Abstract
Cryptochrome (CRY) photoreceptors undergo photoresponsive homo-oligomerization to become physiologically active, and BICs (blue-light inhibitors of CRYs) suppress homo-oligomerization. Structural elucidation of CRY–CRY homo-oligomers and a CRY–BIC heterodimer reveals how the activity of plant CRYs is regulated by alternative protein–protein interactions.
Cryptochromes (CRY) are photolyase-like flavoproteins that act as blue light receptors in plants and as non-photoreceptor components of the circadian clock in mammals1–3. Our understanding of cryptochromes has been greatly improved by crystal structures of cryptochromes, including the photoresponsive Drosophila melanogaster dCRY, photoresponsive pigeon CRY4, and non-photoresponsive mammalian mCRY14–7. Plant cryptochromes undergo reversible photoactivation and inactivation in vivo, whereby blue light prompts homo-oligomerization of CRYs, thus activating the photoreceptors. The photoactivated CRYs may be inactivated by spontaneous reversion to inactive monomers in darkness8 or by the CRY-interacting BIC proteins in blue light8–10. The structural alterations underlying photoactivation and inactivation of plant CRYs are not fully understood. Two studies in this issue, by Shao et al.11 and Ma et al.12, reveal structural insights into the active and inactive states of plant cryptochromes.
Cryptochromes are defined by a common two-domain structure: the highly conserved PHR (photolyase homology region) domain of ~500 residues that noncovalently binds the chromophore FAD (flavin adenine dinucleotide) and the divergent CCE (CRY C-terminal extension), also known as CCT (CRY carboxyl terminus), domain of variable length10,13. The mechanisms of CRY-mediated photoresponses in plants have been extensively studied over the last two decades10,14. Those studies suggest that plant CRYs undergo photoreduction, photo-oligomerization and signal transduction to regulate gene expression and photoresponses (Fig. 1). In darkness, plant CRY contains oxidized FAD in its inactive ground state. CRY then absorbs photons of ~450 nm and FAD is reduced by receiving electrons relayed via three evolutionarily conserved tryptophan residues called the ‘Trp-triad’. Photoreduced CRY molecules undergo conformational changes to form physiologically active homo-oligomers. Photo-oligomerization appears to increase the affinity of CRYs for other proteins, promoting their interaction with one or more of ~30 CRY-interacting proteins, such as the CRY-signaling proteins CIBs (CRY-interacting bHLHs), the COP1 (constitutive photomorphogenic 1)–SPAs (suppressors of phytochrome A) complex or the CRY-inhibitory BICs (Fig. 1). These photoresponsive protein–protein interactions ultimately alter the photoresponsive transcriptome or the stability of photoresponsive proteins to modulate plant growth and development. Key details of this model, including the structural differences that distinguish the physiologically active and inactive CRYs, remain unclear.
Fig. 1 |. Photoactivation and inactivation of plant cryptochromes.
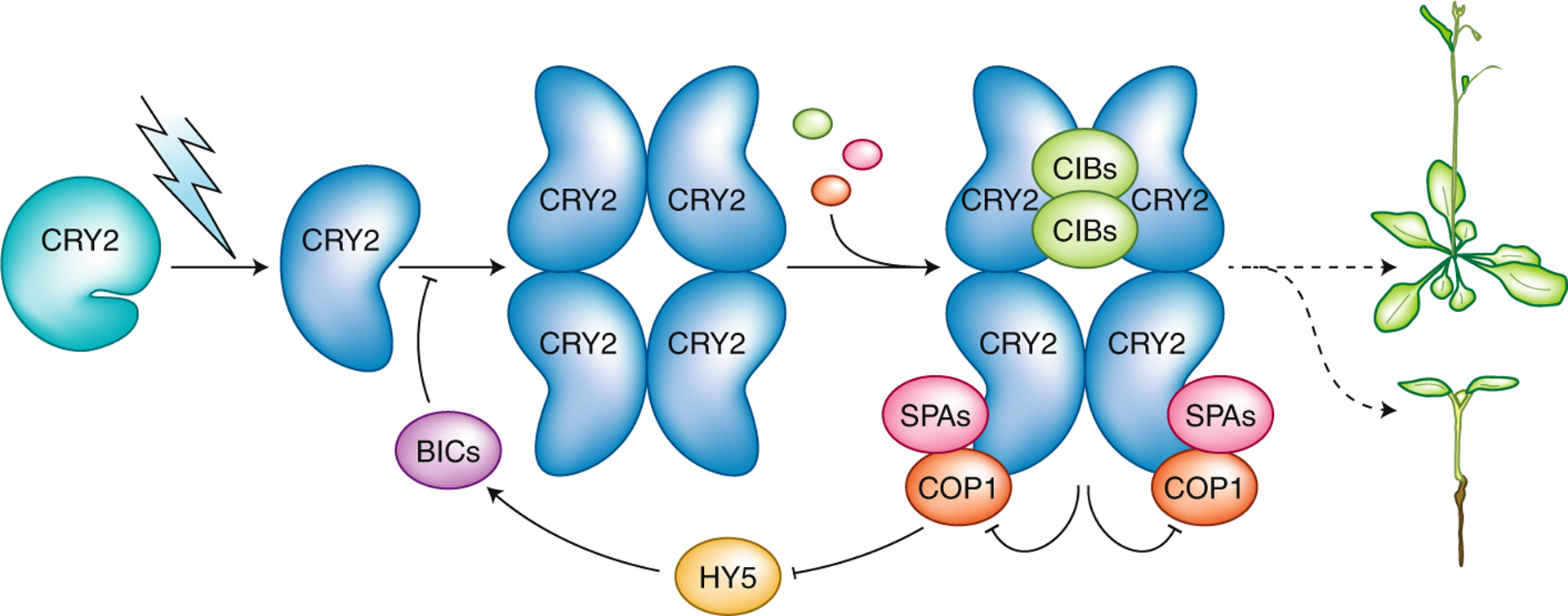
Arabidopsis CRY2 is used as a model to depict the mechanism of photoactivation and inactivation of plant cryptochromes. In darkness, CRY2 exists as an inactive monomer with a closed conformation (depicted as the CCE tail closed) and FAD is oxidized (not shown). In the presence of light, photoexcited CRY2 undergoes photoreduction to adopt an open conformation (with the CCE tail opened), leading to the formation of CRY2 homodimers (not shown) and homotetramers. CRY2 oligomers interact with signaling proteins such as CIB1 and COP1–SPAs to alter gene expression and plant development. CRY2 photoactivation also results in its inactivation by negative feedback inhibition, whereby CRY2 interacts with COP1–SPAs to suppress the E3 ubiquitin ligase activity of COP1, resulting in increased abundance of HY5, which activates transcription of BICs to suppress CRY2 photoactivation. Arrows represent processes or positive effects; T bars represent inhibitory effects.
Genetic studies of the structure–function relationship of plant CRYs have addressed the above questions. Mutation of one or all three Trp-triad residues in Arabidopsis thaliana CRY1 or CRY2 abolished their photoreduction activity in vitro but not their physiological activity in vivo15–17. Counterintuitively, some of the Trp-triad mutants, such as W374A and W397A in Arabidopsis CRY2 (CRY2W374A and CRY2W397A) or their equivalents in Arabidopsis CRY1 (CRY1W377A and CRY1W400A), exhibit constitutive activities, including blue-light-independent inhibition of hypocotyl growth and photoperiod-independent promotion of flowering. Shao et al.11 determined structures of the active CRY homo-oligomers, taking advantage of the constitutively active Trp-triad mutants. In contrast to wild-type CRY2 (which undergoes photo-oligomerization9) or the FAD-deficient CRY2D387A mutant (which fails to homo-oligomerize), the Trp-triad mutant CRY2W374A exhibits constitutive homo-oligomerization activity in vitro11. This result is consistent with reports that photo-oligomerization is necessary for CRY2 function9 and that CRY2W374A is constitutively active in vivo16. Cryo-EM visualization of constitutively active mutants of Arabidopsis CRY2 (AtCRY2W374A) and the equivalent mutant of maize CRY1 (ZmCRY1cW368A) display similar homodimer and homotetramer configurations despite their disordered CCE domains. The PHR domains of these CRYs form homodimers or homotetramers via head-to-head interactions. The constitutively active CRY2W374A mutant migrated as a single homodimeric peak in gel filtration assays. By contrast, the CRY2W374A/W349A and CRY2W374A/R439L mutant proteins, which contain additional mutations of interface residues, eluted as two peaks: a monomer peak and a peak with a molecular mass much greater than that of the homodimer. The CRY2W374A/W349A and CRY2W374A/R439L double mutants exhibited reduced activity in vivo, supporting the interpretation that the CRY2 homodimer is the major active form in vivo. The CRY2 interface mutants also displayed reduced binding affinity for the CRY2-signaling protein, CIB1. These results suggest that CIB1 interacts with CRY2 at the CRY2 homodimer interface (Fig. 1), which explains why CRY homo-oligomers but not CRY monomers are active. Further structural analyses of additional complexes of CRY2 and CRY2-interacting proteins are necessary to determine whether CIBs interact with CRY2 through non-interface structural elements that are generated by conformational changes upon CRY2 homodimerization or whether other CRY2-binding proteins also interact with the CRY2 homodimer at the interface. Interestingly, most of the residues located at one of the two interfaces of the CRY tetramer are conserved in plant CRYs but not in animal CRYs or in DNA photolyases. It remains to be determined whether homo-oligomerization is necessary for the functions of non-plant CRYs, which might have evolved different interface structures, or whether the dimerization-dependent photoactivation mechanism is unique to plant CRYs.
Photoreceptors are commonly inactivated or desensitized by various mechanisms to maintain appropriate photosensitivity of the cell. Plant cryptochromes can be inactivated by at least three mechanisms: spontaneous dark reversion from (reduced) homo-oligomers to (oxidized) monomers, interaction with and inhibition by negative regulators such as BICs, and ubiquitination-dependent proteolysis10. Among these mechanisms, BIC-dependent inhibition of CRYs is itself photoresponsive (Fig. 2), implying that the CRY–BIC interaction may play a more dynamic role in regulating the activity of CRYs in plants grown in light9. BICs are small proteins (usually <200 amino acids) that share a highly conserved CRY-interacting domain (CID) at their carboxyl termini9. BICs are found only in plant lineages, from moss to angiosperm, but not in bacteria or animals. Arabidopsis has two BIC homologs, BIC1 and BIC2, which both bind to the PHR domain of photoexcited CRYs9. The photoresponsive interaction of BICs and CRY2 (at 1:1 stoichiometry) inhibits all photoresponsive activities of CRY2, including photoreduction, photo-oligomerization, light-dependent phosphorylation, light-dependent ubiquitination and CRY-mediated changes of the transcriptome, stem elongation and flowering time10.
Fig. 2 |. Structural insights into cry2 inactivation.
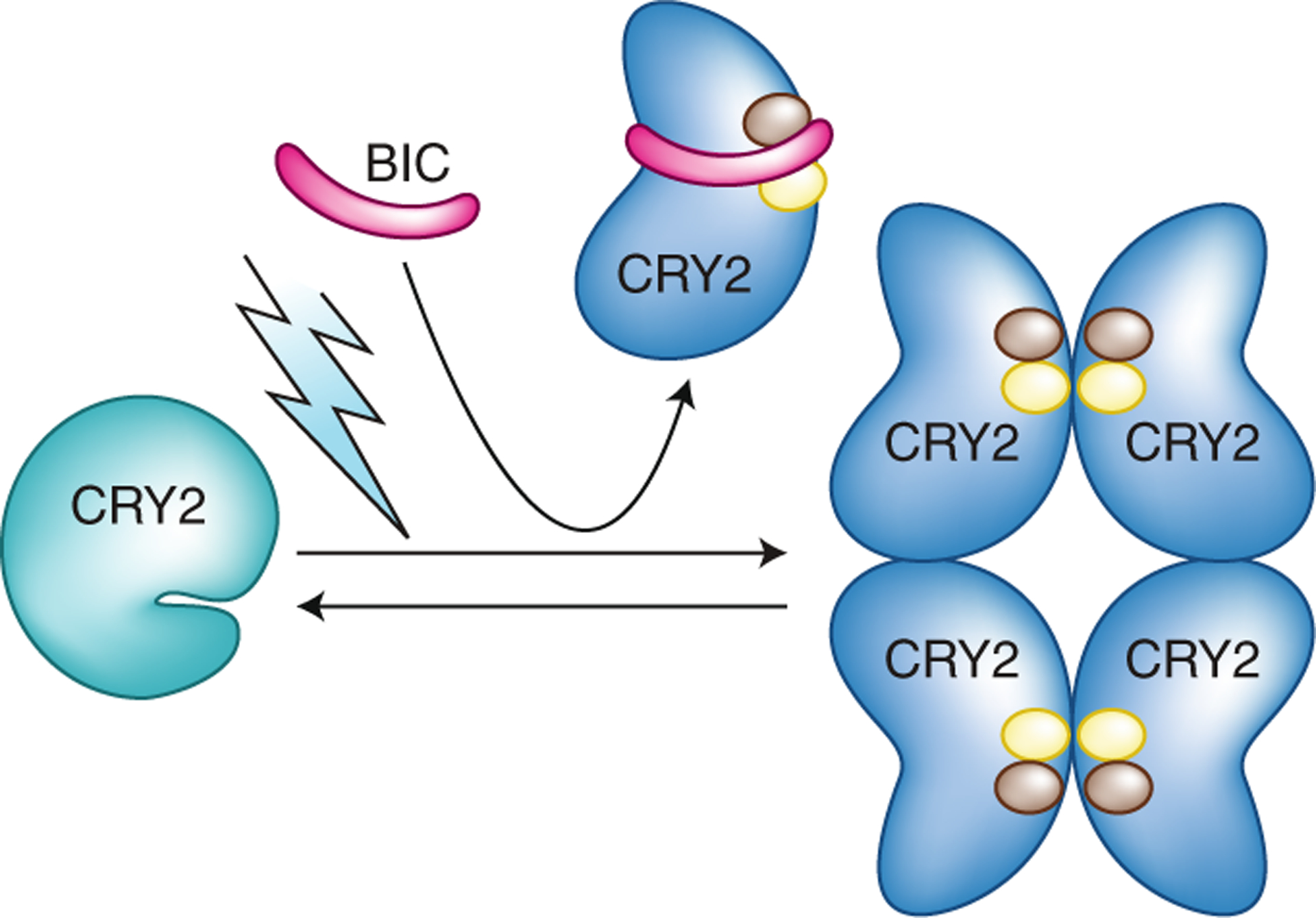
BIC displays a ‘waist belt’ structure, which interacts with photoexcited CRY2 and wraps around the PHR domain to prevent CRY2 dimerization. The present structural analyses11,12 reveal that at least two residues of CRY2, R208 and W349, represented by brown and yellow circles, are located at the interfaces of both the CRY–CRY complex and the CRY–BIC complex. This finding suggests that BIC acts as a competitive inhibitor, binding CRY2 to block CRY photo-oligomerization and photoactivation.
Ma et al. determined X-ray crystal structures of the PHR domain of Arabidopsis CRY2 (1–489) at 2.7 Å resolution and of a CRY2N–BIC2 heterodimer comprising the CRY2 PHR domain (1–506) and the CID fragment of Arabidopsis BIC2 (33–97) from 1 Å to 3 Å resolution12. As anticipated, the PHR domain of Arabidopsis CRY2 has the same α and α/β subdomains, and the structure is almost identical to that of the PHR domain of Arabidopsis CRY1, although the two domains share only 58% amino acid sequence identity. In contrast to DNA photolyases that usually contain pterine or deazaflavin as the antenna chromophore3, the CRY2N (C39–G49) region near the corresponding chromophore-binding antenna pocket of photolyases contains a short helix that extends into the antenna pocket, blocking the entry of a possible second cofactor. This result suggests that Arabidopsis CRY2 may not possess a second chromophore.
Examination of the CRY2N–BIC2 complex structure provides two possible mechanisms to explain how BIC proteins may inactivate plant CRYs. First, BIC may inhibit CRY photoreduction. At least ten residues of CRY2, including the better known D387, contact the FAD chromophore. Binding of BIC2 to CRY2N increases the distance between the electron donor (W397) and the electron acceptor of the isoalloxazine ring of FAD by ~1 Å, which probably makes the electron transfer more difficult. Moreover, binding of BIC2 to CRY2N also results in a rotation of the carboxyl side chain of the presumed proton donor (D393) by 52°, which increases the distance between proton donor and proton acceptor in FAD by ~5 Å, making protonation of FAD by D393 all but impossible. Consequently, BIC2 may block reduction of FAD to FAD• by W397 and protonation of FAD• to FADH• by D393 to prevent CRY2 photoreduction. Second, BICs may act as competitive inhibitors of CRY homo-oligomerization. In the CRY2N–BIC2 complex, the CID fragment of BIC2 displays a ‘waist belt’ structure that wraps around the groove between the α and α/β subdomains of the CRY2 PHR domain (Fig. 2). CRY2N and BIC2 each have at least 16 residues that are involved in forming the CRY2N–BIC2 complex. Individual mutations of six or nine interface residues of CRY2 and BIC2 lowered the apparent affinity of the CRY2N–BIC2 complex in vitro. All CRY2-interacting residues of BIC2 within the CID domain are highly conserved in plants. The CRY–BIC heterodimer and the CRY–CRY homodimer interfaces share at least two residues, CRY2 W349 and R208. In the CRY2N–BIC2 heterodimer, CRY2 residue W349 hydrophobically interacts with I57 of BIC2, whereas CRY2 R208 forms a salt bridge and a hydrogen bond with E50 of BIC212. In the CRY homodimers, CRY2 W349 and R208 and their counterparts in CRY1 are at the interfaces of the CRY2–CRY2 and CRY1-CRY1 homodimers11, respectively. These results argue strongly that binding of BICs to CRYs competitively inhibits CRY photo-oligomerization to block photoactivation (Fig. 2).
The studies by Shao et al. and Ma et al. provide structural insights into how PHR domains are involved in photoactivation and inactivation of plant CRYs. The CCE domain of plant CRYs contains abundant intrinsically disordered regions, which impose a formidable technical barrier to structurally determining the full-length plant CRYs. This leaves us with many unanswered questions. For example, genetic studies argue that that the photoexcited CRYs should adopt an open conformation by opening up the CCE domain, presumably prior to CRY–CRY homo-oligomerization; however, this conformation has yet to be structurally resolved18. Similarly, it has been shown that photoexcited CRYs have an increased affinity for BICs, but the structural changes underlying photoresponsive CRY–BIC heterodimerization remain to be elucidated. Moreover, it has been shown that plant CRYs form not only small homodimers or homotetramers but also larger complexes called photobodies19,20. Because intrinsically disordered regions are known to facilitate liquid–liquid phase separation of many proteins, CRY photobodies may represent condensed liquid droplets of CRY homo-oligomers that could alter the equilibrium of multivalent protein–protein interactions. It would thus be of great interest to investigate how the CCE domain contributes to the photoactivation and inactivation of plant cryptochromes.
Acknowledgements
Work in the authors’ laboratories is supported in part by the National Natural Science Foundation of China (31970265 to Q.W.), the Natural Science Foundation of Fujian Province (2019J06014 to Q.W.) and the National Institutes of Health (GM56265 to C.L.).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Ahmad M & Cashmore AR Nature 366, 162–166 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Cashmore AR Plant Cell Environ. 20, 764–767 (1997). [Google Scholar]
- 3.Sancar A Chem. Rev 103, 2203–2237 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Brautigam CA et al. Proc. Natl Acad. Sci. USA 101, 12142–12147 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zoltowski BD et al. Nature 480, 396–399 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czarna A et al. Cell 153, 1394–1405 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Zoltowski BD et al. Proc. Natl Acad. Sci. USA 116, 19449–19457 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Q et al. Mol. Plant 13, 398–413 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q et al. Science 354, 343–347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q & Lin C Annu. Rev. Plant Biol 10.1146/annurev-arplant-050718-100300 (2020). [DOI] [PMC free article] [PubMed]
- 11.Shao K et al. Nat. Struct. Mol. Biol 10.1038/s41594-020-0420-x (2020). [DOI]
- 12.Ma L et al. Nat. Struct. Mol. Biol 10.1038/s41594-020-0410-z (2020). [DOI]
- 13.Lin C & Shalitin D Annu. Rev. Plant Biol 54, 469–496 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Chaves I et al. Annu. Rev. Plant Biol 62, 335–364 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Gao J et al. Proc. Natl Acad. Sci. USA 112, 9135–9140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X et al. Proc. Natl Acad. Sci. USA 108, 20844–20849 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Su T, He W, Wang Q & Lin C Mol. Biol. Evol 37, 327–340 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang H-Q et al. Cell 103, 815–827 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Más P, Devlin PF, Panda S & Kay SA Nature 408, 207–211 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Yu X et al. Plant Cell 21, 118–130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]


