the chemostat experiments, all incubation experiments were performed in triplicates and the data presented as the average results of the triplicate samples along with their standard deviations. Statistical analyses were performed using R software version 3.6.3 (www.r-project.org), where the one-sample Student’s t test was used to determine the statistical significance of temporal changes in transcript copy numbers or N species concentrations. A P value threshold of 0.05 was applied.
Data availability.
The partial 16S rRNA gene sequences were deposited in NCBI’s GenBank database (accession numbers MT426123 to MT426164). The draft genome sequences of the six confirmed DNRA isolates were deposited in NCBI’s GenBank database (accession numbers JABAIU000000000, JABAIT000000000, JABAIS000000000, JABAIR000000000, JABAIQ000000000, and JABAIP000000000).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (grant number 2020R1C1C1007970) and a USDA-NIFA grant (grant number 2014-67019-21614). The authors were also financially supported by the Brain Korea 21 Plus Project (grant number 21A20132000003).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Williams AG, Audsley E, Sandars DL. 2010. Environmental burdens of producing bread wheat, oilseed rape and potatoes in England and Wales using simulation and system modelling. Int J Life Cycle Assess 15:855–868. doi: 10.1007/s11367-010-0212-3. [DOI] [Google Scholar]
- 2.Erisman JW, Sutton MA, Galloway J, Klimont Z, Winiwarter W. 2008. How a century of ammonia synthesis changed the world. Nat Geosci 1:636–639. doi: 10.1038/ngeo325. [DOI] [Google Scholar]
- 3.Erisman JW, Galloway JN, Seitzinger S, Bleeker A, Dise NB, Petrescu AMR, Leach AM, de Vries W. 2013. Consequences of human modification of the global nitrogen cycle. Philos Trans R Soc Lond B Biol Sci 368:20130116. doi: 10.1098/rstb.2013.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rockström J, Steffen W, Noone K, Persson Å, Chapin FS III, Lambin EF, Lenton TM, Scheffer M, Folke C, Schellnhuber HJ, Nykvist B, de Wit CA, Hughes T, van der Leeuw S, Rodhe H, Sörlin S, Snyder PK, Costanza R, Svedin U, Falkenmark M, Karlberg L, Corell RW, Fabry VJ, Hansen J, Walker B, Liverman D, Richardson K, Crutzen P, Foley JA. 2009. A safe operating space for humanity. Nature 461:472–475. doi: 10.1038/461472a. [DOI] [PubMed] [Google Scholar]
- 5.Canfield DE, Glazer AN, Falkowski PG. 2010. The evolution and future of earth’s nitrogen cycle. Science 330:192–196. doi: 10.1126/science.1186120. [DOI] [PubMed] [Google Scholar]
- 6.Chen D, Suter HC, Islam A, Edis R. 2010. Influence of nitrification inhibitors on nitrification and nitrous oxide (N2O) emission from a clay loam soil fertilized with urea. Soil Biol Biochem 42:660–664. doi: 10.1016/j.soilbio.2009.12.014. [DOI] [Google Scholar]
- 7.Akiyama H, Yan X, Yagi K. 2009. Evaluation of effectiveness of enhanced-efficiency fertilizers as mitigation options for N2O and NO emissions from agricultural soils: meta-analysis. Glob Change Biol 16:1837–1846. doi: 10.1111/j.1365-2486.2009.02031.x. [DOI] [Google Scholar]
- 8.Putz M, Schleusner P, Rütting T, Hallin S. 2018. Relative abundance of denitrifying and DNRA bacteria and their activity determine nitrogen retention or loss in agricultural soil. Soil Biol Biochem 123:97–104. doi: 10.1016/j.soilbio.2018.05.006. [DOI] [Google Scholar]
- 9.Yoon S, Song B, Phillips RL, Chang J, Song MJ. 2019. Ecological and physiological implications of nitrogen oxide reduction pathways on greenhouse gas emissions in agroecosystems. FEMS Microbiol Ecol 95:fiz066. doi: 10.1093/femsec/fiz066. [DOI] [PubMed] [Google Scholar]
- 10.Friedl J, De Rosa D, Rowlings DW, Grace PR, Müller C, Scheer C. 2018. Dissimilatory nitrate reduction to ammonium (DNRA), not denitrification dominates nitrate reduction in subtropical pasture soils upon rewetting. Soil Biol Biochem 125:340–349. doi: 10.1016/j.soilbio.2018.07.024. [DOI] [Google Scholar]
- 11.Shan J, Zhao X, Sheng R, Xia Y, Ti C, Quan X, Wang S, Wei W, Yan X. 2016. Dissimilatory nitrate reduction processes in typical Chinese paddy soils: rates, relative contributions, and influencing factors. Environ Sci Technol 50:9972–9980. doi: 10.1021/acs.est.6b01765. [DOI] [PubMed] [Google Scholar]
- 12.Fitzhugh RD, Lovett GM, Venterea RT. 2003. Biotic and abiotic immobilization of ammonium, nitrite, and nitrate in soils developed under different tree species in the Catskill Mountains, New York, USA. Global Change Biol 9:1591–1601. doi: 10.1046/j.1365-2486.2003.00694.x. [DOI] [Google Scholar]
- 13.Moreno-Vivián C, Flores E. 2007. Nitrate assimilation in bacteria In Bothe H, Ferguson SJ, Newton WE (ed), Biology of the nitrogen cycle, p 263–282. Elsevier B.V., Amsterdam, Netherlands. doi: 10.1016/B978-044452857-5.50018-7. [DOI] [Google Scholar]
- 14.van den Berg EM, Elisário MP, Kuenen JG, Kleerebezem R, van Loosdrecht M. 2017. Fermentative bacteria influence the competition between denitrifiers and DNRA bacteria. Front Microbiol 8:1684. doi: 10.3389/fmicb.2017.01684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonin P. 1996. Anaerobic nitrate reduction to ammonium in two strains isolated from coastal marine sediment: a dissimilatory pathway. FEMS Microbiol Ecol 19:27–38. doi: 10.1111/j.1574-6941.1996.tb00195.x. [DOI] [Google Scholar]
- 16.Cruz-Garcia C, Murray AE, Klappenbach JA, Stewart V, Tiedje JM. 2007. Respiratory nitrate ammonification by Shewanella oneidensis MR-1. J Bacteriol 189:656–662. doi: 10.1128/JB.01194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pandey A, Suter H, He J-Z, Hu H-W, Chen D. 2018. Nitrogen addition decreases dissimilatory nitrate reduction to ammonium in rice paddies. Appl Environ Microbiol 84:e00870-18. doi: 10.1128/AEM.00870-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon S, Cruz-García C, Sanford R, Ritalahti KM, Löffler FE. 2015. Denitrification versus respiratory ammonification: environmental controls of two competing dissimilatory NO3−/NO2− reduction pathways in Shewanella loihica strain PV-4. ISME J 9:1093–1104. doi: 10.1038/ismej.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laverman AM, Van Cappellen P, van Rotterdam-Los D, Pallud C, Abell J. 2006. Potential rates and pathways of microbial nitrate reduction in coastal sediments. FEMS Microbiol Ecol 58:179–192. doi: 10.1111/j.1574-6941.2006.00155.x. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt CS, Richardson DJ, Baggs EM. 2011. Constraining the conditions conducive to dissimilatory nitrate reduction to ammonium in temperate arable soils. Soil Biol Biochem 43:1607–1611. doi: 10.1016/j.soilbio.2011.02.015. [DOI] [Google Scholar]
- 21.Hardison AK, Algar CK, Giblin AE, Rich JJ. 2015. Influence of organic carbon and nitrate loading on partitioning between dissimilatory nitrate reduction to ammonium (DNRA) and N2 production. Geochim Cosmochim Acta 164:146–160. doi: 10.1016/j.gca.2015.04.049. [DOI] [Google Scholar]
- 22.van den Berg EM, van Dongen U, Abbas B, van Loosdrecht M. 2015. Enrichment of DNRA bacteria in a continuous culture. ISME J 9:2153–2161. doi: 10.1038/ismej.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Berg EM, Boleij M, Kuenen JG, Kleerebezem R, van Loosdrecht M. 2016. DNRA and denitrification coexist over a broad range of acetate/N-NO3− ratios, in a chemostat enrichment culture. Front Microbiol 7:1842–1853. doi: 10.3389/fmicb.2016.01842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vuono DC, Read RW, Hemp J, Sullivan BW, Arnone JA, Neveux I, Blank RR, Loney E, Miceli D, Winkler MKH, Chakraborty R, Stahl DA, Grzymski JJ. 2019. Resource concentration modulates the fate of dissimilated nitrogen in a dual-pathway Actinobacterium. Front Microbiol 10:3. doi: 10.3389/fmicb.2019.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tiedje JM, Sexstone AJ, Myrold DD, Robinson JA. 1982. Denitrification: ecological niches, competition and survival. Antonie Van Leeuwenhoek 48:569–583. doi: 10.1007/BF00399542. [DOI] [PubMed] [Google Scholar]
- 26.Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61:533–616. doi: 10.1128/.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgin AJ, Hamilton SK. 2007. Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Front Ecol Environ 5:89–96. doi: 10.1890/1540-9295(2007)5[89:HWOTRO]2.0.CO;2. [DOI] [Google Scholar]
- 28.Murphy AE, Anderson IC, Smyth AR, Song B, Luckenbach MW. 2016. Microbial nitrogen processing in hard clam (Mercenaria mercenaria) aquaculture sediments: the relative importance of denitrification and dissimilatory nitrate reduction to ammonium (DNRA). Limnol Oceanogr 61:1589–1604. doi: 10.1002/lno.10305. [DOI] [Google Scholar]
- 29.Behrendt A, de Beer D, Stief P. 2013. Vertical activity distribution of dissimilatory nitrate reduction in coastal marine sediments. Biogeosciences 10:7509–7523. doi: 10.5194/bg-10-7509-2013. [DOI] [Google Scholar]
- 30.Orellana LH, Chee-Sanford JC, Sanford RA, Löffler FE, Konstantinidis KT. 2017. Year-round shotgun metagenomes reveal stable microbial communities in agricultural soils and novel ammonia oxidizers responding to fertilization. Appl Environ Microbiol 84:e01646-17. doi: 10.1128/AEM.01646-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson MB, Martiny AC, Martiny J. 2016. Global biogeography of microbial nitrogen-cycling traits in soil. Proc Natl Acad Sci U S A 113:8033–8040. doi: 10.1073/pnas.1601070113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandey A, Suter H, He J-Z, Hu H-W, Chen D. 2019. Dissimilatory nitrate reduction to ammonium dominates nitrate reduction in long-term low nitrogen fertilized rice paddies. Soil Biol Biochem 131:149–156. doi: 10.1016/j.soilbio.2019.01.007. [DOI] [Google Scholar]
- 33.Anonymous. 2013. The cultural revolution. Nat Rev Microbiol 11:1–1. doi: 10.1038/nrmicro2948. [DOI] [PubMed] [Google Scholar]
- 34.Welsh A, Chee-Sanford JC, Connor LM, Löffler FE, Sanford RA. 2014. Refined NrfA phylogeny improves PCR-based nrfA gene detection. Appl Environ Microbiol 80:2110–2119. doi: 10.1128/AEM.03443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Gunsalus RP. 2000. The nrfA and nirB nitrite reductase operons in Escherichia coli are expressed differently in response to nitrate than to nitrite. J Bacteriol 182:5813–5822. doi: 10.1128/jb.182.20.5813-5822.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stremińska MA, Felgate H, Rowley G, Richardson DJ, Baggs EM. 2012. Nitrous oxide production in soil isolates of nitrate-ammonifying bacteria. Environ Microbiol Rep 4:66–71. doi: 10.1111/j.1758-2229.2011.00302.x. [DOI] [PubMed] [Google Scholar]
- 37.Mania D, Heylen K, Spanning RJ, Frostegård Å. 2014. The nitrate-ammonifying and nosZ carrying bacterium Bacillus vireti is a potent source and sink for nitric and nitrous oxides under high nitrate conditions. Environ Microbiol 16:3196–3210. doi: 10.1111/1462-2920.12478. [DOI] [PubMed] [Google Scholar]
- 38.Heylen K, Keltjens J. 2012. Redundancy and modularity in membrane-associated dissimilatory nitrate reduction in Bacillus. Front Microbiol 3:371. doi: 10.3389/fmicb.2012.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y, De Vos P, Willems A. 2018. Influence of nitrate and nitrite concentration on N2O production via dissimilatory nitrate/nitrite reduction to ammonium in Bacillus paralicheniformis LMG 6934. MicrobiologyOpen 7:e00592. doi: 10.1002/mbo3.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanford RA, Cole JR, Tiedje JM. 2002. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic Myxobacterium. Appl Environ Microbiol 68:893–900. doi: 10.1128/AEM.68.2.893-900.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baethgen WE, Alley MM. 1989. A manual colorimetric procedure for measuring ammonium nitrogen in soil and plant Kjeldahl digests. Comm Soil Sci Plant Anal 20:961–969. doi: 10.1080/00103628909368129. [DOI] [Google Scholar]
- 42.Mania D, Heylen K, van Spanning RJM, Frostegård Å. 2016. Regulation of nitrogen metabolism in the nitrate-ammonifying soil bacterium Bacillus vireti and evidence for its ability to grow using N2O as electron acceptor. Environ Microbiol 18:2937–2950. doi: 10.1111/1462-2920.13124. [DOI] [PubMed] [Google Scholar]
- 43.Smith MS. 1982. Dissimilatory reduction of NO2− to NH4+ and N2O by a soil Citrobacter sp. Appl Environ Microbiol 43:854–860. doi: 10.1128/AEM.43.4.854-860.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Decleyre H, Heylen K, Colen C, Willems A. 2015. Dissimilatory nitrogen reduction in intertidal sediments of a temperate estuary: small scale heterogeneity and novel nitrate-to-ammonium reducers. Front Microbiol 6:1124. doi: 10.3389/fmicb.2015.01124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sung Y, Fletcher KE, Ritalahti KM, Apkarian RP, Ramos-Hernández N, Sanford RA, Mesbah NM, Löffler FE. 2006. Geobacter lovleyi sp. nov. strain SZ, a novel metal-reducing and tetrachloroethene-dechlorinating bacterium. Appl Environ Microbiol 72:2775–2782. doi: 10.1128/AEM.72.4.2775-2782.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Y, De Vos P, Heylen K. 2016. Nitrous oxide emission by the non-denitrifying, nitrate ammonifier Bacillus licheniformis. BMC Genom 17:1–11. doi: 10.1186/s12864-016-2382-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qu Z, Bakken LR, Molstad L, Frostegård Å, Bergaust LL. 2016. Transcriptional and metabolic regulation of denitrification in Paracoccus denitrificans allows low but significant activity of nitrous oxide reductase under oxic conditions. Environ Microbiol 18:2951–2963. doi: 10.1111/1462-2920.13128. [DOI] [PubMed] [Google Scholar]
- 48.Chang J, Gu W, Park D, Semrau JD, DiSpirito AA, Yoon S. 2018. Methanobactin from Methylosinus trichosporium OB3b inhibits N2O reduction in denitrifiers. ISME J 12:2086–2089. doi: 10.1038/s41396-017-0022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Felgate H, Giannopoulos G, Sullivan MJ, Gates AJ, Clarke TA, Baggs E, Rowley G, Richardson DJ. 2012. The impact of copper, nitrate and carbon status on the emission of nitrous oxide by two species of bacteria with biochemically distinct denitrification pathways. Environ Microbiol 14:1788–1800. doi: 10.1111/j.1462-2920.2012.02789.x. [DOI] [PubMed] [Google Scholar]
- 50.Liu B, Mao Y, Bergaust L, Bakken LR, Frostegard A. 2013. Strains in the genus Thauera exhibit remarkably different denitrification regulatory phenotypes. Environ Microbiol 15:2816–2828. doi: 10.1111/1462-2920.12142. [DOI] [PubMed] [Google Scholar]
- 51.Bengtsson G, Bengtson P, Månsson KF. 2003. Gross nitrogen mineralization-, immobilization-, and nitrification rates as a function of soil C/N ratio and microbial activity. Soil Biol Biochem 35:143–154. doi: 10.1016/S0038-0717(02)00248-1. [DOI] [Google Scholar]
- 52.Luckmann M, Mania D, Kern M, Bakken LR, Frostegård Å, Simon J. 2014. Production and consumption of nitrous oxide in nitrate-ammonifying Wolinella succinogenes cells. Microbiology 160:1749–1759. doi: 10.1099/mic.0.079293-0. [DOI] [PubMed] [Google Scholar]
- 53.Corker H, Poole RK. 2003. Nitric oxide formation by Escherichia coli. Dependence on nitrite reductase, the NO-sensing regulator Fnr, and flavohemoglobin Hmp. J Biol Chem 278:31584–31592. doi: 10.1074/jbc.M303282200. [DOI] [PubMed] [Google Scholar]
- 54.Yoon S, Sanford R, Löffler FE. 2015. Nitrite control over dissimilatory nitrate/nitrite reduction pathways in Shewanella loihica strain PV-4. Appl Environ Microbiol 81:3510–3517. doi: 10.1128/AEM.00688-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolin EA, Wolin MJ, Wolfe RS. 1963. Formation of methane by bacterial extracts. J Biol Chem 238:2882–2886. [PubMed] [Google Scholar]
- 56.Rehr B, Klemme J-H. 1989. Competition for nitrate between denitrifying Pseudomonas stutzeri and nitrate ammonifying enterobacteria. FEMS Microbiol Lett 62:51–57. doi: 10.1016/0378-1097(89)90105-5. [DOI] [Google Scholar]
- 57.Akunna JC, Bizeau C, Moletta R. 1993. Nitrate and nitrite reductions with anaerobic sludge using various carbon sources: glucose, glycerol, acetic acid, lactic acid and methanol. Water Res 27:1303–1312. doi: 10.1016/0043-1354(93)90217-6. [DOI] [Google Scholar]
- 58.Garcia-Robledo E, Corzo A, Papaspyrou S. 2014. A fast and direct spectrophotometric method for the sequential determination of nitrate and nitrite at low concentrations in small volumes. Mar Chem 162:30–36. doi: 10.1016/j.marchem.2014.03.002. [DOI] [Google Scholar]
- 59.Yoshinari T, Hynes R, Knowles R. 1977. Acetylene inhibition of nitrous oxide reduction and measurement of denitrification and nitrogen fixation in soil. Soil Biol Biochem 9:177–183. doi: 10.1016/0038-0717(77)90072-4. [DOI] [Google Scholar]
- 60.Martin M. 2011. Cutadapt removes adapter sequences from high-sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 61.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fish J, Chai B, Wang Q, Sun Y, Brown CT, Tiedje J, Cole J. 2013. FunGene: the functional gene pipeline and repository. Front Microbiol 4:291. doi: 10.3389/fmicb.2013.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maalej S, Denis M, Dukan S. 2004. Temperature and growth-phase effects on Aeromonas hydrophila survival in natural seawater microcosms: role of protein synthesis and nucleic acid content on viable but temporarily nonculturable response. Microbiology 150:181–187. doi: 10.1099/mic.0.26639-0. [DOI] [PubMed] [Google Scholar]
- 66.Miranda KM, Espey MG, Wink DA. 2001. A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5:62–71. doi: 10.1006/niox.2000.0319. [DOI] [PubMed] [Google Scholar]
- 67.Park D, Kim H, Yoon S. 2017. Nitrous oxide reduction by an obligate aerobic bacterium Gemmatimonas aurantiaca strain T-27. Appl Environ Microbiol 83:e00502-17. doi: 10.1128/AEM.00502-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sander R. 2015. Compilation of Henry’s law constants (version 4.0) for water as solvent. Atmos Chem Phys 15:4399–4981. doi: 10.5194/acp-15-4399-2015. [DOI] [Google Scholar]
- 69.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The partial 16S rRNA gene sequences were deposited in NCBI’s GenBank database (accession numbers MT426123 to MT426164). The draft genome sequences of the six confirmed DNRA isolates were deposited in NCBI’s GenBank database (accession numbers JABAIU000000000, JABAIT000000000, JABAIS000000000, JABAIR000000000, JABAIQ000000000, and JABAIP000000000).