Abstract
Rapid and accurate diagnosis is crucial for successful outbreak containment. During the current coronavirus disease 2019 (COVID-19) public health emergency, the gold standard for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection diagnosis is the detection of viral RNA. Additional diagnostic methods õenabling the detection of current or past SARS-CoV-2 infection would be highly beneficial. We assessed 2 immunochromatographic lateral flow assays (LFA-1, LFA-2) and 2 enzyme-linked immunosorbent assay kits (IgA/IgG ELISA-1, IgM/IgG ELISA-2) using 325 samples: serum samples from polymerase chain reaction–confirmed COVID-19 hospitalized patients (n = 55) and healthcare workers (n = 143) and 127 samples from negative controls. Diagnostic performances were assessed according to days after symptom onset (dso) and the antigenic format used by manufacturers. Clinical sensitivities varied greatly among the assays, showing poor mutual agreement. After 15 dso, ELISA-1 (Euroimmun) and LFA-1 (Biosynex) combining IgM and IgG detection showed the best performances. A thorough selection of serological assays for the detection of ongoing or past infections is advisable.
Keywords: COVID-19, SARS-CoV-2, Serological diagnosis, Humoral response
Highlights
-
•
We assessed 2 immunochromatographic lateral flow assays (LFA-1, LFA-2) and two enzyme-linked immunosorbent assay kits (IgA/IgG ELISA-1, IgM/IgG ELISA-2) using 325 well-characterized samples.
-
•
The clinical sensitivity varied greatly according to days after symptom onset, the antigenic format, and the disease severity.
-
•
The assays showed poor mutual agreement.
-
•
A thorough selection of serological assays for the detection of ongoing or past infections is advisable.
1. Introduction
A novel coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causing coronavirus disease 2019 (COVID-19) has emerged as a major healthcare threat (World Health Organization (WHO), n.d.. Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases). At the beginning of the pandemic, the main healthcare objective was to stop the spread of the virus. A key aspect to achieve this goal was to ensure early and accurate infection diagnosis and appropriate quarantine for infected people. The gold standard for identifying SARS-CoV-2 infection relies on the detection of viral RNA by reverse transcription polymerase chain reaction (RT-PCR)–based techniques. However, the large-scale routine implementation of this approach has been hampered by its time-consuming nature (most often 4–6 h) and shortages of materials. Moreover, the presence of sufficient amounts of the viral genome at the site of sample collection is a prerequisite to allow genome detection. Missing the time window of active viral replication or low-quality sampling can lead to false-negative results, which would allow infected patients to spread the virus to their relatives and working environment. In such conditions, additional diagnostic methods would be highly beneficial to ensure timely diagnosis of all infected and recovered patients. Combining RT-PCR with the screening of the onset and strength of the humoral response against SARS-CoV-2 could enhance diagnostic sensitivity and accuracy. There are now several studies describing the kinetics of anti–SARS-CoV-2 IgM and IgG detection using laboratory enzyme-linked immunosorbent assay (ELISA) tests, most reporting that IgM is detectable as early as 5–14 days after the first clinical symptoms (Guo et al., 2020; Liu et al., 2020; Xu et al., 2020; Yong et al., 2020; Zhang et al., 2020; Zhao et al., 2020a). At this stage of the pandemic, many countries are now questioning how to prepare and manage the easing of lockdown. Serological tools have an important place in establishing such strategies. Validated serological assays are crucial for patient contact tracing and epidemiological studies. Several formats of serological methods are beginning to be marketed, i.e., lateral flow assays (LFAs) and ELISAs detecting IgA, IgM, and/or IgG or total antibodies. Data about the analytical and clinical performances of these devices are still lacking, as well as their indication in the diagnosis of SARS-CoV-2 infection. In this context, we evaluated the diagnostic performances of 2 LFAs and 2 commercial ELISA kits detecting IgM, IgA, and IgG based on well-characterized panels of serum samples from PCR-confirmed COVID-19 patients and healthcare workers and from SARS-CoV-2–negative patients. Diagnostic performances of each assay were assessed according to days after symptom onset (dso) and the antigenic format used by manufacturers. This evaluation led us to propose a decisional diagnostic algorithm based on serology, which may be applicable in future seroprevalence studies.
2. Materials and methods
2.1. Patients and serum samples/study design
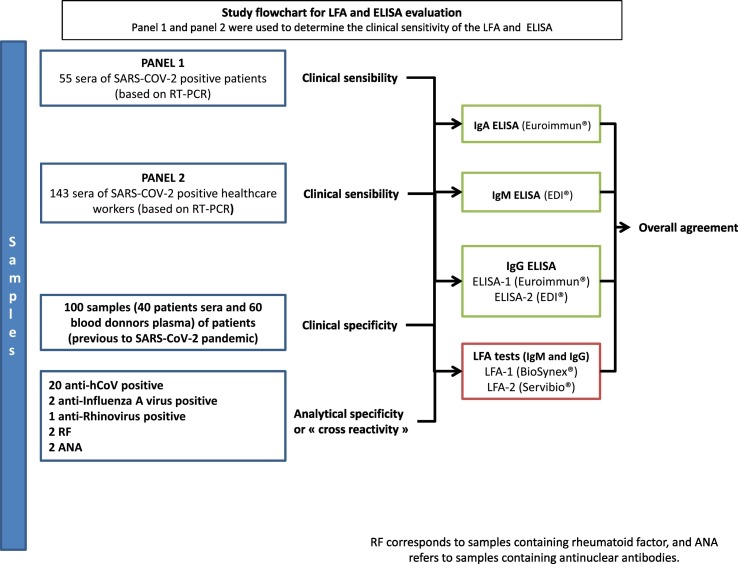
The study design is summarized in Fig. 1 . A total of 325 samples were used, including 55 serum samples from hospitalized patients (panel 1),; 143 serum samples from healthcare workers (panel 2) diagnosed with COVID-19 at Strasbourg University Hospital (Strasbourg, France), recruited in April 2020; and 67 serum and 60 plasma samples from negative controls. All sera of panels 1 and 2 were tested with 2 LFAs and 2 ELISAs (Fig. 1). Patient characteristics were collected for each panel (Table 1 ). Laboratory detection of SARS-CoV-2 was performed by RT-PCR testing of nasopharyngeal swab specimens according to current guidelines (Institut Pasteur, Paris, France; WHO technical guidance). This assay targets 2 regions of the viral RNA-dependent RNA polymerase (RdRp) gene, with a threshold limit of detection of 10 copies per reaction. Serum samples were collected at a median of 7 dso (range, 0–31 dso) for panel 1 and 24 dso (range, 15–39 dso) for panel 2. Serum samples from 40 patients and plasma samples from 60 healthy blood donors collected before the COVID-19 pandemic onset (from March to November 2019) were selected as negative controls to determine clinical specificity. Another 27 serum samples collected before the COVID-19 pandemic onset were used to study cross-reactivity, including 20 samples from patients infected with 4 other human coronaviruses 2 to 3 months before sampling (HCoV-229E, HCoV-HKU1, HCoV-NL63, and HCoV-OC43), 2 from patients previously infected with influenza A virus, 1 from a patient previously infected with human rhinovirus, 2 containing rheumatoid factor, and 2 positive for antinuclear antibodies. All these negative controls were tested with all evaluated assays. Ethical approval was granted by the local institutional review board (CE-2020-34). All patients provided written informed consent.
Fig. 1.
Study flowchart for LFA and ELISA evaluation.
Panel 1 and panel 2 were used to determine the clinical sensitivity of the LFA and ELISA.
RF corresponds to samples containing rheumatoid factor, and ANA refers to samples containing antinuclear antibodies.
Table 1.
Patient characteristics.
| COVID-19 patients (panel 1) | COVID-19 healthcare workers (panel 2) | Total | |
|---|---|---|---|
| Number of patients | 55 | 143 | 198 |
| Median age (years) [range] |
68 [34–93] |
32 [21–62] |
43 [21–93] |
| Sex (female/male) | 17/38 | 96/47 | 113/85 |
| Median dso at RT-PCR analysis [range] |
3 [0–13] |
2 [0–11] |
2 [0–13] |
| Median dso at serum collection [range] |
8 [0–28] |
24 [15–39] |
22 [0–39] |
| Hospitalized in ICU | 23 | NA | NA |
| Hospitalized without ICU admission | 33 | NA | NA |
NA = not applicable.
Samples analyzed within 7 days were stored at 4 °C. The other samples were stored at −20 °C with only a single freeze–thaw cycle.
2.2. Immunochromatographic LFAs
We evaluated 2 commercial CE-marked LFAs: i) LFA-1: Biosynex COVID-19 BSS (Biosynex, Switzerland, Fribourg) and ii) LFA-2: COVID-19 Sign IgM/IgG (Servibio/VEDALAB, France, Alençon). Technical characteristics of the assays are summarized in the Supplementary data (Table S1). Both were tested according to the manufacturer's instructions. Briefly, for each test, 10 μL of serum sample and 2 drops of buffer were added. The strip was placed flat at room temperature for 10 min, and then the results were scored according to the sample and control line intensity only for the tests validated by the appearance of the control line. Interpretation was performed by 2 independent readers using the standardized intensity scoring system that was established previously. The absence of the sample line was scored as 0 (negative), whereas a visible sample line was classified as positive, and the results were scored as follows: a weak line as 1, a clear visible line with an intensity lower than that of the control line as 2, a clear visible line with an intensity similar to that of the control line as 3, and a clear visible line with an intensity higher than that of the control line as 4.
2.3. ELISA (IgA, IgM, and IgG)
The following ELISA diagnostic kits were used for the detection of anti–SARS-CoV-2 IgA, IgM, and IgG antibodies according to the manufacturer's instructions: 1) ELISA-1: ELISA anti–SARS-CoV-2 IgA and IgG (Euroimmun, Lübeck, Germany) and 2) ELISA-2: EDI™ novel coronavirus COVID-19 IgM and IgG (Epitope Diagnostics, San Diego, CA). Technical characteristics of the assays are summarized in the Supplementary data (Table S1). The assessed ELISA kits used as their antigenic source full-length recombinant nucleocapsid protein and the recombinant S1 domain of the spike protein for IgA and IgG in ELISA-1 and for IgM and IgG in ELISA-2, respectively. In brief, the optical density (OD) of the samples and calibrators was detected at 450 nm. Cutoffs for IgG detection were calculated according to the manufacturer's instructions. ELISA-1 results were expressed as a ratio, and a ratio greater than 1.1 was considered positive. For ELISA-2, values greater than the cutoff were considered positive. To allow correlation of the results, the results for IgG ELISA-2 were also expressed as a ratio (OD sample/OD cutoff).
2.4. Statistical analysis
Clinical sensitivity was determined on samples from SARS-CoV-2 RT-PCR–positive patients and healthcare workers (inclusion criterion). Percentages of IgA, IgM, and IgG detection were calculated and compared among all evaluated serological devices according to the dso category in panel 1 (i.e., 0 to 7, 8 to 14, and more than 14 days) an in panel 2 (i.e., 15 to 21, 22 to 28, and more than 28 days). For both LFAs and ELISAs, overall positivity was also evaluated based on positive results for the IgA, IgM, or the IgG test line or ratio (S/Co). Clinical specificity was calculated using the serum samples from 40 patients and the plasma samples from 60 healthy blood donors collected before the COVID-19 pandemic onset (from March to November 2019). Agreement among kits was determined for IgM and IgG parameters using Fleiss' kappa (overall agreement) and Cohen's kappa (agreement between pairs). A kappa value >0.80 was deemed satisfactory. Moreover, the diagnostic performances were estimated by comparing the combined IgM and IgG results according to SARS-CoV-2 infection status for each sample. Performance was considered satisfactory if the diagnostic accuracy exceeded 90%. Analyses were conducted using GraphPad (San Diego, CA) Prism 6 software.
3. Results
3.1. Study population
The general characteristics of the COVID-19 study participants are presented in Table 1. We collected serum samples from a total of 198 patients, including 85 men. Ages ranged from 21 to 93 years, with a median of 43. Serum samples were divided into several panels for evaluation, i.e., panel 1 corresponds to COVID-19 hospitalized patients, and panel 2 corresponds to COVID-19 healthcare workers. Among COVID-19 patients, the median age was 68 years (range: 34–93 years), and the median age was 32 (range: 21–62 years) among COVID-19 healthcare workers.
3.2. LFA and ELISA clinical performances
3.2.1. Clinical sensitivity
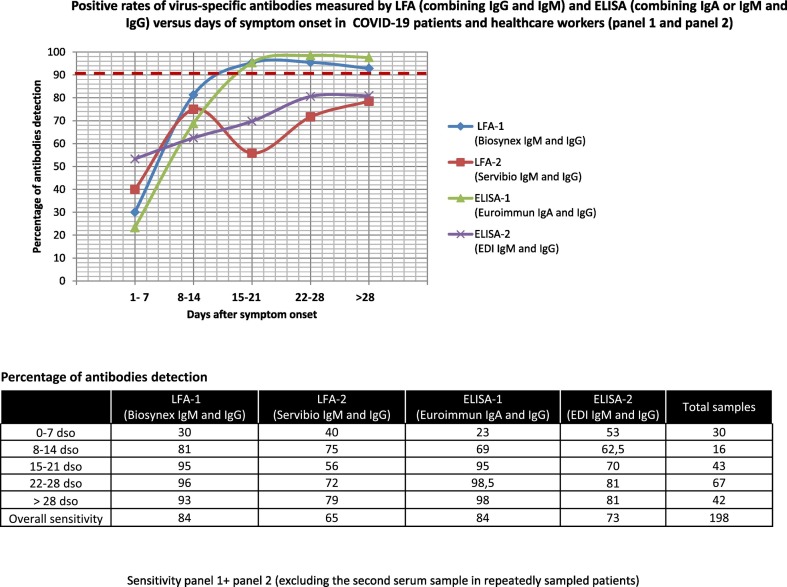
The overall clinical sensitivity evaluated on 198 serum samples (combining panel 1 and panel 2 samples) from COVID-19 patients varied greatly between the 2 LFAs tested, especially for IgM, which was found in 76% and 31% of samples, respectively. A higher percentage of IgM detection (93%) was observed between 15 and 21 dso for LFA-1, whereas the optimal IgM detection rate for LFA-2 was obtained between 8 and 14 dso, with IgM detected in only 56% of cases (Fig. S1). The sensitivity was similar for IgG between both devices, with 62% of samples detected positive using LFA-1 and 63% detected positive using LFA-2. The maximal detection rate for IgG was observed after 28 dso, with 86% and 79% for LFA-1 and LFA-2, respectively (Fig. S2). Combining IgM and IgG detection led to an overall sensitivity of 84% using LFA-1 but only 65% using LFA-2 (Fig. 2 ). The global clinical sensitivity for IgA (ELISA-1) and for IgM (ELISA-2) was evaluated to be 80% and 37%, respectively (Fig. S1). The global clinical sensitivity estimated for IgG detection with ELISA-1 and ELISA-2 on the same 198 serum samples was 75% and 71%, respectively (Fig. S2). Combining IgA or IgM and IgG detection led to an overall sensitivity of 84% (ELISA-1) and 73% (ELISA-2) (Fig. 2). Only LFA-1 and ELISA-1 kits exceeded 90% of antibodies detection after 15 dso. Between 22 and 28 dso, antibodies detection rates peaked at 96% for LFA-1 and 98.5% for ELISA-1.
Fig. 2.
Positive rates of virus-specific antibodies measured by LFA (combining IgG and IgM) and ELISA (combining IgA or IgM and IgG) versus days of symptom onset in COVID-19 patients and healthcare workers (panel 1 and panel 2).
Sensitivity panel 1+ panel 2 (excluding the second serum sample in repeatedly sampled patients).
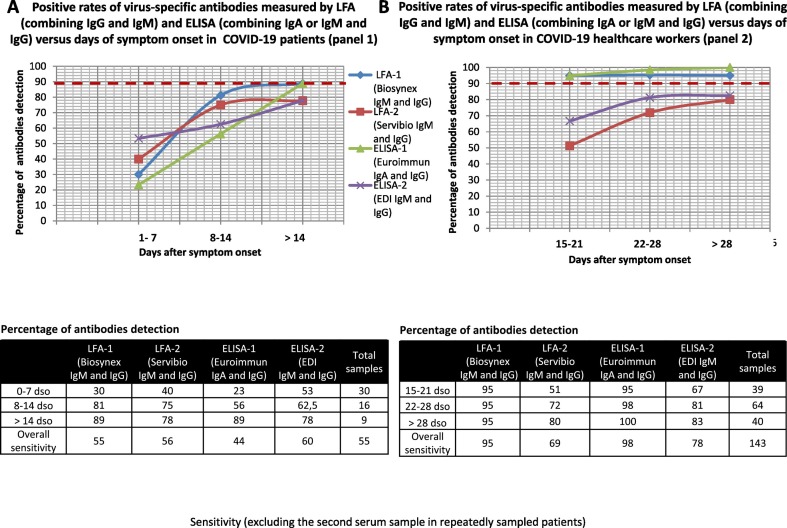
Clinical sensitivity was also evaluated in each panel separately given that specimens were sampled earlier after symptom onset in panel 1 than in panel 2. In panel 1, IgM was detected in 45% and 40% of sera using LFA-1 and LFA-2, respectively (Fig. S3). The optimum IgM detection rate was observed after 14 dso with 67% of IgM detection rate for both devices. Overall sensitivity for IgA (ELISA-1) and IgM (ELISA-2) was 44% and 33%, respectively. However after 14 dso, ELISA-1 showed an IgA detection rate of 89%. In this panel, the overall sensitivity for IgG detection ranged from 33% (LFA-1 and ELISA-1) to 56% (ELISA-2) (Fig. S4). The optimum rate of IgG detection was observed for all assays after 14 dso, with rates ranging from 67% (ELISA-2) to 89% (LFA-1). When combining IgA or IgM and IgG results using LFA and ELISA devices in panel 1, the sensitivity ranged from 44% for ELISA-1 to 60% for ELISA-2 (Fig. 3A). The optimum rate of IgG detection was observed for all assays after 14 dso, with rates ranging from 78% (LFA-2 and ELISA-2) to 89% (LFA-1 and ELISA-1). However, only 9 infected patients were sampled after 14 dso in this panel.
Fig. 3.
(A) Positive rates of virus-specific antibodies measured by LFA (combining IgG and IgM) and ELISA (combining IgA or IgM and IgG) versus days of symptom onset in COVID-19 patients (panel 1).
(B) Positive rates of virus-specific antibodies measured by LFA (combining IgG and IgM) and ELISA (combining IgA or IgM and IgG) versus days of symptom onset in COVID-19 healthcare workers (panel 2).
Sensitivity (excluding the second serum sample in repeatedly sampled patients).
In panel 2, the sensitivity for IgM detection was 87% and 29% for LFA-1 and LFA-2, and 38% for ELISA-2, respectively. The sensitivity for IgA detection was 94% for ELISA-1. LFA-1 was more efficient at detecting IgM from 15 to 21 dso in 92% of the cases, whereas the highest percentage of IgM detection for LFA-2 was measured 28 dso, with only 35% of the cases detected (Fig. S5). The highest IgM detection rate observed for ELISA-2 only reached 41% between 15 and 21 dso. ELISA-1 detected IgA in more than 92% of cases from 15 dso, with the highest rate (97.5%) observed after 28 dso. The percentage of IgG detection ranged from 64% for ELISA-2 to 87% for ELISA-1 (Fig. S6). The optimum rate of IgG detection was observed for all assays after 28 dso [i.e., 68% (ELISA-2), 80% (LFA-2), 88% (LFA-1), and 100% (ELISA-1)]. Combining IgA or IgM and IgG detection in this panel increased the overall sensitivity to 78% for ELISA-2, 95% for LFA-1, and 98% for ELISA-1(Fig. 3B).
3.2.2. Clinical specificity and cross-reactivity between SARS-CoV-2 and other human coronaviruses
IgM clinical specificity was estimated to be 78% (LFA-2), 98% (ELISA-2), and 99% (LFA-1). ELISA-1 showed a specificity of 91% for IgA. IgG specificity was 99% for LFA-1 and ELISA-1 and 96% for ELISA-2, whereas it reached only 83% for LFA-2, corresponding to 17/100 false-positive results with a weak intensity score of 1 to 2 (Table S2).
Analytical specificity reached 89% for IgM and 100% for IgG for both LFAs. LFA-1 cross-reacted with the 2 serum samples containing rheumatoid factor (IgM band intensity scored from 1 to 3). Both LFAs cross-reacted with seasonal human coronaviruses (HCoV-HKU1/NL63, 229E, and OC43) with IgM band intensities scored from 1 to 2 (Table S3). The analytical specificity was 96% for both IgG ELISA devices and reached 93% for IgA ELISA-1 (Euroimmun) and 100% for IgM ELISA-2 (EDI). Both IgG ELISAs cross-reacted with a different seasonal human coronavirus [HCoV-HKU1 for ELISA-2 (EDI) and HCoV-NL63 for ELISA-1 (Euroimmun)].
3.3. Relative performances of serological tools for SARS-CoV-2 (panels 1 and 2)
The relative performance of evaluated assays was assessed on both panels 1 and 2. The overall agreement among the 4 assays was 77.19% [Fleiss' kappa: 0.37; 95% confidence interval (CI): 0.27–0.47]. When comparing the 2 LFAs, the kappa agreement statistic was 0.421 (95% CI: 0.291–0.551) for IgG, 0.085 (95% CI: 0.000–0.173) for IgM, and 0.250 (95% CI: 0.118–0.382) combining IgM and IgG results. Between the 2 ELISAs, the kappa value reached 0.349 (95% CI: 0.205–0.492) for IgG and 0.338 (95% CI: 0.190–0.486) combining IgM or IgA and IgG results. High variability in signal intensities was observed among the tested assays (Fig. S7A). Bland–Altman analysis of the IgG ratio measured by ELISA-1 and ELISA-2 defined a 95% limit of agreement of 4.93 (S/CO), showing a good correlation between the 2 IgG ELISAs with ratios of at least 2 S/CO (Fig. S7B).
3.4. Time to IgM and IgA antibody onset
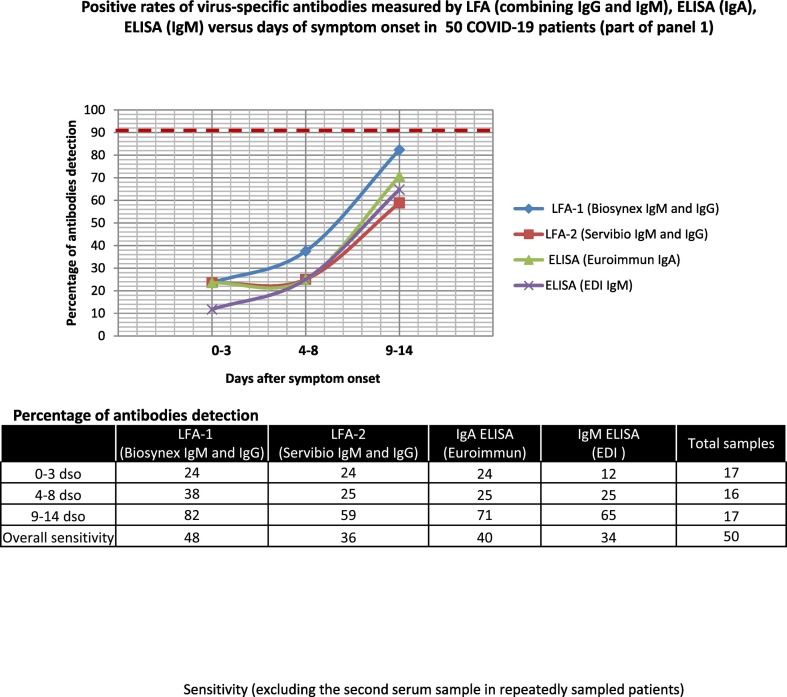
Fifty early serum samples (part of panel 1) of COVID-19 patients were tested with ELISA IgA (Euroimmun) and ELISA-IgM (EDI) assays as well as with both LFA devices. The IgM detection rate ranged from 34% (ELISA-IgM EDI) to 48% (LFA-1), whereas IgA was detected in 40% of samples. The optimum rate of detection for IgM and IgA was observed between 9 and 14 dso (82% for LFA-1 IgM and 71% for IgA ELISA) (Fig. 4 ). We further analyzed the delay of antibody onset in this panel according to the hospitalization unit. When considering samples positive in at least 2 of the 4 assays, we observed a trend towards an earlier detection of antibodies in patients admitted to the intensive care unit (ICU) than in those with milder disease, but the specimen numbers per time interval were low (Fig. S8).
Fig. 4.
Positive rates of virus-specific antibodies measured by LFA (combining IgG and IgM), ELISA (IgA), and ELISA (IgM) versus days of symptom onset in 50 COVID-19 patients (part of panel 1).
Sensitivity (excluding the second serum sample in repeatedly sampled patients).
4. Discussion
In our study, we evaluated test performance for 2 LFAs [i.e., Biosynex (LFA-1) and Servibio (LFA-2)] and 2 ELISA kits (i.e., ELISA-1 Euroimmun IgA/IgG and ELISA-2 EDI IgM/ IgG). We first considered the comparison between tests for the entire cohort and then in each panel separately.
We found a good clinical specificity of 99% for LFA-1 (IgM and IgG) and ELISA-1 (IgG). Except for ELISA-1 IgA and for the IgM test line on both LFA devices, other assays did not cross-react or they poorly cross-reacted. Clinical sensitivity was first calculated on combined panels 1 and 2 according to days after symptom onset. Considering the 198 serum samples, the majority of patients displayed anti–SARS-CoV-2 antibodies only 15 days after symptom onset. For IgM, IgA, and IgG detection, none of the assays we evaluated reached a global sensitivity of 90% even when combining IgA, IgM, and IgG results. The best performances for IgM detection were obtained for LFA-1 with a detection rate of 76%. However between 15 and 21 dso, LFA-1 showed a clinical sensitivity of 95%. The overall clinical sensitivity for IgA detection was evaluated to be 80% for ELISA-1, with the best detection rate exceeding 95% after 15 dso. For IgG detection, only ELISA-1 showed values over 95% from 22 dso. Combining IgA or IgM and IgG results modestly improved sensitivity values for LFA-1 and ELISA-1, and detection rates of more 95% were only observed from 15 dso. The assays we tested showed variable sensitivities and poor mutual agreement (Fig. S7A). However, we found concordant values of overall sensitivity for LFA-1 and ELISA-1 in the entire cohort (84%) (Fig. 2), as well as in panel 2 (95% for LFA-1 and 98% for ELISA-1) (Fig. 3B). The observed differences in terms of sensitivity may reflect the material used as an antigenic source for each assay. Among the 4 coronavirus structural proteins, the spike (S) and nucleocapsid (N) proteins are the main immunogens (Meyer et al., 2014). Specifically, antibodies directed against the viral S protein are expected to appear earlier than those directed against the N protein (Liu et al., 2020; Okba et al., 2020). ELISA-1 (Euroimmun) and LFA-1 (Biosynex) use the recombinant spike protein S1 domain and the receptor binding domain as antigenic sources, respectively, whereas ELISA-2 (EDI) and ELISA-2 (Servibio) are based on a recombinant complete N protein. However, the overall clinical sensitivity calculated in panel 1, corresponding to hospitalized COVID-19 patients, is more discrepant between LFA-1 (55%) and ELISA-1 (44%). In fact, Burbelo et al. (2020) found in 65 patients that antibodies directed against the N protein of SARS-CoV-2 were more sensitive than S protein antibodies for detecting early infection. Sun et al. (2020) investigated IgM and IgG responses against SARS-CoV-2 nucleocapsid (N) and spike (S) protein after symptom onset in the ICU and non-ICU patients. A series of blood samples were collected along the disease course for 11 ICU patients and 27 non-ICU patients for longitudinal analysis. In non-ICU patients, specific IgM and IgG increased after symptom onset, with IgM reaching a peak in the second week in some patients, while IgG continued to increase in the third week. S-IgG was significantly higher in non-ICU patients in the third week, whereas N-IgG was significantly higher in ICU patients. Therefore, another major point explaining the variable results is the choice of the population tested.
Since their development and availability, serological tools have been envisaged to meet 2 different objectives. The first objective was to obtain a faster diagnosis, improve the detection of acute infection by detecting false-negative patients to decrease workloads to central laboratories, and accelerate clinical decision making (Xu et al., 2020; Zhang et al., 2020). We evaluated clinical sensitivity in panel 1, including serum samples from hospitalized patients with COVID-19. The overall sensitivity for all the assays was lower than that calculated on the entire cohort. Only LFA-1 and ELISA-1 reached 89% detection after 14 dso when combining IgA or IgM and IgG detection. For the same time point, the other assays (LFA-2 and ELISA-2) showed a suboptimal sensitivity of 78%, which moderates the interest in their use in the triage of patients with suspected COVID-19. Moreover, because of possible delays in seroconversion, we suggest that rapid serology tests such as LFA cannot replace RT-PCR but should instead be considered complementary tools to enhance access to the screening of symptomatic and asymptomatic patients at the population level.
We further investigated IgM and IgA detection in 50 sera from infected patients sampled early after symptom onset (part of panel 1). If some patients developed anti–SARS-CoV-2 antibodies from 1 to 3 dso, most had detectable IgM (65% with ELISA-2) and IgA (71% with ELISA-1) only between 9 and 14 days (Fig. 4). In addition, symptom severity may affect the rate of seropositivity. A delayed or absent humoral response against SARS-CoV-2 has been reported in some patients (Zhao et al., 2020b) and may result in negative serology results (Yongchen et al., 2020; Zhang et al., 2020). Future studies are required to shed further light on the underlying mechanisms. We observed a trend towards higher seroprevalence, with at least 2 of the 4 assays being positive for patients admitted to the ICU compared to those with milder disease.
The second diagnostic application of a SARS-CoV-2 serological diagnostic tool would be to determine population seroprevalence. At this stage of the pandemic, many countries are now exiting from lockdown and trying to prevent any COVID-19 upsurge. Serological tools have an important place in establishing such strategies. Therefore, we evaluated the 4 assays in panel 2, composed of 143 serum samples from COVID-19 healthcare workers with a diagnosis proven by RT-PCR. ELISA-1 showed a good overall clinical sensitivity of 94% and IgA detection rates higher than 90% from 15 dso. LFA-1 showed the best performances for IgM detection with an overall sensitivity of 87%. For IgG detection, only ELISA-1 showed good performances with an overall sensitivity of 87% and 100% detection rate after 28 dso. When combining IgA or IgM and IgG detection, only LFA-1 and ELISA-1 displayed an excellent clinical sensitivity (≥95%) after 15 days from the onset of symptoms in the range of acceptable values defined by the French National Health Authority (90–100%) (Fig. 3 B).
At this stage of the pandemic, there are no data available about the COVID-19 global seroprevalence in our country (or only partial data obtained in a small specific cohort). It would be interesting in light of future prevalence studies to determine and discuss the positive predictive value of the LFA and ELISA kits we evaluated.
In this study, we first demonstrated that serological tools cannot replace RT-PCR for acute infection diagnosis. Detection rate could be modestly improved by combining IgA or IgM and IgG detection. The bests performances, but not sufficient in this purpose, were obtained only after 14 dso for LFA-1 (Biosynex) and ELISA-1 (Euroimmun). In this study, we did not evaluate assays detecting total antibodies. Recent studies evaluated several serological assays including total anti–SARS-CoV-2 antibodies ELISAs (GeurtsvanKessel et al., 2020; Weidner et al., 2020). GeurtsvanKessel et al. showed that the Wantai ELISA detecting total immunoglobulins against the receptor binding domain of SARS CoV-2 had the best overall characteristics to detect functional antibodies even before 14 dso and regardless of disease severity.
Secondly, a thorough selection of serological assays for detecting ongoing or past infections is advisable following the lifting of lockdowns. Special attention should be paid to antigenic sources and validation against RT-PCR results. After 15 dso, ELISA-1 (Euroimmun) showed the best overall characteristics for detection of IgA and/or IgG, and when combining IgM and IgG detection, only LFA-1 (Biosynex) displayed an excellent clinical sensitivity (≥95%).
Moreover, the reading of sample test lines on LFA devices is still subjective regardless of the manufacturers, especially for weak and/or equivocal bands, requiring a double reading of results. This subjectivity makes it difficult to globalize their use with good reproducibility among healthcare workers. Manufacturers should provide some intensity scale to facilitate the interpretation of these assays. We recommend optimizing antibody detection by combining 1 LFA and 1 IgG ELISA in cases of weak or equivocal signals on the LFA. Long-term studies are required to investigate antibody persistence.
Acknowledgments
We thank Nadège Frey, Cécile Lang, Véronique Sohn, Anne Moncollin, Axelle Grub, Nathalie Durand, and Elise Sühr for their excellent technical assistance. Prof. Fafi-Kremer had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. This study was supported by the Strasbourg University Hospital (COVID-HUS study). We declare no conflicts of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.diagmicrobio.2020.115181.
Appendix A. Supplementary data
Supplementary material
References
- Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J Infect Dis. 2020;222(2):206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., Okba N.M.A., Igloi Z., Bogers S., Embregts C.W.E., Laksono B.M. An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat Commun. 2020;11(1):3436. doi: 10.1038/s41467-020-17317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin infect dis. 2020 doi: 10.1093/cid/ciaa310. Mar 21. Pii: ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W. Evaluation of nucleocapsid and spike protein-based ELISAs for detecting antibodies against SARS-CoV-2. Clin Microbiol. 2020 doi: 10.1128/JCM.00461-20. Mar 30. Pii: JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B, Drosten C, Müller MA. 2014. Serological assays for emerging coronaviruses: challenges and pitfalls virus Res. Dec 19; 194:175–83. 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed]
- Okba N., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis. 2020;26(7) doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Feng Y., Mo X., Zheng P., Wang Q., Li P. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):940–948. doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner L., Gänsdorfer S., Unterweger S., Weseslindtner L., Drexler C., Farcet M. Quantification of SARS-CoV-2 antibodies with eight commercially available immunoassays. J Clin Virol. 2020;129:104540. doi: 10.1016/j.jcv.2020.104540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117 Published March 19, 2020.
- Xu Y., Xiao M., Liu X., Xu S., Du T., Xu J. Significance of serology testing to assist timely diagnosis of SARS-CoV-2 infections: implication from a family cluster. Emerg Microbes Infect. 2020;14:1–12. doi: 10.1080/22221751.2020.1752610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong G., Yi Y., Tuantuan L., Xiaowu W., Xiuyong L., Ang L. Evaluation of the auxiliary diagnostic value of antibody assays for the detection of novel coronavirus (SARS-CoV-2) J Med Virol. 2020 doi: 10.1002/jmv.25919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongchen Z., Shen H., Wang X., Shi X., Li Y., Yan J. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg Microbes Infect. 2020;9(1):833–836. doi: 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Chen C., Shen W., Tang F., Lei H., Xie Y. Emerg Microbes Infect. 2020;22:1–28. doi: 10.1080/22221751.2020.1760143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.J., Liao X.X., Wang H.H., Wei L.L., Xing M.M., Liu L.L. Early virus clearance and delayed antibody response in a case of COVID-19 with a history of co-infection with HIV-1 and HCV. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa408. ciaa408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material