Abstract
Introduction
During the first month of the SARS-CoV-2 outbreak, rapid development of PCR-based diagnostic tests became a global priority so that timely diagnosis, isolation, and contact tracing could minimize the advancing pandemic surge. Designing these tests for broad, long-term detection was complicated by limited information about the novel virus’ genome sequence and how it might mutate during global spread and adaptation to humans.
Methods
We assessed eight widely adopted lab developed PCR tests for SARS-CoV-2 against 15,001 SARS-CoV-2 genome sequences. Using a custom bioinformatic pipeline called PCR_strainer, we identified all mismatches and sequence variants in genome locations targeted by 15 sets of primer/probe oligonucleotides from these assays.
Results
For 12 out of 15 primer/probe sets, over 98 % of SARS-CoV-2 genomes had no mismatches. Two primer/probe sets contained a single mismatch in the reverse primer that was present in over 99 % of genomes. One primer/probe set targeted a location with extensive polymorphisms with 23 sequence observed variants at the forward primer location. One of these variants, which contains three nucleotide mismatches, arose in February as part of the emergence of a viral clade and was present in 18.8 % of the genomes we analyzed.
Discussion
Most early PCR diagnostic tests for SARS-CoV-2 remain inclusive of circulating viral diversity, but three assays with extensive mismatches highlight assay design challenges for novel pathogens and provide valuable lessons for PCR assay design during future outbreaks. Our bioinformatics pipeline is also presented as a useful general-purpose tool for assessing PCR diagnostics assays against circulating strains.
Keywords: SARS-CoV-2, COVID-19, Real-time polymerase chain reaction, Molecular diagnostic techniques, Laboratory developed test, Circulating strains
1. Introduction
In late 2019, the novel coronavirus SARS-CoV-2 emerged into humans. Within the span of two-and-a-half months, it spread from a cluster of pneumonia cases of unknown origin in Wuhan, China to a World Health Organization (WHO)-declared global pandemic emergency impacting 114 countries [1,2]. During this time, rapid development of laboratory diagnostics for the novel virus became a global priority; prompt diagnosis, isolation, and contact tracing became central to controlling the advancing pandemic surge. Several government, hospital, and academic labs designed PCR-based assays which were compiled by the World Health Organization (WHO) for global dissemination. Seven of these laboratory developed tests (LDTs) were shared on the WHO website on Jan 24, 2020, barely a month after the first reported cases [3]. An additional LDT was developed in mid-January 2020 at the British Columbia Centre for Disease Control Public Health Laboratory [4].
PCR-based diagnostic assays rely on the careful design of synthetic oligonucleotide primers and probes. Nucleotide mismatches between primers, probes, and target genetic material result in thermodynamic instabilities that can disrupt PCR chemistry, impair detection, and produce false negative results. This is especially problematic when detecting RNA viruses, like coronaviruses, whose genomes mutate readily compared to DNA-based organisms. Consequently, oligonucleotide design strategies for viral pathogens focus on genomic locations where low mutation rates are crucial to preserving biological function and pathogen viability.
Novel pathogens present unique challenges for oligonucleotide design because their genomes are poorly characterized. Without extensive reference genome sequences, it becomes difficult to discern stable genomic loci to target for oligonucleotide design. In the case of recently emerged zoonotic viruses, these challenges are compounded by hard-to-predict genomic changes accompanying adaptation to the new host. This makes a pandemic caused by a novel zoonotic RNA virus a challenging scenario in which to develop PCR-based diagnostics: oligonucleotide design choices carry high stakes but must be made with incomplete and insufficient information.
Here, we provide an assessment of oligonucleotide designs from those crucial early LDTs for SARS-CoV-2, many of which were widely adopted as the pandemic grew globally. Our analysis benefits from an unprecedented global genomics effort that has generated around 40,000 publicly released SARS-CoV-2 genomes in a mere 5 months. We have used this abundance of genome sequences to evaluate the frequency of mutations in genomic locations targeted by eight early LDTs.
We also provide the bioinformatic pipeline used for our analysis, called PCR_strainer, as a valuable general-purpose tool for clinical microbiology laboratories. It allows straightforward and fast assessment of diagnostic PCR assays against numerous reference genomes representing the circulating strain diversity of a target pathogen.
2. Material and methods
2.1. SARS−COV-2 genome sequences
We downloaded 38,980 SARS-CoV-2 genome sequences and accompanying metadata from the Global Initiative for Sharing All Influenza Data (GISAID). We gratefully acknowledge the work of the originating and submitting laboratories for these sequences, who are listed in Supplement S1. These sequences were submitted before June 8, 2020 from specimens collected between Dec 31, 2019 and May 31, 2020. We selected all available sequences, restricted to full genomes (over 29,000 bp), and excluded sequences with over 5% Ns. We further filtered genomes to remove any containing degenerate bases further than 100 positions from the 5′ or 3’ sequence ends. We manually removed non-human animal and environmental isolates.
2.2. PCR_strainer
The PCR_strainer script was written in Python v3.7.3. It is available from https://github.com/KevinKuchinski/PCR_strainer. It performed in silico PCR simulation using Thermonucleotide BLAST (TNTBLAST) v2.04 [5]. TNTBLAST results were limited to best matches, with 30 °C minimum annealing temperature for primers and probes to generate maximum amplicon diversity. Default salt and oligonucleotide molarity parameters were used. PCR_strainer parsed the output from TNTBLAST to extract the number of gaps and mismatches between each assay oligonucleotide and each reference genome at target locations. PCR_strainer tabulated these data and wrote them to a TSV file. This tabulated data was used to identify all sequence variants, calculate their frequency, and generate PCR_strainer’s final report on the performance of each assay.
2.3. Phylogenetic tree of SARS-CoV-2 genomes
All SARS-CoV-2 genomes sequences in the final dataset from May 2020 were copied into a separate FASTA file (n = 778 sequences), then 100 bases were trimmed from the 5′ and 3’ ends of each sequence to remove low-quality degenerate nucleotides. A multiple sequence alignment was generated using MUSCLE v3.8.1551 using a maximum of 16 iterations [6]. The resulting multiple sequence alignment was used for tree construction, which was carried out using PhyML with default parameters [7]. The resulting tree was visualized using the ETE Toolkit module for Python [8].
3. Results
3.1. Assays analyzed in this study
For this analysis, we focused on 7 SARS-CoV-2 LDTs compiled by the WHO and initially released on Jan 24, 2020. The original document of compiled assays, as provided by the WHO, is included as Supplement S2 [3]. We also analyzed an assay developed by the British Columbia Centre for Disease Control Public Health Laboratory [4]. These LDTs are summarized in Table 1 . The majority of these assays (6 of 8) were multiplexed as 2 or 3 oligonucleotide sets. This gave a total of 15 target locations in the SARS-CoV-2 genome. The most commonly targeted gene was the nucleocapsid (N) gene (n = 8), followed by the RNA-dependent RNA polymerase (RdRP) gene (n = 4). Each of the following was targeted by one set of oligonucleotides: the envelope (E) gene, nonstructural protein 14 (nsp14) gene, and an uncharacterized location in open reading frame 1ab (orf1ab).
Table 1.
Summary of SARS-CoV-2 lab developed tests analyzed in this study.
| Assay developers | Assay name | Assay target |
|---|---|---|
| Charité Virology, Tib-Molbiol, Erasmus MC, Public Health England [9] | Charité group – Na | Nucleocapsid |
| Charité group – RdRPb | RNA-dependent RNA polymerase | |
| Charité group – Ec | Envelope | |
| Chinese Centre for Disease Control | China CDC – N | Nucleocapsid |
| China CDC – orf1ab | Open reading frame 1ab (orf1ab) | |
| Hong Kong University [10] | HKU – N | Nucleocapsid |
| HKU – orf1b-nsp14 | Open reading frame 1b – nonstructural protein 14 (orf1b-nsp14) | |
| National Institute of Infectious Disease, Japan [11] | Japan NIID - N | Nucleocapsid |
| Institut Pasteur France | Pasteur - RdRP-IP2 | RNA-dependent RNA polymerase |
| Pasteur - RdRP-IP4 | RNA-dependent RNA polymerase | |
| National Institute of Health, Thailand | Thailand NIH - N | Nucleocapsid |
| Centres for Disease Control and Prevention, USA | CDC USA - N1 | Nucleocapsid |
| CDC USA - N2 | Nucleocapsid | |
| CDC USA - N3a | Nucleocapsid | |
| British Columbia Centre for Disease Control Public Health Laboratory [4] | BCCDC PHL – RdRP | RNA-dependent RNA polymerase |
Omitted from latest version of assay.
Only the SARS-CoV-2 specific probe is considered in this analysis.
Also used in assays from Institut Pasteur, Japan NIID, and BC CDC PHL assays as confirmatory target.
3.2. Three sets of oligonucleotides had extensive mismatches against SARS-CoV-2 genome sequences
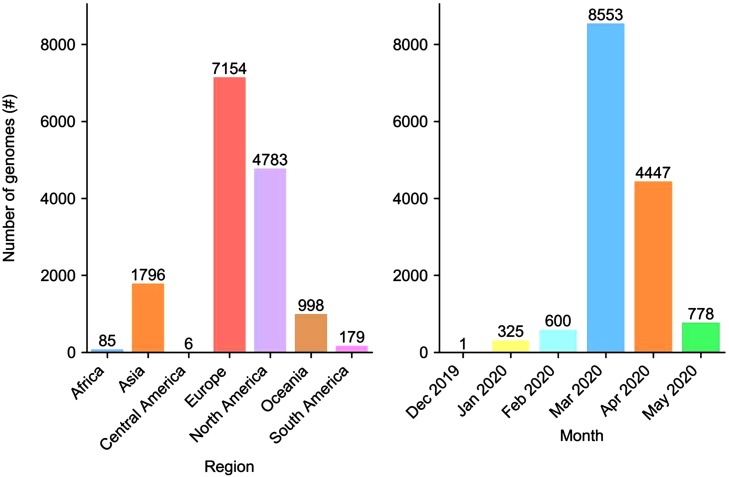
To assess the inclusivity of oligonucleotides from these assays, we curated a dataset of 15,001 SARS-CoV-2 genome sequences from specimens collected between Dec 31, 2019 and May 31, 2020. The composition of the dataset by region and collection month is shown in Fig. 1 .
Fig. 1.
Regional composition and collection month of SARS-CoV-2 genomes analyzed in this study. Region and country information was available for all 15,001 genomes, but collection month information was only available for 14,704 genomes.
Using PCR_strainer, we identified genomes in our dataset with nucleotide mismatches in locations targeted by the early SARS-CoV-2 LDTs described above. The frequency of mismatches for each set of oligonucleotides is reported in Table 2 . Overall, these oligonucleotide designs maintained high inclusivity among circulating SARS-CoV-2 variants throughout the first five months of the pandemic. For 12 out of 15 sets of oligonucleotides, over 98 % of genomes had no mismatches. The remaining 3 sets of oligonucleotides (dark shading in Table 2) had outlying mismatch frequencies and were analyzed further using PCR_strainer.
Table 2.
Frequency of mismatches between 15,001 SARS-CoV-2 genome sequences and 15 sets of oligonucleotides from early lab developed tests. The Charite group - RdRP, Japan NIID - N, and China CDC - N assays were further analyzed due to extensive mismatches..
| Assay | 0 mismatches |
1 mismatches |
2 mismatches |
3+ mismatches |
|---|---|---|---|---|
| Charité group - N | 98.9 % | 0.9 % | 0.0 % | 0.1 % |
| Charité group - RdRP | 0.0 % | 99.6 % | 0.3 % | 0.0 % |
| Charité group - E | 99.6 % | 0.2 % | 0.1 % | 0.0 % |
| China CDC - N | 79.9 % | 1.0 % | 0.0 % | 19.0 % |
| China CDC - orf1ab | 99.1 % | 0.8 % | 0.0 % | 0.0 % |
| HKU - N | 98.9 % | 0.9 % | 0.1 % | 0.1 % |
| HKU - orf1b-nsp14 | 99.5 % | 0.4 % | 0.0 % | 0.0 % |
| Japan NIID - N | 0.0 % | 99.5 % | 0.5 % | 0.0 % |
| Pasteur - RdRP-IP2 | 99.5 % | 0.2 % | 0.3 % | 0.0 % |
| Pasteur - RdRP-IP4 | 99.5 % | 0.4 % | 0.1 % | 0.0 % |
| Thailand NIH - N | 99.1 % | 0.4 % | 0.2 % | 0.3 % |
| CDC USA - N1 | 97.6% | 0.6 % | 0.1 % | 1.7 % |
| CDC USA - N2 | 99.3% | 0.6 % | 0.0 % | 0.0 % |
| CDC USA - N3 | 98.4% | 1.4 % | 0.0 % | 0.1 % |
| BCCDC PHL - RdRP | 99.7 % | 0.3 % | 0.0 % | 0.0 % |
3.3. Pervasive single nucleotide mismatches in assays from Charité Group and Japan NIID
Two sets of oligonucleotides had mismatches against all 15,001 SARS-CoV-2 reference genomes in our dataset: the Charité group’s RdRP gene assay and the Japan NIID’s N gene assay. For both assays, almost all genomes contained a single mismatch: 99.7 % against the RdRP gene assay and 99.5 % against the N gene assay (Table 2). Using PCR_strainer, we obtained all viral genomic sequences at locations targeted by these assays and counted the frequency of each variant at the forward primer, reverse primer, and probe target locations (Table 3 ). For both assays, one sequence variant in the reverse primer location accounted for nearly all mismatched genomes, suggesting a simple update the oligonucleotide sequences for these assays.
Table 3.
SARS-CoV-2 genomic sequence variants at locations targeted by three early lab developed tests with substantial mismatches. Mismatched nucleotides are bolded and underlined. For convenience, genome locations are expressed as DNA sequences from the strand containing the assay oligonucleotide (top strand for forward primers and probes and bottom strand for reverse primers). Only variants present in at least 1% of all genomes in the dataset are shown (table with all variants available as Supplementary Table S1).
| Assay | Oligonucleotide | Variant genome sequence at target location | Mismatches | Frequency among all genomes |
|---|---|---|---|---|
| Charité group - RdRP | Forward primer | 5′ GTGAAATGGTCATGTGTGGCGG 3’ | 0 | 99.7 % |
| Probe | 5′ CAGGTGGAACCTCATCAGGAGATGC 3’ | 0 | 99.9% | |
| Reverse primer | 5′ CAAATGTTAAAAACACTATTAGCATA 3’ | 1 | 100.0 % | |
| Japan NIID - N | Forward primer | 5′ AAATTTTGGGGACCAGGAAC 3’ | 0 | 99.7 % |
| Probe | 5′ ATGTCGCGCATTGGCATGGA 3’ | 0 | 99.9% | |
| Reverse primer | 5′ TGGCACCTGTGTAGGTCAAC 3’ | 1 | 99.8% | |
| China CDC - N | Forward primer | 5′ GGGGAACTTCTCCTGCTAGAAT 3’ | 0 | 80.1 % |
| 5′ AACGAACTTCTCCTGCTAGAAT 3’ | 3 | 18.8 % | ||
| Probe | 5′ TTGCTGCTGCTTGACAGATT 3’ | 0 | 100.0 % | |
| Reverse primer | 5′ CAGACATTTTGCTCTCAAGCTG 3’ | 0 | 99.8% |
3.4. Investigating Polymorphisms in China CDC’S N Gene Assay
The third oligonucleotide set with extensive mismatches was the China CDC’s N gene assay. PCR_strainer reported mismatches against 20.1 % of SARS-CoV-2 genomes analyzed, with 3 or more mismatches in 19.0 % of genomes (Table 2). Using PCR_strainer, we obtained the sequences of the forward primer, reverse primer, and probe target locations in all genomes. We observed 23 different sequence variants with up to 8 mismatches at the forward primer target location. One of these variants, which we will call the ‘AAC’ variant, was present in 18.8 % of genomes in our dataset (Table 3).
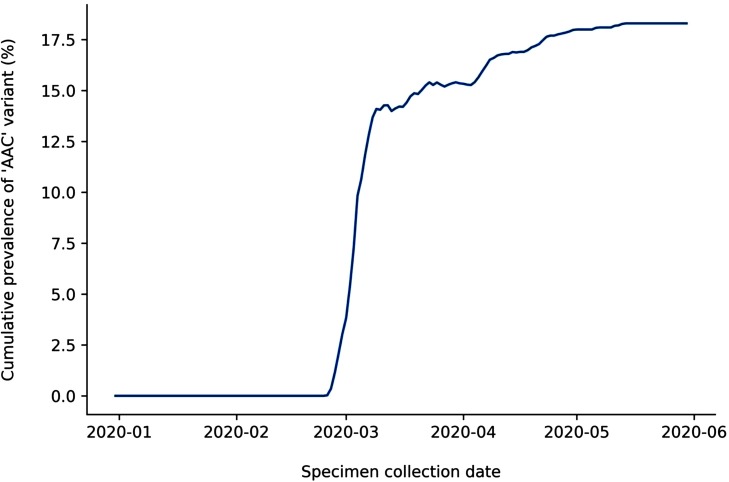
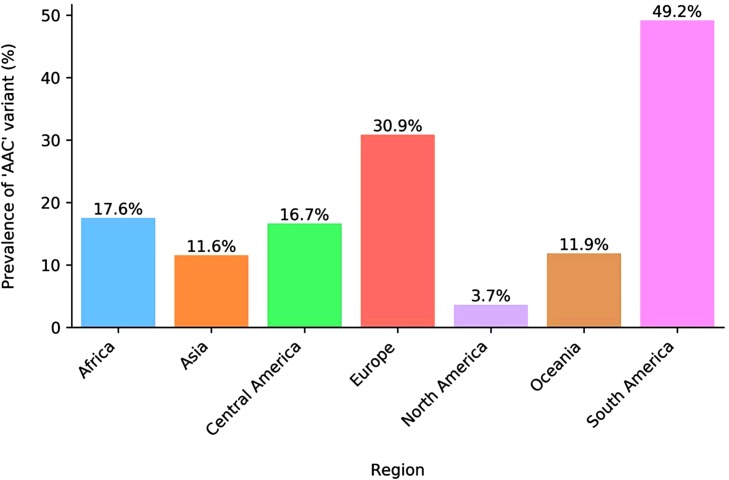
We analyzed the prevalence of the ‘AAC’ variant within our reference genomes over time (Fig. 2 ). Its first appearance was in a specimen collected Feb 25, 2020 in Germany. This suggests that polymorphisms in this location arose during spread outside of China, at least one month after the assay had been designed. We also analyzed the ‘AAC’ variant’s prevalence by region (Fig. 3 ), revealing it was most prevalent in South America and Europe, appearing in 49.2 % and 30.9 % of genomes respectively. In China, where this assay was designed during the initial phase of the outbreak, only 9 genomes (1.5 %) were reported with this variant before June 1, 2020. The earliest of these was not collected until March 15, 2020.
Fig. 2.
Cumulative prevalence of ‘AAC’ variant in SARS-CoV-2 genomes collected between Dec 31, 2019 and May 31, 2020. Cumulative prevalence indicates the cumulative number of genomes with the variant divided by the cumulative total number of genomes up to the given date. Specimen collection date information was available for 14,477 genomes in the dataset.
Fig. 3.
Regional prevalence of ‘AAC’ variant among 15,001 SARS-CoV-2 genomes collected between Dec 31, 2019 and May 31, 2020. A table of prevalence by country is provided as Supplementary Table S2.
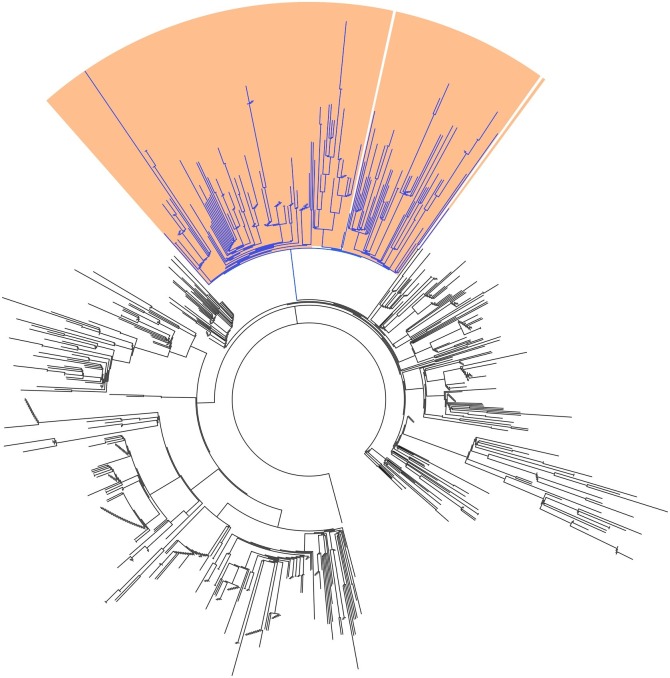
Next, we constructed a phylogeny of all SARS-CoV-2 genomes in our dataset from May 2020, the most recent completed month at the time of writing (Fig. 4 ). We coloured branches by whether all descendent genomes contained ‘AAC’ variant (blue) or original (black) genotypes. This tree showed the variant belonged to a clearly defined clade. We identified the common ancestral node of all variants, and 99.5 % of leaves descending from this node represented genomes containing the ‘AAC’ variant sequence. Furthermore, this clade closely overlapped with the GISAID ‘GR’ clade: 99.5 % of genomes with the ‘AAC’ variant were identified as members of Clade ‘GR’, and 100 % of genomes identified as members of clade ‘GR’ contained the ‘AAC’ variant. It also overlapped less-closely with the B.1.1 lineage proposed by Rambault et al. [12]: 100 % of genomes identified as members of Lineage B.1.1 contained the ‘AAC’ variant, while only 83.0 % of genomes with ‘AAC’ variant were identified as members of Lineage B.1.1.
Fig. 4.
Phylogenetic tree of 778 SARS-CoV-2 genomes from May 2020. Genomes containing the ‘AAC’ variant sequence in the location targeted by the China CDC’s N gene assay are coloured blue. Genomes belonging to the GISAID ‘GR’ clade are shaded in orange (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
When polymorphisms are identified in a PCR target location, a common strategy is to generate a consensus sequence and incorporate degenerate bases into the affected oligonucleotide. PCR_strainer facilitates this task by generating separate FASTA files containing target location sequences from all genomes in the provided dataset. We used this FASTA file to generate a consensus sequence for the forward primer target location of the China CDC’s N gene assay, incorporating degenerate nucleotides into polymorphic positions with a consensus threshold of 10 %. This consensus sequence provides a possible update to the forward primer sequence, changing it from 5′-GGGGAACTTCTCCTGCTAGAAT-3’ to 5′-RRMGAACTTCTCCTGCTAGAAT-3’.
4. Discussion
From our analysis using PCR_strainer, we report that most oligonucleotides in these assays have maintained high inclusivity during the first fourth months of the SARS-CoV-2 pandemic. There were only 3 oligonucleotides with substantial mismatches and polymorphisms: the reverse primer from the Charité group’s RdRP gene assay, the reverse primer from the Japan NIID’s N gene assay, and the forward primer from the China CDC’s N gene assay.
Single mismatches in the Charité group and Japan NIID assays were uniform and pervasive, suggesting they were artefacts of the oligonucleotide design process. The mismatch in the Japan NIID’s assay was due to an error in the first reported SARS-CoV-2 genome assembly, and investigations by the assay’s designers did not find any impact on clinical performance [11]. The mismatch in the Charité group’s assay appears to be the result of using closely related sarbecovirus genomes as reference material in the design process. These sequences contained divergent nucleotides at the mismatched position that swayed the consensus sequence away from the nucleotide present in SARS-CoV-2 [9]. Both of these situations highlight the challenges of designing PCR oligonucleotides for novel pathogens with poorly characterized genomes: assay developers must rely on early, potentially error-prone genome assemblies or supplement their reference sequences with closely related organisms whose genomes may diverge in the target regions selected.
Polymorphisms affecting the China CDC’s assay were extensive and varied, suggesting they were unfortunate consequences of viral evolution. The most common of these variants was present in 14.9 % of genomes in our dataset, but it did not appear until at least one month after the assay was designed and it was not especially prevalent in genomes from China. The clear association of this variant with the emergence of a viral clade highlights another challenge of PCR design for novel pathogens, especially mutable RNA viruses: rapid adaption to new hosts can result in extensive genomic mutations that are difficult to predict and avoid when designing diagnostic assays.
Our analysis cannot determine if these mismatches and polymorphisms would impact clinical performance, but their positions do not obviously suggest PCR failure. The prevalent ‘AAC’ variant in the China CDC’s assay was confined to the 3 least-critical nucleotides at the 5′ end of the primer. Mismatches in the Charité group and Japan NIID assays were both 14 positions from the more-crucial 3’ end of the primers. Indeed, as stated above, the Japan NIID group failed to observe a difference in detection between primers with and without the mismatch [11].
Taken together, these results provide a reassuring assessment of the early SARS-CoV-2 assays, many of which were widely adopted. They also provide instructive lessons about designing PCR assays for the diagnosis of novel viral pathogens. We have highlighted four of these lessons here.
The first lesson is that reference laboratories should maintain a collection of PCR oligonucleotides that are inclusive across higher-level viral taxa. At the beginning of the SARS-CoV-2 outbreak, pan-sarbecovirus oligonucleotides were instrumental for investigating the etiology of the first cases and rapidly developing stop-gap diagnostic assays. These kinds of oligonucleotide collections should focus on higher-level viral taxa that are common sources of novel zoonotic disease and, consequently, pose the most significant pandemic threats. They also allow isolates of accessible, closely related viruses to be used as control and validation material when specimens of the novel etiological pathogen are limited or unavailable.
The second lesson is that public health funders should commit to research characterizing viral diversity in wildlife, livestock, and game species. Crucial laboratory tools that enabled rapid identification of SARS-CoV-2 and prompt design of early diagnostic tests (e.g. pan-sarbecovirus PCR oligonucleotides) existed because of bat coronavirus research arising from the SARS incident in 2003−04. Numerous outbreaks and pandemics in the 20th and 21st centuries originated from the interface between humans, livestock, game, and wildlife. Successful prevention, preparedness, and response to these crises depend on characterizing this viral diversity.
The third lesson is that diagnostic PCR assays for novel pathogens should be multiplexed for redundancy. The SARS-CoV-2 experience reveals that stop-gap assays can be compromised by oligonucleotide design artefacts and unforeseen polymorphisms arising through early viral adaptation and evolution. This risk can be mitigated by including additional targets in the PCR. Assay redundancy can be further enhanced by adding confirmatory targets that are inclusive across higher-level taxa. For instance, the Charité group’s pan-sarbecovirus E gene assay was used for this purpose in half of the assays analyzed in the study.
The fourth lesson is that genomic sequencing of specimens must keep pace with the spread of an outbreak. Polymorphisms accumulate as zoonotic pathogens adapt to new hosts, and the locations where they occur are difficult to predict. Specimens should be sequenced routinely to identify design artefacts, monitor emerging polymorphisms, and promptly update oligonucleotide designs. This is equally true for endemic viruses, where polymorphisms regularly arise through antigenic drift. Laboratories accredited by the College of American Pathologists (CAP) are required to monitor the compatibility of their LDTs with currently circulating strains; routine genomic sequencing of viral isolates and bioinformatic analysis using PCR_strainer can address this requirement.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
We gratefully acknowledge all authors from originating laboratories who obtained specimens and submitting laboratories who generated genomic sequences that were shared via GISAID; a complete listing is provided in the appendix material. We also thank Dr. Inna Sekirov for her comments on the manuscript, and the technologists at the British Columbia Centre for Disease Control Public Health Laboratory for their insight and allowing us to include their assay in our analysis. This study was completed as part of Kevin Kuchinski’s doctoral work, and he thanks the British Columbia Ministry of Agriculture for its financial support.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2020.104581.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.L.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(March (7798)):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . 2020. Coronavirus Disease (COVID-19) Situation Report 111.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200510covid-19-sitrep-111.pdf?sfvrsn=1896976f_2 Accessed on May 1,from. [Google Scholar]
- 3.World Health Organization . 2020. Molecular Assays to Diagnose COVID-19: In-house Developed Molecular Assays.https://www.who.int/docs/default-source/coronaviruse/whoinhouseassays.pdf?sfvrsn=de3a76aa_2&download=true Accessed on May 1, from. [Google Scholar]
- 4.LeBlanc J.J., Gubbay J.B., Li Y., Needle R., Radons Arneson S., Marcino D., Charest H., Desnoyers G., Dust K., Fattouh R., Garceau R., German G., Hatchette T.F., Kozak R.A., Krajden M., Kuschak T., Lang A.L.S., Levett P., Mazzulli T., McDonald R., Mubareka S., Prystajecky N., Rutherford C., Smieja M., Yu Y., Zahariadis G., Zelyas N., Bastien N. COVID-19 pandemic diagnostics investigation team of the canadian public health laboratory network (CPHLN) respiratory virus working group. Real-time PCR-based SARS-CoV-2 detection in Canadian laboratories. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104433. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gans J.D., Wolinsky M. Improved assay-dependent searching of nucleic acid sequence databases. Nucleic Acids Res. 2008;36(July (12)):e74. doi: 10.1093/nar/gkn301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(March 19 (5)):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(October (5)):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 8.Huerta-Cepas J., Serra F., Bork P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016;33(June (6)):1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(January (3)) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66(April 1 (4)):549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., Nakatsu Y., Mori Y., Kageyama T., Matsuyama S., Takeda M. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020:18. doi: 10.7883/yoken.JJID.2020.061. Feb. [DOI] [PubMed] [Google Scholar]
- 12.Rambaut A., Holmes E.C., Hill V., O’Toole A., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. bioRxivorg. 2020 doi: 10.1038/s41564-020-0770-5. https://www.biorxiv.org/content/10.1101/2020.04.17.046086v1 [cited Jun 8, 2020]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.