Abstract
Background
Oral squamous cell carcinoma (OSCC) may develop from a variety of oral potentially malignant disorders, but the mechanism of malignant transformation is still unknown. Among them, oral lichen planus (OLP) has a high prevalence. Previous studies have shown that α-enolase (ENO1) can promote cell proliferation and play an important role in tumorigenesis. In this study, we aim to explore the mechanism of ENO1 regulation in the process of OSCC tumorigenesis from OLP.
Methods
ENO1 expression in tissues was determined by real-time quantitative PCR and immunohistochemistry. ENO1 was knocked down in cal-27 to observe the change in cell proliferation. Then, RNA-seq and bioinformatics analyses were conducted between OLP and OSCC samples. The expression of circ-AMOTL1, miRNA-22-3p, and miRNA-1294 was assessed using the real-time quantitative PCR. With knockdown and overexpression of circ-AMOTL1 in vitro, the change of ENO1 in the mRNA level was also assessed.
Results
ENO1 was enhanced in the OSCC samples in comparison with OLP. Immunohistochemistry and real-time quantitative PCR results showed that ENO1 was significantly higher in OSCC tissue than in the OLP group, with a statistically significant difference (p<0.05). When ENO1 was knocked down in cal-27, cell proliferation was inhibited (p<0.05). The expression of miR-22-3p and miR-1294 was decreased in OSCC tissues, whereas ENO1 and circ-AMOTL1 increased. In an in vitro study, knockdown of circ-AMOTL1 resulted in a decrease of ENO1, while overexpression of circ-AMOTL1 led to an increase of ENO1 in the mRNA level.
Conclusion
We confirmed that ENO1 expression was elevated in OSCC and increased cell proliferation. In an in vitro study, ENO1 expression was promoted by circ-AMOTL1. ENO1 may play a role as a tumor-promoting gene in OSCC through the circ-AMOTL1/miR-22-3p/miR-1294 network. These novel findings may shed further light on the pathogenesis from OLP to OSCC and the potential precursor markers.
Keywords: oral lichen planus, oral squamous cell carcinoma, ENO1, AMOTL1, ceRNA
Introduction
Oral cancer is a malignant neoplasia which arises on the lip or oral cavity. According to the latest published GLOBOCAN report, there were 354,864 new cases of oral cancer (ICD-10 code C00-08: lip, oral cavity) and 177,384 deaths worldwide in 2018.1 Despite advances in diagnosis and treatment, the five-year survival rate is less than 50%, and the recurrence rate is about 65%.2 Traditionally, oral cancer was defined as oral squamous cell carcinoma (OSCC).1 Thus, OSCC is an important component of oral cancer. The primary cause of low survival rate and high recurrence rate lies in that OSCC is diagnosed at advanced stages in most cases. Therefore, research on the pathogenesis of OSCC is crucial to reduce morbidity and mortality. OSCC may develop from a variety of oral potentially malignant disorders (OPMDs). Among them, oral lichen planus (OLP) has a high prevalence3 and the malignancy rate is underestimated.4 OLP is a chronic mucocutaneous disorder with different kinds of clinical forms and may develop into OSCC. Thus, the process of OSCC tumorigenesis from OLP should be fully investigated.
Previous studies suggested that metabolic disorder was part of the causes and even an activator of tumorigenesis.5,6 Under fully oxygenated conditions, cancer cells were characterized by accelerated glycolysis. This metabolic shift was observed by Otto Warburg, and was subsequently named the “Warburg Effect.”7 Considered as a hallmark of cancer, the role of the Warburg Effect has been explored for nearly a century. With a variety of genomic techniques and cancer biology studies, researchers started to realize that the orchestrated performance of oncogenes, metabolic disorder, and tumor suppressors resulted in the increase of glycolysis and its production: lactate acts as a key player in tumorigenesis by upregulating monocarboxylate transporter 1 (MCT1) and monocarboxylate transporter 4 (MCT4) expression in the regulation of tumor growth and tumorigenesis.6 Recently, Dai et al confirmed that α-enolase (ENO1) mediates the metabolic shift to glycolysis.8 Several studies on cancer cells showed that raised ENO1 can result in the promotion of cell proliferation.9,10 However, the mechanism of ENO1 regulation has not been well studied yet.
Proteins are translated from mRNA, and the latest mRNA regulation mechanism holds that competing endogenous RNAs (mRNA, LncRNA, circle RNA, pseudogenes, etc.) can competitively bind to microRNAs, thereby removing the regulation of target genes by microRNAs. Non-coding RNA (ncRNA) is involved in many life processes and plays an important role. Its regulation can extend to almost the whole life process, such as development, proliferation, apoptosis, differentiation, carcinogenesis, etc. Long noncoding RNAs (LncRNAs) and circle RNAs (circRNAs) are the two main types of ncRNAs. MicroRNA (miRNA) is a type of short non-coding RNA, about 22nt in length. By binding to the 3‘UTR region of target gene mRNA, microRNA can directly degrade the target gene or block its translation into proteins with physiological functions, and inhibit the expression of the target gene at the post-transcriptional level. This mechanism widely exists in mRNA regulation. However, the regulation of ENO1 at the mRNA level has not been well understood.
In this research project, we first identified that ENO1 can promote proliferation in cal-27, and its expression increases in OSCC when compared to OLP in clinical samples. Then, we performed RNA-seq and bioinformatics analysis between OLP and OSCC samples to explore the regulation of ENO1. Finally, we verified it at both tissue and cell level.
Materials and Methods
Clinical Samples Collection
This study was approved by the ethics review committee of Huashan Hospital (Fudan University). All biopsy samples of 6 OLP and 6 OSCC tissues were collected from 2017 to 2019 at Huashan Hospital (Table 1). The diagnostic criteria of OLP were from the American Academy of Oral and Maxillofacial Pathology in 2016.11 Samples of normal mucosal tissues were collected in wisdom teeth extraction operations without clinically visible inflammation under sterile conditions at Huashan Hospital, Fudan University (Shanghai, China).
Table 1.
Clinical Characteristics of Patients
| Patient ID | Gender | Age | Site of Lesions | Type of OLP Lesions | TNM Status | Associated Condition | HE |
|---|---|---|---|---|---|---|---|
| 1 | F | 60 | BM | II | OLP | ||
| 2 | F | 40 | BM | I, II | OLP | ||
| 3 | F | 40 | BM | II | OLP | ||
| 4 | F | 58 | BM | II | OLP | ||
| 5 | F | 68 | T | II | OLP | ||
| 6 | F | 61 | BM | II | OLP | ||
| 7 | M | 40 | BM | T1N0M0 | OSCC | ||
| 8 | F | 62 | T-L | T1N0M0 | Hypertension, diabetes | OSCC | |
| 9 | M | 62 | G | T1N0M0 | Hypertension | OSCC | |
| 10 | M | 67 | LU | T1N0M0 | Heart disease | OSCC | |
| 11 | M | 53 | T | T2N1M0 | OSCC | ||
| 12 | F | 68 | T | T2N1M0 | Heart disease | OSCC |
Abbreviations: BM, buccal mucosa; G, gingiva; L, left; LU, under of lip; OLP, oral lichen planus; OLP I, reticular type; OLP II, atrophic-erosive type; OSCC, oral squamous cell carcinoma; T, tongue; TNM, tumor node metastasis.
Cell Culture
Cal-27 and HaCaT cells were presented as a gift from Ninth People’s Hospital, Shanghai Jiaotong University. This was approved by the ethics committee of Huashan Hospital. Cells were cultured in DMEM medium (Gibco, USA) with 10% fetal bovine serum (Hyclone, USA) at 37°C in the presence of 5% CO2. HaCaT cells were stimulated with 10 μg/mL lipopolysaccharide (LPS) for 24 h as the OLP cell model.
Cell Transfection
Small interfering RNAs (siRNAs) of ENO1 were synthesized by Proteintech (Shanghai, China). The aim of cell transfection was to knockdown ENO1 expression. It was transfected with lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions.
Lentivirus Production and Cell Infection
The sh-circ-AMOTL1 and circ-AMOTL1 sequences were, respectively, inserted into GV493 and GV367 vectors containing green fluorescent protein (GENE Company, Shanghai, China). Cells were infected according to the manufacturer’s instructions with MOI=100.
CCK8 Assay
Cell counting kit 8 (CCK8, Dojindo, Japan) assay was used to test cell proliferation according to the manufacturer’s instructions. Cells (2000 cells per well) were seeded into 96-well plates. After knockdown ENO1, OD values were tested at 450 nm at 24, 48, and 72 h.
Immunohistochemical (IHC) Analysis
Tissue sections were deparaffinized with xylene and rehydrated through a series of ethanol solutions with descending concentrations. The sections were further treated following the procedure of UltraSensitiveTM SP IHC Kit (KIT-9710, MXB Biotechnologies, China). Antigen retrieval was performed by heating in citrate buffer (pH 6.0). Then, the sections were incubated in Rabbit ENO1 antibody (1:2000, Abcam, Cambridge, MA, USA, cat# ab227978) overnight at 4°C. Fresh DAB solution (Sheng-Gong Technologies, Shanghai, China, cat# E670033) was used to stain sections.
Real-Time Quantitative PCR (qPCR)
Total RNA was extracted by using a TRIzol reagent (Invitrogen, cat# 15,596–026). cDNA was synthesized by using PrimeScriptTM RT Master Mix Kit (TaKaRa, cat# RR036A). The qPCR was performed on the StepOne™ Real-Time PCR System (Applied Biosystems) with a SYBR Premix Ex Taq Kit (TaKaRa, cat# RR420A). All the reactions were performed in triplicate. According to the manufacturer’s instructions, the PCRs were conducted at 95°C for 30 s, followed by 40 cycles of 95°C for 3 s, and 60°C for 30 s in the StepOne™ Real-Time PCR System (Applied Biosystems). The expression value was calculated by the comparative Ct method with formula 2−ΔΔCt. Human β-Actin RNA was amplified as an endogenous control for ENO1. Human 18SrRNA was amplified as an endogenous control for circ-AMOTL1. Human U6 RNA was amplified as an endogenous control for miR-22-3p/miR-1294. The primers used are listed in Table 2.
Table 2.
The Primers and Oligonucleotides Used in This Work
| Name | Sequence (5ʹ-3ʹ) |
|---|---|
| ENO1 forward primer | 5′- GTTCACAGCCAGTGCAGGAA-3′ |
| ENO1 reverse primer | 5′- GGAGGCAGTTGCAGGACTTC-3′ |
| cric-AMOTL1 forward primer | 5′- ATGCTCCACGAGATGGTCAAG-3′ |
| cric-AMOTL1 reverse primer | 5′- ACTACTGGGGCTATAACAAGCAG-3′ |
| miR-1294 RT primer | 5′-GTCGTATCCAGTGCAGGGTCCGAGGT ATTCGCACTGGATACGACAGACAA-3′ |
| miR-1294 forward primer | 5′-GCGTGTGAGGTTGGCATTG-3′ |
| miR-1294 reverse primer | 5′-AGTGCAGGGTCCGAGGTATT-3′ |
| miR-22-3p RT primer | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTAT TCGCACTGGATACGACACAGTT-3′ |
| miR-22-3p forward primer | 5′- GCGAAGCTGCCAGTTGAAG-3′ |
| miR-22-3p reverse primer | 5′- AGTGCAGGGTCCGAGGTATT-3′ |
| β-actin forward primer | 5′-CCTGGCACCCAGCACAAT-3′ |
| β-actin reverse primer | 5′-GCTGATCCACATCTGCTGGAA-3′ |
| U6 RT primer | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATT CGCACTGGATACGACAAAATA-3′ |
| U6 forward primer | 5′-AGAGAAGATTAGCATGGCCCCTG-3′ |
| U6 reverse primer | 5′-ATCCAGTGCAGGGTCCGAGG-3′ |
| 18S rRNA forward primer | 5′- GGAGTATGGTTGCAAAGCTGA-3′ |
| 18S rRNA reverse primer | 5′- ATCTGTCAATCCTGTCCGTGT-3′ |
RNA Extraction and Sequencing
Total RNAs were extracted by using a TRIzol reagent according to the manufacturer’s manual. The cDNA library was sequenced on Illumina HiSeq™ (Illumina Inc., San Diego, CA, USA). For each sample, raw data quality was analyzed and visually evaluated with FastQC. Clean data sequences were then aligned with the reference human genome in HISAT2 software. Expression statistics were performed by using the transcripts per million (TPM) means. The annotated mRNA gene expression fold (Shift versus Normal) and p-value were acquired by DEGseq software. Transcripts or genes were considered differentially expressed when the fold change (FC) >2 (Log2FC>1) or <0.5, and p-value or FDR were less than 0.05.
GO Functional and KEGG Enrichment Analysis
Based on differentially expressed genes, Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG), pathway analyses were performed by using the DAVID Database. A threshold of p<0.05 was set for enrichment identification. Routinely, GO analysis includes biological process (BP), cellular component (CC), and molecular function (MF) terms.
The Protein–Protein Interaction
The protein–protein interaction (PPI) network was constructed by using the STRING database. Only mRNAs that were expressed differentially and associated with ENO1 were screened for network construction.
Statistical Analysis
All continuous data were described as mean ± standard and compared by using either Student’s t test (for normally distributed variables) or Mann–Whitney U-test (not normally distributed). All statistical tests were two-sided, and a p-value <0.05 is considered statistically significant. SAS 9.4 software was used to analyze the distribution of data and perform statistical tests.
Results
ENO1 Facilitating Cell Proliferation Was Increased in OSCC
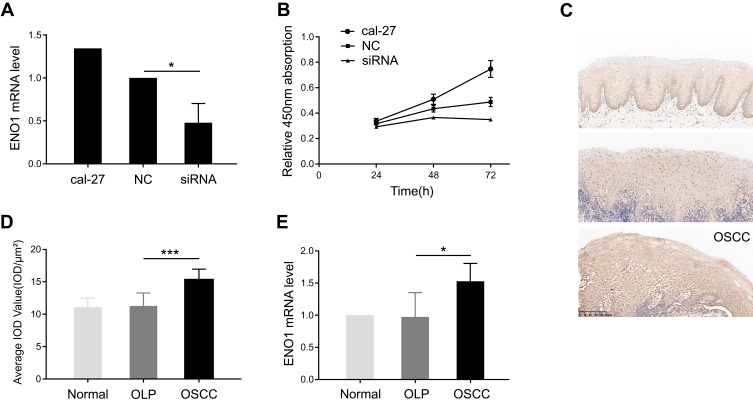
First, the effect of ENO1 on cell proliferation was investigated in cal-27. We knocked down ENO1 expression in cal-27, and the cell proliferation was inhibited (Figure 1A and B), which suggested that ENO1 promoted cell proliferation in OSCC. Second, we also noticed that the expression of ENO1 was enhanced in the OSCC samples in comparison with the OLP ones (Figure 1C and D). The increase in ENO1 expression was further validated through qPCR with three groups of samples (3 normal, 3 OLP, and 3 OSCC) (p < 0.05) (Figure 1E). Real-time quantitative PCR results showed that the mRNA level of ENO1 was changed. Recently, many scholars have reported that mRNA with protein-coding function can be regulated by competing endogenous RNAs (mRNA, LncRNA, circle RNA, pseudogenes, etc.).12,13 At the same time, researchers have constructed the transcriptome expression profile of the OLP-OSCC disease process, and suggested that noncoding RNAs could play an important role in this disease process.14 Therefore, we explored the noncoding RNAs which acted as a competing endogenous RNA to regulate ENO1 in this disease process through RNA-seq. Collectively, ENO1 may participate in tumorigenesis of OSCC by promoting cell proliferation.
Figure 1.
ENO1 facilitating cell proliferation was increased in oral squamous cell carcinoma (OSCC).
Notes: (A) ENO1 was knocked down, which was confirmed by real-time quantitative PCR. (B) Cell proliferation was inhibited by knocking down ENO1. (C) The expression of ENO1 by immunohistochemistry (IHC) staining in normal, OLP, and OSCC groups. (D) Semi-quantitative analysis demonstrated increased expression level of ENO1 in OSCC tissue compared to OLP. (E) Validation of increase in ENO1 expression through real-time quantitative PCR. t test: *p < 0.05, ***p < 0.001.
Abbreviations: NC, negative control; OLP, oral lichen planus; OSCC, oral squamous cell carcinoma; PCR, polymerase chain reaction.
RNA-Seq to Explore the Regulation Network Associated with ENO1
To identify the regulation network associated with ENO1 in tumorigenesis of OSCC, we further conducted transcriptome sequencing from 12 samples (6 OLP and 6 OSCC samples) and bioinformatics analysis to determine the miRNA targets and candidate competing endogenous RNAs. There were 6734 differentially expressed mRNA genes (3327 upregulateregulated and 3408 downregulateregulated) and 1985 differentially expressed ceRNAs (608 upregulateregulated and 1377 downregulateregulated) (Figure 2A–D). With this result, we found that the circRNA AMOTL1 was significantly increased in OSCC samples (fold change = 2.05, p < 0.01) and was the most correlated with ENO1 expression. In our study, the influence of the circ-AMOTL1/ENO1 axis on OSCC was analyzed as well. With the TCGA head and neck squamous cell carcinoma database, we observed a positive relationship between the expression of circ-AMOTL1 and ENO1 (Figure 2E). The potential binding miRNAs were screened through data from starBase according to the following standard: clip data strict stringency ≥5 and degradome high stringency ≥3. There were 69 binding miRNAs towards circ-AMOTL1 and 45 towards ENO1, and among the intersection factors, several miRNAs were included: hsa-miR-22-3p, hsa-miR-1294, hsa-miR-330-3p, hsa-miR-485-5p, and hsa-miR-193a-5p, etc. Next, these miRNAs were sorted by the clipExpNum, and it was found that hsa-miR-22-3p and hsa-miR-1294 were highly likely to target both ENO1 and circ-AMOTL1 (Figure 2F–I).
Figure 2.
RNA-seq to explore the regulation network associated with ENO1.
Notes: Heat map showing differentially expressed noncoding RNAs (A) and mRNAs (B), when comparing OSCC with OLP. Red denotes high expression and blue denotes low expression. The transcriptome sequencing reveals 6734 differentially expressed mRNA genes (C), 3327 upregulateregulated and 3408 downregulateregulated; and 1985 differentially expressed ncRNAs (D), 608 upregulateregulated and 1377 downregulateregulated. (E) There is a highly positive correlation between the expression of circ-AMOTL1 and ENO1 in TCGA head and neck squamous cell carcinoma database. (F) Binding sites of ENO1 and miR-22-3p. (G) Binding sites of ENO1 and miR-1294. (H) Binding sites of circ-AMOTL1 and miR-22-3p. (I) Binding sites of circ-AMOTL1 and miR-1294.
Abbreviations: OLP, oral lichen planus; OSCC, oral squamous cell carcinoma.
Circ-AMOTL1 Increased in OSCC and Regulated ENO1 Positively
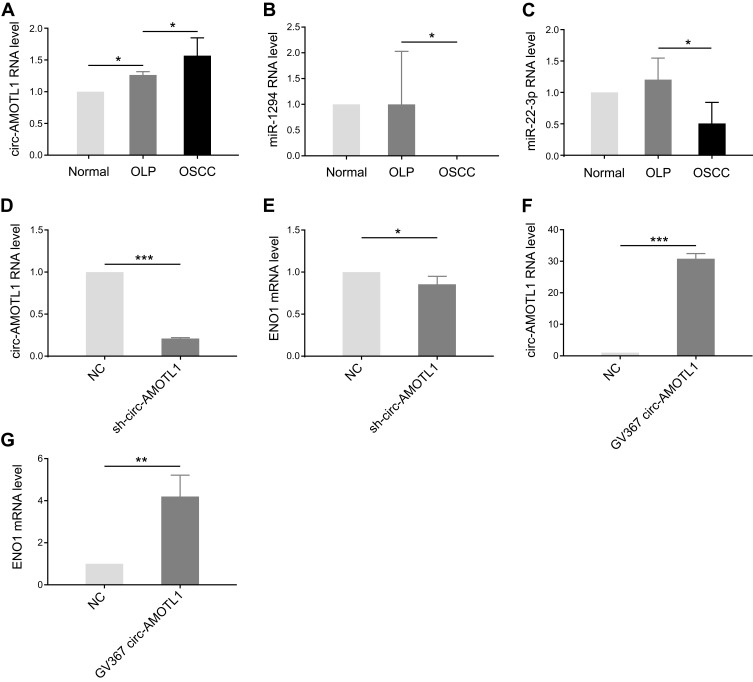
With the results of bioinformatic analysis, circ-AMOTL1, miR-22-3p, and miR-1294 were selected for identification in clinical samples through qPCR. As expected, circ-AMOTL1 expression was enhanced in OSCC samples in comparison with OLP (Figure 3A), and miR-22-3p and miR-1294 expression were decreased (Figure 3B and C). In the OLP cell model, we found that the knocking down of circ-AMOTL1 caused a reduction of ENO1 in the mRNA level (Figure 3D and E). Furthermore, overexpressing circ-AMOTL1 caused an increase in ENO1 in the mRNA level (Figure 3F and G). Collectively, these results suggested that circ-AMOTL1 positively regulated ENO1, and this regulation may be conducted through miR-22-3p/miR-1294.
Figure 3.
Circ-AMOTL1 increased in OSCC and regulated ENO1 positively.
Notes: (A) Knockdown efficiency of circ-AMOTL1 by sh-circ-AMOTL1 in the OLP cell model. (B) The expression of ENO1 in mRNA level after knocking down circ-AMOTL1. (C) Overexpression efficiency of circ-AMOTL1 by GV367 circ-AMOTL1 in the OLP cell model. (D) The expression of ENO1 in mRNA level after overexpression of circ-AMOTL1. (E) The circ-AMOTL1 expression is enhanced in OSCC samples in comparison with OLP in real-time quantitative PCR validation. (F–G) miR-22-3p and miR-1294 expression levels are decreased in OSCC. t test: *p < 0.05, **p < 0.01, ***p < 0.001.
Abbreviations: NC, negative control; OLP, oral lichen planus; OSCC, oral squamous cell carcinoma; PCR, polymerase chain reaction.
KEGG/GO Enrichments and PPI Analysis Focusing on ENO1
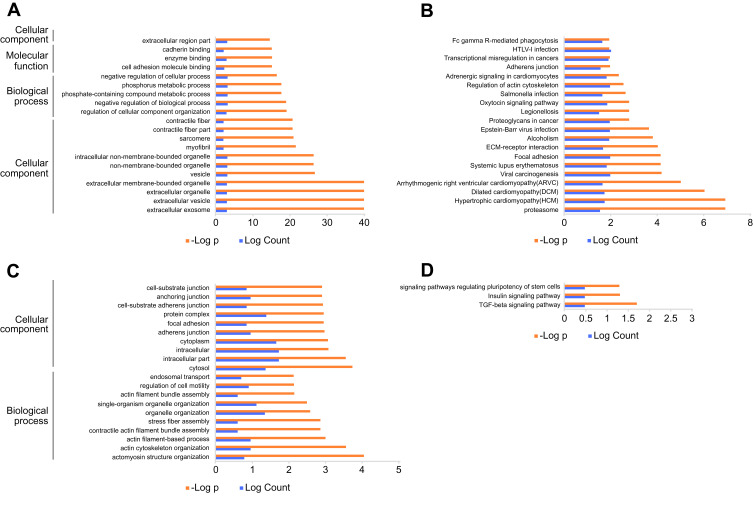
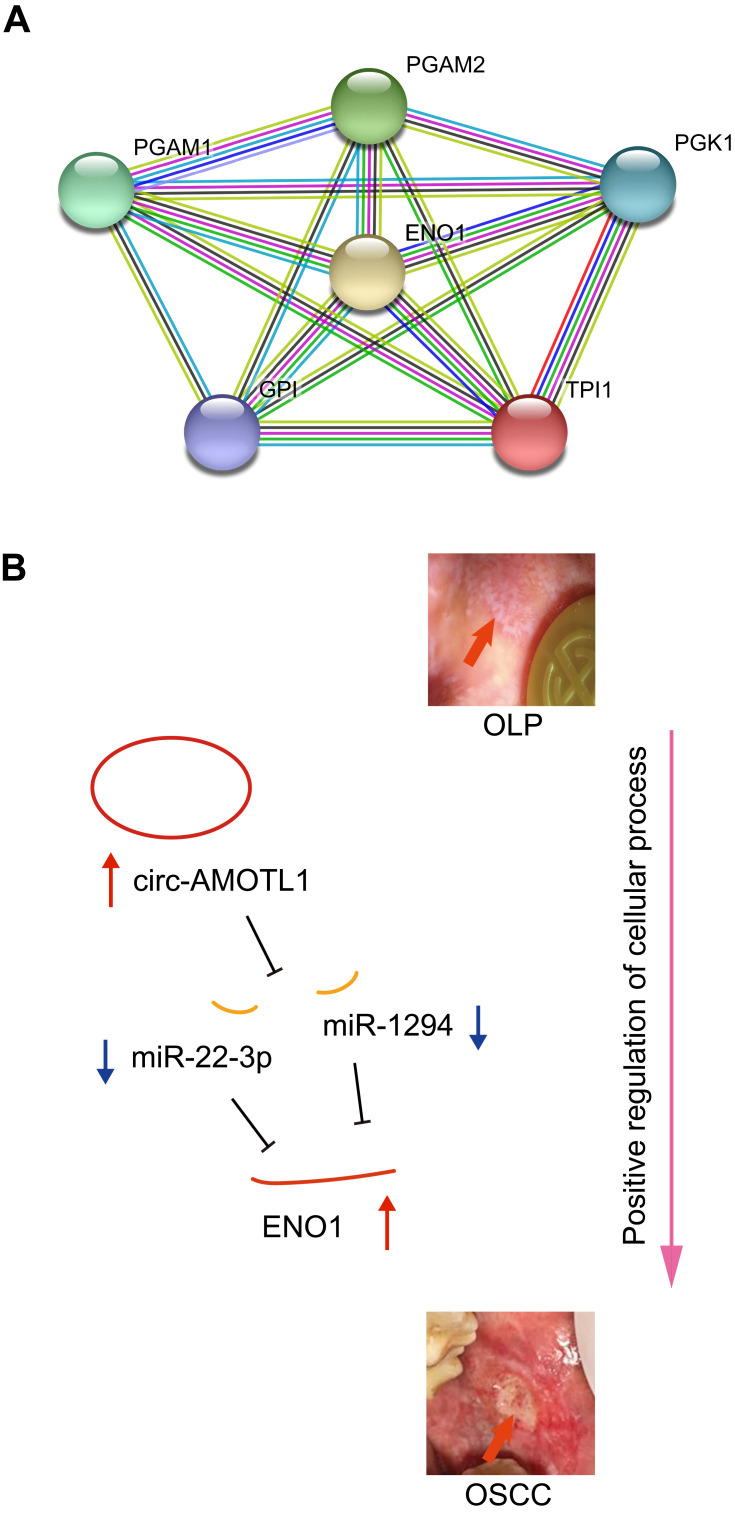
Again, we utilized the sequencing results to probe the potential KEGG and GO enrichments, as well as the PPI network. Based on differential mRNAs, there were 1772 significantly enriched GO terms, among which 150 terms included ENO1. The Top 20 GO functional terms are listed in Figure 4A, including extracellular exosome, extracellular vesicle, extracellular organelle, and extracellular membrane-bounded organelle. For KEGG enrichment analysis, 58 significantly enriched pathways were found (top 20 listed in Figure 4B). Based on differential circRNAs, GO and KEGG enrichment were analyzed. Together, 85 GO terms were significantly enriched, and 28 terms included circ-AMOTL1. The top 20 GO functional terms are listed in Figure 4C. With these results, two common enriched GO terms shared by both circ-AMOTL1 and ENO1 were: positive regulation of multicellular organismal process and positive regulation of the cellular process. Similarly, three KEGG pathways were enriched based on differential circRNAs (Figure 4D). Finally, an ENO1 associated PPI network was generated with the STRING database, and six genes (including ENO1) were screened out as key nodes (Figure 5A): PGAM1, PGAM2, PGK1, GP1, and TPI1. These genes might play the essential roles in the circ-AMOTL1/ENO1 regulation net in tumorigenesis of OSCC.
Figure 4.
KEGG/GO enrichments.
Notes: The top 20 GO functional terms (A) and KEGG enrichments (B) are based on differential mRNAs of transcriptome sequencing using OSCC and OLP samples. The top 20 GO functional terms (C) and KEGG enrichments (D) are based on differential noncoding RNAs of transcriptome sequencing.
Abbreviations: GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; OLP, oral lichen planus; OSCC, oral squamous cell carcinoma.
Figure 5.
An ENO1 associated PPI network generated using the STRING database (A) and the hypothesis schematic diagram of the circ-AMOTL1/miR-22-3p/miR-1294/ENO1 ceRNA network during OSCC tumorigenesis (B).
Abbreviations: OLP, oral lichen planus; OSCC, oral squamous cell carcinoma; PPI, protein–protein interaction.
Discussion
Our study suggested that the expression of ENO1 was increased in the tumorigenesis of OSCC and promoted cell proliferation. Using sequencing and bioinformatics analysis, as well as qPCR and IHC confirmation, we depicted a novel mechanism of tumorigenesis of OSCC (from OLP). The hypothesis schematic diagram is shown in Figure 5B: circ-AMOTL1, miR-22-3p, miR-1294 and ENO1 construct a CeRNA network, in which increased circ-AMOTL1 upregulateregulates ENO1 expression through miR-22-3p/miR-1294, and finally, ENO1 drives OSCC onset via enriched functions like positive regulation of cellular process and associated proteins such as PGAM1, PGAM2, PGK1, GP1, and TPI1.
As shown by a number of studies on cancer, ENO1 displayed a higher expression level in sputum supernatant of early-stage lung cancer patients,15 as well as in the tumor samples of lung adenocarcinomas.16 Meanwhile, it is noticed that the ENO1 gene was most frequently amplified in squamous cell lung carcinoma.17 This implied that increased ENO1 expression can also be a marker of squamous cell carcinoma (SCC) formation. So far, there is one study that has reported its relationship with head and neck cancer, indicating that ENO1 promotes transformation partly via chemokine CCL20 induction and can be regarded as a potential prognostic marker.18 Moreover, ENO1 participates in the process of lymphoma cell adhesion mediated drug resistance (CAM-DR), besides promoting tumor proliferation.19 ENO1 overexpression was also observed in pancreatic cancer,20 and ENO1 silencing could sensitize hypoxia-induced chemoresistance.21 Some Italian scholars used a genetic model of pancreatic carcinoma and observed the effects that vaccination with ENO1 DNA elicits. The result showed that the vaccinated mice had increased serum levels of anti-ENO1 immunoglobulin G and induced complement-dependent cytotoxicity, which delayed tumor progression and significantly extended survival.22 It was also noticed that ENO1 promoted cancer progression in hepatocellular carcinoma, breast cancer, and gastric cancer.10,23–25 Generally, most studies support the idea that ENO1 expression is elevated and promotes the Warburg effect and cancer proliferation and invasion; ENO1 overexpression and modifications have diagnostic and prognostic value.26
However, it may play distinctive roles in different cancer types. Regarding neuroblastoma, ENO1 has a tumor-suppressor activity and a high level of ENO1 expression has significant growth inhibitory effects.27 Meanwhile, another lung cancer study reported that overexpressed ENO1 inhibited the mobility of A549, as well as the expression of the mesenchymal markers N-cadherin and vimentin, but it upregulated the epithelial marker E-cadherin, and suppressed ERK1/2 phosphorylation in the EGF-stimulating assay.28
Our results supported the suggestion that positive regulation of the cellular process could be a crucial function which mediated the procarcinogenic role of the circ-AMOTL1/ENO1 axis in OSCC. Additionally, PPI analysis suggested that PGAM1, PGAM2, PGK1, GPI, and TPI1 were important factors associated with ENO1. PGAM1 (phosphoglycerate mutase 1) encodes a protein that catalyzes the reversible reaction of 3-phosphoglycerate (3-PGA) to 2-phosphoglycerate (2-PGA) in the glycolytic pathway. It strongly impacts the glycolysis and gluconeogenesis pathways, which are essential for oncogenesis, as is shown in studies focusing on pancreatic ductal adenocarcinoma,29 renal clear cell carcinoma,30 osteosarcoma,31 urothelial bladder cancer,32 etc. PGAM2 (phosphoglycerate mutase 2) encodes the muscle-specific PGAM subunit.33 This gene has seldom been studied but is already known to be a factor in tumor formation, e.g. in hepatocellular carcinoma and lung cancer.34–36 PGK1 is also a glycolytic enzyme that catalyzes the conversion of 1.3-diphosphoglycerate to 3-phosphoglycerate; it has been widely reported as a marker of tumorigenesis including gastric cancer,37 endometrioid adenocarcinoma,38 lung cancer,39 breast cancer,40 hepatocellular carcinoma,41 etc. Consistent with the circ-AMOTL1/ENO1 axis, it enhances the metabolic process and hence drives tumor onset.41,42 GPI (glucose-6-phosphate isomerase) functions as a glycolytic enzyme (glucose-6-phosphate isomerase) that interconverts glucose-6-phosphate and fructose-6-phosphate. In addition, mammalian GPI can function as a tumor-secreted cytokine and an angiogenic factor that stimulates endothelial cell motility. Overexpression of GPI was noticed in different cancer types, such as breast cancer,43 hepatocellular carcinoma,44 and gastric cancer.45 TPI1 (triosephosphate isomerase 1) catalyzes the isomerization of glyceraldehydes 3-phosphate (G3P) and dihydroxy-acetone phosphate (DHAP) in glycolysis and gluconeogenesis. As well, it contributes largely to tumorigenesis, as documented in lung cancer,46 gastrointestinal cancer,47 and colon cancer.48
We here proposed a ceRNA network constructed mainly by circ-AMOTL1/miR-22-3p/miR-1294/ENO1. The circRNA AMOTL1 is a clear oncogene, which is highly expressed in patient tumor samples and cancer cell lines. Underlying its procarcinogenic effects, different mechanisms have been proposed, including increasing the affinity of c-myc binding to a number of promoters49 and serving as a sponge for binding miR-193a-5p and relieving miR-193a-5p repression of some oncogene clusters.50 In addition, the gene symbol AMOTL1 (angiomotin like 1) encodes a peripheral membrane protein that is a component of tight junctions. This protein is also elevated in tumors.51–53 Moreover, the potent tumorigenicity might be triggered through interactions between circ-AMOTL1 and c-myc;49 and the alternative transcription form of the ENO1, MBP1, also interacts with c-myc P2 promoter, which explains the positive relationship between circ-AMOTL1 and ENO1 in another view. MiR-22-3p promotes cancer progression in cervical squamous carcinoma cells,54 papillary thyroid cancer,55 and pancreatic cancer;56 meanwhile, it also acts as a tumor suppressor in colorectal cancer57 and hepatocellular carcinoma.58 MiR-1294 acts as a tumor suppressor in gastric cancer,59 clear cell renal cell carcinoma,60 pancreatic ductal adenocarcinoma,61 and esophageal cancer;62 in parallel, it confers cisplatin resistance in ovarian cancer cells by targeting IGF1R.59 So far, there has been no evidence showing the relationship between circ-AMOTL1/miR-22-3p/miR-1294 and OSCC. In addition, ENO1, miR-22-3p, miR-1294, and circ-AMOTL1 could serve as new biomarkers in the tumorigenesis of OSCC. The decreased expression of these two miRNAs in our results deserves further experiments to support the findings.
We also wanted to point out the limitations of our work. First, the experiments were not blinded and only a few cell lines were involved. More cell lines need to be applied to blinded experiments to clarify the molecular mechanism. Second, we did not prove the direction binding of miR-22-3p/miR-1294 and circ-AMOTL1 or ENO1. Third, confounding factors were not taken into consideration in this study, like alcohol, tobacco, and hepatitis C infection history, which may have an effect on the carcinogenesis of OSCC. In addition, not all OSCC specimens are from OLP malignant transformation and the sample size used in this study was limited, so to include more specimens related to this disease process could help to get more reliable conclusions. Furthermore, our study is an observational study. A causal relationship between the circ-AMOTL1/ENO1 axis and carcinogenesis of OSCC should be studied in longitudinal studies in the future.
Conclusions
Although OLP-Associated Oral Squamous Cell Carcinoma had been studied previously at the RNA level,63 we confirmed that ENO1 expression was elevated in OSCC, and ENO1 might function as a tumor-promoting gene in OSCC through the circ-AMOTL1/miR-22-3p/miR-1294 network in this study. The carcinogenesis mechanism of ENO1 may include the enrichment of GO terms such as regulation of cellular component organization, negative regulation of the biological process, phosphate-containing compound metabolic process, phosphorus metabolic process, etc., and enrichment of KEGG pathways such as glycolysis/gluconeogenesis, HIF-1 signaling pathway, and RNA degradation. Positive regulation of cellular process is a crucial function that mediates the procarcinogenic role of the circ-AMOTL1/ENO1 axis in OSCC. ENO1 interacted proteins such as PGAM1, PGAM2, PGK1, GPI, and TPI1 might contribute to OSCC development. These novel findings may shed further light on the pathogenesis of OSCC and the potential precursor markers.
Acknowledgments
This study was funded by grants from the National Natural Science Foundation of China (81470736 and 81901008). Jin Liu and Qiaozhen Yang are co-first authors for this study.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee of Huashan Hospital and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All patients provided written informed consent.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Chow LQM. Head and Neck Cancer. N Engl J Med. 2020;382(1):60–72. doi: 10.1056/NEJMra1715715 [DOI] [PubMed] [Google Scholar]
- 3.González-Moles MÁ, Warnakulasuriya S, González-Ruiz I, et al. Worldwide prevalence of oral lichen planus: A systematic review and meta-analysis. Oral Dis. 2020. doi: 10.1111/odi.13323 [DOI] [PubMed] [Google Scholar]
- 4.González-Moles MÁ, Ruiz-Ávila I, González-Ruiz L, Ayén Á, Gil-Montoya JA, Ramos-García P. Malignant transformation risk of oral lichen planus: A systematic review and comprehensive meta-analysis. Oral Oncol. 2019;96:121–130. doi: 10.1016/j.oraloncology.2019.07.012 [DOI] [PubMed] [Google Scholar]
- 5.Xing Y, Zhao S, Zhou BP, Mi J. Metabolic reprogramming of the tumour microenvironment. FEBS J. 2015;282(20):3892–3898. doi: 10.1111/febs.13402 [DOI] [PubMed] [Google Scholar]
- 6.San-Millan I, Brooks GA. Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis. 2017;38(2):119–133. doi: 10.1093/carcin/bgw127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asgari Y, Zabihinpour Z, Salehzadeh-Yazdi A, Schreiber F, Masoudi-Nejad A. Alterations in cancer cell metabolism: the Warburg effect and metabolic adaptation. Genomics. 2015;105(5–6):275–281. doi: 10.1016/j.ygeno.2015.03.001 [DOI] [PubMed] [Google Scholar]
- 8.Dai J, Zhou Q, Chen J, Rexius-Hall ML, Rehman J, Zhou G. Alpha-enolase regulates the malignant phenotype of pulmonary artery smooth muscle cells via the AMPK-Akt pathway. Nat Commun. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan GR, Xu SH, Tan ZL, et al. Proteomics characterization of gastrokine 1-induced growth inhibition of gastric cancer cells. Proteomics. 2011;11(18):3657–3664. doi: 10.1002/pmic.201100215 [DOI] [PubMed] [Google Scholar]
- 10.Yu S, Li N, Huang Z, et al. A novel lncRNA, TCONS_00006195, represses hepatocellular carcinoma progression by inhibiting enzymatic activity of ENO1. Cell Death Dis. 2018;9(12):1184. doi: 10.1038/s41419-018-1231-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng YS, Gould A, Kurago Z, et al. Diagnosis of oral lichen planus: a position paper of the American Academy of Oral and Maxillofacial Pathology. Oral Surg Oral Med Oral Pathol Oral Radiol. 2016;122(3):332–354. doi: 10.1016/j.oooo.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 12.Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Q, Xu B, Sun H, et al. A genome-wide association scan of biological processes involved in oral lichen planus and oral squamous cell carcinoma. Medicine. 2017;96(25):e7012. doi: 10.1097/MD.0000000000007012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu L, Shen J, Mannoor K, et al. Identification of ENO1 as a potential sputum biomarker for early-stage lung cancer by shotgun proteomics. Clin Lung Cancer. 2014;15(372–378):e1. doi: 10.1016/j.cllc.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okudela K, Mitsui H, Matsumura M, et al. The potential significance of alpha-enolase (ENO1) in lung adenocarcinomas - A utility of the immunohistochemical expression in pathologic diagnosis. Pathol Int. 2017;67(12):602–609. doi: 10.1111/pin.12607 [DOI] [PubMed] [Google Scholar]
- 17.Racz A, Brass N, Hofer M, Sybrecht GW, Remberger K, Meese EU. Gene amplification at chromosome 1pter-p33 including the genes PAX7 and ENO1 in squamous cell lung carcinoma. Int J Oncol. 2000;17(1):67–73. doi: 10.3892/ijo.17.1.67 [DOI] [PubMed] [Google Scholar]
- 18.Tsai ST, Chien IH, Shen WH, et al. ENO1, a potential prognostic head and neck cancer marker, promotes transformation partly via chemokine CCL20 induction. Eur J Cancer. 2010;46(9):1712–1723. doi: 10.1016/j.ejca.2010.03.018 [DOI] [PubMed] [Google Scholar]
- 19.Zhu X, Miao X, Wu Y, et al. ENO1 promotes tumor proliferation and cell adhesion mediated drug resistance (CAM-DR) in Non-Hodgkin’s Lymphomas. Exp Cell Res. 2015;335(2):216–223. doi: 10.1016/j.yexcr.2015.05.020 [DOI] [PubMed] [Google Scholar]
- 20.Yin H, Wang L, Liu HL. ENO1 Overexpression in Pancreatic Cancer Patients and Its Clinical and Diagnostic Significance. Gastroenterol Res Pract. 2018;2018:3842198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Bi R, Yin H, et al. ENO1 silencing impairs hypoxia-induced gemcitabine chemoresistance associated with redox modulation in pancreatic cancer cells. Am J Transl Res. 2019;11(7):4470–4480. [PMC free article] [PubMed] [Google Scholar]
- 22.Cappello P, Rolla S, Chiarle R, et al. Vaccination with ENO1 DNA prolongs survival of genetically engineered mice with pancreatic cancer. Gastroenterology. 2013;144(5):1098–1106. doi: 10.1053/j.gastro.2013.01.020 [DOI] [PubMed] [Google Scholar]
- 23.Zhu X, Yu H, Li B, et al. Targeting a LncRNA P5848-ENO1 axis inhibits tumor growth in hepatocellular carcinoma. Biosci Rep. 2018;2:548. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Cancemi P, Buttacavoli M, et al. Expression of Alpha-Enolase (ENO1), Myc Promoter-Binding Protein-1 (MBP-1) and Matrix Metalloproteinases (MMP-2 and MMP-9) Reflect the Nature and Aggressiveness of Breast Tumors. Int J Mol Sci. 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao H, Wang YF, Yuan WZ, et al. Silencing of ENO1 by shRNA Inhibits the Proliferation of Gastric Cancer Cells. Technol Cancer Res Treat. 2018;17:1533033818784411. doi: 10.1177/1533033818784411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cappello P, Principe M, Bulfamante S, Novelli F. Alpha-Enolase (ENO1), a potential target in novel immunotherapies. Front Biosci. 2017;22:944–959. doi: 10.2741/4526 [DOI] [PubMed] [Google Scholar]
- 27.Ejeskar K, Krona C, Caren H, et al. Introduction of in vitro transcribed ENO1 mRNA into neuroblastoma cells induces cell death. BMC Cancer. 2005;5(1):161. doi: 10.1186/1471-2407-5-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou X, Zhang Y, Han N, et al. [alpha-Enolase (ENO1) inhibits epithelial-mesenchymal transition in the A549 cell line by suppressing ERK1/2 phosphorylation]. Zhongguo Fei Ai Za Zhi. 2013;16(5):221–226. doi:10.3779/j.issn.1009-3419.2013.05.01. Chiense. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Weng Y, Liu P, et al. Identification of PGAM1 as a putative therapeutic target for pancreatic ductal adenocarcinoma metastasis using quantitative proteomics. Onco Targets Ther. 2018;11:3345–3357. doi: 10.2147/OTT.S162470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Shu F, Lei B, et al. Expression of PGAM1 in renal clear cell carcinoma and its clinical significance. Int J Clin Exp Pathol. 2015;8(8):9410–9415. [PMC free article] [PubMed] [Google Scholar]
- 31.Shen Y, Zhao S, Wang S, et al. S1P/S1PR3 axis promotes aerobic glycolysis by YAP/c-MYC/PGAM1 axis in osteosarcoma. EBioMedicine. 2019;40:210–223. doi: 10.1016/j.ebiom.2018.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng XC, Gong FM, Chen Y, et al. Proteomics identification of PGAM1 as a potential therapeutic target for urothelial bladder cancer. J Proteomics. 2016;132:85–92. doi: 10.1016/j.jprot.2015.11.027 [DOI] [PubMed] [Google Scholar]
- 33.Yang H, He J, Wei W, et al. The c.-360 T>C mutation affects PGAM2 transcription activity and is linked with the water holding capacity of the longissimus lumborum muscle in pigs. Meat Sci. 2016;122:139–144. doi: 10.1016/j.meatsci.2016.07.023 [DOI] [PubMed] [Google Scholar]
- 34.Geng J, Sun J, Lin Q, et al. Methylation status of NEUROG2 and NID2 improves the diagnosis of stage I NSCLC. Oncol Lett. 2012;3(4):901–906. doi: 10.3892/ol.2012.587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knobloch TJ, Ryan NM, Bruschweiler-Li L, et al. Metabolic Regulation of Glycolysis and AMP Activated Protein Kinase Pathways during Black Raspberry-Mediated Oral Cancer Chemoprevention. Metabolites. 2019;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarathi A, Palaniappan A. Novel significant stage-specific differentially expressed genes in hepatocellular carcinoma. BMC Cancer. 2019;19(1):663. doi: 10.1186/s12885-019-5838-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zieker D, Konigsrainer I, Traub F, et al. PGK1 a potential marker for peritoneal dissemination in gastric cancer. Cell Physiol Biochem. 2008;21(5–6):429–436. doi: 10.1159/000129635 [DOI] [PubMed] [Google Scholar]
- 38.Zhou JW, Tang JJ, Sun W, et al. PGK1 facilities cisplatin chemoresistance by triggering HSP90/ERK pathway mediated DNA repair and methylation in endometrial endometrioid adenocarcinoma. Mol Med. 2019;25(1):11. doi: 10.1186/s10020-019-0079-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu T, Zhao Y, Hu Z, et al. MetaLnc9 Facilitates Lung Cancer Metastasis via a PGK1-Activated AKT/mTOR Pathway. Cancer Res. 2017;77(21):5782–5794. doi: 10.1158/0008-5472.CAN-17-0671 [DOI] [PubMed] [Google Scholar]
- 40.Xu D, Aka JA, Wang R, Lin SX. 17beta-hydroxysteroid dehydrogenase type 5 is negatively correlated to apoptosis inhibitor GRP78 and tumor-secreted protein PGK1, and modulates breast cancer cell viability and proliferation. J Steroid Biochem Mol Biol. 2017;171:270–280. doi: 10.1016/j.jsbmb.2017.04.009 [DOI] [PubMed] [Google Scholar]
- 41.Xie H, Tong G, Zhang Y, Liang S, Tang K, Yang Q. PGK1 Drives Hepatocellular Carcinoma Metastasis by Enhancing Metabolic Process. Int J Mol Sci. 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarnopolsky MA. Myopathies Related to Glycogen Metabolism Disorders. Neurotherapeutics. 2018;15(4):915–927. doi: 10.1007/s13311-018-00684-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu G, Guo Z, Chatterjee A, et al. Overexpression of glycosylphosphatidylinositol (GPI) transamidase subunits phosphatidylinositol glycan class T and/or GPI anchor attachment 1 induces tumorigenesis and contributes to invasion in human breast cancer. Cancer Res. 2006;66(20):9829–9836. doi: 10.1158/0008-5472.CAN-06-0506 [DOI] [PubMed] [Google Scholar]
- 44.Lyu Z, Chen Y, Guo X, Zhou F, Yan Z, Xing J. Genetic variants in glucose-6-phosphate isomerase gene as prognosis predictors in hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2016;40(6):698–704. doi: 10.1016/j.clinre.2016.05.001 [DOI] [PubMed] [Google Scholar]
- 45.Ma YT, Xing XF, Dong B, Cheng XJ, Guo T, Du H. Higher autocrine motility factor/glucose-6-phosphate isomerase expression is associated with tumorigenesis and poorer prognosis in gastric cancer. Cancer Manag Res. 2018;10:4969–4980. doi: 10.2147/CMAR.S177441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puzone R, Savarino G, Salvi S, et al. Glyceraldehyde-3-phosphate dehydrogenase gene over expression correlates with poor prognosis in non small cell lung cancer patients. Mol Cancer. 2013;12(1):97. doi: 10.1186/1476-4598-12-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Husi H, Fernandes M, Skipworth RJ, Miller J, Cronshaw AD. Identification of diagnostic upper gastrointestinal cancer tissue type-specific urinary biomarkers. Biomed Rep. 2019;10(3):165–174. doi: 10.3892/br.2019.1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumamoto K, Nakachi Y, Mizuno Y, et al. Expressions of 10 genes as candidate predictors of recurrence in stage III colon cancer patients receiving adjuvant oxaliplatin-based chemotherapy. Oncol Lett. 2019;18(2):1388–1394. doi: 10.3892/ol.2019.10437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Q, Du WW, Wu N, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24(9):1609–1620. doi: 10.1038/cdd.2017.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Z, Qu CB, Zhang Y, et al. Dysregulation of p53-RBM25-mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L-miR-193a-5p-Pcdha pathway. Oncogene. 2019;38(14):2516–2532. doi: 10.1038/s41388-018-0602-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wan HY, Li QQ, Zhang Y, Tian W, Li YN, Liu M. MiR-124 represses vasculogenic mimicry and cell motility by targeting amotL1 in cervical cancer cells. Cancer Lett. 2014;355(1):148–158. doi: 10.1016/j.canlet.2014.09.005 [DOI] [PubMed] [Google Scholar]
- 52.Couderc C, Boin A, Fuhrmann L, et al. AMOTL1 Promotes Breast Cancer Progression and Is Antagonized by Merlin. Neoplasia. 2016;18(1):10–24. doi: 10.1016/j.neo.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng Y, Zhang Y, Barutello G, Chiu K, Arigoni M, Giampietro C. Angiomotin like-1 is a novel component of the N-cadherin complex affecting endothelial/pericyte interaction in normal and tumor angiogenesis. Sci Rep. 2016;6(1):30622. doi: 10.1038/srep30622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lv KT, Liu Z, Feng J, Zhao W, Hao T. MiR-22-3p Regulates Cell Proliferation and Inhibits Cell Apoptosis through Targeting the eIF4EBP3 Gene in Human Cervical Squamous Carcinoma Cells. Int J Med Sci. 2018;15:142–152. doi: 10.7150/ijms.21645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang M, Chen B, Ru Z, Cong L. CircRNA circ-ITCH suppresses papillary thyroid cancer progression through miR-22-3p/CBL/beta-catenin pathway. Biochem Biophys Res Commun. 2018;504(1):283–288. doi: 10.1016/j.bbrc.2018.08.175 [DOI] [PubMed] [Google Scholar]
- 56.Hussein NA, Kholy ZA, Anwar MM, Ahmad MA, Ahmad SM. Plasma miR-22-3p, miR-642b-3p and miR-885-5p as diagnostic biomarkers for pancreatic cancer. J Cancer Res Clin Oncol. 2017;143(1):83–93. doi: 10.1007/s00432-016-2248-7 [DOI] [PubMed] [Google Scholar]
- 57.Sha QK, Chen L, Xi JZ, Song H. Long non-coding RNA LINC00858 promotes cells proliferation, migration and invasion by acting as a ceRNA of miR-22-3p in colorectal cancer. Artif Cells Nanomed Biotechnol. 2019;47(1):1057–1066. doi: 10.1080/21691401.2018.1544143 [DOI] [PubMed] [Google Scholar]
- 58.Chen J, Wu FX, Luo HL, Liu JJ, Luo T, Bai T. Berberine upregulates miR-22-3p to suppress hepatocellular carcinoma cell proliferation by targeting Sp1. Am J Transl Res. 2016;8(11):4932–4941. [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Huang S, Guo Y, Li L. MiR-1294 confers cisplatin resistance in ovarian Cancer cells by targeting IGF1R. Biomed Pharmacother. 2018;106:1357–1363. doi: 10.1016/j.biopha.2018.07.059 [DOI] [PubMed] [Google Scholar]
- 60.Pan W, Pang LJ, Cai HL, Wu Y, Zhang W, Fang J-C. MiR-1294 acts as a tumor suppressor in clear cell renal cell carcinoma through targeting HOXA6. Eur Rev Med Pharmacol Sci. 2019;23(9):3719–3725. doi: 10.26355/eurrev_201905_17797 [DOI] [PubMed] [Google Scholar]
- 61.Xu Y, Yao Y, Gao P, Cui Y. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun. 2019;509(1):138–142. doi: 10.1016/j.bbrc.2018.12.088 [DOI] [PubMed] [Google Scholar]
- 62.Zhang Z, Lin W, Gao L, et al. Hsa_circ_0004370 promotes esophageal cancer progression through miR-1294/LASP1 pathway. Biosci Rep. 2019;39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Q, Guo B, Sun H, et al. Identification of the key genes implicated in the transformation of OLP to OSCC using RNA-sequencing. Oncol Rep. 2017;37(4):2355–2365. doi: 10.3892/or.2017.5487 [DOI] [PubMed] [Google Scholar]