Understanding the persistence of enveloped viruses helps inform infection control practices and procedures in health care facilities and community settings. These data convey to public health investigators that enveloped viruses can persist and remain infective on surfaces, thus demonstrating a potential risk for transmission. Under these laboratory-simulated Western indoor hospital conditions, we assessed the suitability of phi 6 as a surrogate for environmental persistence research related to enveloped viruses, including EBOV and coronaviruses.
KEYWORDS: Ebola virus, phi 6 bacteriophage, surface persistence, enveloped viruses, surrogate, coronavirus, environmental microbiology, public health, virology, health care transmission
ABSTRACT
The infection of health care workers during the 2013 to 2016 Ebola outbreak raised concerns about fomite transmission. In the wake of the coronavirus disease 2019 (COVID-19) pandemic, investigations are ongoing to determine the role of fomites in coronavirus transmission as well. The bacteriophage phi 6 has a phospholipid envelope and is commonly used in environmental studies as a surrogate for human enveloped viruses. The persistence of phi 6 was evaluated as a surrogate for Ebola virus (EBOV) and coronaviruses on porous and nonporous hospital surfaces. Phi 6 was suspended in a body fluid simulant and inoculated onto 1-cm2 coupons of steel, plastic, and two fabric curtain types. The coupons were placed at two controlled absolute humidity (AH) levels: a low AH of 3.0 g/m3 and a high AH of 14.4 g/m3. Phi 6 declined at a lower rate on all materials under low-AH conditions, with a decay rate of 0.06-log10 PFU/day to 0.11-log10 PFU/day, than under the higher AH conditions, with a decay rate of 0.65-log10 PFU/h to 1.42-log10 PFU/day. There was a significant difference in decay rates between porous and nonporous surfaces at both low AH (P < 0.0001) and high AH (P < 0.0001). Under these laboratory-simulated conditions, phi 6 was found to be a conservative surrogate for EBOV under low-AH conditions in that it persisted longer than Ebola virus in similar AH conditions. Additionally, some coronaviruses persist longer than phi 6 under similar conditions; therefore, phi 6 may not be a suitable surrogate for coronaviruses.
IMPORTANCE Understanding the persistence of enveloped viruses helps inform infection control practices and procedures in health care facilities and community settings. These data convey to public health investigators that enveloped viruses can persist and remain infective on surfaces, thus demonstrating a potential risk for transmission. Under these laboratory-simulated Western indoor hospital conditions, we assessed the suitability of phi 6 as a surrogate for environmental persistence research related to enveloped viruses, including EBOV and coronaviruses.
INTRODUCTION
Ebola virus (EBOV) is an enveloped RNA virus in the family Filoviridae, which also includes Marburg virus and Cueva virus. Ebola virus disease (EVD) is a rare but deadly disease with a case fatality rate of around 50% (ranging from 25% to 90% based on outbreaks from 1976 to 2015) according to the World Health Organization (WHO) (https://www.who.int/en/news-room/fact-sheets/detail/ebola-virus-disease). The ongoing outbreak that started in 2018, mainly occurring in Democratic Republic of Congo (DRC) and spilling over into Uganda, is the second largest, with 2,997 cases and 1,998 deaths as of 27 August 2019 (https://www.who.int/news-room/detail/29-08-2019-as-ebola-cases-reach-3000-in-drc-who-calls-on-partners-to-fulfill-promises-to-communities). The largest and deadliest EBOV outbreak on record (2013 to 2016) had more than 28,000 cases and over 11,000 deaths globally (1). The outbreak directly impacted the United States when, in 2014, a health care worker in Texas tested positive for EBOV after caring for an infected patient (2; https://www.cdc.gov/media/releases/2014/s1015-texas-second-health-care-worker.html). Recorded cases from past and present outbreaks show that EBOV hospital transmission is a global concern as highlighted in a review of health care worker (HCW) infections by Selvaraj et al. (3), which details occupational exposure increasing transmission risk to health care workers.
Coronaviruses are also enveloped RNA viruses, belonging to the family Coronaviridae. Human coronaviruses are responsible for some common colds, but in 2002, severe acute respiratory syndrome coronavirus 1 (SARS)-CoV-1 emerged from Guangdong Province, China, as the first known deadly coronavirus (4), resulting in 8,422 illnesses and 916 deaths. In 2012, the Middle East respiratory syndrome coronavirus (MERS-CoV) outbreak, believed to have originated from Saudi Arabia, caused 858 deaths across 27 countries (https://www.who.int/emergencies/mers-cov/en/). Coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, emerged from Hubei Province, China (late 2019), and spread rapidly around the world, being declared a pandemic by the WHO in March 2020 (https://www.who.int/news-room/detail/27-04-2020-who-timeline---covid-19). Fomites were thought to have played a role in the transmission of SARS-CoV-1 (5, 6), and though SARS-CoV-2 is thought to be primarily spread by aerosols, investigations are underway to determine if fomites also contribute to transmission. Emerging data suggest that SARS-CoV-2 can persist for days on surfaces (7), though the influence of environmental factors still needs to be explored.
The Centers for Disease Control and Prevention (CDC) recommends a combination of measures to prevent transmission of EVD in hospitals, and these recommendations have been adapted for SARS-CoV-2 as well. The recommendations include patient isolation and record keeping, proper personal protection equipment (PPE) and correct use of PPE, dedicated equipment, limited use of sharps, avoiding aerosol-generating procedures, hand hygiene, and monitoring potentially exposed personnel and visitors for signs and symptoms (https://www.cdc.gov/vhf/ebola/clinicians/evd/infection-control.html; https://www.cdc.gov/coronavirus/2019-ncov/hcp/infection-control-recommendations.html). Further guidance from the CDC covers environmental infection control beyond PPE to include disinfectant use, routine cleaning, how to handle soiled surfaces and textiles, and how to transport or dispose of contaminated items and waste (https://www.cdc.gov/vhf/ebola/clinicians/cleaning/hospitals.html; https://www.cdc.gov/coronavirus/2019-ncov/hcp/infection-control-recommendations.html).
The WHO offers similar guidance on PPE (including proper donning and doffing), infection prevention and control, hand hygiene, and management of wastes (8, 9; https://www.who.int/en/news-room/fact-sheets/detail/ebola-virus-disease). Despite implementation of these best practices, transmission of Ebola from patient to health care worker continued, as documented by at least four reported cases in the fall of 2018 (http://www.cidrap.umn.edu/news-perspective/2018/08/drc-ebola-cases-surpass-earlier-outbreak-total-virus-infects-4-more-health) in the United States. There has also been documented health care worker transmission of SARS-CoV-1 and MERS (10). These transmission events highlight the need to understand the persistence of EBOV and other pathogenic enveloped viruses on fomites and the role of fomites in transmission, especially in the presence of body fluids (11, 12).
EBOV is a select agent and requires a biosafety level 4 (BSL-4) laboratory and specialized PPE to prevent potential life-threatening exposure. SARS-CoV-1 and SARS-CoV-2 are also labeled as select agents and require a BSL-3 laboratory in order to culture and conduct environmental persistence, sampling, or disinfection studies in which working with live virus is required. For safety concerns, researchers have incorporated other methods to avoid handling and propagating this virus. Some have opted for the molecular detection of viral RNA to demonstrate potential transmission in the hospital environment (12, 13). Surrogate viruses have historically been used for transmission studies related to health care practices and can be employed as surrogates for viral persistence. Bacteriophage phi 6, a member of the family Cystoviridae, was previously used as a surrogate for EBOV, influenza virus, coronavirus (SARS-1), Venezuelan equine encephalitis virus, and other pathogenic enveloped viruses (14–19). Casanova et al. (20, 21) used phi 6 as an Ebola surrogate to demonstrate transference to health care worker hands and scrubs during the PPE doffing procedures. However, data regarding the persistence of EBOV or its surrogates in the health care environment are limited. This study evaluated the persistence of phi 6 in the presence of artificial test soil (ATS) as a potential surrogate for EBOV or coronaviruses at two absolute humidity (AH) conditions on four potential fomites: nonporous stainless steel (SS) and plastic (PL) and two types of porous hospital curtain fabrics.
RESULTS
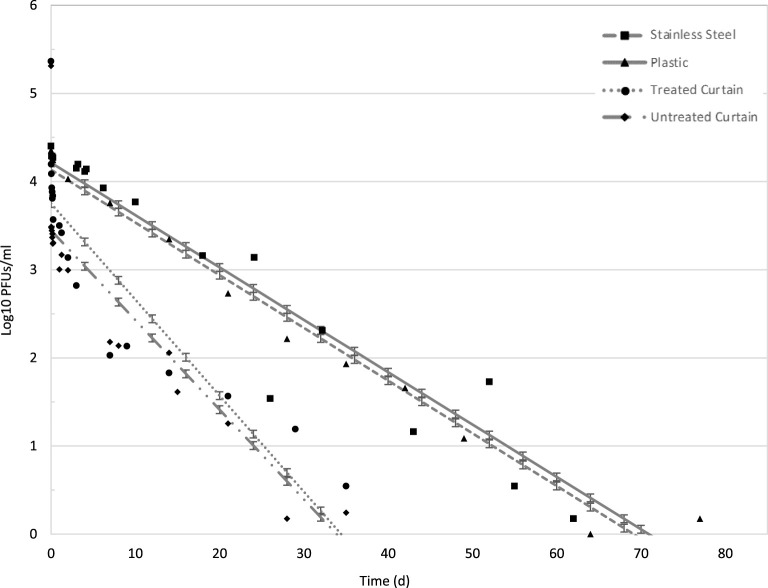
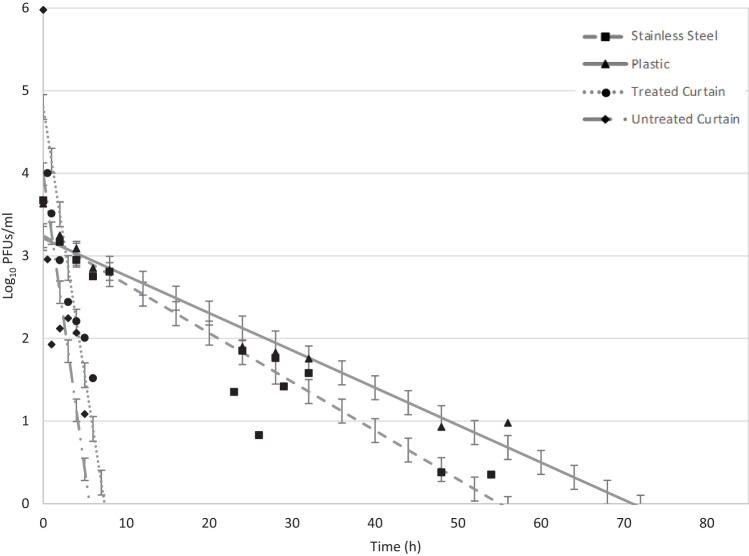
At the lower AH (3.0 g/m3) on SS, phi 6 persisted >76 days with 4.08-log10 reduction (inoculum of 2.5 × 104 plaque-forming units [PFU]), and there was no detectable infective phage by 102 days (Table 1). Phi 6 persisted on PL at the same AH up to 77 days with 4.20-log10 reduction (inoculum of 2.4 × 104 PFU), and there was no detectable phage by 78 days. The statistical model, using a least-squares method in SAS v9.4 (Cary, NC), projected phi 6 persistence until 72 days on SS and 70 days on PL (Fig. 1). The model predicted a decay rate of 0.06 log10/day for both SS and PL, with r2 values of 0.88 and 0.96, respectively, at AH 3.0 g/m3 (Table 1). The projected data have a uniform standard error of ±0.04-log10 PFU/ml. There was no difference, by analysis of variance and the F statistic, in decay rates between SS and PL at the low-AH (3.0 g/m3) conditions.
TABLE 1.
Persistence of bacteriophage phi 6 compared to reports of the Ebola Makona variant and human coronavirus on nonporous surfacesa
| Virus | Surface | Matrix | AH (g/m3) (°C, % RH) | Decay rateb | r2 | D value | Time to no detection (reduction at last sampling point detected [log10]) | Source or reference no. |
|---|---|---|---|---|---|---|---|---|
| Phi 6 | Steel | ATS | 3.0 (18, 20) | 0.06/day | 0.88 | 18 days | 102 days (4.08 at 76 days) | Current data |
| Phi 6 | Plastic | ATS | 3.0 (18, 20) | 0.06/day | 0.96 | 14 days | 78 days (4.20 at 77 days) | Current data |
| Phi 6 | Steel | ATS | 14.4 (26, 57) | 1.42/day | 0.90 | 6 h | > 3 days (77 h) (3.33 at 54 h) | Current data |
| Phi 6 | Plastic | ATS | 14.4 (26, 57) | 1.09/day | 0.91 | 6 h | > 3 days (73 h) (3.68 at 72 h) | Current data |
| Phi 6 | Steel | Blood | 2.7–11.3 (21.1–21.7, 15–59) | N/Ac | N/A | 7–88 hd | N/A | 43 |
| Phi 6 | Steel | PBS | 2.7–11.3 (21.1–21.7, 15–59) | N/A | N/A | 5 h | N/A | 43 |
| Ebola | Steel | Organic soil load | 5.7 (21.5, 30) | 0.22/day | 0.90 | 30 h | N/A (∼3 at 8 days) | 30 |
| Ebola | 25-well plate/plastic | Human blood | 7.4 (21, 40) | 0.69/day | 0.90 | 2 days | 6 days (∼4.5) | 22 |
| Ebola | 25-well plate/plastic | Human blood | 21.5 (27, 80) | 0.68/day | 0.87 | 2 days | 7 days (5) | 22 |
| Ebola | Steel | Human blood | 3.3 (22, 17) | 0.79/h | N/A | <12 h | <3 days (∼3) | 32 |
| Ebola | Plastic | Human blood | 3.3 (22, 17) | 0.79/h | N/A | <12 h | 3 days (N/A) | 32 |
| Ebola | Steel | Human blood | 8.0 (22, 41) | 0.63/h | N/A | <12 h | 4 days (∼ 3.5) | 32 |
| Ebola | Plastic | Human blood | 8.0 (22, 41) | 0.63/h | N/A | <12 h | 4 days (N/A) | 32 |
| Ebola | Steel | Human blood | 25.8 (28, 90) | 0.29/h | N/A | ∼72 h | 10 days (∼3) | 32 |
| Ebola | Plastic | Human blood | 25.8 (28, 90) | 0.29/h | N/A | ∼72 h | 10 days (∼2.5) | 32 |
| CoV-2 | Steel and plastic | Artificial Saliva | 14.4e (26, 57) | N/A | N/A | N/A | 4.4 days for 99.99% reduction (4) | https://www.dhs.gov/science-and-technology/sars-calculator |
| SARS-CoV-2 | Plastic and steel | N/A | 7.4–8.4 (21–23, 40) | N/A | N/A | 8–24 h | 3–4 days (∼3.7) | 7 |
| SARS-CoV-2 | Plastic | Culture medium | 12.7 (22, 65) | N/A | N/A | ∼6 h | 7 days (5.8) | 33 |
| SARS-CoV-2 | Steel | Culture medium | 12.7 (22, 65) | N/A | N/A | 1 day | 7 days (5.8) | 33 |
| SARS-CoV-1 | Plastic and steel | N/A | 7.4–8.4 (21–23, 40) | N/A | N/A | 8–24 h | 3–4 days (∼3.4) | 7 |
| SARS CoV-1 | Plastic | Culture medium | 7.8–11.9 (22–25, 40–50) | N/A | N/A | 6 d | 28 days (∼5) | 50 |
| MERS-COV | Plastic and steel | Culture medium | 6.9 (20, 40) | N/A | N/A | 8–10 h | 3 days (∼5) | 29 |
| MERS-COV | Plastic and steel | Culture medium | 26.1 (30, 80) | N/A | N/A | 4–6 h | 1 day (∼5) | 29 |
| HCV OC43 | Latex | Culture medium | 10.1–13.8 (21, 55–75) | N/A | N/A | <1 h | 1 h (∼3) | 34 |
| HCV 229E | Aluminum and latex | Culture medium | 10.1–13.8 (21, 55–75) | N/A | N/A | ∼4–5 h | 12 h (∼3) | 34 |
| HCV OC43 | Aluminum | Culture medium | 10.1–13.8 (21, 55–75) | N/A | N/A | 1–2 h | 3 h (∼3) | 34 |
Surfaces include stainless steel, aluminum, latex, and plastic.
Decay rates for current data based upon model predictions graphed in Fig. 1 and 2 and expressed in log10 per day. Other decay rates taken from literature.
N/A means that no data were provided or that the experiment was not extended until there was no detection.
Medians of two experiments.
Conditions not actually tested but estimated from the model.
FIG 1.
Model projection of low temperature and low humidity conditions (LTLH; AH = 3.0 g/m3) on all surfaces: stainless steel (SS), plastic (PL), treated curtains (TC), and untreated curtains (UC). Projected SS is represented by a dashed line, and observed mean is a square. Projected PL is represented by a solid line, and observed mean is a triangle. The projected TC is represented by a dotted line, and the observed mean is a circle. The UC projected persistence is represented by a dotted/dashed line, and the observed mean is a diamond shape. Data points and error bars indicate mean and standard deviation of the projected data. Refer to Table 1 for r2 values.
At the higher AH (14.4 g/m3) on SS, phi 6 persisted >54 h with 3.33-log10 reduction (inoculum of 5.2 × 103 PFU), and there was no infective phage by 77 h (Table 1). Phi 6 persisted on PL at the same AH up to 72 h with 3.68-log10 reduction (inoculum of 1.9 × 104 PFU), and there was no detectable infective phage by 73 h. The model projected phi 6 persisting until 56 h on SS and 71 h on PL (Fig. 2). The model predicted a decay rate of 1.42 log10/day (r2 = 0.90) for SS and 1.09 log10/day (r2 = 0.91) for PL (Table 1). The projected data have a uniform standard error of ±0.14-log10 PFU/ml. There was a significant difference in decay rates between SS and PL (P = 0.001) at the higher AH. Overall, phi 6 persisted longer on SS and PL surfaces at the lower AH of 3.0 g/m3 (no detectable phi 6 at 78 days to 102 days) than at the higher AH of 14.4 g/m3 (no detectable phi 6 at 73 h to 77 h).
FIG 2.
Model projection of high temperature and high humidity conditions (HTHH; AH = 14.4 g/m3 unit) on all surfaces: stainless steel (SS), plastic (PL), treated curtains (TC), and untreated curtains (UC). Projected SS is represented by a dashed line, and observed mean is a square. Projected PL is represented by a solid line, and observed mean is a triangle. The projected TC is represented by a dotted line, and the observed mean is a circle. The UC projected persistence is represented by a dotted/dashed line, and the average observed mean is a diamond shape. Data points and error bars indicate mean and standard deviation of the projected data. Refer to Table 1 for r2 values.
Phi 6 persistence on porous treated and untreated curtains (TC and UC) at the low-AH (3.0 g/m3) conditions was >35 days with 5.40-log10 reduction (inoculum of 2.3 × 105 PFU) and 5.27-log10 reduction (inoculum of 2.2 × 105 PFU), respectively, and no phi 6 was detectable at 42 d for either curtain type (Table 2). The model projected phi 6 persisting until 35 days in the low-AH conditions (Fig. 1). The model of persistence on TC and UC predicted decay rates of 0.11 log10/day for TC (r2 = 0.72) and 0.10 log10/day (r2 = 0.71) for UC (Table 2). The projected data have a uniform standard error of ±0.04-log10 PFU/ml. There was no significant difference in the decay rates of the curtain types in low-AH conditions (P = 0.200).
TABLE 2.
| Virus | Surface | Matrix | AH (g/m3) (°C, % RH) | Decay ratec | r2 | D value (min [log10]) | Time to no detection (reduction at last sampling point detected [log10]) | Source or reference no. |
|---|---|---|---|---|---|---|---|---|
| Phi 6 | TC | ATS | 3.0 (18, 20) | 0.11/day | 0.72 | 30 | 42 days (5.40 at 35 days) | Current data |
| Phi 6 | UC | ATS | 3.0 (18, 20) | 0.10/day | 0.71 | 30 | 42 days (5.27 at 35 days) | Current data |
| Phi 6 | TC | ATS | 14.4 (26, 57) | 0.65/h | 0.71 | <30 (2) | N/Ad (4.91 at 6 h) | Current data |
| Phi 6 | UC | ATS | 14.4 (26, 57) | 0.71/h | 0.49 | <30 (3) | N/A (5.19 at 5 h) | Current data |
| HCoV 229 | Cotton gauze | Growth medium | 10.1–12.9 (21, 55–70) | N/A | N/A | N/A | 12 h (3) | 34 |
| HCoV OC43 | Cotton gauze | Growth medium | 10.1–12.9 (21, 55–70) | N/A | N/A | N/A | 1 h (3) | 34 |
| SARS-CoV-1 | Cotton gown | Growth medium | N/A (20, N/A)e | N/A | N/A | N/A | 1 day (6) | 44 |
One treated with zinc pyrithione (TC) and one untreated (UT).
Human coronavirus studies on other porous surfaces are presented for comparison. Limited comparison data from the literature for three coronaviruses are also presented.
Decay rates for current data based upon model predictions graphed in Fig. 1 and 2 and expressed in log10.
N/A means that no data were provided or that the experiment was not extended until there was no detection.
AH was not calculated because RH was not provided.
Phi 6 persistence on porous curtains (TC and UC) at high-AH (14.4 g/m3) conditions decreased drastically compared to the low-AH results for TC and UC. Time to no detection decreased from days to hours, where persistence on TC was 6 h with a 4.91-log10 reduction (inoculum of 3.3 × 106 PFU), and, on UC, it was 5 h with a 5.19-log10 reduction (inoculum of 1.7 × 106 PFU) (Table 2). The model projected phi 6 persisting until 9 h on TC and until 7 h on UC in the high-AH conditions (Fig. 2). The model predicted decay rates of 0.65 log10/h for TC (r2 = 0.71) and 0.71 log10/h (r2 = 0.49) for UC (Table 2). The projected data have a uniform standard error of ±0.15 for TC and ±0.14-log10 PFU/ml for UC. The difference between curtain types was not significant (P = 0.500) under high-AH conditions.
Overall, bacteriophage phi 6 in body fluid simulant persisted longer when held at the low AH of 3.0 g/m3 (35 days to 102 days) than at the higher AH of 14.4 g/m3 (5 h to 77 h) regardless of surface and material type (Table 1 and 2). With respect to decay rates, phi 6 declined more slowly on all materials under low-AH (3.0 g/m3) conditions (0.06 log10/day to 0.11 log10/day) than under the higher AH (14.4 g/m3; 0.65 log10/h to 1.42 log10/day; Table 1 and 2). There were significant differences in decay rates between porous and nonporous surfaces at both low-AH (P < 0.0001) and high-AH (P < 0.0001) conditions.
DISCUSSION
Phi 6 is an enveloped bacteriophage and was chosen for this study because it has been used as a surrogate for the persistence of other enveloped viruses such as influenza virus, coronavirus, and Venezuelan equine encephalitis virus (14, 16, 18). Using a nonpathogenic surrogate removes the need for resources associated with a BSL-3 or BSL-4 agent and makes the research procedures accessible to more laboratories. This current work demonstrated that the persistence of phi 6 was similar to the published reports of EBOV (22–26), human respiratory viruses (15, 27), and coronavirus (28, 29) in that the phage persisted longer in colder temperatures and at lower relative and absolute humidity.
To date, two studies have evaluated EBOV persistence and found variability based on temperature, humidity, and substrate (22, 30). Persistence was also shown to vary between species of EBOV, Sudan EBOV and Zaire EBOV, and between variants Makona-C05 and Yambuku-Mayinga (11, 31, 32). Bausch et al. (11) found the risk of infection from fomites to be low when working with the Sudan EBOV, where only 2 of 33 surface swab samples taken daily in an Ebola isolation ward in Uganda were positive by PCR detection only; no samples were culture positive. In contrast, Bibby et al. found that the Zaire EBOV survived for over 14 days on glass and plastic in guinea pig sera held at 4°C (31), demonstrating the role of both temperature and strain in survival. Schuit et al. found differences between Makona-C05 and Yambuku-Mayinga variants, with the Makona-C05 variant remaining viable for longer in hospital room conditions (32).
Phi 6 was shown here in the current study to be a conservative surrogate for EBOV in a laboratory-simulated Western hospital room condition of 3.0 g/m3 AH, persisting longer than the Makona-C05 variant (AH = 3.3 g/m3), with decay rates of 0.06 log10/d and 0.79 log10/h, respectively (Table 1). Due to different conditions, persistence comparisons between the current phi 6 study and the Schuit et al. 2016 Ebola work at the higher AH were not possible; phi 6 was evaluated at an AH of 14.4 g/m3, and EBOV was evaluated at an AH of 8 g/m3 or 25.8 g/m3 (32). Interestingly, contrary to the trend we report with phi 6 and studies seen with MERS-CoV (29), Schuit et al. demonstrated increased survival of Ebola at the higher AH of 25.8 g/m3 (28°C; 90% RH) when deposited in dried blood compared to his lower-AH conditions tested.
A controlled laboratory study of SARS-CoV-1 and SARS-CoV-2 investigated persistence on steel and plastic and found that both viruses behaved similarly (7). When conditions were held at AH 7.4 to 8.4 (21 to 23°C and 40% RH), no infective virus was detected by 72 and 96 h, respectively (Table 1). Chin et al. (33) demonstrated that SARS-CoV-2 persisted for 7 days at AH 12.7 (22°C; 65% RH). These conditions are close to our AH of 14.4, in which persistence of phi 6 was observed for only 3 days, suggesting phi 6 may not be a suitable surrogate for SARS-CoV-2. Additionally, a model based on testing at several humidity and temperature conditions predicted that SARS CoV-2 would persist for 4.4 days on steel and plastic, as suspended in artificial saliva, and held at an AH of 14.4 (26°C; 57% RH), one of the same conditions we tested phi 6, though the model would not extend to the lower-AH condition (https://www.cdc.gov/coronavirus/2019-ncov/hcp/infection-control.html). Our work reported phi 6 persistence for 3 days under these conditions, a significantly shorter period, though the matrices were different. This adds evidence to the Chin et al. (33) results that phi 6 may not be a suitable surrogate for SARS-CoV-2 when persistence is being studied.
MERS-CoV was less persistent than phi 6 under close but not exactly the same conditions, surviving only to days 3 and 1 at low (6.9 g/m3) and high (26.1 g/m3) AH, respectively (29). Our study showed significantly greater persistence of phi 6 (78 to 102 days) at lower AH (3.0 g/m3) than was tested for MERS-CoV. At the highest-AH conditions tested, 14.4 g/m3 for phi 6 and 26.1 g/m3 for MERS-CoV, persistence for phi 6 was 6 h, while MERS-CoV was 24 h. Whether these differences can be explained by environmental test conditions, surface characteristics, or organism structure differences remains to be explored, but little parallel is seen between the studies. Other coronaviruses tested at AH levels of 10.1 to 13.8 g/m3 on aluminum persisted for even less time; human coronavirus (HCV) 229 persisted for 12 h and HCV OC43 for 3 h (34) (Table 1). Persistence declines with increasing AH for phi 6, but the literature does not reveal this trend for coronaviruses.
An integral data gap for enveloped viruses is the risk of transmission under various humidity and temperature conditions. The two temperature and humidity combinations applied in this study were upper and lower health care facility extremes. As related to EBOV and coronaviruses, the upper extreme might be found in a tropical setting without adequate air conditioning, as seen in Liberia and Sierra Leone during the 2014 to 2016 epidemic, which was 27.4°C (35). The lower extreme was chosen as a setting common in Western health care facilities. The environmental temperature, along with relative humidity (RH), was used to calculate the AH frequently referenced in the literature. AH is the measure of the water vapor in the air regardless of temperature, while RH is the ratio of the concentration of water vapor to the maximum possible concentration at a given temperature. Research on both phi 6 and influenza virus has shown that AH may be more important than RH in virus infectivity (26, 36, 37) and the role of AH may be linked to changes in the viral envelope (28, 38–40). Shaman and Kohn reported that influenza virus survival is more dependent on the water vapor in the air (AH) than how close the air is to saturation (RH) (37). Prussin et al. came to the same conclusion for phi 6 when using a multiple regression analysis, showing that AH is a better predictor of virus infectivity (26). The role of AH in survival and infectivity may be linked to changes in the viral envelope during desiccation (28, 38–40) and the protective effects of proteins upon concentration (41). One study noted a drop in infectivity between 60 and 80% RH at 25° and 37°C (AH, 14.4 to 40 g/m3) (26), and similar findings were seen in an influenza virus persistence study (41), though these RH and AH conditions were higher than those tested here. Health care environments vary around the world, and more information is needed to understand what influences persistence of enveloped viruses.
Damage from environmental conditions (e.g., AH, surface type, and matrices) to the viral envelope impact infectivity and persistence. Mateu et al. (42) suggest that repeated disruption of the capsid envelope can cause irreparable damage. Casanova et al. (28) suggest that viral inactivation is due to structural damage during desiccation, occurring at the air-water interface of the viral envelope. Persistence and infectivity of EBOVs have been shown to be influenced by the suspension liquid (blood, serum, or cell culture media) and surface type, such as plastic and metal (nonporous), as well as curtain material (porous) (23, 25). Blood, mucus, stool, and other body fluids can provide protection from envelope structural damage due to desiccation (14, 31). In the hospital environment, Bausch et al. (11) detected EBOV by reverse transcription PCR (RT-PCR) only upon visibly contaminated surfaces that were bloodstained. The transmission pathway of infected blood, and the protective nature of the blood matrix as shown by Fischer et al. (22), led to the use, in the current study, of ATS as a blood stimulant, as it contains proteins, hemoglobin, and carbohydrates. Though testing only continued for 72 h, Wood et al. demonstrated better persistence of phi 6 when suspended in blood diluent than in phosphate-buffered saline (PBS) (Table 1) and that the influence of the matrix overshadowed the influence of the fomite material (43). The current data show that phi 6 infectivity persists longer in dried ATS (18°C, 20% RH, 3.04 g/m3 AH) than the Ebola Makona-WPGC07 strain persisted in blood under similar, but not exact, conditions (21°C, 40% RH, 7.35 g/m3 AH) (22). Overall, variations in log10 reductions have been seen between species and strains of filoviruses (23) as previously mentioned. However, this comparison to the Makona variant in blood under similar conditions as used in this work suggests phi 6 may be a conservative surrogate for Ebola at lower-AH conditions (Table 1).
Differences in persistence for both Lake Victoria marburgvirus (MARV) and Zaire Ebola virus (ZEBOV) were seen between metal (316 stainless steel) and plastic by Piercy et al. (23). Both Lake Victoria MARV and ZEBOV suspended in guinea pig sera could not be recovered from metal surfaces at any time point regardless of the temperature and humidity. When placed on plastic, however, persistence improved, with infectivity lasting up to 50 days at 4°C (23).
Nonporous surfaces appear to be a greater risk in the transmission of EBOV in hospital settings than porous surfaces. Like EBOV and coronavirus, phi 6 did not persist as long on porous surfaces (TC and UC) as it did on nonporous surfaces (SS and PL) (Table 1 and 2). Cook et al. (30) found the Makona variant of EBOV suspended in organic soil load was completely inactivated on cotton gowns within 24 h and persisted longer on steel carriers than PL (21.5°C, 30% RH, 5.69 g/m3 AH).
Sizun et al. (34) investigated the human coronavirus (HCV) 229E on cotton sponges and found that the virus declined rapidly as well, with no infective virus found after 12 h, and HCV OC43 was not infective after 1 h. Lai et al. (44) inoculated cotton gown fabric with SARS-CoV-1 and found survival for only 24 h when held at 20°C (RH not reported). No other controlled laboratory studies were found in the literature describing survival of other enveloped viruses on porous surfaces, though Abad et al. (45) demonstrated that persistence for some nonenveloped viruses such as hepatitis A virus and bacteriophage B40-8 was lower if placed on cotton fabric than on hard surfaces, while other nonenveloped viruses (human rotavirus [HRV], poliovirus, and enteric adenovirus) persisted as well or longer on cotton than on hard surfaces.
One limitation to this evaluation of phi 6 bacteriophage as a suitable surrogate on porous materials is the difficulty in elucidating if there is a true lack of persistence or if the phage adhered to the fabric, making it difficult to elute. Adsorption to the curtains could potentially explain some of the declines in recoverable infective phage, though the adsorption would most likely inhibit touch-transfer as well. Viral adsorption to fabrics is influenced by whether a fabric is tightly or loosely knit and the surface charge of the virus and the fabric (46). These data inform public health investigators that enveloped viruses are likely to persist longer on surfaces in modern climate-controlled health care facilities than in tropical field stations. Additional precautions and disinfection strategies must be taken to prevent transmission when treating infected patients. This work also provides additional evidence that phi 6 may be considered a conservative surrogate for EBOV when conducting persistence investigations in that it persisted longer. We also report that phi 6 may not be a suitable surrogate for coronaviruses in all environmental conditions, though there seems to be a wide range of persistence reported in the literature. The model for SARS-CoV-2 (https://www.dhs.gov/science-and-technology/sars-calculator) predicted persistence of 4.4 days (99.99% inactivation), which is considerably more than our experimental finding of 73 h for phi 6. As reported by Aquino de Carvalo et al. (14), when evaluating phi 6 as a surrogate for persistence in water matrices, phi 6 cannot be considered a universal surrogate for persistence of all enveloped viruses on surfaces. Instead, multiple surrogate viruses should be considered if the virus of interest itself cannot be investigated.
MATERIALS AND METHODS
Test materials.
Four test materials were chosen to simulate surfaces typically found within the U.S. health care environment and cut into 1-cm2 coupons. The two nonporous surfaces were stainless steel (SS) (24 gauge, type 304; Stewart Stainless Supply, Suwanee, GA) and plastic (PL) (polyvinyl chloride [PVC] acrylic alloy, Kydex T [0.08-in. thickness, P1 haircell texture; Kydex, LLC, Bloomsburg, PA]). The two porous surfaces were polyester curtain fabric with (treated curtain [TC]) and without (untreated curtain [UC]) a zinc pyrithione treatment (ModoMed, Grand Rapids, MI). Nonporous (SS and PL) coupons were prepared by washing three times with dilute detergent Fisherbrand Versa-Clean (Unica Canada, Inc., Boucherville, Quebec, Canada), rinsing with reverse osmosis water (3 times), and spraying with 70% ethanol. The SS coupons were sterilized by autoclaving at 121°C (30 lb/in2) for 20 min. The PL, TC, and UC coupons were sterilized by UV (UVC wavelength of 254 nm) treatment for 30 min on each side.
Phi 6 propagation.
Bacteriophage phi 6 and host Pseudomonas syringae were obtained from Laboratoire de Sylvain Moineau in Québec, Canada. Using prepared 18-h growth of P. syringae (HER1102) in 50 ml tryptic soy broth (TSB), bacteriophage phi 6 (HER102) was propagated from lyophilized stock by reconstituting with 1 ml of prewarmed (37°C) TSB. Reconstituted phi 6 (500 μl) was transferred into 50 ml fresh TSB with 100 μl of overnight host P. syringae, followed by gentle agitation using a vortex and incubating with agitation (100 to 110 rpm) for 18 h at 22°C. The phi 6 lysate was then filter sterilized using a 0.22 μm syringe filter (polyvinylidene difluoride [PVDF]; Millex-GV; Millipore, Burlington, MA) into a sterile 50-ml tube. The filtrate (phi 6) was protected from light and stored in the dark at 4°C until experiments were performed. The phi 6 stock titer was approximately 108 PFU/ml.
Inoculation and environmental exposure.
A 20% solution of artificial test soil (ATS; Healthmark Industries Company Inc., Fraser, MI) was prepared in phage buffer (SM buffer) according to the formulation in Cold Spring Harbor Protocols (47) (NaCl, MgSO4·7H2O, Tris-Cl [1 M, pH 7.5]), and used as a body fluid simulant, with ATS containing proteins (albumin), hemoglobin, carbohydrates, cellulose, and lipids. The stock phi 6 was diluted in series to obtain a 106 PFU/ml suspension in ATS. The coupons were inoculated with 10 μl of 106 PFU/ml suspension in ATS, resulting in an inoculum of 104 PFU/cm2. Negative controls consisted of ATS (20%) without phi 6 inoculated onto coupons. The test coupons were placed in open petri dishes in triplicate, along with the negative coupons, in a Caron model 6030 environmental chamber (Marietta, OH) at two controlled temperature (T) and relative humidity (RH) levels: 26°C and 57% RH (AH = 14.4 g/m3) and 18°C and 20% RH (AH = 3.0 g/m3). Three inoculated coupons and a negative control were removed and processed immediately (T0) and at designated time points until phi 6 PFU could no longer be detected (limit of detection from coupons was ≤2 PFU/cm2). Each experimental condition was repeated twice with triplicate coupons for a total of 6 coupons at each time point. One exception is SS, which had a total of 5 samples for each time point for the high-AH (14.4 g/m3 AH) environmental condition.
Recovery.
The phage(s) were dislodged from the coupons in 5 ml phosphate-buffered saline with Tween 80 (PBST; 0.02%) (0.01 M PBS [7.2 to 7.4 pH] and Tween 80 [0.02%]) by alternating between vortex and sonicating bath for 30 s each, repeating the rotation 3 times (48). The eluate was diluted in series in SM buffer. One ml of sample was plated with 500 μl of host P. syringae in tryptic soy agar (Becton, Dickinson and Company, Franklin Lakes, NJ), using a double agar overlay method (49). The overlay plates incubated for 18 to 24 h (22°C) before we counted the PFU. Coupon removal, processing, and plating continued until no infective phages (as determined by PFU) were observed. PFU per milliliter were calculated based on dilution factors and log10 transformed. Results were calculated in both RH and AH to compare to other studies.
Statistical analysis.
SAS v9.4 (SAS, Cary, NC) was used to create linear models that assessed the potential relationships between the mean log10 change of virus concentration on SS, PL, TC, and UC at two environmental conditions (3.0 g/m3, 14.4 g/m3). For these analyses, the data points were used individually and not averaged. Least-squares methods were used to fit the model and determine the rate of log10 reductions written as decay rate (log10 reduction per day or hour) in Table 1 and 2 for these data; r2 was used to assess goodness of fit of the model. Analysis of variance and the F statistic were used to test the differences between various materials (i.e., SS versus PL, TC versus UC, and porous versus nonporous) under the same absolute humidity, where significance was set at a P value of <0.05.
ACKNOWLEDGMENTS
The authors acknowledge Matthew Arduino for his help in the design of this experiment and Amanda Lyons for her technical assistance.
REFERENCES
- 1.Agua-Agum J, Allegranzi B, Ariyarajah A, Aylward R, Blake IM, Barboza P, Bausch D, Brennan RJ, Clement P, Coffey P, Cori A, Donnelly CA, Dorigatti I, Drury P, Durski K, Dye C, Eckmanns T, Ferguson NM, Fraser C, Garcia E, Garske T, Gasasira A, Gurry C, Hamblion E, Hinsley W, Holden R, Holmes D, Hugonnet S, Jaramillo Gutierrez G, Jombart T, Kelley E, Santhana R, Mahmoud N, Mills HL, Mohamed Y, Musa E, Naidoo D, Nedjati-Gilani G, Newton E, Norton I, Nouvellet P, Perkins D, Perkins M, Riley S, Schumacher D, Shah A, Tang M, Varsaneux O, Van Kerkhove MD, WHO Ebola Response Team. 2016. After Ebola in West Africa–unpredictable risks, preventable epidemics. N Engl J Med 375:587–596. doi: 10.1056/NEJMsr1513109. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy M. 2014. Texas healthcare worker is diagnosed with Ebola. Br Med J 349:6200. doi: 10.1136/bmj.g6200. [DOI] [PubMed] [Google Scholar]
- 3.Selvaraj SA, Lee KE, Harrell M, Ivanov I, Allegranzi B. 2018. Infection rates and risk factors for infection among health workers during Ebola and marburg virus outbreaks: a systematic review. J Infect Dis 218:S679–S689. doi: 10.1093/infdis/jiy435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drosten C, Günther S, Preiser W, van der Werf S, Brodt H-R, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RAM, Berger A, Burguière A-M, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra J-C, Müller S, Rickerts V, Stürmer M, Vieth S, Klenk H-D, Osterhaus ADME, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y-C, Huang L-M, Chan C-C, Su C-P, Chang S-C, Chang Y-Y, Chen M-L, Hung C-C, Chen W-J, Lin F-Y, Lee Y-T, SARS Research Group of National Taiwan University College of Medicine and National Taiwan University Hospital. 2004. SARS in hospital emergency room. Emerg Infect Dis 10:782–788. doi: 10.3201/eid1005.030579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowell SF, Simmerman JM, Erdman DD, Wu J-SJ, Chaovavanich A, Javadi M, Yang J-Y, Anderson LJ, Tong S, Ho MS. 2004. Severe acute respiratory syndrome coronavirus on hospital surfaces. Clin Infect Dis 39:652–657. doi: 10.1086/422652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL, Thornburg NJ, Gerber SI, Lloyd-Smith JO, de Wit E, Munster VJ. 2020. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med 382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization. 2014. Interim infection prevention and control guidance for care of patients with suspected or confirmed filovirus haemorrhagic fever in health-care settings, with focus on Ebola: interim guidance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 9.World Health Organization. 2020. Infection prevention and control during health care when novel coronavirus (nCoV) infection is suspected: interim guidance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 10.Chowell G, Abdirizak F, Lee S, Lee J, Jung E, Nishiura H, Viboud C. 2015. Transmission characteristics of MERS and SARS in the healthcare setting: a comparative study. BMC Med 13:210. doi: 10.1186/s12916-015-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bausch DG, Towner JS, Dowell SF, Kaducu F, Lukwiya M, Sanchez A, Nichol ST, Ksiazek TG, Rollin PE. 2007. Assessment of the risk of Ebola virus transmission from bodily fluids and fomites. J Infect Dis 196(Suppl 2):S142–S147. doi: 10.1086/520545. [DOI] [PubMed] [Google Scholar]
- 12.Palich R, Irenge LM, de Sainte Fare EB, Augier A, Malvy D, Gala J-L. 2017. Ebola virus RNA detection on fomites in close proximity to confirmed Ebola patients; N’Zerekore, Guinea, 2015. PLoS One 12:e0177350. doi: 10.1371/journal.pone.0177350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poliquin PG, Vogt F, Kasztura M, Leung A, Deschambault Y, Van den Bergh R, Dorion C, Maes P, Kamara A, Kobinger G, Sprecher A, Strong JE. 2016. Environmental contamination and persistence of Ebola virus RNA in an Ebola treatment center. J Infect Dis 214:S145–S152. doi: 10.1093/infdis/jiw198. [DOI] [PubMed] [Google Scholar]
- 14.Aquino de Carvalho N, Stachler EN, Cimabue N, Bibby K. 2017. Evaluation of phi6 persistence and suitability as an enveloped virus surrogate. Environ Sci Technol 51:8692–8700. doi: 10.1021/acs.est.7b01296. [DOI] [PubMed] [Google Scholar]
- 15.Adcock NJ, Rice EW, Sivaganesan M, Brown JD, Stallknecht DE, Swayne DE. 2009. The use of bacteriophages of the family Cystoviridae as surrogates for H5N1 highly pathogenic avian influenza viruses in persistence and inactivation studies. J Environ Sci Health A Tox Hazard Subst Environ Eng 44:1362–1366. doi: 10.1080/10934520903217054. [DOI] [PubMed] [Google Scholar]
- 16.Phillpotts RJ, Thomas RJ, Beedham RJ, Platt SD, Vale CA. 2010. The cystovirus phi6 as a simulant for Venezuelan equine encephalitis virus. Aerobiologia 26:301–309. doi: 10.1007/s10453-010-9166-y. [DOI] [Google Scholar]
- 17.Bearden RL, Casanova LM. 2016. Survival of an enveloped virus on toys. Pediatr Infect Dis J 35:923–924. doi: 10.1097/INF.0000000000001193. [DOI] [PubMed] [Google Scholar]
- 18.Casanova LM, Weaver SR. 2015. Evaluation of eluents for the recovery of an enveloped virus from hands by whole-hand sampling. J Appl Microbiol 118:1210–1216. doi: 10.1111/jam.12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casanova LM, Waka B. 2013. Survival of a surrogate virus on N95 respirator material. Infect Control Hosp Epidemiol 34:1334–1335. doi: 10.1086/673994. [DOI] [PubMed] [Google Scholar]
- 20.Casanova LM, Teal LJ, Sickbert-Bennett EE, Anderson DJ, Sexton DJ, Rutala WA, Weber DJ, CDC Prevention Epicenters Program. 2016. Assessment of self-contamination during removal of personal protective equipment for Ebola patient care. Infect Control Hosp Epidemiol 37:1156–1161. doi: 10.1017/ice.2016.169. [DOI] [PubMed] [Google Scholar]
- 21.Casanova L, Alfano-Sobsey E, Rutala WA, Weber DJ, Sobsey M. 2008. Virus transfer from personal protective equipment to healthcare employees' skin and clothing. Emerg Infect Dis 14:1291–1293. doi: 10.3201/eid1408.080085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer R, Judson S, Miazgowicz K, Bushmaker T, Prescott J, Munster VJ. 2015. Ebola virus stability on surfaces and in fluids in simulated outbreak environments. Emerg Infect Dis 21:1243–1246. doi: 10.3201/eid2107.150253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piercy TJ, Smither SJ, Steward JA, Eastaugh L, Lever MS. 2010. The survival of filoviruses in liquids, on solid substrates and in a dynamic aerosol. J Appl Microbiol 109:1531–1539. doi: 10.1111/j.1365-2672.2010.04778.x. [DOI] [PubMed] [Google Scholar]
- 24.Bibby K, Fischer RJ, Casson LW, Stachler E, Haas CN, Munster VJ. 2015. Persistence of Ebola virus in sterilized wastewater. Environ Sci Technol Lett 2:245–249. doi: 10.1021/acs.estlett.5b00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sagripanti JL, Rom AM, Holland LE. 2010. Persistence in darkness of virulent alphaviruses, Ebola virus, and Lassa virus deposited on solid surfaces. Arch Virol 155:2035–2039. doi: 10.1007/s00705-010-0791-0. [DOI] [PubMed] [Google Scholar]
- 26.Prussin AJ, Schwake DO, Lin K, Gallagher DL, Buttling L, Marr L. 2018. Survival of the enveloped virus Phi6 in droplets as a function of relative humidity, absolute humidity, and temperature. Appl Environ Microbiol 84:e00551-18. doi: 10.1128/AEM.00551-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan J, Chan M, VanderZaag A. 2017. Inactivation of avian influenza viruses on porous and non‐porous surfaces is enhanced by elevating absolute humidity. Transbound Emerg Dis 64:1254–1261. doi: 10.1111/tbed.12499. [DOI] [PubMed] [Google Scholar]
- 28.Casanova LM, Jeon S, Rutala WA, Weber DJ, Sobsey MD. 2010. Effects of air temperature and relative humidity on coronavirus survival on surfaces. Appl Environ Microbiol 76:2712–2717. doi: 10.1128/AEM.02291-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Doremalen N, Bushmaker T, Munster V. 2013. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Emerg Infect Dis 18:20590. doi: 10.2807/1560-7917.ES2013.18.38.20590. [DOI] [PubMed] [Google Scholar]
- 30.Cook BW, Cutts TA, Nikiforuk AM, Poliquin PG, Court DA, Strong JE, Theriault SS. 2015. Evaluating environmental persistence and disinfection of the Ebola virus Makona variant. Viruses 7:1975–1986. doi: 10.3390/v7041975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bibby K, Casson LW, Stachler E, Haas CN. 2015. Ebola virus persistence in the environment: state of the knowledge and research needs. Environ Sci Technol Lett 2:2–6. doi: 10.1021/ez5003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuit M, Miller DM, Reddick-Elick MS, Wlazlowski CB, Filone CM, Herzog A, Colf LA, Wahl-Jensen V, Hevey M, Noah JW. 2016. Differences in the comparative stability of Ebola virus Makona-C05 and Yambuku-Mayinga in blood. PLoS One 11:e0148476. doi: 10.1371/journal.pone.0148476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chin A, Chu J, Perera M, Hui K, Yen H-L, Chan M, Peiris M, Poon L. 2020. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe 1:e10. doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sizun J, Yu MW, Talbot PJ. 2000. Survival of human coronaviruses 229E and OC43 in suspension and after drying on surfaces: a possible source of hospital-acquired infections. J Hosp Infect 46:55–60. doi: 10.1053/jhin.2000.0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters JL, Cho DK, Aluisio AR, Kennedy SB, Massaquoi MBF, Sahr F, Perera SM, Levine AC. 2019. Environmental temperature and case fatality of patients with Ebola virus disease in Sierra Leone and Liberia, 2014–2015: a retrospective cohort study. Trop Med Int Health 24:23–30. doi: 10.1111/tmi.13166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coulliette A, Perry K, Edwards J, Noble-Wang J. 2013. Persistence of the 2009 pandemic influenza A (H1N1) virus on N95 respirators. Appl Environ Microbiol 79:2148–2155. doi: 10.1128/AEM.03850-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaman J, Kohn M. 2009. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc Natl Acad Sci U S A 106:3243–3248. doi: 10.1073/pnas.0806852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson SS, Flury M, Yates MV, Jury WA. 1998. Role of the air-water-solid interface in bacteriophage sorption experiments. Appl Environ Microbiol 64:304–309. doi: 10.1128/AEM.64.1.304-309.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson SS, Yates MV. 1999. Bacteriophage inactivation at the air-water-solid interface in dynamic batch systems. Appl Environ Microbiol 65:1186–1190. doi: 10.1128/AEM.65.3.1186-1190.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trouwborst T, Kuyper S, De Jong J, Plantinga A. 1974. Inactivation of some bacterial and animal viruses by exposure to liquid-air interfaces. J Gen Virol 24:155–165. doi: 10.1099/0022-1317-24-1-155. [DOI] [PubMed] [Google Scholar]
- 41.Yang W, Elankumaran S, Marr LC. 2012. Relationship between humidity and influenza A viability in droplets and implications for influenza’s seasonality. PLoS One 7:e46789. doi: 10.1371/journal.pone.0046789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mateu MG. 2012. Mechanical properties of viruses analyzed by atomic force microscopy: a virological perspective. Virus Res 168:1–22. doi: 10.1016/j.virusres.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Wood JP, Richter W, Sunderman M, Calfee MW, Serre S, Mickelsen L. 2020. Evaluating the environmental persistence and inactivation of MS2 bacteriophage and the presumed Ebola virus surrogate phi6 using low concentration hydrogen peroxide vapor. Environ Sci Technol 54:3581–3590. doi: 10.1021/acs.est.9b06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai MY, Cheng PK, Lim WW. 2005. Survival of severe acute respiratory syndrome coronavirus. Clin Infect Dis 41:e67–e71. doi: 10.1086/433186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abad FX, Pinto RM, Bosch A. 1994. Survival of enteric viruses on environmental fomites. Appl Environ Microbiol 60:3704–3710. doi: 10.1128/AEM.60.10.3704-3710.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeargin T, Buckley D, Fraser A, Jiang X. 2016. The survival and inactivation of enteric viruses on soft surfaces: a systematic review of the literature. Am J Infect Control 44:1365–1373. doi: 10.1016/j.ajic.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 47.Cold Spring Harbor Laboratory. 2006. SM buffer. Cold Spring Harb Protoc 2006. doi: 10.1101/pdb.rec8111. [DOI] [Google Scholar]
- 48.Goeres DM, Loetterle LR, Hamilton MA, Murga R, Kirby DW, Donlan R. 2005. Statistical assessment of a laboratory method for growing biofilms. Microbiology 151:757–762. doi: 10.1099/mic.0.27709-0. [DOI] [PubMed] [Google Scholar]
- 49.Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. 2009. Enumeration of bacteriophages by double agar overlay plaque assay, p 69–76. In Clokie MRJ, Kropinski AM (ed), Bacteriophages: methods and protocols, volume 1: isolation, characterization, and interactions. Humana Press, Totowa, NJ. doi: 10.1007/978-1-60327-164-6_7. [DOI] [PubMed] [Google Scholar]
- 50.Chan K, Peiris J, Lam S, Poon L, Yuen K, Seto W. 2011. The effects of temperature and relative humidity on the viability of the SARS coronavirus. Adv Virol 2011:734690. doi: 10.1155/2011/734690. [DOI] [PMC free article] [PubMed] [Google Scholar]