Abstract
Crustaceans are commonly used model organisms to study neuromodulation. Despite numerous reported crustacean neuropeptide families and their functions, there has been no report on neuropeptide glycosylation. This is in part due to a lack of sensitive method that enables deciphering this intricate low-abundance post-translational modification (PTM), even though glycosylation has been shown to play an important role in neuromodulation. Here, we describe the discovery of glycosylated neuropeptides with an enrichment-free approach taking advantage of signature oxonium ions produced in higher-energy collision dissociation (HCD) MS/MS spectra. The detection of the oxonium ions in the HCD scans suggests glycan attachment to peptides, allowing electron-transfer/higher-energy collision dissociation (EThcD) to be performed to selectively elucidate structural information of glycosylated neuropeptides that are buried in non-glycosylated peptides. Overall, 4 N-linked and 14 O-linked glycosylated neuropeptides have been identified for the first time in the crustacean nervous system. In addition, 91 novel putative neuropeptides have been discovered based on the collected HCD scans. This hybrid approach, coupling shotgun method for neuropeptide discovery and targeted strategy for glycosylation characterization enables the first report on glycosylated neuropeptides in crustacean and the discovery of additional neuropeptides simultaneously. The elucidation of novel glycosylated neuropeptides sheds light on the crustacean peptidome and offers novel insights into future neuropeptide functional studies.
Keywords: glycosylation, neuropeptide, EThcD, crustacean nervous system, LC-MS/MS
For TOC Only
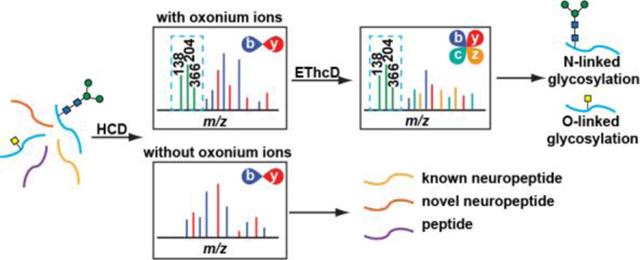
Introduction
Crustacea represents a vast group of arthropod animals, including crabs, shrimps and lobsters. Their relatively simple and well-defined neural networks1 have made themselves attractive model organisms in the neuromodulation studies2–4. The crustacean nervous system where active intercellular interactions take place includes the central nervous system (brain), the stomatogastric nervous system, the pericardial organs (POs), the sinus glands (SGs) and the thoracic ganglia (TG). Neuropeptides are secreted signaling peptides from neurons that play critical roles in signal transmission5–7 and physiological regulations8–10. Over the decades, there have been increasing interests in crustacean neuropeptides because many of them are functional homologues with human neuropeptides11–13. Also, the well-characterized anatomy enables isolation of certain neuronal tissues for circuit-specific studies triggered by stimuli14–15. The neuropeptides inside different organs can react in completely different ways under experimental conditions, as reported in previous studies16–18. Thus, the analysis of crustacean neuropeptides typically involves characterizing various tissue organs to promote thorough understanding toward crustacean nervous system.
Unlike small molecule neurotransmitters, neuropeptides are synthesized by ribosome, further modified in endoplasmic reticulum, transported to Golgi apparatus for necessary processing, then secreted in dense core vesicles and experience local diffusion between axons and dendrites for intercellular communication19–20. Before a mature neuropeptide can be generated, the inactive precursor prohormone undergoes extensive enzymatic cleavages and post-translational modifications (PTMs). The occurrence of PTMs could change neuropeptide structure and function21. To obtain a better understanding toward neuromodulation mechanism, the characterization of neuropeptides and PTMs are of great significance. The large sequence variance brought by amino acids (building blocks of peptides) and dynamic PTMs generated by diverse enzymes have imposed great challenges to neuropeptide discovery and identification. Mass spectrometry (MS) has become an effective tool to study biomolecules22–41, including neuropeptides and PTMs, because it enables production of structure-specific fragments that can be integrated to elucidate chemical composition and peptide sequence. Although there have been prosperous advancements of MS analytical methods, the characterization of one important PTM, glycosylation, on neuropeptides is still underdeveloped, largely due to the heterogeneity of glycans and extremely low abundance of both neuropeptides and glycan modification.42–43 To fully characterize glycosylated neuropeptides, peptide sequence, glycosylation site as well as glycan composition should be reported. In commonly-used MS fragmentation techniques, collision-induced dissociation (CID) mainly produces extensively fragmented glycan and peptide pieces masking glycan site information whereas electron-transfer dissociation (ETD) generates limited glycan side chain composition with reasonable peptide backbone fragmentation.44 To achieve better identification of glycosylated peptides, electron-transfer/higher-energy collision dissociation (EThcD) has been utilized to combine advantages of both CID and ETD and has become an emerging tool for glycopeptide and glycoprotein analysis.27, 45–50 In an EThcD scan, both b/y and c/z ion series are produced,51 uncovering peptide sequence, glycan site position and glycan composition in a single spectrum.27, 45–48
Even with a proper MS fragmentation approach, detection of glycosylated neuropeptides in the crustacean nervous system is still compromised by biological system complexity. The nervous system consists of a variety of molecules, including neurotransmitters, metabolites, lipids, proteins and peptides among which neuropeptides often exist in low abundance and small percentage. The glycosylated neuropeptides, as a subset of neuropeptides, usually occurs with extremely low abundance that makes its confident characterization increasingly difficult. To unravel the sample complexity, a commonly applied strategy is glycosylation enrichment through reversible affinity assays, including hydrazide chemistry52, lectin affinity chromatography53, HILIC54–56, boronic acid57–59, and titanium dioxide60 enrichment. While these methods demonstrated their effectiveness in glycosylation analysis, they do suffer from various disadvantages such as sample loss and unsatisfactory reproducibility61. Therefore, a selective and facile method that can differentiate glycosylated and non-glycosylated peptides in a complex sample mixture is highly desirable. Previous studies62–66 showcase an effective strategy utilizing predominant glycan low-mass fragments (e.g. m/z 204.08, 366.14) produced in HCD/CID as diagnostic ions to probe glycopeptides in the subsequent ETD/EThcD events that allows structural elucidation of glycopeptide and the successful application in glycopeptides, glycoproteins and glycoproteomics without prior knowledge of the glycan structure and peptide sequences.
Hence, here we demonstrate the use of an MS/MS fragment-dependent mass spectrometric workflow for simultaneous targeted glycosylated neuropeptide characterization without the need of sample enrichment and global neuropeptide discovery. In this data-dependent acquisition (DDA) method, top-intensity peaks are first fragmented with HCD. Due to the labile nature of glycan, a peptide bearing glycosylation is prone to produce high abundance oxonium ions in the HCD spectra. Once the signature oxonium ions are identified in the HCD scans, the very same precursor ion will be further selected for additional EThcD fragmentation. In this decision-tree driven MS/MS approach, the simultaneous screening of non-glycosylated neuropeptides and glycosylated neuropeptides can be achieved in a single LC-MS/MS experiment to maximize instrument throughput. The identified N-linked glycosylation and O-linked glycosylation together with novel non-glycosylated neuropeptides offers deeper coverage of the neuropeptidome and novel insights into the crustacean nervous system.
Experimental Procedures
Chemicals.
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified.
Animal Experiments.
All animal experiments were performed following institutional guidelines (University of Wisconsin-Madison IACUC). Rock crabs Cancer irroratus were obtained from Ocean Resources Inc. (Sedgwick, ME) for our pilot study. Subjected to rock crab availability, all other experiments were carried out using blue crabs Callinectes sapidus purchased at a local grocery store. The crabs were maintained in artificial saline tank at 12–13 °C. Upon experiments, crabs were sacrificed for tissue collection after 15 min anesthetization in ice. 4 rock crabs and 20 blue crabs were dissected following previously described protocol67. The isolated neuronal tissues were immediately transferred into acidified methanol68 (MeOH/H2O/HAc = 90/9/1, v/v/v) and stored in dry ice. All tissues were then kept in a −80 °C freezer until further handling.
Sample Processing.
Tissues were probe sonicated in 1 mL acidified methanol and the extracts were centrifuged (4 °C, 18000g, 20min) to collect supernatant. The peptides were collected in the flow-through after spinning (4 °C, 14000g, 4×15min) the supernatant using Microcon YM-30 cutoff filters (pre-rinsed with 80 μL ACN/MeOH/H2O=20/30/50, v/v/v, 4 °C, 13000g, for two times). After drying in vacuum, the peptide extracts were reconstituted in 100 μL 50 mM ammonium bicarbonate. The samples were further reduced by 5 mM DTT (room temperature, 1h) then alkylated by 15 mM IAA (room temperature, in the dark, 30 min). The alkylation reaction was stopped by 5 min DTT incubation at room temperature. An offline high pH RP HPLC was utilized to fractionate complex sample contents before online LC-MS/MS analysis using a C18 column (Phenomenex Kinetex 5u EVO, 150 mm × 2.1 mm, 100 Å) with a 90 min gradient. Mobile phase A is 10 mM NH4HCOO in H2O, pH = 10 and mobile phase B contains 10 mM NH4HCOO in ACN/H2O (90/10, v/v) with pH = 10. Crustacean tissue extracts were separated at column temperature 30 °C with a flow rate of 0.2 mL/min. The gradient stayed at 1% B for 3 min then was linearly ramped to 35% B in 3–50 min, then was ramped to 60% in 4 min and continued to 70% in another 4 min then ramped to 100% in 1 min and stayed at 100% B for 15 min, followed by a 15.5 min equilibrium at the end with 1% B. A fraction collector was utilized to collect sample every 2 min during the time period of 6–62 min. The fractionated samples were pooled into 4~5 final fractions which were then dried down under vacuum.
LC-MS/MS Analysis.
LC-MS/MS experiments were performed on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) coupled with a Dionex Ultimate 3000 UPLC system (Thermo Fisher Scientific). Mobile phase A is 0.1% formic acid in H2O with mobile phase B as 0.1% formic acid in ACN. Chromatography separation was achieved using a 75 μm × 15 cm homemade column packed with C18 material (1.7 μm, 150 Å, BEH) from a Waters UPLC column (part no. 186004661). Separation gradient started from 3% B, linearly ramped to 30% B in 90 min, then ramped to 75% in another 20 min. The full MS spectra (m/z 400–1800) were acquired in the Orbitrap with 120K resolution in positive polarity mode with AGC target 2e5 and maximum injection time 100 ms. A Top Speed method was utilized to make sure full MS spectra were acquired every 3 s. Data-dependent HCD was used to fragment most abundant precursor ions at a resolution of 30K with 35% normalized collision energy, dynamic exclusion 20 s, AGC target 1e5 and maximum injection time 60 ms. If oxonium ions at m/z 138.0545, m/z 204.0867, or m/z 366.1396 (± m/z 0.01) were within the top 30 most abundant peaks, an EThcD MS/MS fragmentation would be further applied to the precursor at a resolution of 60K in the Orbitrap. In EThcD scans, precursors with different charges underwent different ETD reaction times (z = 2, 50 ms; z = 3–4, 20 ms; z = 5, 10 ms; z = 6–15, use instrument charge-dependent ETD parameters) then subject to supplemental activation energy of 33%.
Data Processing.
The raw data files were first processed by PEAKS Studio 7.0 with following parameters: no enzymatic cleavage, parent mass error tolerance 10.0 ppm, fragment mass error tolerance 0.02 Da, carbamidomethylation on cysteine as fixed modification, and variable modifications including methionine oxidation, C-terminal amidation, dehydration, pyro-Glu from E and Q, sulfation on Y, methylation, glycans on serine/threonine including HexNAc(1), HexNAc(1)Hex(1), HexNAc(1)Hex(1)NeuAc(1), HexNAc(1)Hex(1)NeuAc(2), HexNAc(2), HexNAc(2)Hex(1) and glycan on asparagine including HexNAcylation. The generated de novo sequences were then combined with the Li Lab crustacean neuropeptide database to produce a new peptide database. The raw data were analyzed by Proteome Discoverer 2.1 embedded with Byonic. EThcD spectra were searched against the new peptide database and mammalian glycan database using precursor mass tolerance 10.0 ppm and fragment mass tolerance 0.01 Da. Common dynamic modifications include C-terminal amidation, pyro-Glu from N terminal Q and methionine oxidation, with cysteine residue carbamidomethylation as static modification. The glycosylation sites were assigned based on N-glycosylation occurring on asparagine with a sequence pattern of N-X-S/T where X cannot be proline and O-glycosylation occurring on serine or threonine together with accurate mass matching between observed peptide/glycan fragment masses and theoretical masses. For novel neuropeptide discovery, the de novo sequences were searched by a homebuilt Python program against consensus motif (the shared peptide sequence pattern within a certain neuropeptide family) in common neuropeptide families69, including A-type allatostatin (AST-A), B-type allatostatin, (AST-B), C-type allatostatin (AST-C), allatostatin-combos, GSEFLamide, kinin, orcokinin, orcomyotropin, pyrokinin, RFamide, RYamide, YRamide, SIFamide, tachykinin, and WXXXnRamide where X denotes any amino acid and Xn is a glycine or nothing.
Results and Discussion
Discovery of N-linked and O-linked glycosylated neuropeptides
Mass spectrometry has become an attractive technique to detect and identify crustacean neuropeptides owing to its sensitivity and high throughput. Even though crustacean neuropeptides have been extensively characterized, the identification of glycosylation on neuropeptides remains to be challenging without proper analytical tools. The naturally occurring extremely low abundance has limited discovery of glycosylated neuropeptides as enrichment is typically difficult. Meanwhile, confident structural elucidation is challenged by glycan heterogeneity and site-specific modification. In our analysis workflow, a straightforward and enrichment-free method was developed to enable identification of both glycosylated neuropeptides and novel neuropeptides without glycosylation via a signature-ion driven approach with MS. To overcome signal suppression from non-glycosylated peptides, the Orbitrap Fusion Lumos mass spectrometer only selectively applies EThcD hybrid fragmentation to those precursors with diagnostic oxonium ions present in prior HCD scans. As the entire crustacean genome has not been sequenced, there is no complete neuropeptide database available for data analysis. To reduce missed detection, the de novo sequences with average local confidence (ALC) >=50% generated from PEAKS Studio 7 software70–71 were combined with in-house crustacean neuropeptide database as a new database for glycosylation screening in Proteome Discoverer 2.1 embedded with Byonic module. The thousands of de novo sequences were also searched by a homebuilt Python program for identification of novel neuropeptides based on neuropeptide family consensus sequence motifs. The discovered glycosylated neuropeptides and novel neuropeptides were further manually inspected to eliminate false discovery.
Summarized in Table 1 are all glycosylated neuropeptides identified by analyzing 4 different neuronal tissue organs (brain, PO, SG and TG) isolated from blue crab Callinectes sapidus. And the corresponding EThcD spectra are shown in Supporting Information Figure S1 through Figure S17, respectively. Overall, 4 N-linked glycoforms on 3 neuropeptides and 14 O-linked glycoforms on 11 neuropeptides were discovered in the crustacean nervous system, covering 5 distinct neuropeptide families. To the best of our knowledge, this study presents the first investigation and characterization of glycosylation on crustacean neuropeptides. No glycosylated neuropeptide was detected from blue crab brain sample while an O-glycosylated orcomyotropin (Figure 1a) was discovered in rock crab brain sample in our pilot study. This might be caused by signal suppression of other biomolecules in the blue crab brain sample or biological differences among crustacean species.
Table 1.
List of glycosylated neuropeptides detected from blue crab Callinectes sapidus neuronal tissues (brain, PO, SG and TG). The square bordered character X indicates glycan position verified from EThcD spectra and underlined character X denotes putatively identified glycan position based upon Byonic search results. The neuropeptide family labeled with asterisk (X*) shows peptide sequences matched with neuropeptide consensus. AST-B: B-type allatostatin; CPRP: crustacean hyperglycemic hormone precursor-related peptide.
| Type | Tissue | Neuropeptide Family |
Neuropeptide Sequence | Glycan | Observed mass (M+H+) |
Calculated mass (M+H+) |
GlcNAc to GalNAc Ratio |
|---|---|---|---|---|---|---|---|
| N | PO | AST-B | NNNWTKFQGSWa | HexNAc(2)Hex(2)Fuc(1) | 2256.967 | 2256.967 | N/A |
| N | PO | AST-B | NNNWTKFQGSWa | HexNAc(2)Hex(3)Fuc(2) | 2565.082 | 2565.077 | N/A |
| N | PO | AST-B* | NNWSKFQGSWa | HexNAc(2)Hex(3)Fuc(2) | 2437.022 | 2437.019 | N/A |
| N | PO | AST-B* | DNNWTKFQGSWa | HexNAc(2)Hex(3)Fuc(2) | 2566.064 | 2566.061 | N/A |
| O | PO | AST-B* | NNWSKFQGSWa | HexNAc(2)Hex(2)Fuc(2) | 2274.969 | 2274.966 | 5.32 |
| O | PO | AST-B* | DNNWTKFQGSWa | HexNAc(2)Hex(1)Fuc(2) | 2241.961 | 2241.956 | 6.81 |
| O | PO | AST-B* | DNNWTKFQGSWa | HexNAc(2)Hex(2)Fuc(2) | 2404.012 | 2404.009 | 5.76 |
| O | PO | AST-B* | PDYPAVSPRSTNWSSLRGTWa | HexNAc(1) | 2479.205 | 2479.200 | 0.77 |
| O | PO | CPRP | SLKSDTVTPLRGFEGETGHPLE | HexNAc(1) | 2573.273 | 2573.273 | 0.56 |
| O | SG | CPRP | SDTVTPLRGFEGETGHPLE | HexNAc(1) | 2245.069 | 2245.062 | 0.63 |
| O | SG | CPRP | SDTVTPLRGFEGETGHPLE | HexNAc(1)Hex(1) | 2407.121 | 2407.115 | 0.83 |
| O | SG | CPRP | GFEGETGHPLE | HexNAc(1)Hex(1) | 1537.656 | 1537.654 | 0.58 |
| O | TG | Orcomyotropin | FDAFTTGFGHS | HexNAc(1)Hex(1) | 1551.656 | 1551.649 | 0.96 |
| O | TG | RFamide | DARTAPLRLRFa | HexNAc(1)Hex(1) | 1679.918 | 1679.908 | 0.74 |
| O | TG | RFamide | SMPTLRLRFa | HexNAc(1)Hex(1) | 1484.783 | 1484.778 | 0.97 |
| O | TG | RFamide | SMPTLRLRFa | HexNAc(1)Hex(1) | 1484.783 | 1484.778 | 0.72 |
| O | PO | RYamide* | QVTERSGFYANRYa | HexNAc(1) | 1792.861 | 1792.861 | 0.84 |
| O | PO | RYamide* | TERSGFYANRYa | HexNAc(1) | 1565.736 | 1565.734 | 0.99 |
Figure 1.
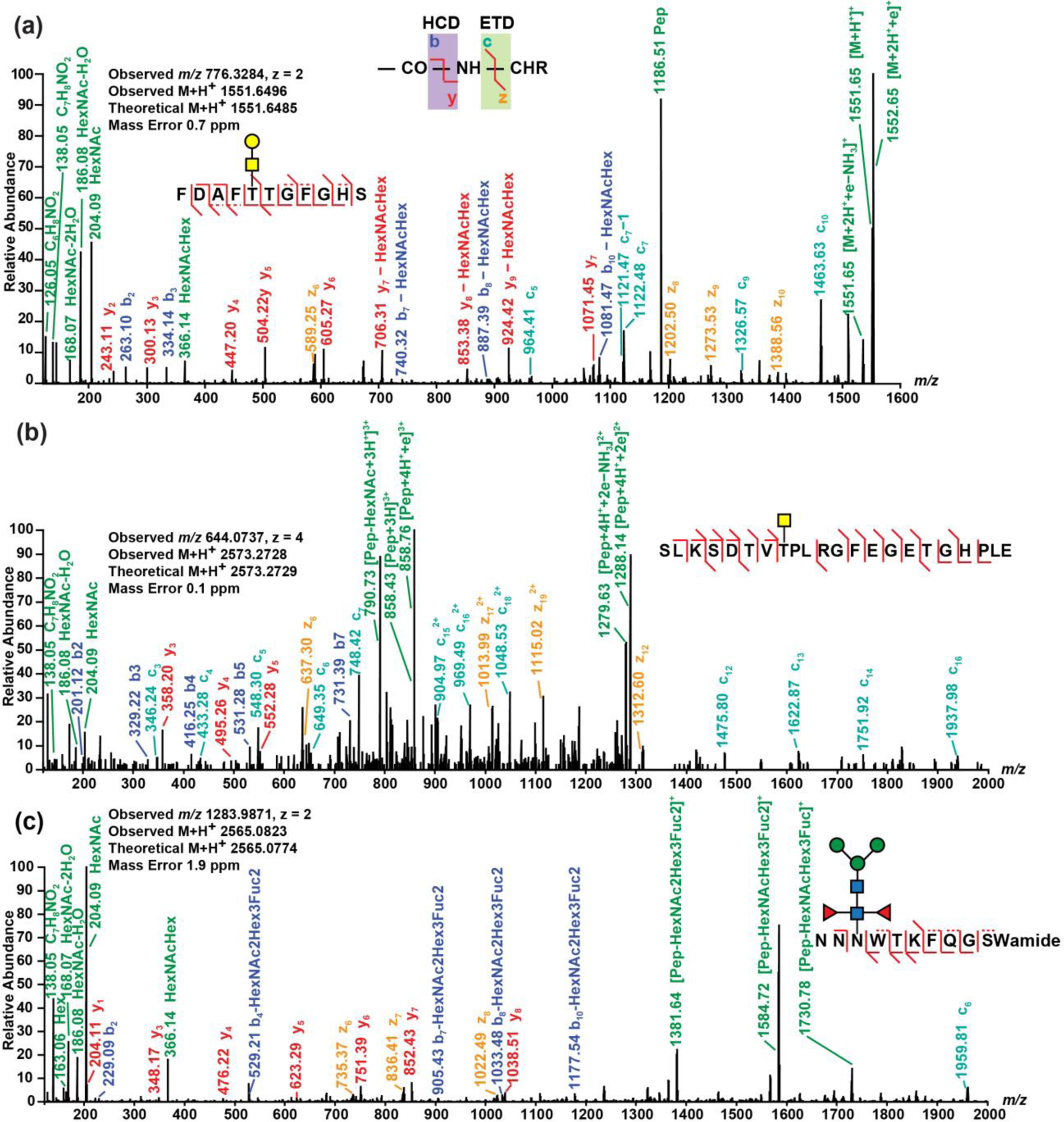
EThcD spectra of (a) an O-linked orcomyotropin neuropeptide discovered in rock crab Cancer irroratus nervous system, (b) an O-linked truncated crustacean hyperglycemic hormone precursor-related (CPRP) neuropeptide discovered in blue crab Callinectes sapidus nervous system, and (c) an N-linked B-type allatostatin (AST-B) neuropeptide discovered in blue crab Callinectes sapidus nervous system. The spectra were analyzed using Proteome Discoverer 2.1 embedded with Byonic with following parameters: precursor mass tolerance 10.0 ppm, fragment mass tolerance 0.01 Da, dynamic modifications C-terminal amidation, N-terminal pyro-Glu from Q and methionine oxidation, and static modification cysteine residue carbamidomethylation.
Structural elucidation of N-linked and O-linked glycosylated neuropeptides
The direct characterization of glycan linkage is difficult solely based on the masses detected from the spectra, due to structural complexity and isomerization of glycans. However, the oxonium ion groups generated by HCD enabled differentiation of O-GlcNAc and O-GalNAc. According to Halem et al.72, the GlcNAc/GalNAc ratio calculated from oxonium ion intensities in the HCD spectra is a useful tool to distinguish GlcNAc and GalNAc, with the ratio <1 indicating GalNAc and ratio >1 suggesting GlcNAc. The GlcNAc and GalNAc ratio is calculated as shown below:
where m/z 126.06, 138.06, 144.07 and 168.07 generated from HexNAc neutral losses represent [C6H7NO2]+, [C7H8NO2]+, [C6H10NO3]+ and [C8H10NO3]+, respectively. According to GlcNAc/GalNAc ratios, the predominant population of neuropeptides consist of GalNAc with the ratio <1, except for 3 B-type allatostatin (AST-B) O-glycosylated neuropeptides with relatively large glycan attached to amino acid side chain. It is interesting to note that even within the same neuropeptide family AST-B, there are significant glycosylation heterogeneity, reflected by different types of glycosylation (N-linked and O-linked) and glycan composition variance. For example, there are 4 N-linked AST-B peptides and 4 other O-linked AST-B peptides. And among the O-linked glycosylated AST-B peptides, PDYPAVSPRSTNWSSLRGTWa has glycan modification of only one GalNAc and other 3 AST-B peptides are linked to larger glycans with at least 5 monosaccharide units, including 2 GlcNAc. The difference of glycan composition and saccharide heterogeneity could be associated with broadly distributed functions of AST-B neuropeptides as autocrine/paracrine factors and circulating hormones69. Although EThcD generates rich fragmentation ions, it does not necessarily provide enough determinant evidence (c/z- ion series) of the glycosylation site for every neuropeptide, especially O-linked ones, so a few neuropeptides in Table 1 with underlined characters have putatively assigned glycosylation sites.
Shown in Figure 1 are three representative EThcD fragmentation spectra of the discovered glycosylated neuropeptides. Figure 1a and Figure 1b display the intact O-linked orcomyotropin and O-linked truncated neuropeptide in the crustacean hyperglycemic hormone precursor-related peptide (CPRP) family. In addition, EThcD spectrum of an N-linked AST-B neuropeptide is shown in Figure 1c. EThcD provides rich sequence and structural information of the glycosylated neuropeptides, including glycan fragments and both peptide b/y- and c/z- ion series. The orcomyotropin neuropeptide detected in the brain of rock crab Cancer irroratus (Figure 1a and Figure S18) has a sequence of FDAFTTGFGHS where O-glycosylation could possibly occur at Thr5, Thr6 and Ser11. Since the EThcD spectrum contains c5 ion with the glycan preserved, the glycosylation site has been located at Thr5. On the truncated CPRP neuropeptide (Figure 1b), the glycosylation site has been assigned at Thr8 because the c7 ion does not contain glycan and the c12 ion includes the glycan, indicating Thr8 is the only position for glycan attachment. Fewer N-linked glycosylated neuropeptides have been discovered compared with O-linked glycosylation, as only part of the neuropeptides has the special N-glycosylation sequence pattern (N-X-S/T where X cannot be proline) being localized. In addition to confirmation of O-linked glycosylation, Figure 1c demonstrates how EThcD can benefit identification of N-glycosylation based on produced c/z- ions. Just relying on this AST-B neuropeptide sequence NNNWTKFQGSWamide, the glycosylation could occur at Asn3 (N-linked), Thr5 (O-linked) and Ser10 (O-linked). The produced c/z- ions bring more insights into the assignment of the glycosylation site. As the spectrum contains z6, z7 and z8 ions, the possibility at Thr5 and Ser10 have been excluded as there are no glycan attached on the generated z ions, meaning the glycosylation is N-linked and thus occurs at Asn3. Using the GlcNAc/GalNAc ratio, it is determined that the O-glycosylation cases in Figure 1a and Figure 1b are both O-GalNAc as the ratios are less than 1. The assignment of glycan composition in Figure 1c is slightly different. The Byonic generated N-glycan sequence HexNAc(2)Hex(3)Fuc(2) was searched in UniCarbKB73, and three N-glycan compositions were returned with two of them given known species origins. Among the two N-glycan compositions, only one was selected as our putative N-glycan assignment to the arthropod crustacean neuropeptide because the same composition is present in two other arthropod animals, European honey bee Androctonus australis and scorpion Androctonus australis hector, while the other possibility excluded was detected in a mollusca animal Japanese flying squid Todarodes pacificus. By integrating information from various perspectives, the interpretation of complicated EThcD spectra is possible and has strengthened our understanding toward crustacean neuropeptide glycosylation.
Glycosylation occurring on peptides with neuropeptide consensus sequence motif
In addition to glycosylation occurrence on known neuropeptides, there are other glycosylated peptides whose sequences match with neuropeptide consensus sequence motif. Although currently more evidence is still needed to ultimately confirm their roles as neuropeptides, the findings have expanded the coverage and broadened the understanding toward crustacean peptidome. Figure 2 illustrates two O-linked glycosylated peptides that are potentially neuropeptides. The MS/MS spectra for confident identification of these two peptides (without glycosylation) in blue crab sample are shown in Supporting Information Figure S19 and Figure S20, respectively. EThcD enables glycan site verification on a 20-amino acid peptide, as demonstrated in Figure 2a. The peptide PDYPAVSPRSTNWSSLRGTWa was discovered in rock crab Cancer irroratus as a putative neuropeptide candidate so it was included in database during data analysis. The O-glycosylation could occur at Ser7, Ser10, Thr11, Asn12, Ser14, Ser15 and Thr19 on this peptide with AST-B consensus sequence (WX6Wamide). As suggested by the determinant c11 ion (glycan included) and z12 ion (not containing glycan), the O-glycosylation modification on this relatively long peptide has been assigned at Ser7. Figure 2b shows the glycosylation on a de novo peptide sequence QVTERSGFYANRYa with neuropeptide RYamide consensus sequence. As illustrated in the spectrum, the abundant c3 ion leads to a confident assignment that the O-glycosylation is attached to Thr3. The glycosylation occurrence on peptides with neuropeptide consensus sequence motif requires the combination of de novo sequences with known crustacean neuropeptide database when probing glycosylated (neuro)peptides. An interesting question to ask is what is the ratio of glycosylated to non-glycosylated forms for the same peptide sequence. The absolute quantitation requires synthesis of glycosylated peptide standards which presented technical challenges to several vendors. Although glycosylated and non-glycosylated peptides have different ionization efficiencies, a rough estimation of glycosylated/non-glycosylated ratio can be made by comparing the extracted ion chromatogram (EIC) intensity/area of the glycosylated and non-glycosylated forms. For instance, Figure S21 shows the EIC of glycosylated and non-glycosylated forms of PDYPAVSPRSTNWSSLRGTWa. The O-glycosylated/non-glycosylated estimated ratio is ~10%. Another example is NNWSKFQGSWa, as shown in Figure S22. The N-glycosylated/non-glycosylated estimated ratio is ~0.1%. Although the glycosylated forms only consist of a small portion, they may play critical and indispensable roles in the nervous system which were less studied due to lack of sensitive methods for discovery and identification. Future studies will include electrophysiological experiments to explore the functions of these glycosylated peptides.
Figure 2.
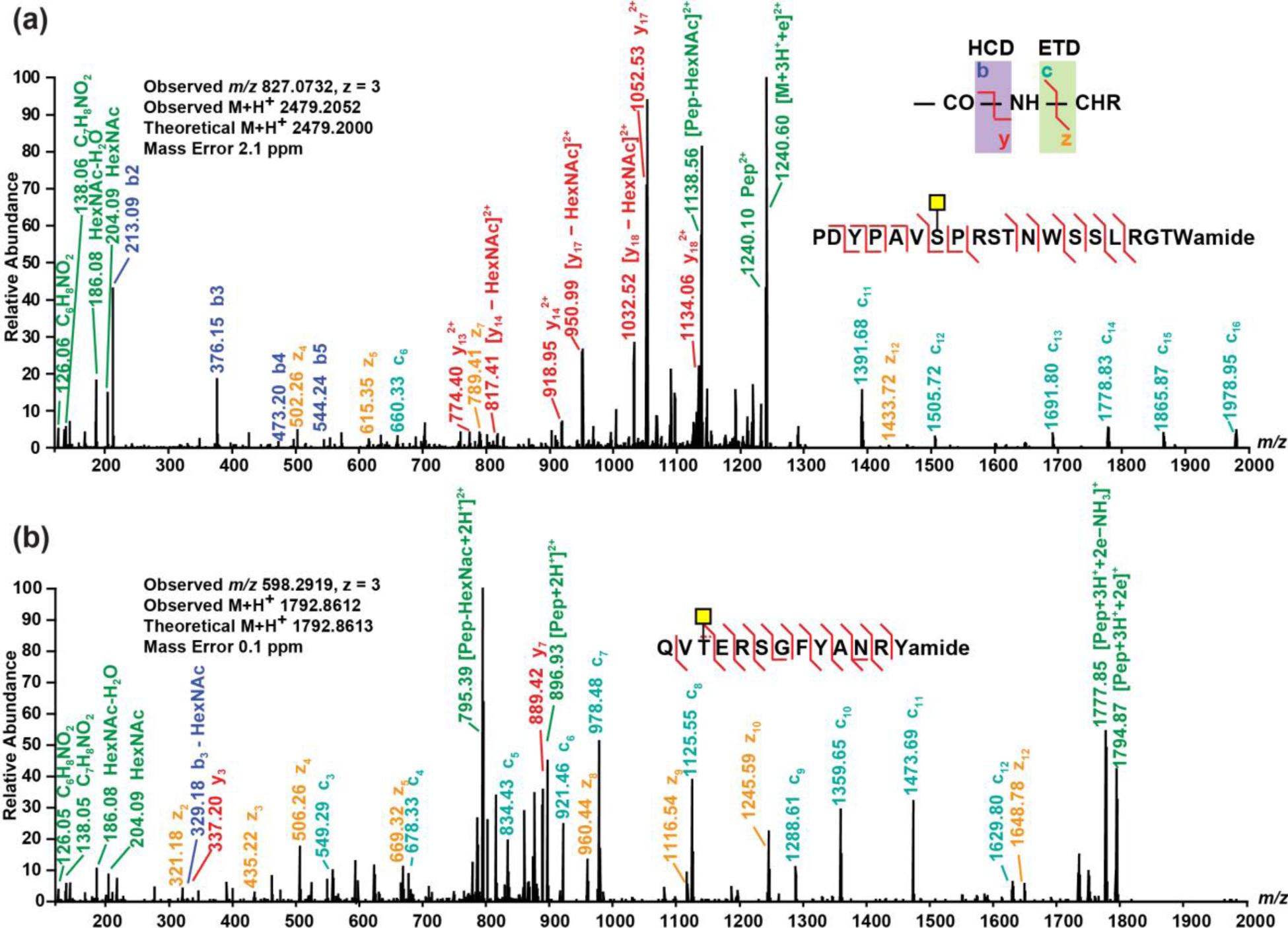
EThcD spectra of (a) an O-linked peptide with crustacean hyperglycemic hormone precursor-related (CPRP) neuropeptide consensus and (b) an O-linked peptide with neuropeptide RYamide consensus discovered from blue crab Callinectes sapidus nervous system. Spectra were analyzed using Proteome Discoverer 2.1 embedded with Byonic with following parameters: precursor mass tolerance 10.0 ppm, fragment mass tolerance 0.01 Da, dynamic modifications C-terminal amidation, N-terminal pyro-Glu from Q and methionine oxidation, and static modification cysteine residue carbamidomethylation.
Discovery of novel neuropeptide sequences
In our oxonium ion-triggered MS/MS fragmentation approach, numerous HCD spectra were acquired for novel neuropeptide discovery in addition to those identified glycosylated neuropeptides. The de novo sequences generated from HCD with ALC>70% were matched to neuropeptide consensus sequence motif from a variety of neuropeptide families, including A-type allatostatin (AST-A), AST-B, pyrokinin, RFamide, RYamide, tachykinin, WXXXnRamide and YRamide. Representative MS/MS spectra of de novo peptides with an AST-B consensus motif, a WXXXnRamide consensus and an RFamide consensus sequence are illustrated in Figure 3a, Figure 3b and Figure 3c, respectively. Figure 3d shows the number of novel non-glycosylated neuropeptides and neuropeptide family variance of de novo sequences detected in different tissue organs, which could be related to neuronal organ function diversities. The full list of the 91 putative novel neuropeptides can be found in Supporting Information Table S1. Although additional experiments will need to be performed to confirm the sequences and test their functional roles, this extended list of putative novel signaling peptides offers new knowledge and insights into the crustacean peptidome and helps to further advance our understanding toward the peptidergic signaling in the crustacean nervous system.
Figure 3.
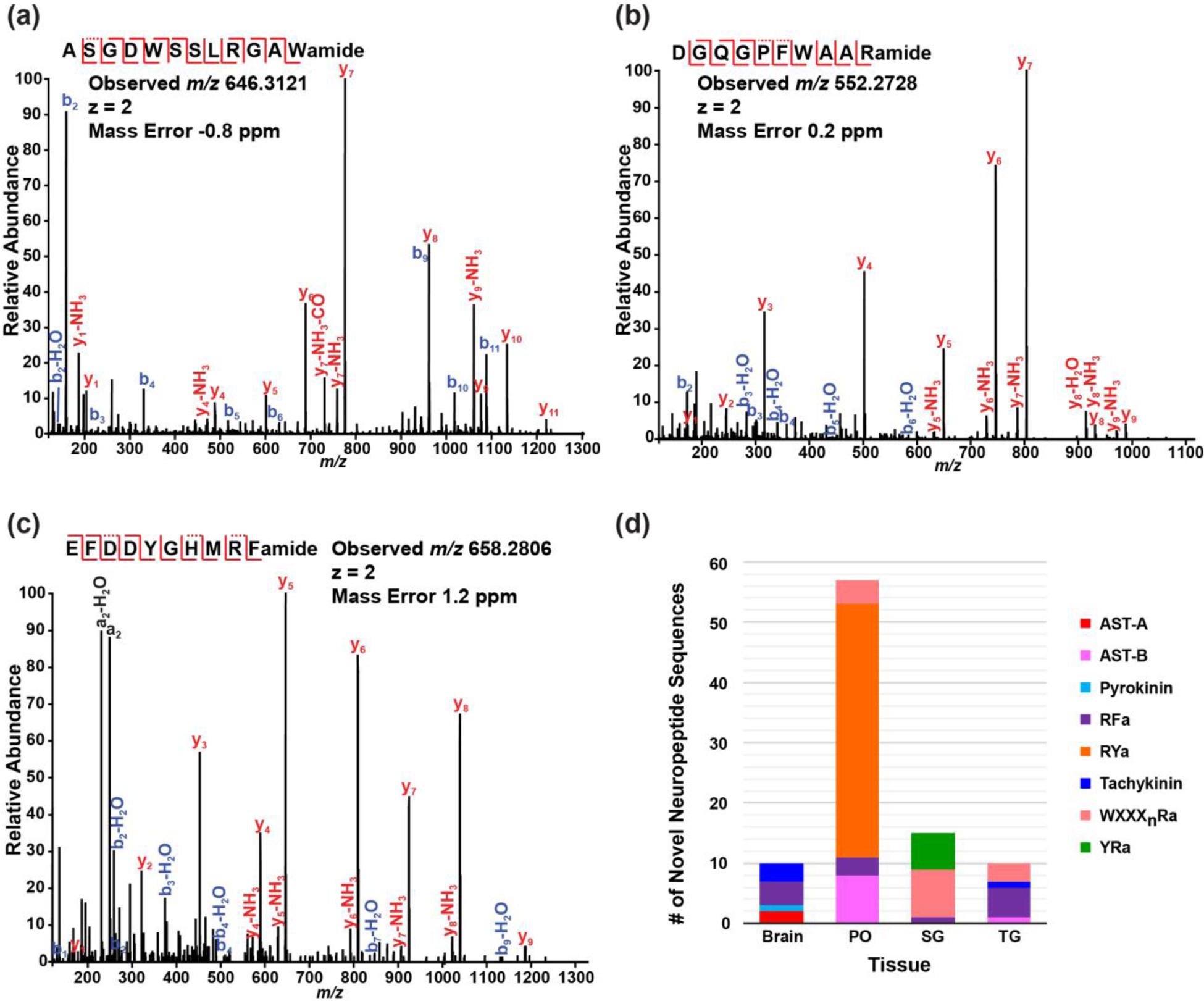
HCD spectra detected in blue crab Callinectes sapidus: (a) a de novo peptide sequence (ALC=96%) with B-type allatostatin neuropeptide consensus sequence motif detected in pericardial organs, (b) a de novo peptide sequence (ALC=99%) with WXXXnRamide (Xn is a glycine or nothing) neuropeptide consensus detected in sinus glands and (c) a de novo peptide sequence (ALC=99%) with RFamide (subfamily sulfakinin) neuropeptide consensus detected in sinus glands. (d) the breakdown of de novo peptide sequences with various neuropeptide consensus distributed in different neuronal organs. PO: pericardial organ; SG: sinus gland; TG: thoracic ganglia. Spectra were analyzed by PEAKS Studio 7.0 with following parameters: no enzymatic cleavage, parent mass error tolerance 10.0 ppm, fragment mass error tolerance 0.02 Da, fixed modification cysteine carbamidomethylation, and variable modifications methionine oxidation, C-terminal amidation, dehydration, pyro-Glu from E and Q, sulfation on Y, methylation, glycans on serine/threonine including HexNAc(1), HexNAc(1)Hex(1), HexNAc(1)Hex(1)NeuAc(1), HexNAc(1)Hex(1)NeuAc(2), HexNAc(2), HexNAc(2)Hex(1) and glycan on asparagine including HexNAcylation.
Conclusions
To summarize, our study established a facile strategy to simultaneously characterize glycosylated neuropeptides and novel neuropeptide sequences in the crustacean nervous system, resulting in a deeper understanding of the peptidome. The employed HCD-triggered EThcD workflow achieved complementary characterization of peptides with and without glycosylation with optimal throughput and coverage. Multiple glycosylated neuropeptides, in the form of N-linked and O-linked, were discovered in crustacean neuronal tissues for the first time. Also, a plethora of novel neuropeptides were sequenced to generate a comprehensive view of the complex biological system. The newly identified glycosylated neuropeptides together with novel neuropeptides have greatly expanded our knowledge about in vivo neuropeptide synthesis processes and are anticipated to offer novel insights into cellular signaling mechanisms and unique opportunities to explore the role of glycosylation in neuromodulation and peptidergic signaling.
Supplementary Material
Figures S1-S17. EThcD spectra of crustacean neuropeptides modified with glycans discovered from blue crab Callinectes sapidus nervous system.
Figure S18. EThcD spectrum of a crustacean neuropeptide modified with glycans discovered from rock crab Cancer irroratus nervous system.
Figures S19-S20. MS/MS spectra of two confidently identified neuropeptides without glycan attached.
Figures S21-S22. EIC comparison of glycosylated and non-glycosylated forms of the peptide.
Table S1. Putative novel neuropeptide sequences identified using HCD via de novo peptide sequencing.
Acknowledgements
This work was supported in part by the National Science Foundation (NSF) grant CHE-1710140, National Institutes of Health grants R01 DK071801 and U01 CA231081. The Orbitrap instruments were purchased through the support of an NIH shared instrument grant (NIH-NCRR S10RR029531) and Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison. We thank Dr. Marshall Bern from the Protein Metrics for providing 1-year access to Byonic software package. L. L. acknowledges a Vilas Distinguished Achievement Professorship and the Charles Melbourne Johnson Distinguished Chair Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
The authors declare no competing financial interest. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE74 partner repository with the dataset identifier PXD014269.
Supporting Information
The following supporting information is available free of charge at ACS website http://pubs.acs.org
References
- 1.Wiese K, The crustacean nervous system. Springer Science & Business Media: 2013. [Google Scholar]
- 2.NUSBAUM MP; Marder E, A neuronal role for a crustacean red pigment concentrating hormone-like peptide: neuromodulation of the pyloric rhythm in the crab, Cancer borealis. Journal of experimental biology 1988, 135 (1), 165–181. [Google Scholar]
- 3.BELTZ BS; KRAVITZ EA, Aminergic and Peptidergic Neuromodulation in Crustacea. Journal of Experimental Biology 1986, 124 (1), 115–141. [Google Scholar]
- 4.Marder E, Neuromodulation of Neuronal Circuits: Back to the Future. Neuron 2012, 76 (1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hökfelt T; Broberger C; Xu Z-QD; Sergeyev V; Ubink R; Diez M, Neuropeptides — an overview. Neuropharmacology 2000, 39 (8), 1337–1356. [DOI] [PubMed] [Google Scholar]
- 6.Crown A; Clifton DK; Steiner RA, Neuropeptide signaling in the integration of metabolism and reproduction. Neuroendocrinology 2007, 86 (3), 175–182. [DOI] [PubMed] [Google Scholar]
- 7.Grimmelikhuijzen CJ; Hauser F, Mini-review: the evolution of neuropeptide signaling. Regulatory peptides 2012, 177, S6–S9. [DOI] [PubMed] [Google Scholar]
- 8.Sakurai T; Amemiya A; Ishii M; Matsuzaki I; Chemelli RM; Tanaka H; Williams SC; Richardson JA; Kozlowski GP; Wilson S; Arch JRS; Buckingham RE; Haynes AC; Carr SA; Annan RS; McNulty DE; Liu W-S; Terrett JA; Elshourbagy NA; Bergsma DJ; Yanagisawa M, Orexins and Orexin Receptors: A Family of Hypothalamic Neuropeptides and G Protein-Coupled Receptors that Regulate Feeding Behavior. Cell 1998, 92 (4), 573–585. [DOI] [PubMed] [Google Scholar]
- 9.Gäde G, REGULATION OF INTERMEDIARY METABOLISM AND WATER BALANCE OF INSECTS BY NEUROPEPTIDES. Annual Review of Entomology 2004, 49 (1), 93–113. [DOI] [PubMed] [Google Scholar]
- 10.Barrios A; Ghosh R; Fang C; Emmons SW; Barr MM, PDF-1 neuropeptide signaling modulates a neural circuit for mate-searching behavior in C. elegans. Nature Neuroscience 2012, 15, 1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandeman RE; Sandeman DC; Watson AHD, Substance P antibody reveals homologous neurons with axon terminals among somata in the crayfish and crab brain. Journal of Comparative Neurology 1990, 294 (4), 569–582. [DOI] [PubMed] [Google Scholar]
- 12.Nässel DR; Wegener C, A comparative review of short and long neuropeptide F signaling in invertebrates: Any similarities to vertebrate neuropeptide Y signaling? Peptides 2011, 32 (6), 1335–1355. [DOI] [PubMed] [Google Scholar]
- 13.Yu N; Smagghe G, CCK(-like) and receptors: Structure and phylogeny in a comparative perspective. General and Comparative Endocrinology 2014, 209, 74–81. [DOI] [PubMed] [Google Scholar]
- 14.Skiebe P, Neuropeptides are ubiquitous chemical mediators: Using the stomatogastric nervous system as a model system. Journal of Experimental Biology 2001, 204 (12), 2035–2048. [DOI] [PubMed] [Google Scholar]
- 15.Nusbaum MP; Beenhakker MP, A small-systems approach to motor pattern generation. Nature 2002, 417, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen R; Hui L; Cape SS; Wang J; Li L, Comparative Neuropeptidomic Analysis of Food Intake via a Multifaceted Mass Spectrometric Approach. ACS Chemical Neuroscience 2010, 1 (3), 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen R; Xiao M; Buchberger A; Li L, Quantitative Neuropeptidomics Study of the Effects of Temperature Change in the Crab Cancer borealis. Journal of Proteome Research 2014, 13 (12), 5767–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y; Buchberger A; Muthuvel G; Li L, Expression and distribution of neuropeptides in the nervous system of the crab Carcinus maenas and their roles in environmental stress. PROTEOMICS 2015, 15 (23–24), 3969–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Den Pol AN, Neuropeptide transmission in brain circuits. Neuron 2012, 76 (1), 98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Q; Liang Z; OuYang C; Li L In Biologically Active Peptides in Invertebrates: Discovery and Functional Studies, Colloquium Series on Neuropeptides, Morgan & Claypool Life Sciences: 2015; pp 1–182. [Google Scholar]
- 21.Fricker LD In Neuropeptides and other bioactive peptides: from discovery to function, Colloquium Series on Neuropeptides, Morgan & Claypool Life Sciences: 2012; pp 1–122. [Google Scholar]
- 22.Baggerman G; Clynen E; Huybrechts J; Verleyen P; Clerens S; De Loof A; Schoofs L, Peptide profiling of a single Locusta migratoria corpus cardiacum by nano-LC tandem mass spectrometry. Peptides 2003, 24 (10), 1475–1485. [DOI] [PubMed] [Google Scholar]
- 23.Dowell JA; Vander Heyden W; Li L, Rat Neuropeptidomics by LC–MS/MS and MALDI–FTMS: Enhanced Dissection and Extraction Techniques Coupled with 2D RP-RP HPLC. Journal of Proteome Research 2006, 5 (12), 3368–3375. [DOI] [PubMed] [Google Scholar]
- 24.Fu Q; Goy MF; Li L, Identification of neuropeptides from the decapod crustacean sinus glands using nanoscale liquid chromatography tandem mass spectrometry. Biochemical and biophysical research communications 2005, 337 (3), 765–778. [DOI] [PubMed] [Google Scholar]
- 25.Jia C; Lietz CB; Ye H; Hui L; Yu Q; Yoo S; Li L, A multi-scale strategy for discovery of novel endogenous neuropeptides in the crustacean nervous system. Journal of Proteomics 2013, 91, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Secher A; Kelstrup CD; Conde-Frieboes KW; Pyke C; Raun K; Wulff BS; Olsen JV, Analytic framework for peptidomics applied to large-scale neuropeptide identification. Nature Communications 2016, 7, 11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Q; Canales A; Glover MS; Das R; Shi X; Liu Y; Keller MP; Attie AD; Li L, Targeted Mass Spectrometry Approach Enabled Discovery of O-Glycosylated Insulin and Related Signaling Peptides in Mouse and Human Pancreatic Islets. Analytical Chemistry 2017, 89 (17), 9184–9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Q; Ouyang C; Zhong X; Li L, Profiling of small molecule metabolites and neurotransmitters in crustacean hemolymph and neuronal tissues using reversed-phase LC-MS/MS. Electrophoresis 2018, 39 (9–10), 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Q; Wang Y; Chen B; Ma F; Hao L; Li G; Ouyang C; Li L, Visualization and Identification of Neurotransmitters in Crustacean Brain via Multifaceted Mass Spectrometric Approaches. ACS Chemical Neuroscience 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang T; Yu N; Kim J; Murgo J-R; Kissai M; Ravichandran K; Miracco EJ; Presnyak V; Hua S, Oligonucleotide Sequence Mapping of Large Therapeutic mRNAs via Parallel Ribonuclease Digestions and LC-MS/MS. Analytical Chemistry 2019. [DOI] [PubMed] [Google Scholar]
- 31.Håkansson K; Cooper HJ; Emmett MR; Costello CE; Marshall AG; Nilsson CL, Electron Capture Dissociation and Infrared Multiphoton Dissociation MS/MS of an N-Glycosylated Tryptic Peptide To Yield Complementary Sequence Information. Analytical Chemistry 2001, 73 (18), 4530–4536. [DOI] [PubMed] [Google Scholar]
- 32.Adamson JT; Håkansson K, Electron Capture Dissociation of Oligosaccharides Ionized with Alkali, Alkaline Earth, and Transition Metals. Analytical Chemistry 2007, 79 (7), 2901–2910. [DOI] [PubMed] [Google Scholar]
- 33.Axelsson J; Palmblad M; Håkansson K; Håkansson P, Electron capture dissociation of substance P using a commercially available Fourier transform ion cyclotron resonance mass spectrometer. Rapid Communications in Mass Spectrometry 1999, 13 (6), 474–477. [DOI] [PubMed] [Google Scholar]
- 34.Amicucci MJ; Galermo AG; Guerrero A; Treves G; Nandita E; Kailemia MJ; Higdon SM; Pozzo T; Labavitch JM; Bennett AB; Lebrilla CB, Strategy for Structural Elucidation of Polysaccharides: Elucidation of a Maize Mucilage that Harbors Diazotrophic Bacteria. Analytical Chemistry 2019, 91 (11), 7254–7265. [DOI] [PubMed] [Google Scholar]
- 35.Li Q; Kailemia MJ; Merleev AA; Xu G; Serie D; Danan LM; Haj FG; Maverakis E; Lebrilla CB, Site-Specific Glycosylation Quantitation of 50 Serum Glycoproteins Enhanced by Predictive Glycopeptidomics for Improved Disease Biomarker Discovery. Analytical Chemistry 2019, 91 (8), 5433–5445. [DOI] [PubMed] [Google Scholar]
- 36.Xiong Y; Karuppanan K; Bernardi A; Li Q; Kommineni V; Lebrilla CB; Dandekar AM; Faller R; McDonald KA; Nandi S, Effects of N-glycosylation on the structure, function, and stability of a plant-made Fc-fusion anthrax decoy protein. Frontiers in Plant Science 2019, 10, 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin H; Chen Y; Mao J; Cheng K; Sun D; Dong M; Wang L; Wang L; Ye M, Proteomics analysis of site-specific glycoforms by a virtual multistage mass spectrometry method. Analytica Chimica Acta 2019, 1070, 60–68. [DOI] [PubMed] [Google Scholar]
- 38.Go EP; Moon H-J; Mure M; Desaire H, Recombinant Human Lysyl Oxidase-like 2 Secreted from Human Embryonic Kidney Cells Displays Complex and Acidic Glycans at All Three N-Linked Glycosylation Sites. Journal of Proteome Research 2018, 17 (5), 1826–1832. [DOI] [PubMed] [Google Scholar]
- 39.Saunders KO; Nicely NI; Wiehe K; Bonsignori M; Meyerhoff RR; Parks R; Walkowicz WE; Aussedat B; Wu NR; Cai F; Vohra Y; Park PK; Eaton A; Go EP; Sutherland LL; Scearce RM; Barouch DH; Zhang R; Von Holle T; Overman RG; Anasti K; Sanders RW; Moody MA; Kepler TB; Korber B; Desaire H; Santra S; Letvin NL; Nabel GJ; Montefiori DC; Tomaras GD; Liao H-X; Alam SM; Danishefsky SJ; Haynes BF, Vaccine Elicitation of High Mannose-Dependent Neutralizing Antibodies against the V3-Glycan Broadly Neutralizing Epitope in Nonhuman Primates. Cell Reports 2017, 18 (9), 2175–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Y; Li Z; Felder MA; Yu Q; Shi X; Peng Y; Cao Q; Wang B; Puglielli L; Patankar MS; Li L, Mass Spectrometry Imaging of N-Glycans from Formalin-Fixed Paraffin-Embedded Tissue Sections Using a Novel Subatmospheric Pressure Ionization Source. Analytical Chemistry 2019, 91 (20), 12942–12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li G; Ma F; Cao Q; Zheng Z; DeLaney K; Liu R; Li L, Nanosecond photochemically promoted click chemistry for enhanced neuropeptide visualization and rapid protein labeling. Nature Communications 2019, 10 (1), 4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Craig AG; Bandyopadhyay P; Olivera BM, Post-translationally modified neuropeptides from Conus venoms. European Journal of Biochemistry 1999, 264 (2), 271–275. [DOI] [PubMed] [Google Scholar]
- 43.Gerwig G; Hocking H; Stöcklin R; Kamerling J; Boelens R, Glycosylation of conotoxins. Marine drugs 2013, 11 (3), 623–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dalpathado DS; Desaire H, Glycopeptide analysis by mass spectrometry. Analyst 2008, 133 (6), 731–738. [DOI] [PubMed] [Google Scholar]
- 45.Yu Q; Wang B; Chen Z; Urabe G; Glover MS; Shi X; Guo L-W; Kent KC; Li L, Electron-transfer/higher-energy collision dissociation (EThcD)-enabled intact glycopeptide/glycoproteome characterization. Journal of the American Society for Mass Spectrometry 2017, 28 (9), 1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Z; Yu Q; Hao L; Liu F; Johnson J; Tian Z; Kao WJ; Xu W; Li L, Site-specific characterization and quantitation of N-glycopeptides in PKM2 knockout breast cancer cells using DiLeu isobaric tags enabled by electron-transfer/higher-energy collision dissociation (EThcD). Analyst 2018, 143 (11), 2508–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glover MS; Yu Q; Chen Z; Shi X; Kent KC; Li L, Characterization of intact sialylated glycopeptides and phosphorylated glycopeptides from IMAC enriched samples by EThcD fragmentation: Toward combining phosphoproteomics and glycoproteomics. International Journal of Mass Spectrometry 2018, 427, 35–42. [Google Scholar]
- 48.Zhu J; Chen Z; Zhang J; An M; Wu J; Yu Q; Skilton SJ; Bern M; Ilker Sen K; Li L, Differential Quantitative Determination of Site-Specific Intact N-Glycopeptides in Serum Haptoglobin between Hepatocellular Carcinoma and Cirrhosis Using LC-EThcD-MS/MS. Journal of proteome research 2018, 18 (1), 359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y; Liu F; Franc V; Halim LA; Schellekens H; Heck AJR, Hybrid mass spectrometry approaches in glycoprotein analysis and their usage in scoring biosimilarity. Nature Communications 2016, 7, 13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reiding KR; Bondt A; Franc V; Heck AJR, The benefits of hybrid fragmentation methods for glycoproteomics. TrAC Trends in Analytical Chemistry 2018, 108, 260–268. [Google Scholar]
- 51.Frese CK; Altelaar AFM; van den Toorn H; Nolting D; Griep-Raming J; Heck AJR; Mohammed S, Toward Full Peptide Sequence Coverage by Dual Fragmentation Combining Electron-Transfer and Higher-Energy Collision Dissociation Tandem Mass Spectrometry. Analytical Chemistry 2012, 84 (22), 9668–9673. [DOI] [PubMed] [Google Scholar]
- 52.Zhang H; Li X.-j.; Martin DB; Aebersold R, Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nature biotechnology 2003, 21 (6), 660. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y; Wu S.-l.; Hancock WS, Approaches to the study of N-linked glycoproteins in human plasma using lectin affinity chromatography and nano-HPLC coupled to electrospray linear ion trap—Fourier transform mass spectrometry. Glycobiology 2006, 16 (6), 514–523. [DOI] [PubMed] [Google Scholar]
- 54.Hägglund P; Bunkenborg J; Elortza F; Jensen ON; Roepstorff P, A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. Journal of proteome research 2004, 3 (3), 556–566. [DOI] [PubMed] [Google Scholar]
- 55.Ma S; Zhang L; Wang S; Zhang H; You X; Ou J; Ye M; Wei Y, Preparation of epoxy-functionalized hierarchically porous hybrid monoliths via free radical polymerization and application in HILIC enrichment of glycopeptides. Analytica Chimica Acta 2019, 1058, 97–106. [DOI] [PubMed] [Google Scholar]
- 56.Zhang L; Ma S; Chen Y; Wang Y; Ou J; Uyama H; Ye M, Facile Fabrication of Biomimetic Chitosan Membrane with Honeycomb-Like Structure for Enrichment of Glycosylated Peptides. Analytical Chemistry 2019, 91 (4), 2985–2993. [DOI] [PubMed] [Google Scholar]
- 57.Xu Y; Wu Z; Zhang L; Lu H; Yang P; Webley PA; Zhao D, Highly specific enrichment of glycopeptides using boronic acid-functionalized mesoporous silica. Analytical chemistry 2008, 81 (1), 503–508. [DOI] [PubMed] [Google Scholar]
- 58.Zhang L; Xu Y; Yao H; Xie L; Yao J; Lu H; Yang P, Boronic Acid Functionalized Core–Satellite Composite Nanoparticles for Advanced Enrichment of Glycopeptides and Glycoproteins. Chemistry – A European Journal 2009, 15 (39), 10158–10166. [DOI] [PubMed] [Google Scholar]
- 59.Xu Y; Zhang L; Lu H; Yang P, On-plate enrichment of glycopeptides by using boronic acid functionalized gold-coated Si wafer. PROTEOMICS 2010, 10 (5), 1079–1086. [DOI] [PubMed] [Google Scholar]
- 60.Larsen MR; Jensen SS; Jakobsen LA; Heegaard NH, Exploring the sialiome using titanium dioxide chromatography and mass spectrometry. Molecular & cellular proteomics 2007, 6 (10), 1778–1787. [DOI] [PubMed] [Google Scholar]
- 61.Pap A; Medzihradszky KF; Darula Z, Using “spectral families” to assess the reproducibility of glycopeptide enrichment: human serum O-glycosylation revisited. Analytical and Bioanalytical Chemistry 2017, 409 (2), 539–550. [DOI] [PubMed] [Google Scholar]
- 62.Jebanathirajah J; Steen H; Roepstorff P, Using optimized collision energies and high resolution, high accuracy fragment ion selection to improve glycopeptide detection by precursor ion scanning. Journal of the American Society for Mass Spectrometry 2003, 14 (7), 777–784. [DOI] [PubMed] [Google Scholar]
- 63.Saba J; Dutta S; Hemenway E; Viner R, Increasing the productivity of glycopeptides analysis by using higher-energy collision dissociation-accurate mass-product-dependent electron transfer dissociation. International journal of proteomics 2012, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mechref Y, Use of CID/ETD mass spectrometry to analyze glycopeptides. Current protocols in protein science 2012, 68 (1), 12.11. 1–12.11. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh C; Zampronio CG; Creese AJ; Cooper HJ, Higher energy collision dissociation (HCD) product ion-triggered electron transfer dissociation (ETD) mass spectrometry for the analysis of N-linked glycoproteins. Journal of proteome research 2012, 11 (9), 4517–4525. [DOI] [PubMed] [Google Scholar]
- 66.Parker BL; Thaysen-Andersen M; Solis N; Scott NE; Larsen MR; Graham ME; Packer NH; Cordwell SJ, Site-specific glycan-peptide analysis for determination of N-glycoproteome heterogeneity. Journal of proteome research 2013, 12 (12), 5791–5800. [DOI] [PubMed] [Google Scholar]
- 67.Kutz KK; Schmidt JJ; Li L, In situ tissue analysis of neuropeptides by MALDI FTMS in-cell accumulation. Analytical chemistry 2004, 76 (19), 5630–5640. [DOI] [PubMed] [Google Scholar]
- 68.Sturm RM; Greer T; Woodards N; Gemperline E; Li L, Mass spectrometric evaluation of neuropeptidomic profiles upon heat stabilization treatment of neuroendocrine tissues in crustaceans. Journal of proteome research 2013, 12 (2), 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Christie AE; Stemmler EA; Dickinson PS, Crustacean neuropeptides. Cellular and Molecular Life Sciences 2010, 67 (24), 4135–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma B; Zhang K; Hendrie C; Liang C; Li M; Doherty‐Kirby A; Lajoie G, PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid communications in mass spectrometry 2003, 17 (20), 2337–2342. [DOI] [PubMed] [Google Scholar]
- 71.Zhang J; Xin L; Shan B; Chen W; Xie M; Yuen D; Zhang W; Zhang Z; Lajoie GA; Ma B, PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Molecular & Cellular Proteomics 2012, 11 (4), M111. 010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Halim A; Westerlind U; Pett C; Schorlemer M; Rüetschi U; Brinkmalm G; Sihlbom C; Lengqvist J; Larson G; Nilsson J, Assignment of Saccharide Identities through Analysis of Oxonium Ion Fragmentation Profiles in LC–MS/MS of Glycopeptides. Journal of Proteome Research 2014, 13 (12), 6024–6032. [DOI] [PubMed] [Google Scholar]
- 73.Campbell MP; Hayes CA; Struwe WB; Wilkins MR; Aoki-Kinoshita KF; Harvey DJ; Rudd PM; Kolarich D; Lisacek F; Karlsson NG; Packer NH, UniCarbKB: Putting the pieces together for glycomics research. PROTEOMICS 2011, 11 (21), 4117–4121. [DOI] [PubMed] [Google Scholar]
- 74.Perez-Riverol Y; Csordas A; Bai J; Bernal-Llinares M; Hewapathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; Pérez E; Uszkoreit J; Pfeuffer J; Sachsenberg T; Yılmaz Ş; Tiwary S; Cox J; Audain E; Walzer M; Jarnuczak AF; Ternent T; Brazma A; Vizcaíno JA, The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Research 2018, 47 (D1), D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S17. EThcD spectra of crustacean neuropeptides modified with glycans discovered from blue crab Callinectes sapidus nervous system.
Figure S18. EThcD spectrum of a crustacean neuropeptide modified with glycans discovered from rock crab Cancer irroratus nervous system.
Figures S19-S20. MS/MS spectra of two confidently identified neuropeptides without glycan attached.
Figures S21-S22. EIC comparison of glycosylated and non-glycosylated forms of the peptide.
Table S1. Putative novel neuropeptide sequences identified using HCD via de novo peptide sequencing.


