Abstract
Introduction
The SARS-CoV-2 pandemic of 2020 is a prime example of the omnipresent threat of emerging viruses that can infect humans. A protocol for the identification of novel coronaviruses by viral metagenomic sequencing in diagnostic laboratories may contribute to pandemic preparedness.
Aim
The aim of this study is to validate a metagenomic virus discovery protocol as a tool for coronavirus pandemic preparedness.
Methods
The performance of a viral metagenomic protocol in a clinical setting for the identification of novel coronaviruses was tested using clinical samples containing SARS-CoV-2, SARS-CoV, and MERS-CoV, in combination with databases generated to contain only viruses of before the discovery dates of these coronaviruses, to mimic virus discovery.
Results
Classification of NGS reads using Centrifuge and Genome Detective resulted in assignment of the reads to the closest relatives of the emerging coronaviruses. Low nucleotide and amino acid identity (81% and 84%, respectively, for SARS-CoV-2) in combination with up to 98% genome coverage were indicative for a related, novel coronavirus. Capture probes targeting vertebrate viruses, designed in 2015, enhanced both sequencing depth and coverage of the SARS-CoV-2 genome, the latter increasing from 71% to 98%.
Conclusion
The model used for simulation of virus discovery enabled validation of the metagenomic sequencing protocol. The metagenomic protocol with virus probes designed before the pandemic, can assist the detection and identification of novel coronaviruses directly in clinical samples.
Keywords: SARS-CoV-2, virus discovery, metagenomics, bioinformatics, COVID-19
1. Introduction
The Severe Acute Respiratory Syndrome Coronavirus type 2 (SARS-CoV-2) pandemic of 2020 demonstrates the devastating effect an emerging virus can have. Although previous pandemics such as the Spanish Flu (1918) and Asian Flu (1957) resulted in a multitude of fatal cases, the SARS-CoV-2 pandemic exhibits an unprecedented impact on public health, the economy and society as a whole. In 2002 and 2012 respectively, the Severe Acute Respiratory Syndrome (SARS) [1] and Middle Eastern Respiratory Syndrome (MERS) Coronavirus [2] have emerged as zoonotic infections causing severe respiratory disease, with continued introductions of MERS-CoV remaining a public health threat up to now [3].
Pandemic preparedness comprises strategies and measures to protect human health and lives in anticipation of the worldwide spread of (re)emerging pathogens. Pandemic preparedness plans [4] focus on measures to contain and control the spread of emerging pathogens. Early detection of the pathogen is the mainstay of initiating infection control measures. Global surveillance as a component of the International Health Regulations (IHR) aims at early detection and monitoring of human cases of zoonotic diseases with pandemic potential [5]. Pandemic surveillance plans commonly focus on specific viruses, such as influenza, and depend on targeted detection of these specific viral threats, limiting the detection of unanticipated and novel viruses. The current SARS-CoV-2 pandemic shows the need for unbiased identification of potential pathogens.
Metagenomic Next-Generation Sequencing (mNGS) enables hypothesis-free sequencing of all nucleic acids in a given sample, including genomes of pathogens. All sequences are amplified, followed by classification of sequences based on a reference database. While research applications are more common, mNGS is being introduced in clinical diagnostic laboratories as indicated by recently diagnosed cases of encephalitis [6]. Implementation of mNGS in clinical diagnostics requires validation of metagenomic protocols. Metagenomic protocols and pipelines have been successfully used for detection of known pathogens [6,7,8]. However, detection and identification of novel, previously unknown emerging viruses presents a challenge due to the absence of their genome sequences in reference databases.
In this study, we validated the identification of emerging coronaviruses by a viral metagenomic protocol, using clinical samples with SARS-CoV-2, and samples spiked with cultivated isolates SARS-CoV Frankfurt-1 (SARS-CoV) and MERS-CoV EMC/2012 (MERS-CoV). The validation included analysis of the performance of both an in-house and a commercially available data analysis pipeline, Genome Detective [9]. Identification of coronaviruses was tested using modified databases lacking SARS-CoV-2, SARS-CoV, and MERS-CoV, mimicking the situation at the time of virus discovery. Additionally, the efficacy of detection of novel coronaviruses using capture probes targeting vertebrate viruses [10,11] known before the current pandemic was analyzed using a SARS-CoV-2 clinical sample.
2. Methods
2.1. Sample selection and preparation
Nasopharyngeal swabs were obtained from two patients who tested positive for SARS-CoV-2 by real-time PCR targeting the SARS-CoV-2 E-gene [12] with Cq values of 20 and 30, respectively. These PCRs were performed as part of routine diagnostics at the Clinical Microbiological Laboratory (CML) of the Leiden University Medical Center.
For the SARS-CoV and MERS-CoV analyses, nasopharyngeal material that had tested negative for all respiratory viruses addressed by in-house multiplex PCRs (coronaviruses 229E, HKU1, NL63, OC43, influenza A, B, human metapneumovirus, parainfluenza 1-4, respiratory syncytial virus and rhinovirus) was spiked in with the cultivated isolates SARS-CoV Frankfurt-1 [1,13] and MERS-CoV EMC/2012 [14] with viral load per sample being 1.3 × 105 PFU and 2.4 × 105 PFU and Cq values of 23 and 22, respectively.
2.2. Metagenomic Next-Generation Sequencing (mNGS)
Library preparation and sequencing were performed using a previously validated protocol [15,16]. Briefly, 200 μl of patient samples were spiked with equine arteritis virus (EAV) and phocid herpesvirus-1 (PhHV-1) prior to NA extraction using the Magnapure 96 DNA and Viral NA Small volume extraction kit on the MagnaPure 96 system (Roche, Basel, Switzerland) resulting in 100 μL nucleic acid-containing eluate. Of this eluate, 50 μl per sample was used as input for the library prep, utilizing the NEBNext Ultra II Directional RNA Library prep kit for Illumina (New England Biolabs, Ipswich, MA, USA), dual indexed NEBNext Multiplex Oligos for Illumina (1.5μM), and a protocol optimized for processing RNA and DNA simultaneously in a single tube [15].
Library preps of the samples where processed both with and without enrichment for viruses using sequence capture probes (see below). Subsequent sequence analysis was performed using a NovaSeq6000 sequencing system (Illumina, San Diego, CA, USA) at GenomeScan BV to obtain approximately 10 million 150bp reads per sample.
2.3. Viral capture probe enrichment
Enrichment of viral sequences from the sample library pools was performed using the SeqCap EZ HyperCap kit according to the manufacturer’s instructions (Roche, Basel, Switzerland). This kit uses a vertebrate virus SeqCap EZ probe pool designed to target a set of sequences from vertebrate viruses that were available in 2015 [10], including the following: Coronaviridae NCBI:txid11118), Coronavirinae (NCBI:txid693995), Alphacoronavirus (NCBI:txid693996), Betacoronavirus (NCBI:txid694002), Gammacoronavirus (NCBI:txid694013), and Deltacoronavirus (NCBI:txid1159901). Amplified DNA libraries from two SARS-CoV-2 samples and one negative control, with a combined mass of 1 μg, were pooled in equal amounts in a single enrichment experiment. Some adaptions were made: human Cot DNA and blocking oligos (Integrated DNA Technologies, Coralville, IA, USA) were added to each enrichment pool to prevent nonspecific binding and binding of human DNA to the probes. Subsequently, hybridization to the probe pool was performed for 40 hours. Next, the Hyber Cap Bead kit was used for washing the captured DNA, followed by post capture PCR amplification using the KAPA HiFi HotStart ReadyMix (2×) (Roche, Basel, Switzerland) and Illumina NGS primers (5 μM). The final washing step was performed using AMPure XP beads (Beckman Coulter, Inc., Brea, CA, USA) after which quality and quantity of the enriched libraries were assessed by Qubit analysis (Thermo Fisher, Waltham, MA, USA) and Bioanalyzer (Agilent, Santa Clara, CA, USA).
2.4. Sequence read classification: Centrifuge
After quality pre-processing using an in-house QC pipeline, Biopet version 0.9.0 [17] and removal of human reads after mapping them to human reference genome GRCh38 [18] with Bowtie2 version 2.3.4 [19], the remaining sequencing reads were taxonomically classified using Centrifuge 1.0.2-beta [20] with the databases prepared by taking all 12,302 Refseq viral genomes (as of Juny 16th, 2020) and extracting the GenBank records annotated before the dates of the existence of the MERS-CoV and SARS-CoV index patients in 2012 and 2002, respectively. Reads with multiple matches were assigned to the lowest common ancestor (k = 1). Taxonomic assignments of reads by Centrifuge were visualized with Krona version 2.0 [21].
2.5. In-house virus discovery protocol
Pre-processed short reads were de novo assembled into contigs using SPAdes version 3.10.1 [22]. All contigs were analyzed using the NCBI Basic Local Alignment Search Tool (BLAST 2.8.1) [23] using the BLAST NCBI’s nucleotide (nt) database (accessed April 2018). Only viral hits for contigs with a length of ≥500bp were selected to identify the best shared homology to viruses. A length of 500bp was taken to ensure coverage of the built contigs by at least 3 reads, to rule out any possible contamination. Only hits dated prior to the date of emergence of the viruses were considered to mimic the virus discovery setting for SARS-CoV, MERS-CoV and SARS-CoV-2.
2.6. Genome Detective: commercial classification and discovery tool
After extraction of human reads, FASTQ files generated for SARS-CoV-2 samples (with and without viral enrichment) were uploaded for classification and de novo assembly by the commercial web-based tool Genome Detective v1.120 (www.genomedetective.com, accessed 2020-05-11) [9], using a reference database (generated 2019-09-21). In brief, after removal of low-quality reads and trimming by Trimmomatic [24], candidate viral reads were identified using the protein-based alignment method DIAMOND [25] in combination with the Swissprot UniRef90 protein database followed by de novo assembly using metaSPAdes [26]. Blastx and Blastn [23] were used to search for candidate reference sequences using the NCBI RefSeq virus database (accessed 2019-09-21). Consensus sequences were produced by joining de novo contigs using Advanced Genome Aligner [27].
3. Results
3.1. Classification of SARS-CoV-2, SARS-CoV, and MERS-CoV using databases created before the emergence of these viruses
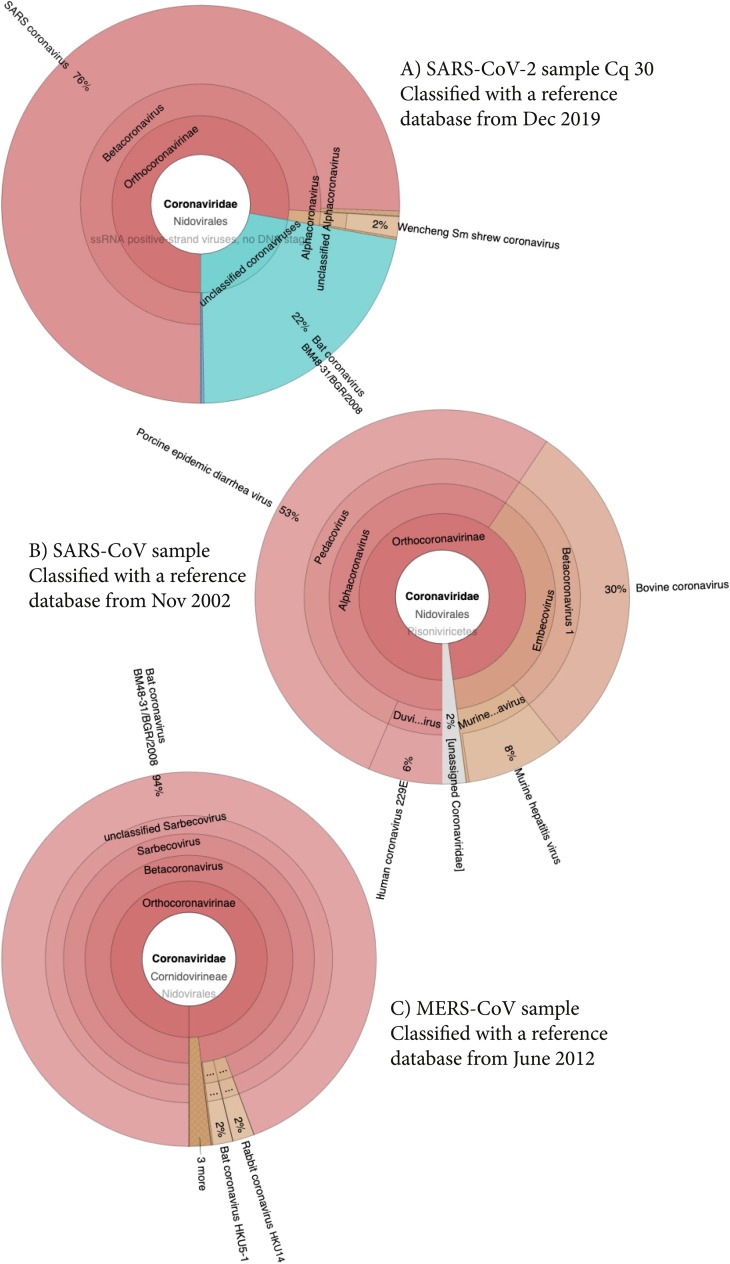
To mimic the classification conditions present in the setting of virus discovery, viral metagenomic reference genome databases created before the emergence of SARS-CoV-2, SARS-CoV and MERS-CoV were used for the classification of sequence reads (December 2019 for the two SARS-CoV-2 positive samples, November 2002 for the SARS-CoV and June 2012 for the MERS-CoV positive samples). Classification results of viral reads are shown in Fig. 1 and Table 1 . Sequence reads obtained for SARS-CoV-2 samples were classified as belonging to SARS coronavirus and Bat coronavirus BM48-31/BGR/2008. Sequence reads of the SARS-CoV sample were classified as belonging to Porcine epidemic diarrhoea virus and bovine coronavirus, and reads of the MERS-CoV sample as Bat coronavirus BM48-31/BGR/2008, belonging to the Betacoronavirus genus (Table 1).
Fig. 1.
Centrifuge classification results of viral reads of SARS-CoV-2, SARS-CoV, and MERS-positive samples, using viral metagenomic databases created before the emergence of these viruses. A) SARS-CoV-2, B) SARS-CoV, C) MERS.
Table 1.
Classification of SARS-CoV-2, SARS-CoV, and MERS sequence reads using reference databases created before their emergence, using metagenomic classifier Centrifuge.
| Sample | Untargeted mNGS, or viral enrichment by capture probes | Total number of non-human reads | Number of reads classified as Coronaviridae (% of total non-human) |
Coronaviridae assignment of >10% classified Coronaviridae reads |
|---|---|---|---|---|
| SARS-CoV-2 Patient A (Cq 20) |
Untargeted | 3,488,842 | 2,166 (0.06) | SARS-CoV Bat coronavirus BM48-31/BGR/2008 |
| Viral capturea | 9,582,942 | 3,518,798 (36.72) | SARS-CoV Bat coronavirus BM48-31/BGR/2008 |
|
| SARS-CoV-2 Patient B (Cq 30) |
Untargeted | 919,930 | 604 (0.07) | SARS-CoV Bat coronavirus BM48-31/BGR/2008 |
| Viral capturea | 9,894,246 | 572,061 (5.78) | SARS-CoV Bat coronavirus BM48-31/BGR/2008 |
|
| SARS-CoV Frankfurt-1 (Cq 23) |
Untargeted | 6,936,399 | 436 (0.006) | Bovine coronavirus Porcine epidemic diarrhea virus |
| MERS-CoV EMC/2012 (Cq 22) |
Untargeted | 8,201,535 | 8,748 (0.1) | Bat coronavirus BM48-31/BGR/2008 |
Enrichment by capture probes targeting vertebrate viruses designed in 2015
3.2. Virus discovery: de novo assembly
Results of de novo assembly of all samples for contigs longer than 500bp are shown in Table 2 . BLASTn was used to search for hits with sequence homology. Only viral hits with the lowest E-value of all matches identified that were submitted before the publication of SARS-CoV-2 genomes were considered. BLASTn search results of the contigs with Coronaviridae hits are listed in Table 2 including the length of the longest contig for each sample. Identity data of the hits with the lowest E-value are listed in Supplementary Table 1. Additional BLAST alignment figures of the longest contigs of both the SARS-CoV and MERS-CoV samples can be found in Supplementary Fig. 1 and 2, respectively.
Table 2.
Classification of SARS-CoV-2, SARS-CoV, and MERS de novo assembled contigs using BLAST.
| Sample | Untargeted mNGS, or viral enrichment by capture probes | Total contigs ≥ 500bp |
Viral contigs ≥ 500bp |
Coronaviridae contig ≥ 500bp | Length of the longest Coronaviridae contig, bp | BLAST alignment length, bp | BLAST identity match, % | Subject taxonomy name | Release year of sequence of the species | Release year of sequence of the subject found |
|---|---|---|---|---|---|---|---|---|---|---|
| SARS-CoV-2 Patient A (Cq 20) |
Untargeted | 8,606 | 15 | 3 | 19,654 | 12,069 | 87.141 | Bat SARS SL CoVZC45 | 2003 | 2018 |
| Viral capturea | 8,232 | 51 | 31 | 5,811 | 5,820 | 90.567 | Bat SARS SL CoVZC45 | 2003 | 2018 | |
| SARS-CoV-2 Patient B (Cq 30) |
Untargeted | 2,815 | 31 | 16 | 2,503 | 2,456 | 91.450 | Bat SARS SL CoVZXC21 | 2003 | 2018 |
| Viral capturea | 2,110 | 39 | 13 | 4,866 | 4,856 | 92.360 | Bat SARS SL CoVZC45 | 2003 | 2018 | |
| SARS-CoV Frankfurt-1 (Cq 23) |
Untargeted | 3,836 | 10 | 1 | 29,692 | 1,236 | 72.411 | Bovine coronavirus isolate 4-17-03 | 2001 | 2018 |
| MERS-CoV EMC/2012 (Cq 22) |
Untargeted | 4,074 | 9 | 1 | 30,097 | 14,856 | 77.248 | Bat coronavirus HKU4-1 | 2006 | 2006 |
Table showing the total number of built contigs with a length > = 500bp, the number of these contigs where the hit with the lowest E-value would be a hit to viruses, the number of contigs where the hit with the lowest E-value would be a hit to Coronaviridae and of this last group the length of the longest contig, the alignment length, identity match, taxonomic name of BLAST result and the release years of sequences belonging to the species and subjects found by BLAST.
Enrichment by capture probes targeting vertebrate viruses designed in 2015
3.3. Virus discovery of SARS-CoV-2 by GenomeDetective
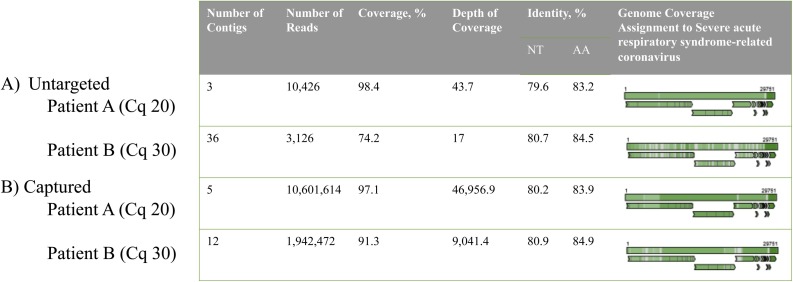
GenomeDetective results of identification of SARS-CoV-2 sequences using a database created before the emergence of SARS-CoV-2 are shown in Fig. 2 . SARS-CoV-2 sequences were identified as SARS-CoV, with nucleotide and amino acid identity of 80-81% and 83-85% respectively in combination with up to 98% genome coverage, being indicative for a novel finding.
Fig. 2.
Discovery performance using metagenomic sequencing (A) without and (B) with enrichment by capture probes targeting vertebrate viruses, designed in 2015. Genome Detective classification of reads, coverage and aligment against the genome of Severe acute respiratory syndrome-related coronavirus are shown.
3.4. Virus discovery using capture probes
The efficacy of a metagenomic sequencing protocol using capture probes targeting vertebrate virus sequences designed before the emergence of SARS-CoV-2, was studied in the context of virus discovery. We analyzed metagenomic data from the two SARS-CoV-2 positive samples prepared both with and without viral enrichment. The total amount of contigs and the number of contigs matching genomes of viruses from Coronaviridae are shown in Table 2 and Fig. 2. For the clinical sample with higher SARS-CoV-2 load (Cq 20), genome coverage was comparable (98% vs. 97% genome coverage), and for the sample with lower load (Cq 30), genome coverage was markedly higher (74% vs. 91% genome coverage) when the metagenomic protocol with viral capture probes was used.
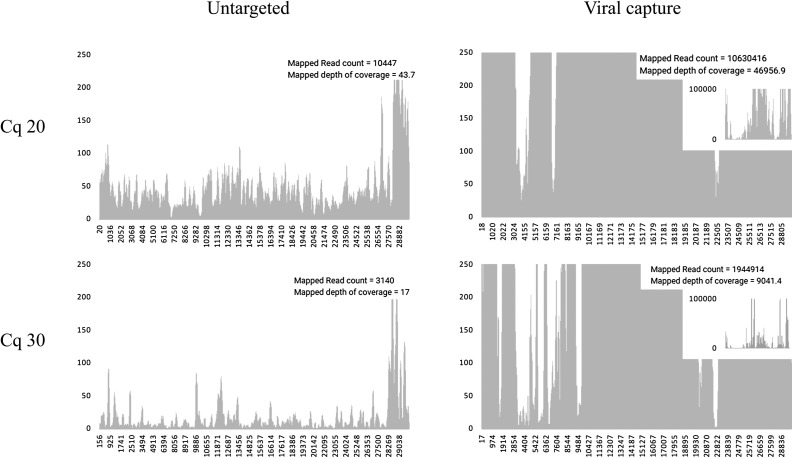
Reads mapping to the SARS-CoV-2 reference genome were used to visualize the difference in using capture probes as depicted in Fig. 3 , where the SARS-CoV-2 genome is almost completely covered. The two largest contigs built by SPAdes that had a hit with the lowest E-value when BLASTed against genomes from Coronaviridae, were 4,866bp and 5,811bp in length for the two SARS-CoV-2 samples enriched using probes.
Fig. 3.
Coverage map of alignment against SARS-CoV-2 reference sequence NC_004718.2, without (left) and with (right) viral capture probes designed in 2015 after metagenomic sequencing of patient samples with respectively Cq 20 (upper graphs) and Cq 30 (lower graphs).
4. Discussion
In this study, we evaluated the performance of a metagenomic sequencing protocol for the identification of emerging viruses using clinical samples in combination with a simulated reference database. High and low loads of SARS-CoV-2, SARS-CoV, and MERS-CoV in clinical samples could be detected as ‘novel’ viruses, using only reference sequences created before these viruses emerged. Sequence reads were assigned to the closest relatives of these viruses available at that time and assembled with heterologous sequences to ‘novel’ consensus genomes. Low identity of these consensus genomes with genomes of closely related ones indicated a novel virus. Additionally, probes targeting sequences of vertebrate viruses, available prior to the coronavirus pandemic of 2020, succeeded in the capture of nearly the full genome of SARS-CoV-2. It must be noted that the validation was performed using emerging viruses with nucleotide identity of over 76% to their closest known relatives and conclusions cannot be extended to novel viruses which are less closely related. Nucleotide (and amino acid) identities reported in literature with regard to novel human pathogenic viruses vary, for example 50% for older viruses like SARS-CoV [1], 80% for MERS-CoV [14], 88% for parts of the Human Metapneumovirus [28] and up to 97.2% for parts of SARS-CoV-2 [29].
Several reports have shown an increase of 100-10,000 fold in sensitivity for detection of known viruses when using capture probes [10], [30] and here we report the potential of using capture probes in the detection of novel viruses. Sequence variation was addressed in the probe design by retaining mutant or variant sequences if sequences diverged by more than 90% [10]. Lipkin and colleagues describe the capture of conserved regions of a rodent hepacivirus isolate with 75% identity using VirSeqCap VERT, and even 40% for detection rather than whole genome sequencing is suggested [10]. The capture probes used in this study targeted sequences of several isolates of alpha-,beta-,gamma-,and deltacoronaviruses. In this study the whole genome of SARS-CoV-2, with 76-100% overall nucleotide identity to the probe targets, was detected using these probes.
Metagenomic sequencing is increasingly being used in diagnostic laboratories as a hypothesis-free approach for suspected infectious diseases in undiagnosed cases. Metagenomic sequencing in diagnostic laboratories has resulted in the detection of pathogens present in the reference database but either not tested for by routine methods due to rare or unknown associations with a specific disease, or for which routine testing failed (e.g., due to primer mismatches). Additionally, mNGS enables the detection of novel pathogens not (yet) present in the databases. Common bioinformatic classifiers are usually not designed for discovery purposes, so additional algorithms including a separate validation to assess the performance in a discovery setting are needed. Reports on specific bioinformatic discovery tools typically describe the algorithm and an in silico analysis and here we present validation studies on the performance of virus discovery tools using clinical samples.
Implementation of virus discovery protocols in diagnostic laboratories may contribute to increased vigilance for emerging viruses and therefore aids in surveillance and pandemic preparedness.
CRediT authorship contribution statement
Ellen C. Carbo: Visualization, Writing - original draft, Formal analysis, Investigation. Igor A. Sidorov: Writing - review & editing, Formal analysis. Jessika C. Zevenhoven-Dobbe: Investigation. Eric J. Snijder: Writing - review & editing. Eric C. Claas: Writing - review & editing. Jeroen F.J. Laros: Methodology, Writing - review & editing. Aloys C.M. Kroes: Resources, Writing - review & editing. Jutte J.C. de Vries: Conceptualization, Writing - original draft, Writing - review & editing, Visualization, Investigation.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
We would like to thank Joost van Harinxma thoe Slooten and Alhena Reyes for the library preparations and viral probe enrichments. Additionally, we would like to thank Lopje Höcker, Margriet Kraakman and Tom Vreeswijk for all their technical assistance in the lab, and Leon Mei and Michel Villerius for their bioinformatic support.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2020.104594.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Drosten C. ‘Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome’. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. May. [DOI] [PubMed] [Google Scholar]
- 2.Ksiazek T.G. ‘A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome’. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. May. [DOI] [PubMed] [Google Scholar]
- 3.http://www.who.int/csr/don/02-jul-2020-mers-saudi-arabia/en/ (Accessed Jul. 04, 2020).
- 4.http://www.euro.who.int/en/health-topics/communicable-diseases/influenza/pandemic-influenza/pandemic-preparedness/national-preparedness-plans (Accessed Jun. 30, 2020).
- 5.‘http://www.who.int/csr/disease/swineflu/global_pandemic_influenza_surveilance_apr09.pdf’. Accessed: Jun. 30, 2020.
- 6.Miller S. ‘Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid’. Genome Res. 2019;29(5):831–842. doi: 10.1101/gr.238170.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y. ‘VIP: an integrated pipeline for metagenomics of virus identification and discovery’. Sci. Rep. 2016;6(1):23774. doi: 10.1038/srep23774. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller R.R., Montoya V., Gardy J.L., Patrick D.M., Tang P. Metagenomics for pathogen detection in public health’. Genome Med. 2013;5(9):81. doi: 10.1186/gm485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vilsker M. ‘Genome Detective: an automated system for virus identification from high-throughput sequencing data’. Bioinformatics. 2019;35(5):871–873. doi: 10.1093/bioinformatics/bty695. Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briese T. ‘Virome Capture Sequencing Enables Sensitive Viral Diagnosis and Comprehensive Virome Analysis’. mBio. 2015;6(5):e01491–15. doi: 10.1128/mBio.01491-15. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wylie T.N., Wylie K.M., Herter B.N., Storch G.A. Enhanced virome sequencing using targeted sequence capture. Genome Res. 2015;25(12):1910–1920. doi: 10.1101/gr.191049.115. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corman V.M. ‘Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR’. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiel V. ‘Mechanisms and enzymes involved in SARS coronavirus genome expression’. J. Gen. Virol. 2003;84(9):2305–2315. doi: 10.1099/vir.0.19424-0. Sep. [DOI] [PubMed] [Google Scholar]
- 14.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. ‘Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia’. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. Nov. [DOI] [PubMed] [Google Scholar]
- 15.van Boheemen S. ‘Retrospective Validation of a Metagenomic Sequencing Protocol for Combined Detection of RNA and DNA Viruses Using Respiratory Samples from Pediatric Patients’. J. Mol. Diagn. 2020;22(2):196–207. doi: 10.1016/j.jmoldx.2019.10.007. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Rijn A.L. ‘The respiratory virome and exacerbations in patients with chronic obstructive pulmonary disease’. PLOS ONE. 2019;14(10):e0223952. doi: 10.1371/journal.pone.0223952. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.http://biopet-docs.readthedocs.io/en/stable/ (Accessed Jul. 03, 2020).
- 18.https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.26/ (Accessed Jul. 10, 2020).
- 19.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D., Song L., Breitwieser F.P., Salzberg S.L. ‘Centrifuge: rapid and sensitive classification of metagenomic sequences’. Genome Res. 2016;26(12):1721–1729. doi: 10.1101/gr.210641.116. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ondov B.D., Bergman N.H., Phillippy A.M. ‘Interactive metagenomic visualization in a Web browser’. BMC Bioinformatics. 2011;12(1):385. doi: 10.1186/1471-2105-12-385. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bankevich A. ‘SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing’. J. Comput. Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. ‘Basic local alignment search tool’. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. Oct. [DOI] [PubMed] [Google Scholar]
- 24.Bolger A.M., Lohse M., Usadel B. ‘Trimmomatic: a flexible trimmer for Illumina sequence data’. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchfink B., Xie C., Huson D.H. ‘Fast and sensitive protein alignment using DIAMOND’. Nat. Methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176. Jan. [DOI] [PubMed] [Google Scholar]
- 26.Nurk S., Meleshko D., Korobeynikov A., Pevzner P.A. ‘metaSPAdes: a new versatile metagenomic assembler’. Genome Res. 2017;27(5):824–834. doi: 10.1101/gr.213959.116. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deforche K. ‘An alignment method for nucleic acid sequences against annotated genomes’. Bioinformatics. 2017 doi: 10.1101/200394. preprint, Oct. [DOI] [Google Scholar]
- 28.van den Hoogen B.G., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. ‘Analysis of the Genomic Sequence of a Human Metapneumovirus’. Virology. 2002;295(1):119–132. doi: 10.1006/viro.2001.1355. Mar. [DOI] [PubMed] [Google Scholar]
- 29.Zhou H. ‘A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein’. Curr. Biol. 2020;30(11):2196–2203. doi: 10.1016/j.cub.2020.05.023. e3, Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carbo E.C. ‘Improved diagnosis of viral encephalitis in adult and pediatric hematological patients using viral metagenomics’. J. Clin. Virol. 2020:104566. doi: 10.1016/j.jcv.2020.104566. Jul. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.