Abstract
Objective
Early detection and treatment are particularly important to epithelial ovarian cancer (EOC). Studies have shown that circular RNA (circRNA) dysregulation is associated with the proliferation and metastasis of ovarian cancer cells. This study focused on the role of serum exosomal circular forkhead box protein P1 (circFoxp1) on survival outcome and cisplatin (DDP) resistance in patients with EOC.
Methods
Quantitative polymerase chain reaction, 5-ethynyl-2′-deoxyuridine (EdU) staining, CCK-8, luciferase reporter assay, RNA immunoprecipitation, tumor xenograft in nude mice, and bioinformatic analysis were performed.
Results
Circulating exosomal circFoxp1 was significantly increased in patients with EOC, especially in DDP-resistant EOC patients. circFoxp1 expression was positively associated with International Federation of Gynecology and Obstetrics stage, primary tumor size, lymphatic metastasis, distant metastasis, residual tumor diameter, and clinical response. Exosomal circFoxp1 also was an independent factor predicting survival and disease recurrence in patients with EOC. Overexpression of circFoxp1 could promote cell proliferation and confer DDP resistance, while knockdown of circFoxp1 could inhibit cell proliferation and enhance DDP sensitivity in vitro and in vivo. In addition, miR-22 and miR-150-3p mimic treatment attenuated circFoxp1-meadiated DDP resistance, while miR-22 and miR-150-3p inhibitor treatment enhanced DDP resistance that mitigated by circFoxp1 knockdown. Furthermore, circFoxp1 positively regulated the expression of CCAAT enhancer binding protein gamma (CEBPG) and formin like 3 (FMNL3) through miR-22 and miR-150-3p.
Conclusions
circFoxp1 is an oncogene in EOC cells and can confer DDP resistance to EOC cells. Circulating exosomal circFoxp1 can be used as a biomarker and potential therapeutic target for EOC.
Keywords: Ovarian Cancer, Exosomes, circRNA, microRNA, Biomarkers, Chemotherapy
INTRODUCTION
Epithelial ovarian cancer (EOC) has the highest mortality rate in gynecological tumors [1]. Many newly diagnostic patients with EOC have reached the advanced stage and extensive intraperitoneal metastasis because of its insidious onset, lack of typical symptoms at an early stage [2]. The prognosis of EOC is extremely poor that 5-year survival rate is only approximately 30%. However, early detection and treatment are particularly important because the 5-year survival rate of early-stage ovarian cancer can reach approximately 90% [3]. Liquid biopsy, as a new method, avoids the harm to the patient caused by surgical operation and puncture. The current methods in the field of liquid biopsy mainly include circulating tumor DNA, circulating tumor cell, extracellular free nucleic acids (RNA and DNA), and exosomes [4]. Exosomes are nano-sized extracellular lipid bilayer vesicles released to the outside by exocytosis, with a diameter of 30 to 150 nm. They can be secreted to the extracellular microspheres by almost all types of cells under various physiological and pathological conditions. Exosomes carry a variety of molecules that have become the focus of tumor biomarker research [5].
Circular RNAs (circRNAs) are a type of endogenous non-coding RNA (ncRNA). circRNAs are formed by back splicing and is a covalently closed single-stranded circular molecule. At present, the potential clinical value of circRNA in tumor diagnosis, treatment and prognosis evaluation is being closely monitored [6]. The expression of circRNA in tumor tissues, plasma and saliva is stable, conservative, and specific, suggesting that they have good clinical application value in disease diagnosis and therapeutic target for tumor [7]. Studies have shown that circRNA dysregulation is associated with the proliferation and metastasis of ovarian cancer cells [8]. Ahmed et al found that circRNAs in primary ovarian tumors and metastatic tumors can be matched with corresponding miRNAs, and that circRNAs have the potential to change the activity of miRNAs, and can be used as targets for ovarian cancer treatment and markers for judging prognosis [9]. Recently, Liu N et al found that circHIPK3, which was highly expressed in EOC tissues, was associated with lymph node invasion, International Federation of Gynecology and Obstetrics (FIGO) staging and prognosis, and may be a new biomarker for predicting the prognosis of patients with EOC [10].
However, the relationship between circRNA in serum exosomes of patients with EOC and the prognosis value is less studied [11]. Whether exosomal circRNAs play a role in EOC is unclear. Circular forkhead box protein P1 (circFoxp1) is a newly identified circular RNA [12]. It has been reported that circFOXP1 expression is significantly upregulated in gallbladder cancer tissues and positively associated with lymph node metastasis, advanced TNM stage and poor prognosis in patients [13]. There are no reports about the expression of circFoxp1 in serum exosomes of patients with EOC and its role in prognostic evaluation. This study focused on serum exosomal circFoxp1 and its relationship with survival outcome and cisplatin (DDP) resistance in patients with EOC.
MATERIALS AND METHODS
1. Human serum sample collection
This study collected serum samples from 112 patients with EOC and 82 healthy people at the time of diagnosis before starting chemotherapy from September 2014 to September 2019. The clinical characteristics of patients with EOC were obtained through electronic medical records, including age, histological type, histological grade, tumor size, metastasis, FIGO staging, and therapeutic intervention, recurrence and survival. All patients were confirmed by pathological diagnosis. DDP resistance of EOC was determined using the adenosine triphosphate-based tumor chemosensitivity assay as previous reported [14]. Based on the criteria, 112 patients with EOC included 46 DDP resistant and 76 DDP sensitive ones. This study was approved by the Institutional Review Board of Third Xiangya Hospital, Central South University (2018-S103). Written informed consents were obtained from the study participants.
2. Exosomal extraction and identification
Approximately 5 mL of serum was centrifuged at 3,000 g for 15 minutes at 4°C. The centrifuged cell supernatant was filtered through a 0.22 μm filter, and exosomes were extracted according to the instructions of the ExoEasy Maxi Kit (catalog No.76064, Qiagen). The BCA kit (Thermo Scientific, Rockford, IL, USA) was used to quantify the exosome protein concentration, and the samples were aliquoted (100 μL/vial) and stored in a −80°C refrigerator. Transmission electron microscopy was used to observe the exosome morphology, and Western Blotting was used to identify the expression of exosome-specific proteins (CD63, TSG101). The particle size distribution and concentration of exosome were analyzed using ZetaView (Particle Metrix, Ammersee, Germany) according to the manufacturer's instructions.
3. Western blotting
Twenty μg of each protein sample was electrophoresed on 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane. The membrane was blocked with 5% skimmed milk at room temperature for 2 hours, and then incubated with monoclonal primary antibody CD63 (1:1,000) and TSG101 (1:1,000), overnight at 4°C. After washing 3 times with Tris-buffered saline with Tween 20, the membranes were incubated with goat anti-rabbit immunoglobulin G antibody (1:2,000) for 2 hours. Images were developed with AP Chromogenic developer (Invitrogen, Waltham, MA, USA). Each of the above experiments was repeated 3 times.
4. Cell culture
EOC cell lines (COC1, OVCAR3, SKOV3, SKOV3/DDP) and normal human ovarian epithelial cells (IOSE-80) were purchased from China Type Culture Collection (Wuhan, China). All cells were cultured in RPMI-1640 medium, supplementary with 10% fetal bovine serum at 37°C, 95% humidity, and 5% CO2. The medium was changed every 2 days and passaged every 4 days.
5. Viral constructions and infection
The plasmid expressing circFoxp1 was synthesized by Sangon (Shanghai, China). The target sequence of circFoxp1 small interfering RNA (siRNA) (siRNA sequence 1#, 5′-TTTTCCCTTTCCAAGGGCACAG-3′; siRNA sequence 2#, 5′-TGACACGGGAACTTTAGAAATGATT-3′) was synthesized by Sangon. The plasmids were packaged into adenoviruses using the AAVPrime AAV System (GeneCopoeia, Inc., Rockville, MD, USA) according to the manufacturer's protocol. The cells were infected with viral at multiplicity of infection = 50 for 48 hours.
6. Quantitative polymerase chain reaction (qPCR)
qPCR was performed as previously described [15]. Total RNA was extracted from serum or exosomes using Plasma/Serum Exosome Purification and RNA Isolation Mini Kit (catalog No. 58300, Thorold, Canada) following the manufacturer recommended protocol. SYBR Premix EX Taq II (TaKaRa, Kyoto, Japan) was used for real-time quantitative PCR on ABI 7500 Real-Time PCR System (SeqGen, Inc., Torrance, CA, USA). The primers were used as: circFoxp1, forward: CTCCTCTGCACCTTCCAAGA, reverse: ATCATAGCCACTGACACGGG; Foxp1, forward: AACGGCAAAGAGGGAGCC, reverse: GGCATGCATAATGCCACAGG; CCAAT enhancer binding protein gamma (CEBPG), forward: ACAGCCAGCGGAGGTACA, reverse: CACTGTTTCGATCCATGGGC. formin like 3 (FMNL3), forward: GATGGGCAACCTGGAGAGC, reverse: ATGCACCATCGTCACCACTT. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), forward: GAAAGCCTGCCGGTGACTAA, reverse: TTCCCGTTCTCAGCCTTGAC. GAPDH was used as an internal control.
7. 5-ethynyl-2′-deoxyuridine (EdU) staining
EdU staining was performed to measure the cell proliferation using the BeyoClick EdU Cell Proliferation Kit with Alexa Fluor 594 (catalog No. C00788L, Beyotime Biotechnology, Shanghai, China) according to the manufacturer's instructions. Briefly, after indicated treatment, the cells were incubated with Edu (10 μM) for 3 hours, and then fixed with fixative solution (P0098) for 15 minutes at room temperature. The cells were washed with phosphate-buffered saline (PBS) for 5 minutes, and incubated with permeate (P0097) for 15 minutes at room temperature. After washed with PBS for 5 minutes each, cells were incubated with Click Additive Solution for 30 minutes at room temperature in the dark. Nuclei were stained with DAPI, and Edu staining-positive cells were observed under a fluorescence microscope. EdU positive cells were counted at 5 different areas per sample and reported as a percentage of EdU-positive cells.
8. Cell Counting Kit-8 (CCK-8) assay
Cell survival rates were measured using CCK-8 (Beyotime, Hangzhou, China). Briefly, after transfection or exosome treatment (1×107 particles/mL), the cells were treated with a series concentration of DDP or paclitaxel for 24 hours. Then 0.5×104 cells were seeded in each 96-well plate for 24 hours. CCK-8 reagents (10 μL) were added to each well at 1 hour before the endpoint of incubation. Optical density 570 nm value in each well was determined by a microplate reader.
9. Tumor xenograft in nude mice
Animal experiments were approved by the Ethical Committee for Animal Research of Central South University. The mice were purchased from Slac Laboratory Animal (Shanghai, China). The animal experiments were performed as our previous reported with some modifications [16]. The SKOV3/DDP cells were transfected with circFopx1 siRNA as abovementioned for 48 hours prior to injections. To assess tumor growth and DDP sensitization, 100 μL of the transfected SKOV3/DDP cells (1×106) was subcutaneously injected into nude mice (n=5 mice/group). On day 12 following the injection of cells, the mice were intraperitoneally injected with DDP every 3 days, with 3 injections administered in total (3 mg/kg/injection). The control mice were injected with an equal volume of saline. The tumor sizes were measured regularly and calculated using the following formula: 0.5×L×W2, where L and W refer to the length and width of the tumor, respectively. On day 26 following cell injections, the mice were sacrificed by injecting an overdose of pentobarbital sodium. The tumor tissues were removed and fixed with 4% paraformaldehyde and paraffin-embedded for the immunohistochemistry (IHC).
10. IHC
The IHC experiments were performed as our previous reported [16]. The tumor tissues were fixed and paraffin-embedded. The tissues were cut into 4 μm. After retrieved in citric acid buffer, the slices were blocked with 100% normal goat serum (Boster Biological Technology, Pleasanton, CA, USA), and with the following primary antibody (anti-Ki67, catalog No. Ab92742, 1:500 dilution, Abcam) overnight at 4°C. The slices were then incubated with a horseradish peroxidase-conjugated anti-rabbit secondary antibody (1:1 dilution; catalog No. KIT-5905; Fuzhou Maixin Biotech Co., Ltd., Fujian, China) for 2 hours at 37°C. Finally, the sections were counterstained with hematoxylin.
11. Luciferase reporter assay
The wild type sequence and mutants in binding sites of circFoxp1, CEBPG, and FMNL3 was cloned downstream of FL reporter vector. COC1 cells were seeded and cultured in 96-well plates for 24 hours. The cells were co-transfected with FL reporter, Renilla luciferase reporter and miR-22 mimic or miR-150-3p mimic (RiBo Biotechnology, Guangzhou, China) for 48 hours. Luciferase activity was measured using a Luciferase Reporter Assay Kit (BioVision Technologies, Inc., Exton, PA, USA) Relative luciferase activity was normalized to Renilla activity.
12. RNA immunoprecipitation (RIP)
A Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Merck Life Science Co., Ltd. Shanghai, China) was used in this assay as previously described [15]. Briefly, COC1 cells were fixed with formaldehyde, lysed with lysis buffer and sonicated. The supernatant was collected and linked with a circFoxp1 probes and dynabeads mixture overnight at 30°C. On the next day, the dynabeads were washed with 200 μL of lysis buffer. Finally, RNA was extracted from these complexes and was used for qPCR.
13. Bioinformatic analysis
Potential miRNA binding sites within circFoxp1 was predicted by miRanda and RNAhybrid as previously described [17]. The potential targets of miR-22 and miR-150-3p was predicted by miRwalk as previously described [18].
14. Statistical analysis
All experiments were repeated at least three times, and data are expressed as the mean±standard error of the mean. Graphpad prism software (version 8; GraphPad Software, Inc., San Diego, CA, USA) was used to perform statistical analysis. Differences between two groups were compared by student t-test. Differences among three or more groups were compared by one-way analysis of variance with a post hoc Bonferroni test. χ2 test and Kruskal-Wallis test pair Compare categorical and continuous variables. Kaplan-Meier method was used to analyze the overall survival (OS) rate and time to relapse. The area under the curve (AUC) values for exosomal circFoxp1 were determined with a receiver operating characteristic (ROC) curve. The p<0.05 was considered to indicate a statistically significant difference.
RESULTS
1. circFoxp1 expression in serum exosomes of patients with EOC
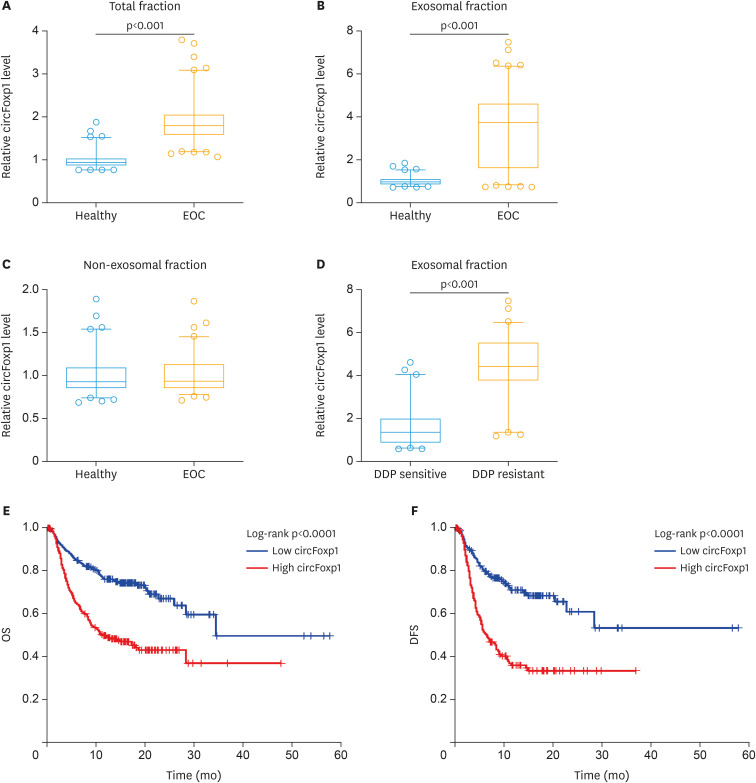
In order to test the role of exosomal circFoxp1 in EOC, we firstly identified whether the collected extracellular vesicles were exosomes. We observed the morphology of extracellular vesicles by scanning electron microscopy (Supplementary Fig. 1A), detected the diameter and concentration of the vesicles by ZetaView, which were about 50–110 nm (Supplementary Fig. 1B), and examined the exosomal markers TSG101 and CD63 (Supplementary Fig. 1C) by Western blot. These results confirmed that the collected extracellular vesicles were the exosomes. We then measured the expression of circFoxp1 in total fraction, exosomal fraction and non-exosomal fraction of serum samples. We found that the expression of circFoxp1 in total fraction was significantly higher than that in healthy people (p<0.001, Fig. 1A). In addition, the expression of circFoxp1 in exosomal fraction, but not in non-exosomal fraction, was significantly upregulated compared with healthy controls (Fig. 1B and C), suggesting that circFoxp1 was enriched in exosomes. It is worth noting that in patients with EOC, the expression of exosomal circFoxp1 in DDP-resistant patients was significantly higher than in DDP-sensitive patients (p<0.001) (Fig. 1D).
Fig. 1. The expression of circFoxp1 in exosomes. (A-C) The expression of circFoxp1 in total fraction of serum (A), in exosomal fraction (B), and in non-exosomal fraction (C). (D) the expression of exosomal circFoxp1 in DDP-sensitive and -resistant EOC patients. (E, F) Kaplan-Meier curve estimated OS rate (E) and DFS rate (F).
circFoxp1, circular forkhead box protein P1; EOC, epithelial ovarian cancer; DDP, cisplatin; OS, overall survival; DFS, disease-free survival.
We further analyzed the correlation between the expression of exosomal circFoxp1 and the clinical characteristics of patients with EOC. Based on the ROC curve (AUC=0.914; 95% confidence interval [CI]=0.867–0.960; Supplementary Fig. 2), the best cut-off was 3.28 that was very close to the mean (3.49) of circFoxp1 expression in exosomes of EOC patients. Therefore, we used the mean value of circFoxp1 expression as the cutoff value to divide the patients into high circFoxp1 group and low circFoxp1 group. As shown in Supplementary Table 1, the expression of serum exosomal circFoxp1 was significantly correlated with FIGO stage (p=0.0312), primary tumor size (p<0.0001), lymph node metastasis (p=0.0009), distant metastasis (p=0.0394), residual tumor diameter (p=0.0275), and clinical response (p<0.0001). There was no significant correlation between the expression of exosome circFoxp1 and age (p=0.3729), histological type (p=0.5548), and histological grade (p=0.6750).
In addition, through survival analysis, we found that patients with EOC with high circFoxp1 expression had lower OS (p<0.0001) and disease-free survival (p<0.0001) (Fig. 1E and F). Univariate and multivariate Cox regression analysis found that exosomal circFoxp1 was an independent factor affecting the survival of patients with EOC (univariate analysis: hazard ratio [HR]=3.21; 95% CI=2.64–5.73; p=0.0035; multivariate analysis: HR=2.79; 95% CI=1.65–4.83; p=0.0073) (Supplementary Tables 2 and 3), suggesting that exosomal circFoxp1 can be used as a biomarker for EOC.
2. circFoxp1 knockdown promotes the sensitivity of EOC cells to DDP
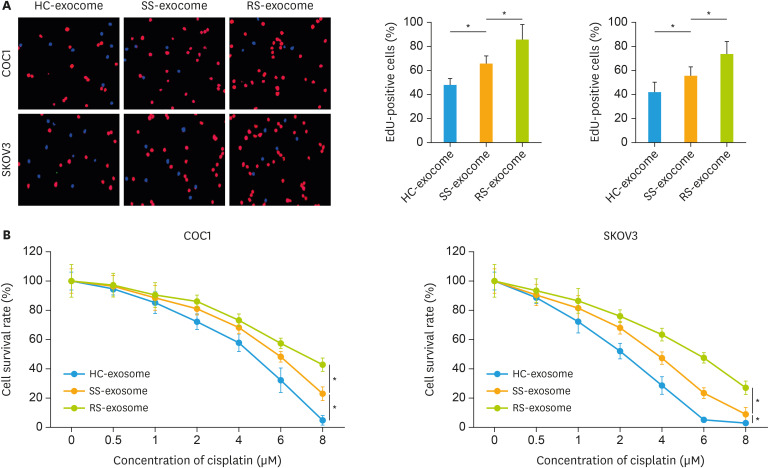
We next tested these circFoxp1-enriched exosomes whether affected the cell survival and proliferation. The results showed that exosome-derived from EOC patients significantly increased the percentage of Edu-positive cells compared with the healthy control (HC-exosome), while the percentage of Edu-positive cells in RS-exosome group (exosome derived from DDP resistant EOC patients)was higher than in SS-exosome group (exosome derived from DDP sensitive EOC patients) (Fig. 2A). In addition, under the condition of DDP stimulation, the treatment of exosome-derived from EOC patients increased the survival rates in COC1 and SKOV3 cells compared with the HC-exosome treatment, while the survival rate in RS-exosome group was higher than in SS-exosome group (Fig. 2B). These results suggest that circFoxp1-enriched exosomes can enhance EOC cell survival and proliferation.
Fig. 2. The effects of exosomes on cell survival and proliferation. (A) Representative images of EdU staining, and quantification of the EdU-positive cells (right). (B) The survival rate in cells treated with exosomes (1 × 107 particles/mL) and cisplatin.
EdU, 5-ethynyl-2′-deoxyuridine; HC, healthy control; SS, cisplatin-sensitive epithelial ovarian cancer patients; RS, cisplatin-resistant epithelial ovarian cancer patients.
*p<0.05.
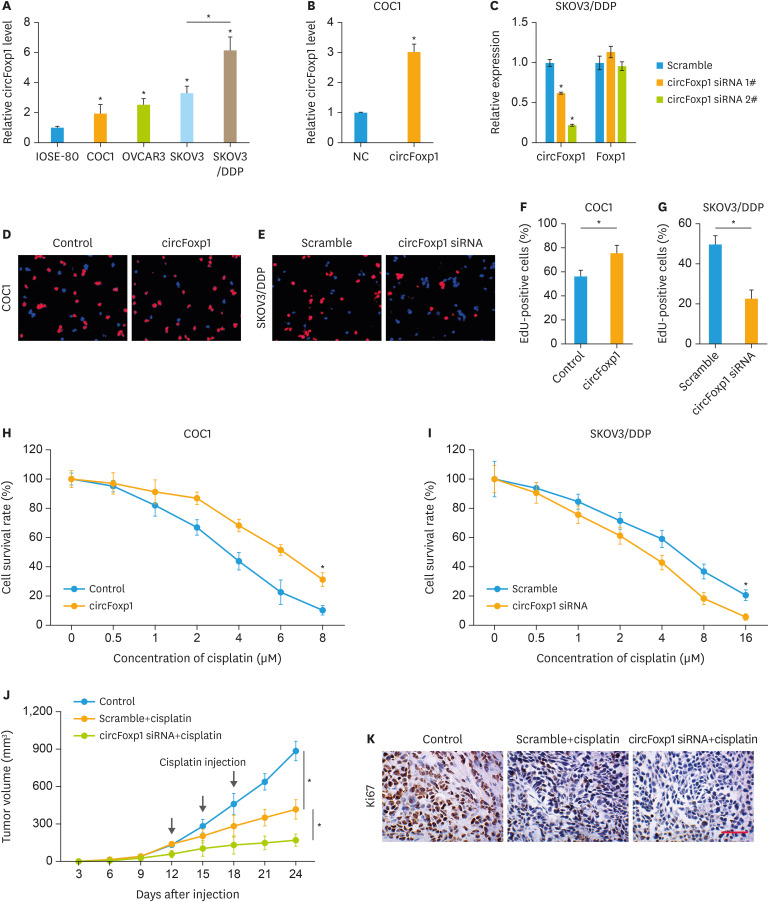
To further study the role of circFoxp1 in EOC cells, we examined the expression of circFoxp1 in different EOC cell lines. Compared to IOSE-80 cells, the expression of circFoxp1 in EOC cells was significantly increased (Fig. 3A). Importantly, we found that the expression of circFoxp1 in SKOV3/DDP was significantly higher than that of its parent cells (Fig. 3A). We overexpressed circFoxp1 in COC1 cells (Fig. 3B). In addition, circFoxp1 was knocked down by siRNA in SKOV3/DDP cells. Both siRNA 1# and siRNA2# transfection decreased the expression of circFoxp1 without significant alterations on the expression of linear mRNA Foxp1, while the efficiency of siRNA 2# was better than siRNA 1# (Fig. 3C). We therefore chose siRNA 2# for next experiments. The effects of circFoxp1 on EOC cell proliferation and DDP resistance were studied by Edu staining and CCK-8 assay. The results showed that overexpression of circFoxp1 in COC1 cells significantly increased the percentage of Edu-positive cells (Fig. 3D and F), while knockdown of circFoxp1 in SKOV3/DDP cells significantly reduced the percentage of Edu-positive cells (Fig. 3E and G). In addition, overexpression of circFoxp1 enhanced DDP and paclitaxel resistance in COC1 cells (Fig. 3H, Supplementary Fig. 3A); in contrast, knockdown of circFoxp1 promoted cell sensitivity to DDP and paclitaxel in SKOV3/DDP and SKOV3 cells, respectively (Fig. 3I, Supplementary Fig. 3B). We further verified the role of circFoxp1 in DDP resistance in EOC cells through in vivo experiments. The results showed that circFoxp1 knockdown significantly inhibited tumor growth and promoted the sensitivity of SKOV3/DDP cells to DDP (Fig. 3J). IHC analysis showed that knockdown of circFoxp1 significantly reduced the expression of tumor cell growth marker Ki67 (Fig. 3K).
Fig. 3. circFoxp1 knockdown attenuates cisplatin-resistance in EOC cells. (A) The expression of circFoxp1 in EOC cell lines. (B) The expression of circFoxp1 in COC1 cells after transfection. (C) The expression of circFoxp1 and Foxp1 in SKOV3/DDP cells after siRNA transfection. (D, E) Representative images of EdU staining in COC1 cells (D) and SKOV3/DDP cells (E). (F, G) quantification of the EdU-positive cells in (D) and (E). (H, I) The survival rate in COC1 cells (H) and SKOV3/DDP cells (I). (J) The tumor volumes of each group. (K) The expression of Ki67 in xenografted tumor tissues (bar, 200 μm).
circFoxp1, circular forkhead box protein P1; EOC, epithelial ovarian cancer; DDP, cisplatin; EdU, 5-ethynyl-2′-deoxyuridine.
*p<0.05.
3. circFoxp1 sponges miR-22 and miR-150-3p
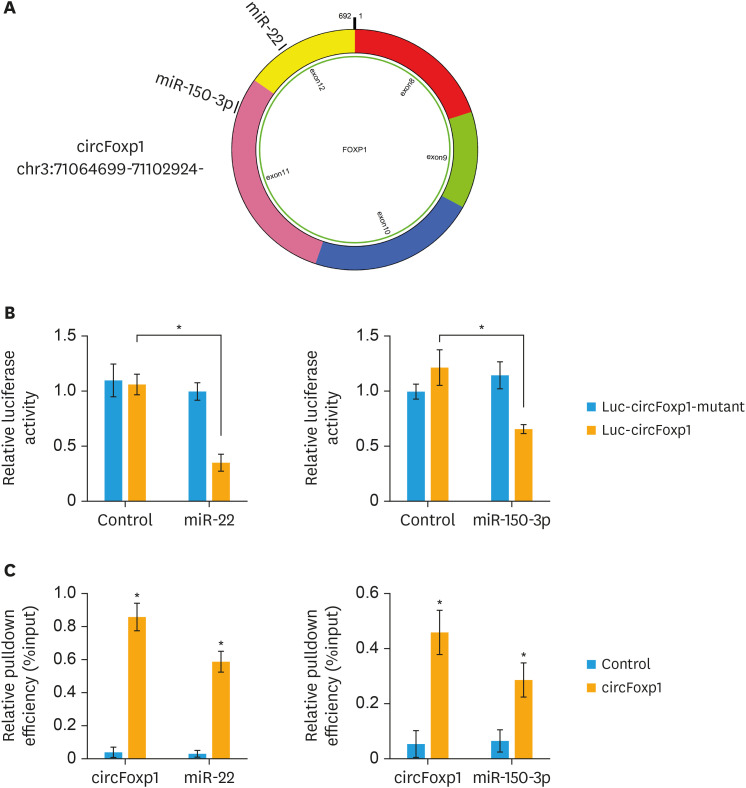
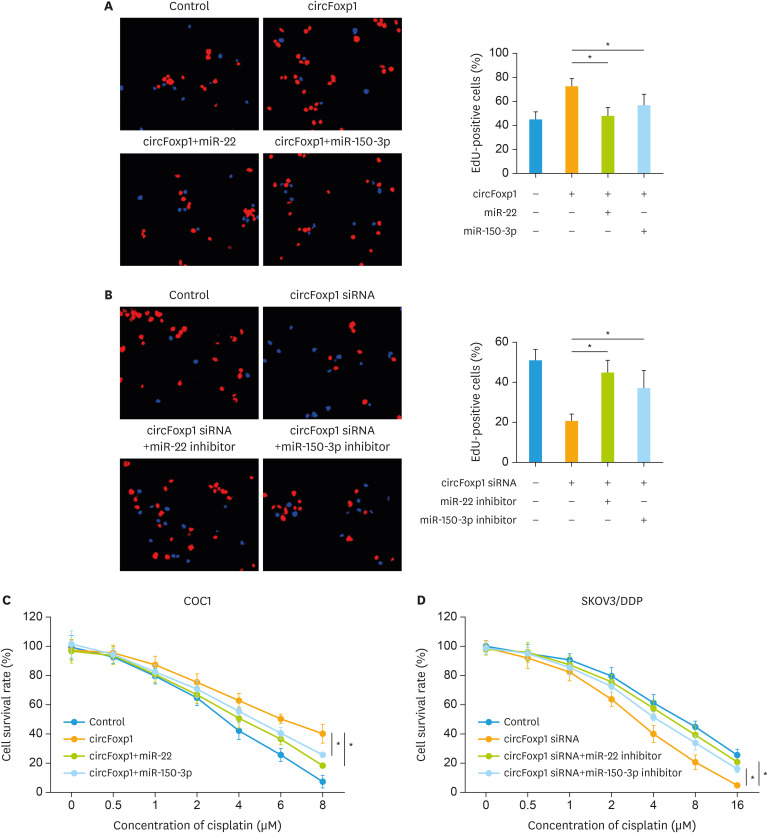
Previous studies have shown that circRNAs exhibit their functions mainly through sponging miRNA. Bioinformatics analysis revealed that circFoxp1 interacted with miR-22 and miR-150-3p simultaneously (Fig. 4A). Further luciferase assay confirmed that circFoxp1 targeted to miR-22 and miR-150-3p (Fig. 4B), suggesting that circFoxp1 may function as a sponge to miR-22 and miR-150-3p. In addition, RIP assay revealed that miR-22 and miR-150-3p directly interacted with circFoxp1 (Fig. 4C). To investigate the downstream mechanism by which circFoxp1 exerted its functions in EOC cells, the mimic of miR-22 and miR-150-3p was co-transfected with circFoxp1. MiR-22 and miR-150-3p mimic could significantly reversed circFoxp1-promoted proliferation in COC1 cells (Fig. 5A). While co-transfection of miR-22 and miR-150-3p inhibitor increased the percentage of EdU-positive cells that inhibited by circFoxp1 knockdown in SKOV3/DDP cells (Fig. 5B). In addition, miR-22 and miR-150-3p mimic treatment attenuated circFoxp1-meadiated DDP resistance in COC1 cells (Fig. 5C). However, miR-22 and miR-150-3p inhibitor treatment enhanced DDP resistance that mitigated by circFoxp1 knockdown in SKOV3/DDP cells (Fig. 5D).
Fig. 4. circFoxp1 interacts with miR-22 and miR-150-3p. (A) Diagram showing the binding site for miR‐22 and miR-150-3p in circFoxp1. (B) Luciferase reporter gene assay was performed to measure luciferase activity. (C) RNA pull-down assay for circFoxp1, miR-22 and miR-150-3p in COC1 cells.
circFoxp1, circular forkhead box protein P1.
*p<0.05.
Fig. 5. circFoxp1 serves as a sponge for miR-22 and miR-150-3p in EOC cells. (A) Representative images of EdU staining in COC1 cells (left), and quantification of the positive cells (right). (B) Representative images of EdU staining in SKOV3/DDP (left), and quantification of the positive cells (right). (C) The survival rate in COC1 cells after treatment. (D) The survival rate in SKOV3/DDP cells after treatment.
circFoxp1, circular forkhead box protein P1; EOC, epithelial ovarian cancer; EdU, 5-ethynyl-2′-deoxyuridine; DDP, cisplatin; siRNA, small interfering RNA.
*p<0.05.
4. MiR-22 and miR-150-3p target to CEBPG and FMNL3
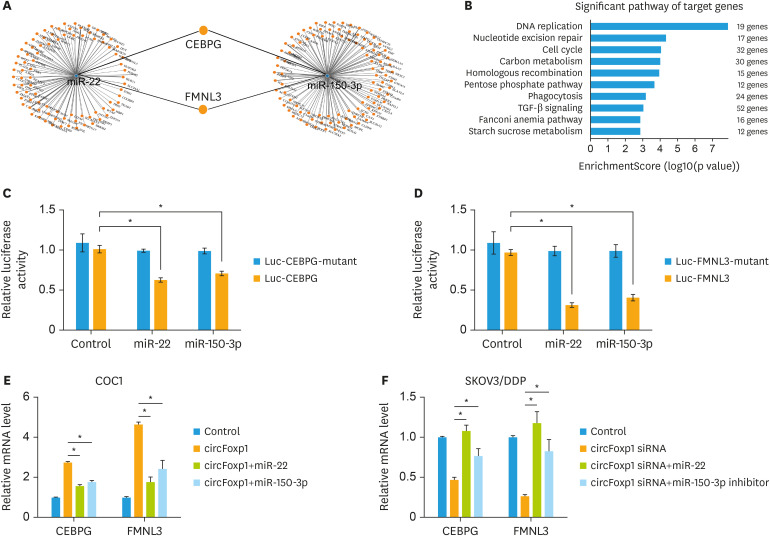
Bioinformatics analysis predicted two potential targets of miR-22 and miR-150-3p, CEBPG, and FMNL3 (Fig. 6A). KEGG analysis showed that DNA replication, nucleotide excision repair, cell cycle and TGF-β signaling pathway were the greatest enriched-pathways that regulated by the targets of miR-22 and miR-150-3p (Fig. 6B). Further luciferase assay confirmed that miR-22 and miR-150-3p targeted to CEBPG and FMNL3 (Fig. 6C and D). In addition, we found that circFoxp1 positively regulated the expression of CEBPG and FMNL3, which were negatively controlled by miR-22 and miR-150-3p in COC1 and SKOV3/DDP cells (Fig. 6E and F).
Fig. 6. miR-22 and miR-150-3p targets to CEBPG and FMNL3 in EOC cells. (A) The mRNA--miRNA gene co‐expression network. (B) KEGG analyzed the significant pathway of target genes of miR-22 and miR-150-3p. (C, D) Luciferase reporter gene assay was performed to measure luciferase activity in COC1 cells after treatment. (E, F) The expression of CEBPG and FMNL3 in COC1 cells (E) and SKOV3/DDP cells (F) after transfection.
CEBPG, CCAAT enhancer binding protein gamma; FMNL3, formin like 3; EOC, epithelial ovarian cancer; DDP, cisplatin.
*p<0.05.
DISCUSSION
Exosomes have high stability and biocompatibility, low immunogenicity and low toxicity. The lipid membrane structure of exosomes plays a good role in protecting the contained nucleic acid molecules [19]. The higher concentration of exosomes in body fluids is conducive to the early diagnosis of tumors. circRNAs are often enriched in exosomes and have high stability and tissue specificity [20]. Therefore, exosomal circRNAs may become special markers for diagnosis and therapeutic target of cancers [11]. This study found that circulating exosomal circFoxp1 was significantly increased in patients with EOC. circFoxp1 expression was positively correlated with FIGO stage, primary tumor size, lymph node metastasis, distant metastasis, residual tumor diameter and clinical response, and was an independent factor predicting survival and disease recurrence in patients with EOC. Confirming the role of circRNA as biomarkers in early diagnosis, prognosis of EOC requires more clinical trials.
Platinum-containing chemotherapy is the first chemotherapy option for patients with advanced ovarian cancer after the first tumor cell depletion [21]. However, after chemotherapy, the tumor-free survival time has gradually shortened, from being sensitive to platinum to being resistant to platinum [22]. Therefore, resistance to chemotherapy drugs is a leading cause of death in patients with advanced ovarian cancer. Studies on resistance to chemotherapy in patients with ovarian cancer have found that exosomes can transmit resistant phenotypes [23]. Crow et al. [24] found that exosomes derived from carboplatin-resistant ovarian cancer cell lines increased the chemotherapy resistance of ovarian cancer cell lines. In addition, stromal cell-derived exosomal miRNA can cause ovarian cancer cells to resist chemotherapeutic drugs [25]. However, the role of exosomal circRNA in ovarian cancer patients' resistance to chemotherapy has not been fully elucidated. By gain- and loss- functional experiments, we found that overexpression of circFoxp1 could promote cell proliferation and reduce DDP sensitivity, while knockdown of circFoxp1 inhibited cell proliferation and enhanced chemotherapy effects.
By bioinformatics analysis, we predicted that circFoxp1 could sponge miR-22 and miR-150-3p. Previous studies have found that miR-22 and miR-150-3p are tumor suppressor genes in a variety of cancers [26,27,28]. The expression of miR-22 is downregulated in EOC, and associated with OS as well as progression-free survival [29]. miR-22 also shows an inhibitory effect on cell migration and invasion in SKOV3 cells [30]. In addition, miR-150 is associated with paclitaxel-resistance in ovarian cancer, and treatment with pre-miR-150 re-sensitizes cancer cells to paclitaxel [31]. Pertuzumab induced miRNA-150 expression in SKOV3 and SNU119 cells, while suppression of miRNA-150 resulted in decreased drug sensitivity to pertuzumab [32]. These results suggest that circFoxp1 may play a role by affecting the function of downstream target genes by sponging miR-22 and miR-150-3p. Further analysis revealed that miR-22 and miR-150-3p targeted to the two genes CEBPG and FMNL3 together. C/EBPs are a family of transcription factors including CEBPG that regulate growth and differentiation of various tissues and control redox homeostasis in normal and cancerous cells [33,34]. FMNL3 is an oncogene in tongue squamous cell carcinoma, nasopharyngeal carcinoma, and colorectal carcinoma [35,36]. This study found that circFoxp1 positively regulated the expression of CEBPG and FMNL3 through miR-22 and miR-150-3p, suggesting that CEBPG and FMNL3 are oncogenes in ovarian cancer, but the specific regulatory mechanism still needs more experimental proof.
In summary, this study finds that upregulation of circulating exosomal circFoxp1 is an independent predictor of survival outcome and DDP resistance in EOC patients. circFoxp1 confers DDP resistance in EOC cells. Circulating exosomal circFoxp1 can be used as a biomarker and potential therapeutic target for EOC.
Footnotes
Funding: This work was supported by National Natural Science Foundation of China (No. 81802668) and Natural Science Foundation of Hunan Province (No. 2018JJ3776).
Conflict of Interest: No potential conflict of interest relevant to this article was reported.
- Conceptualization: Luo Y, Gui R.
- Data curation: Luo Y, Gui R.
- Formal analysis: Luo Y, Gui R.
- Funding acquisition: Luo Y.
- Investigation: Luo Y, Gui R.
- Methodology: Luo Y, Gui R.
- Project administration: Luo Y, Gui R.
- Resources: Luo Y, Gui R.
- Software: Luo Y, Gui R.
- Supervision: Luo Y, Gui R.
- Validation: Luo Y, Gui R.
- Visualization: Luo Y, Gui R.
- Writing - original draft: Luo Y, Gui R.
- Writing - review & editing: Luo Y, Gui R.
SUPPLEMENTARY MATERIALS
Association of tumor characteristics and circFoxp1 expression
Univariate analysis of prognostic factors of EOC
Multivariate analysis of independent prognostic factors of EOC
Identification of circulating exosomes. (A) The morphology of circulating exosomes (indicated by arrow) was observed by transmission electron microscopy. (B) The concentration and size of particles isolated from serum samples. (C) Western blot was performed to test the markers of exosomes, TSG101 and CD63. PBS was used as negative control.
ROC curve of the exosomal circFoxp1 signature.
circFoxp1 knockdown attenuates paclitaxel-resistance in EOC cells. (A) CCK-8 was performed to measure the survival rate after circFoxp1 overexpression and paclitaxel treatment in COC1 cells. (B) CCK-8 was performed to measure the survival rate after circFoxp1 knockdown and paclitaxel treatment in SKOV3 cells.
References
- 1.Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. 2019;393:1240–1253. doi: 10.1016/S0140-6736(18)32552-2. [DOI] [PubMed] [Google Scholar]
- 2.Armbruster S, Coleman RL, Rauh-Hain JA. Management and treatment of recurrent epithelial ovarian cancer. Hematol Oncol Clin North Am. 2018;32:965–982. doi: 10.1016/j.hoc.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Pignata S, Pisano C, Di Napoli M, Cecere SC, Tambaro R, Attademo L. Treatment of recurrent epithelial ovarian cancer. Cancer. 2019;125 Suppl 24:4609–4615. doi: 10.1002/cncr.32500. [DOI] [PubMed] [Google Scholar]
- 4.Van Berckelaer C, Brouwers AJ, Peeters DJ, Tjalma W, Trinh XB, van Dam PA. Current and future role of circulating tumor cells in patients with epithelial ovarian cancer. Eur J Surg Oncol. 2016;42:1772–1779. doi: 10.1016/j.ejso.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Giannopoulou L, Zavridou M, Kasimir-Bauer S, Lianidou ES. Liquid biopsy in ovarian cancer: the potential of circulating miRNAs and exosomes. Transl Res. 2019;205:77–91. doi: 10.1016/j.trsl.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Li D, Luo H, Zhu X. Circular RNAs: the star molecules in cancer. Mol Aspects Med. 2019;70:141–152. doi: 10.1016/j.mam.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, et al. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu KS, Pan F, Mao XD, Liu C, Chen YJ. Biological functions of circular RNAs and their roles in occurrence of reproduction and gynecological diseases. Am J Transl Res. 2019;11:1–15. [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmed I, Karedath T, Andrews SS, Al-Azwani IK, Mohamoud YA, Querleu D, et al. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget. 2016;7:36366–36381. doi: 10.18632/oncotarget.8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu N, Zhang J, Zhang LY, Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22:3713–3718. doi: 10.26355/eurrev_201806_15250. [DOI] [PubMed] [Google Scholar]
- 11.Shi X, Wang B, Feng X, Xu Y, Lu K, Sun M. circRNAs and exosomes: a mysterious frontier for human cancer. Mol Ther Nucleic Acids. 2020;19:384–392. doi: 10.1016/j.omtn.2019.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherubini A, Barilani M, Rossi RL, Jalal MM, Rusconi F, Buono G, et al. FOXP1 circular RNA sustains mesenchymal stem cell identity via microRNA inhibition. Nucleic Acids Res. 2019;47:5325–5340. doi: 10.1093/nar/gkz199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S, Zhang Y, Cai Q, Ma M, Jin LY, Weng M, et al. Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. Mol Cancer. 2019;18:145. doi: 10.1186/s12943-019-1078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao D, Wu LY, Li XG, Wang XB, Li M, Li YF, et al. Application of ATP-tumor chemosensitivity assay in recurrent epithelial ovarian cancer. Zhonghua Zhong Liu Za Zhi. 2010;32:855–858. [PubMed] [Google Scholar]
- 15.Luo Y, Fu Y, Huang R, Gao M, Liu F, Gui R, et al. CircRNA_101505 sensitizes hepatocellular carcinoma cells to cisplatin by sponging miR-103 and promotes oxidored-nitro domain-containing protein 1 expression. Cell Death Dis. 2019;5:121. doi: 10.1038/s41420-019-0202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma J, Niu W, Wang X, Zhou Y, Wang H, Liu F, et al. Bromodomain‑containing protein 7 sensitizes breast cancer cells to paclitaxel by activating Bcl2‑antagonist/killer protein. Oncol Rep. 2019;41:1487–1496. doi: 10.3892/or.2018.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao M, Xu J, Zhong S, Liu Y, Xiao H, Geng L, et al. Expression profiles and potential functions of circular RNAs in extracellular vesicles isolated from radioresistant glioma cells. Oncol Rep. 2019;41:1893–1900. doi: 10.3892/or.2019.6972. [DOI] [PubMed] [Google Scholar]
- 18.Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One. 2018;13:e0206239. doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18:116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fanale D, Taverna S, Russo A, Bazan V. Circular RNA in exosomes. Adv Exp Med Biol. 2018;1087:109–117. doi: 10.1007/978-981-13-1426-1_9. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Wang X. The emerging roles and therapeutic potential of exosomes in epithelial ovarian cancer. Mol Cancer. 2017;16:92. doi: 10.1186/s12943-017-0659-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Namee NM, O'Driscoll L. Extracellular vesicles and anti-cancer drug resistance. Biochim Biophys Acta Rev Cancer. 2018;1870:123–136. doi: 10.1016/j.bbcan.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Shen X, Lyu W. Research advances on the role of exosomes in chemotherapy resistance of ovarian cancer. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2019;48:116–120. doi: 10.3785/j.issn.1008-9292.2019.02.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crow J, Atay S, Banskota S, Artale B, Schmitt S, Godwin AK. Exosomes as mediators of platinum resistance in ovarian cancer. Oncotarget. 2017;8:11917–11936. doi: 10.18632/oncotarget.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Chen X, Cheng R, Yang F, Yu M, Wang C, et al. The Jun/miR-22/HuR regulatory axis contributes to tumourigenesis in colorectal cancer. Mol Cancer. 2018;17:11. doi: 10.1186/s12943-017-0751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K, Li XY, Wang ZM, Han ZF, Zhao YH. MiR-22 inhibits lung cancer cell EMT and invasion through targeting Snail. Eur Rev Med Pharmacol Sci. 2017;21:3598–3604. [PubMed] [Google Scholar]
- 27.Zhao YJ, Song X, Niu L, Tang Y, Song X, Xie L. Circulating exosomal miR-150-5p and miR-99b-5p as diagnostic biomarkers for colorectal cancer. Front Oncol. 2019;9:1129. doi: 10.3389/fonc.2019.01129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan WN, Zhang YQ, Wang XM, Liu YJ, Zhang YX, Que YH, et al. Down-regulated miR-22 as predictive biomarkers for prognosis of epithelial ovarian cancer. Diagn Pathol. 2014;9:178. doi: 10.1186/s13000-014-0178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Liang S, Yu H, Zhang J, Ma D, Lu X. An inhibitory effect of miR-22 on cell migration and invasion in ovarian cancer. Gynecol Oncol. 2010;119:543–548. doi: 10.1016/j.ygyno.2010.08.034. [DOI] [PubMed] [Google Scholar]
- 30.Kim TH, Jeong JY, Park JY, Kim SW, Heo JH, Kang H, et al. miR-150 enhances apoptotic and anti-tumor effects of paclitaxel in paclitaxel-resistant ovarian cancer cells by targeting Notch3. Oncotarget. 2017;8:72788–72800. doi: 10.18632/oncotarget.20348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wuerkenbieke D, Wang J, Li Y, Ma C. miRNA-150 downregulation promotes pertuzumab resistance in ovarian cancer cells via AKT activation. Arch Gynecol Obstet. 2015;292:1109–1116. doi: 10.1007/s00404-015-3742-x. [DOI] [PubMed] [Google Scholar]
- 32.Huggins CJ, Mayekar MK, Martin N, Saylor KL, Gonit M, Jailwala P, et al. C/EBPγ is a critical regulator of cellular stress response networks through heterodimerization with ATF4. Mol Cell Biol. 2015;36:693–713. doi: 10.1128/MCB.00911-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alberich-Jordà M, Wouters B, Balastik M, Shapiro-Koss C, Zhang H, Di Ruscio A, et al. C/EBPγ deregulation results in differentiation arrest in acute myeloid leukemia. J Clin Invest. 2012;122:4490–4504. doi: 10.1172/JCI65102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Chen S, Chen Y, Geng N, Feng C. High expression of FMNL3 associates with cancer cell migration, invasion, and unfavorable prognosis in tongue squamous cell carcinoma. J Oral Pathol Med. 2019;48:459–467. doi: 10.1111/jop.12857. [DOI] [PubMed] [Google Scholar]
- 35.Wu Y, Shen Z, Wang K, Ha Y, Lei H, Jia Y, et al. High FMNL3 expression promotes nasopharyngeal carcinoma cell metastasis: role in TGF-β1-induced epithelia-to-mesenchymal transition. Sci Rep. 2017;7:42507. doi: 10.1038/srep42507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng YF, Xiao YS, Liu Y, Luo XJ, Wen LD, Liu Q, et al. Formin-like 3 regulates RhoC/FAK pathway and actin assembly to promote cell invasion in colorectal carcinoma. World J Gastroenterol. 2018;24:3884–3897. doi: 10.3748/wjg.v24.i34.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Association of tumor characteristics and circFoxp1 expression
Univariate analysis of prognostic factors of EOC
Multivariate analysis of independent prognostic factors of EOC
Identification of circulating exosomes. (A) The morphology of circulating exosomes (indicated by arrow) was observed by transmission electron microscopy. (B) The concentration and size of particles isolated from serum samples. (C) Western blot was performed to test the markers of exosomes, TSG101 and CD63. PBS was used as negative control.
ROC curve of the exosomal circFoxp1 signature.
circFoxp1 knockdown attenuates paclitaxel-resistance in EOC cells. (A) CCK-8 was performed to measure the survival rate after circFoxp1 overexpression and paclitaxel treatment in COC1 cells. (B) CCK-8 was performed to measure the survival rate after circFoxp1 knockdown and paclitaxel treatment in SKOV3 cells.