Recent studies highlight the importance of mucins, in particular Muc2, in intestinal homeostasis. Our functional study demonstrates that an alternative secreted mucin, MUC5AC/Muc5ac, is induced in colitis to protect the colonic barrier by limiting host-bacterial interaction.
Keywords: ulcerative colitis, epithelium, mucin, bacteria
Abstract
Background
The mucus gel layer (MGL) lining the colon is integral to exclusion of bacteria and maintaining intestinal homeostasis in health and disease. Some MGL defects allowing bacteria to directly contact the colonic surface are commonly observed in ulcerative colitis (UC). The major macromolecular component of the colonic MGL is the secreted gel-forming mucin MUC2, whose expression is essential for homeostasis in health. In UC, another gel-forming mucin, MUC5AC, is induced. In mice, Muc5ac is protective during intestinal helminth infection. Here we tested the expression and functional role of MUC5AC/Muc5ac in UC biopsies and murine colitis.
Methods
We measured MUC5AC/Muc5ac expression in UC biopsies and in dextran sulfate sodium (DSS) colitis. We performed DSS colitis in mice deficient in Muc5ac (Muc5ac-/-) to model the potential functional role of Muc5ac in colitis. To assess MGL integrity, we quantified bacterial-epithelial interaction and translocation to mesenteric lymph nodes. Antibiotic treatment and 16S rRNA gene sequencing were performed to directly investigate the role of bacteria in murine colitis.
Results
Colonic MUC5AC/Muc5ac mRNA expression increased significantly in active UC and murine colitis. Muc5ac-/- mice experienced worsened injury and inflammation in DSS colitis compared with control mice. This result was associated with increased bacterial-epithelial contact and translocation to the mesenteric lymph nodes. However, no change in microbial abundance or community composition was noted. Antibiotic treatment normalized colitis severity in Muc5ac-/- mice to that of antibiotic-treated control mice.
Conclusions
MUC5AC/Muc5ac induction in the acutely inflamed colon controls injury by reducing bacterial breach of the MGL.
INTRODUCTION
The mucus gel layer (MGL) lines the epithelial surface of the gastrointestinal tract, where it has a myriad of physiologic functions. The MGL is both a physical and a chemical barrier, controlling the passage of nutrients and regulating host interactions with the intestinal luminal environment.1, 2 Mucins are the major protein constituent of mucus and are broadly classified into 2 groups: transmembrane and secreted mucins. Transmembrane mucins are tethered to the intestinal epithelium to form the glycocalyx, and secreted gel-forming mucins are predominantly released by goblet cells.3 In the healthy small and large intestine the secreted gel-forming mucin MUC2 (human)/Muc2 (murine) is the predominant mucin expressed.4, 5 MUC2/Muc2 is continuously secreted by goblet cells in the intestine, where it incorporates water to form the MGL that lines the intestinal epithelium. In the colon, the MGL consists of an inner adherent layer devoid of intestinal bacteria and an outer loosely adherent layer in which intestinal bacteria reside.5 Mouse models have illustrated the importance of an intact MGL to intestinal homeostasis, with the deletion of Muc2 resulting in perturbation of the normal MGL and spontaneous colitis with progression to colon cancer in these mice.6, 7 Indeed, it has been known for some time that the MGL is thinner in ulcerative colitis (UC), with a reduction in MGL thickness correlating with more severe colonic inflammation.8-11 Furthermore, reduced thickness of the MGL in UC is associated with increased bacterial interaction with the colonic mucosal surface.8, 12 These findings suggest that perturbations in the MGL may lead to intestinal inflammation as observed in UC.
Although MUC2 is the predominant secreted mucin in the human colon,4, 5, 13 studies using UC biopsies have been inconsistent as to the expression levels of MUC2 in UC. Indeed, MUC2 mRNA has been shown to be unchanged,14-17 increased,18 or decreased19 in UC, depending on the study. Greater consensus has existed in analyses of MUC2 protein in UC biopsies, with studies showing decreased MUC2 levels in UC. However, whether these results are because of decreased MUC2 synthesis or sulfation is not clear.15, 17, 20-23 Whereas the exact expression pattern of MUC2 in UC appears uncertain, the expression of an alternative secreted mucin, MUC5AC (human), has been consistently observed to be increased during active inflammation in UC.16, 18, 20, 24 Moreover, recent RNA seq16 and microarray data18 have shown that MUC5AC expression is significantly increased in treatment-naive UC patients and that reduced MUC5AC expression is associated with endoscopic improvement in UC.25 These studies suggested that MUC5AC expression is associated with colonic inflammation. However, whether the induction of MUC5AC in UC plays a functional role in colitis is not yet known.
Although its role in UC is unknown, previous studies in mice have shown a protective role for Muc5ac (murine) in helminth infection, suggesting that Muc5ac expression in the gastrointestinal tract is a tissue-protective response.26 Given these findings, our study sought to investigate if MUC5AC expression during colitis may protect the colon during tissue inflammation as observed in UC. We used expression analysis in active UC patient biopsies and experimental murine colitis to determine the expression pattern of MUC5AC/Muc5ac during intestinal inflammation. Mice deficient in Muc5ac (Muc5ac-/-) were used in a model of experimental colitis to test the potential functional role of this mucin in disease. Our study demonstrates for the first time that an alternative secreted mucin plays a protective role in colonic inflammation as observed in UC.
MATERIALS AND METHODS
Ethical Statement
Patient biopsies were harvested at the time of colonoscopy under a biobank protocol approved by the Colorado Multiple Institutional Review Board (COMIRB #14–2012). All human patients provided informed consent to participate. All murine experiments were performed in accordance with protocols approved by the University of Colorado Denver Institutional Animal Care and Use Committee, reference numbers B97511(10)1D, B97513(10)1D, and B97516(10)1E.
Human Biopsies and Reverse Transcription-Polymerase Chain Reaction
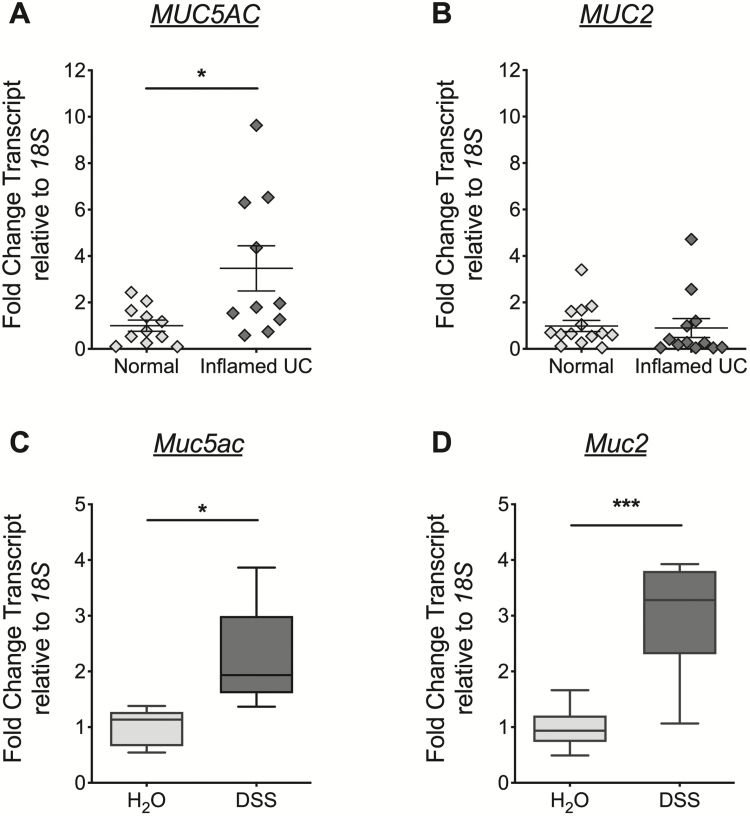
Colonic biopsies were harvested at the time of colonoscopy under a biobank protocol approved by the Colorado Multiple Institutional Review Board (COMIRB #14–2012). All human patients provided informed consent to participate. Biopsies were taken from healthy areas of colonic mucosa at the time of colonoscopy from healthy patients undergoing colon cancer screening (normal). Biopsies of inflamed colonic mucosa were taken at the time of colonoscopy from patients with UC undergoing standard-of-care endoscopic evaluation. Alternative causes for active colitis were excluded. The RNA was extracted from frozen tissue biopsies by phenol:chloroform extraction followed by RNA elution using an RNeasy minElute column (Qiagen, Thermo Fisher Scientific, Waltham, MA). The cDNA was synthesized as previously described,27, 28 and reverse transcription-polymerase chain reaction (RT-PCR) was performed using specific primers for MUC5AC, MUC2, and 18S. Relative change in the gene of interest was calculated by the 2-ΔΔCT method using 18S as the control gene. While the same number of control patient and UC samples were tested for 18S, MUC5AC, and MUC2 expression, the RT-PCR failed to detect the control gene and/or the mucin gene in some samples, likely because of a technical failure. Consequently, there are a different number of data points for each patient group in each graph (Figs. 1A, B).
FIGURE 1.
MUC5AC/Muc5ac gene expression increased at the mucosal surface in active UC and in murine colitis. A and B, Colonic mucosal biopsies collected from healthy control patients (normal) or from sites of endoscopically visible inflammation in patients with UC (inflamed UC). Total RNA extracted and Taqman RT-PCR for MUC5AC (A) and MUC2 (B) performed. Transcript levels calculated relative to 18S and expressed as fold change compared to normal. Data displayed as mean ± SEM. Results represent n = 11 to 14 normal samples and n = 10 to 12 inflamed UC. Statistical difference assessed by 2-tailed Mann-Whitney U test. C and D, Administered DSS (3%) or water to C57 Black 6 mice. After 6 days, mucosal scrapings from the distal colon harvested and total RNA extracted. TaqMan RT-PCR for Muc5ac (C), Muc2 (D), and 18S performed. Transcript levels calculated relative to 18S and expressed as fold change compared to water control. Data displayed as mean ± SEM. Results represent n = 5 to 8 mice per group. Statistical differences calculated using 2-tailed unpaired Student t test. SEM indicates standard error of the mean. *P< 0.05, ***P< 0.0001.
Experimental Animals
All experimental animals were bred and housed at the University of Colorado Anschutz Medical Campus under institutional approved protocols. Muc5ac-/- mice were obtained from Dr. Christopher Evans (University of Colorado, Denver) and crossed in-house with C57 Black 6 mice (Jackson Laboratory, Bar Harbor, ME). Genotyping PCR performed on tails was used to determine the specific deletion of Muc5ac (GeneTyper, Inc., New York, NY).
Dextran Sulfate Sodium Colitis
In the majority of colitis studies that were performed, co-housed, male and female, adult Muc5ac-/- mice and their wildtype controls, of similar age and weight were used. For studies where 16S rRNA gene sequencing was planned (microbiome analysis method described below), male and female adult Muc5ac-/- mice and their littermate control mice bred using a heterozygous breeding strategy were used. We added DSS (40,000–50,000 molecular weight; Affymetrix, Cleveland, OH) to drinking water for 6 days followed by a return to normal drinking water for 1 or 3 days (3%-4.5% DSS; details on DSS percentage used given in each figure legend within the manuscript). Mice were returned to normal drinking water for 3 days in some studies to allow for successful performance of 16S RT-PCR (further details in 16S RT-PCR method section). Research has shown that DSS inhibits the polymerase chain reaction.29 Our pilot studies (not shown) demonstrated that 3 days of normal drinking water post-DSS was optimal to allow for washout of DSS and performance of reliable 16S RT-PCR. Therefore, in studies where tissue collection for 16S RT-PCR and sequencing were planned, this colitis protocol was used.
Body weight was monitored as described in earlier research,27, 28, 30 with changes in body weight calculated relative to initial body weight. Postmortem whole colons and in some cases intact mesenteric lymph nodes (MLN) or ceca were harvested by blunt dissection and colon length measured. For expression studies, strips of whole colon or scrapings from the proximal or distal colon were snap-frozen in liquid nitrogen for future processing. Serial segments of transverse colon were fixed in 10% buffered-formalin (Sigma Aldrich, St. Louis, MI) before staining of paraffin sections with hemotoxylin and eosin and histological scoring by a board-certified pathologist blinded to the study. The histological scoring system used has been described in earlier work.27, 28, 30
The inflammation index gave 0 points for no lamina propria inflammation, 1 point for increased lamina propria inflammatory cells, 2 points for confluence of inflammatory cells extending into the submucosa, and 3 points for transmural inflammation. The injury index gave 0 points for no crypt damage, 1 point for partial (up to 50%) crypt dropout, 2 points for partial to complete (50%-100%) crypt dropout, and 3 points for complete (100%) crypt dropout. The score from each category was then multiplied by 1 for 1% to 25% of intestinal length involved, 2 for 25 to 50% of intestinal length involved, 3 for 50% to 75% of intestinal length involved, or 4 for 75% to 100% of intestinal length involved. It was also noted whether the section was from the proximal or the distal colon. The resulting products for each category (injury and inflammation) were then added for an overall score (total) of 0 to 24 for each mouse. Data were displayed as the total histological score, which represented the summation of the injury and inflammation scores from all sections scored, both proximal and distal, and was therefore a histological score representing the whole colon. Representative images were selected based on histologic scores. Images were acquired at 10x using an Olympus BX51 with an Olympus DP72 camera and cellSens imaging software (V1.6).
Mouse RT-PCR
We performed RNA isolation, cDNA synthesis, and RT-PCR from whole colonic tissue as described in earlier research.27, 28 Specific primers for Muc2, Muc5ac, Muc5b, Muc6, trefoil factor 3, and the internal control 18S (Taqman, Thermo Fisher Scientific, Waltham, MA) were used. Relative change in the gene of interest was calculated by the 2-ΔΔCT method using 18S as the control gene.
Tissue Cytokine Measurements
Following DSS or water administration, flash-frozen whole colon was homogenized in Tris lysis buffer containing a protease and phosphatase inhibitor cocktail (Thermo Fisher), using a handheld tissue homogenizer. The concentration of cytokines in tissue lysates was measured using the V-Plex Pro-Inflammatory Panel Mouse kit (Meso Scale Discovery, Rockville, MD) per the manufacturer’s instructions. Signal detection was performed using a Sector Imager 2400 (Meso Scale Discovery). A Bicinchoninic Acid (BCA) Protein Assay Kit (Thermo Fisher) was used to determine the amount of protein loaded for each sample. Cytokine concentrations in tissue homogenate were normalized to total protein.
Flow Cytometry
Colons were excised, opened longitudinally, washed with cold phosphate-buffered saline (PBS), and then incubated with intraepithelial leukocyte solution to remove epithelial cells (PBS, 15 mmol/L HEPES, and 1 mmol/L EDTA) as described elsewhere.28, 31 Tissues were cut into ∼1 cm pieces and enzymatically digested (Roswell Park Memorial Institute medium, 2% fetal bovine serum, 15 mmol/L HEPES, 200 U/mL collagenase VIII [Sigma-Aldrich], and 1 µg/mL deoxyribonuclease [Thermo Fisher]) at 37°C with gentle agitation for ~20 minutes before being passed through a 100 µm filter. The resulting cell suspension was washed twice with cold Roswell Park Memorial Institute medium/10% fetal bovine serum, and FC receptors were blocked with incubation with an anti-CD16/32 antibody (eBioscience, San Diego, CA). Cells from indicated compartments were then incubated with fluorescently labeled antimouse antibodies against MHCII (M5/114.15.2), CD11b (M1/70), CD11c (N418), Ly6G (1A8), Ly6C (HK1.4), SiglecF (12-1702-82), and CD45 (30-F11) (eBioscience, San Diego, CA) or corresponding isotype controls. Dead cells were excluded using a Live/Dead Fixable Aqua Stain (Invitrogen, Carlsbad, CA), and flow cytometry was performed using a BD FACSCanto II (BD) and analyzed using FLOWJo software (Tree Star, Ashland, OR). The percentage of live total cells of each population was calculated. Cell populations were defined as follows: neutrophils, % live CD45.2+ MHCIINeg SiglecFNeg Ly6G+ cells; eosinophils, % live CD45.2+ MHCIINeg Ly6GNeg SiglecF+ cells; dendritic cells, % live CD45.2+ Ly6GNeg SiglecFNeg MHCII+ CD11c+ cells; resident macrophages, % live CD45.2+ Ly6GNeg SiglecFNeg MHCII+ Ly6CNeg cells; monocytes, % live CD45.2+ Ly6GNeg SiglecFNeg MHCIINeg Ly6C+ cells; and differentiating monocytes, % live CD45.2+ Ly6GNeg SiglecFNeg MHCII+ Ly6C+ cells.
16S Fluorescence In Situ Hybridization
We administered DSS to Muc5ac-/- mice and their wildtype control mice for 6 days before withdrawal of DSS and administration of normal drinking water for 3 days. Mice were sacrificed and intact colon was fixed overnight in 60% methanol (Thermo Fisher), 30% chloroform (MP Biomedicals, Santa Ana, CA), and 10% glacial acetic acid (Thermo Fisher), followed by 100% methanol (Thermo Fisher) before embedding in paraffin. Transverse sections of tissue were baked onto glass slides and deparaffinized with xylene. Slides were moved through a descending series of ethanol/water baths to rehydrate. Cross-linking was achieved by incubation of slides under an ultraviolet light for 20 minutes while submerged in a PBS bath. Tissues were incubated overnight with a fluorescent 16S eubacterial probe (5 ng/uL, GCTGCCTCCCGTAGGAGT with a 5’ ALEXA 555 modification) in a hybridization chamber with humidity. Slides were washed in PBS 3 times for 10 minutes each. Tissue was counterstained with 4′,6-diamidino-2-phenylindole (1:50,000, MP Biomedicals) for 5 minutes. Slides were washed with PBS 3 times for 5 minutes and then with deionized water for 5 minutes. Tissues were dehydrated and mounted in ProLong Gold Antifade (Thermo Fisher Scientific). Representative images were acquired at 40x using an Olympus BX51 with an Olympus DP72 camera and cellSens imaging software (V1.6). To determine bacterial contact with the intestinal epithelium at least 3 images positive for 16S staining were acquired for each mouse. The percentage of epithelium in contact with bacteria was determined by measuring the total length of the intestinal epithelium and the length of epithelium in contact with bacteria in each image.
16S RT-PCR
We administered DSS to Muc5ac-/- mice and their wildtype control mice for 6 days before withdrawal of DSS and administration of normal drinking water for 3 days. Mice were sacrificed, and intact MLN were snap-frozen in liquid nitrogen. Total DNA was isolated using the QIAamp PowerFecal DNA kit (Qiagen, Venlo, Netherlands). We performed RT-PCR of the 16S rRNA gene with a custom 16S primer mix (forward: TCCTACGGAGGCAGCAGT and reverse: GACTACCAGGGTATCTAATCCTGTT, Invitrogen) and the internal control 18S (QuantiTect Primer assays, Qiagen) using the Power SYBR Green PCR Master Mix (Thermo Scientific, Warrington, United Kingdom). Relative change in 16S was calculated by the 2-ΔΔCT method using 18S as the control gene.
Antibiotic Administration
An antibiotic cocktail of ampicillin (1 mg/mL, Sigma-Aldrich), gentamicin (1 mg/mL, Sigma-Aldrich), metronizadole (1 mg/mL, Sigma-Aldrich), neomycin (1 mg/mL, Sigma-Aldrich), and vancomycin (0.5 mg/mL, Sigma-Aldrich), or vehicle (HyPure Cell Culture Grade Water, HyClone, Logan, UT), was administered daily via oral gavage to Muc5ac-/- mice and their wildtype control mice for 10 days before delivery of DSS or normal drinking water and for 5 days during DSS.32 Changes in body weight were assessed and tissue collected following the sacrifice of animals on day 7 post-DSS as described in the Dextran Sulfate Sodium Colitis method section above.
Microbiome Analysis
We administered DSS or normal drinking water to Muc5ac-/- mice and their wildtype littermate control mice for 6 days. We replaced DSS with normal drinking water for 3 days, after which mice were sacrificed. As discussed in the Dextran Sulfate Sodium Colitis method section above, a 3-day washout period was needed post-DSS to limit the possibility that DSS might interfere with the planned PCR-based study. Mice were weighed regularly. Given the influence of parentage on the microbiome,33 mice were taken from multiple separate breeding pairs. Following sacrifice, the whole colon was excised, measured, and filleted open. Large fecal matter was gently removed if present, and a brushing of the colonic mucosal surface was performed to test for the mucosally associated microbiome. The DNA was extracted using the QIAamp PowerFecal DNA kit (Qiagen Inc, Carlsbad, CA), which employs the chemical and mechanical disruption of biomass. For sequencing, PCR amplicons were generated using bar-coded primers that targeted approximately 450 base pairs of the V3V4 variable region of the 16S rRNA gene (338F: 5’ACTCCTACGGGAGGCAGCAG and 806R: 5’ GGACTACHVGGGTWTCTAAT), fused to Illumina adapter sequences. Illumina paired-end sequencing was performed following the manufacturer’s protocol on the MiSeq platform using a 600-cycle version 3 reagent kit and version v2.4 of the MiSeq Control Software.
Illumina MiSeq paired-end reads were aligned to the mouse reference genome mm10 with bowtie2,34 and matching sequences were discarded. The remaining paired-end sequences were demultiplexed and then assembled using phrap; 35 read pairs that did not assemble were discarded. Assembled sequence ends were trimmed over a moving window of 5 nucleotides until average quality met or exceeded 20 nucleotides. Trimmed sequences with more than 1 ambiguity or that were shorter than 350 nucleotides were discarded. Potential chimeras identified with Uchime (usearch6.0.203_i86linux32)36 were removed from subsequent analyses. Assembled sequences were aligned and classified with SINA (1.3.0-r23838)37 using the 418,497 bacterial sequences in Silva 115NR9938 as a reference configured to yield the Silva taxonomy. Operational taxonomic units were produced by clustering sequences with identical taxonomic assignments. The software package Explicet (v2.10.5)39 was used for microbial diversity analysis.
Statistics
Data analysis was performed using GraphPad Prism Analysis software (version 6.0). For analysis of patient biopsies, a 2-tailed Mann-Whitney U test was used. When comparing more than 2 groups, a 2-way analysis of variance with a posthoc Bonferroni or Sidak t test was used. When comparing 2 groups, an unpaired 2-tailed Student t test was used. Statistical significance was set at P< 0.05. For 16S rRNA gene sequencing data, nonparametric permutation-based multiple analysis of variance tests of Morisita-Horn dissimilarities were used to compare groups. The specific statistical test used for each experimental analysis is stated in all figure legends throughout the manuscript. Measurements were considered to be outliers if they were greater than 3 standard deviations from the mean and were identified by the Grubbs test.40 These observations were investigated further to ensure the correct data collection, processing, and handling of these samples.
RESULTS
MUC5AC/Muc5ac Expression Is Increased in Active UC and Experimental Colitis
To establish a potential role for MUC5AC in UC, we first measured the expression of MUC5AC mRNA in colonic biopsies taken from the visibly inflamed colon of patients with UC (Fig. 1 and Supplementary Table 1). MUC2 mRNA expression was also determined because it is the major mucin expressed in the colon. In line with previous studies we observed a significant increase in MUC5AC mRNA expression in active UC (inflamed UC) compared to healthy control patient colons (normal, Fig. 1A).16, 18, 20, 24 There was no statistical difference observed in MUC2 expression between inflamed UC and normal colon, which corresponded to findings in the majority of studies in UC.14-17
Analysis of mRNA expression in colonic mucosal samples taken from the actively inflamed colon during experimental (DSS) colitis in mice demonstrated significantly increased Muc5ac expression (Fig. 1C), mirroring our findings in active UC. Notably, Muc2 mRNA was also increased in DSS colitis relative to that in control mice (Fig. 1D), which differed from our findings in active UC. This result suggests a potential difference in the expression pattern of MUC2/Muc2 between active UC and the DSS colitis model. Nonetheless, these findings showed that MUC5AC/Muc5ac expression is induced during colonic inflammation as observed in UC and experimental colitis.
Genetic Deletion of Muc5ac Significantly Exacerbates Experimental Colitis
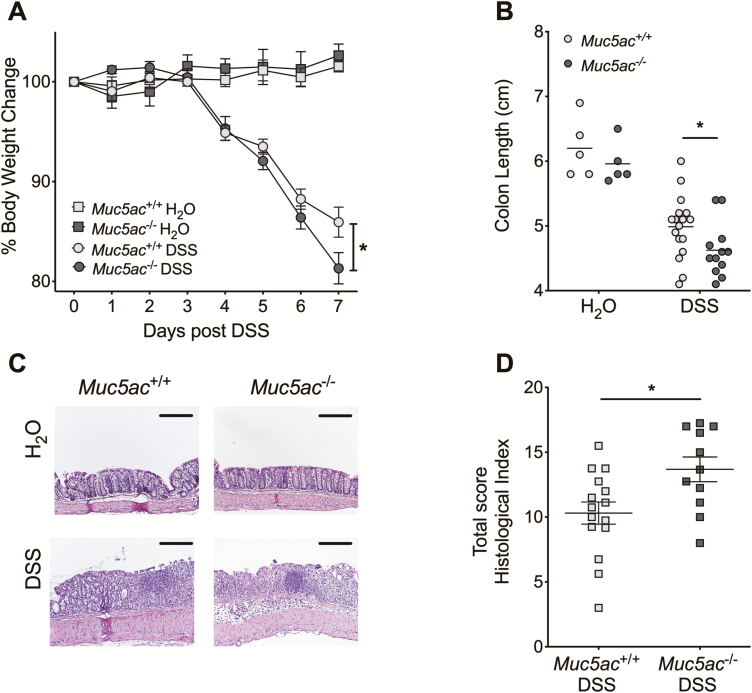
Having observed that MUC5AC/Muc5ac expression in the colon during active UC and experimental colitis was increased (Fig. 1), we utilized mice with genetic deletion of Muc5ac to determine its possible functional role in colitis.26 An initial study with 4.5% DSS resulted in increased mortality in Muc5ac-/- mice compared with control mice (Supplementary Fig. 1). This suggested that Muc5ac may be protective in colitis and prompted us to use a reduced concentration of DSS (3%) in our studies. In these subsequent studies, Muc5ac-/- mice demonstrated greater weight loss, colon shortening, and histologic disease during DSS colitis than did the wildtype control mice (Muc5ac+/+, Figs. 2A-D). Taken together, our data suggest that induction of Muc5ac in the inflamed colon is protective during acute intestinal inflammation as is observed in colitis.
FIGURE 2.
Muc5ac deficiency is detrimental in experimental colitis. Administered DSS (3%) or water to Muc5ac-deficient mice (Muc5ac-/-) and their wildtype control mice (Muc5ac+/+). A, Daily weight measurements displayed as percentage of body weight at day 0. B, On day 7, post-DSS colons harvested and measured. C, Representative histological sections from colon harvested at day 7 post-DSS (bar represents 200 µm; images acquired at 10x). D, Histological analysis of whole colon (proximal and distal combined scores) harvested at day 7 post-DSS provided by pathologist blinded to the groups and the study. Results representative of at least 3 independently performed experiments and displayed as mean ± SEM; n = 5 mice/water group, n = 11 to 17 mice/DSS group. Two-way ANOVA with posthoc Bonferroni t test used to determine statistical weight change; in all other cases unpaired Student t test used. ANOVA indicates analysis of variance. SEM indicates standard error of the mean. *P< 0.05.
To determine the mechanism of Muc5ac-mediated colonic protection, we first asked if the observed exacerbation in intestinal injury and inflammation in Muc5ac-/- mice during DSS coincided with the reduced expression of other gel-forming mucins in these mice (Supplementary Fig. 2). There was no significant decrease in the expression of Muc2, Muc5b, or Muc6 mRNA in the colon of Muc5ac-/- mice compared with that of control mice at baseline (water) or during DSS. In addition, there was no significant difference in the goblet cell marker trefoil factor 3 between the 2 groups, suggesting that greater severity of disease in Muc5ac-/- mice did not result from greater loss of goblet cells. The mRNA expression data were supported by staining for mucins that revealed no major difference in the pattern of Alcian Blue-Periodic acid Schiff positive staining in Muc5ac-/- mice at baseline or during DSS (Supplementary Fig. 3). Therefore, we conclude that the greater severity of DSS colitis in Muc5ac-/- mice does not result from the loss of other gel-forming mucins or a deficit in goblet cells.
Muc2 deficient mice develop spontaneous colitis that is associated with increased colonic epithelial proliferation in the early phase of disease.7 To determine if alterations in colonic epithelial proliferation might be responsible for the increased severity of DSS colitis in Muc5ac-/- mice, we measured the proliferation of E-cadherin-positive intestinal epithelia using Ki67 immunofluorescent staining of colonic tissue at baseline and following DSS (Supplementary Fig. 4). We noted a significant decrease in proliferating cells in the distal colon following DSS, as has been previously described.41 However, we failed to observe any differences in the percentage of proliferating intestinal epithelia in Muc5ac-/- mice compared with Muc5ac+/+ mice in either the distal or proximal colon at baseline or following DSS. Therefore, differences in the proliferative capacity of intestinal epithelial cells does not appear to be responsible for the increased severity of DSS colitis in Muc5ac-/- mice.
Loss of Muc5ac Results in Increased Neutrophil Recruitment and Inflammatory Cytokine Expression in the Injured Colon
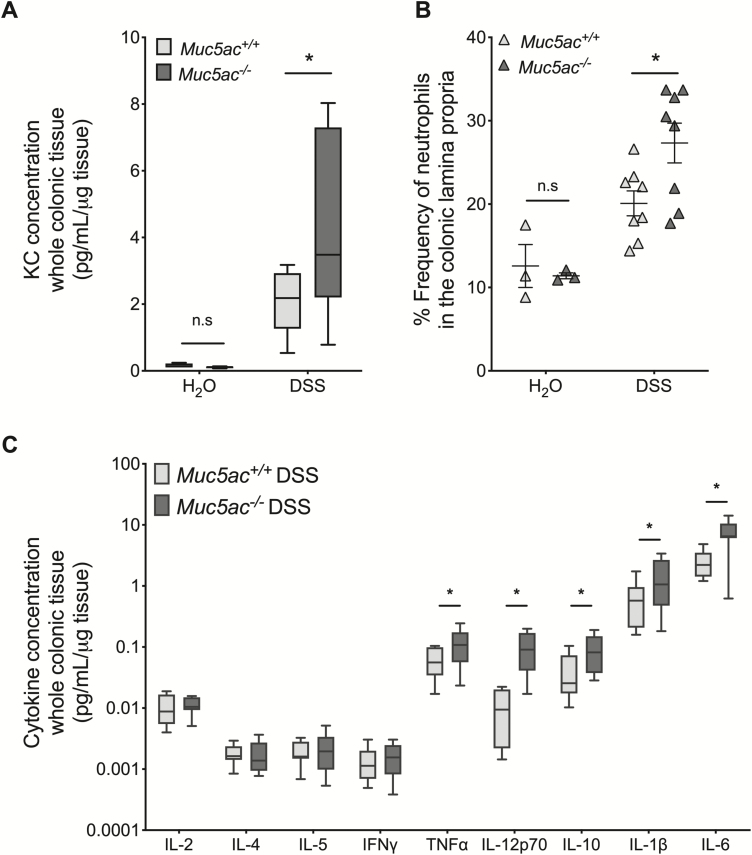
Our previous findings excluded decreased mucin expression (Supplementary Figs. 2, 3) or alterations in intestinal epithelial proliferation (Supplementary Fig. 4) as mechanisms by which the loss of Muc5ac is detrimental in DSS colitis. We therefore undertook studies to quantify colonic inflammation in Muc5ac-/- and Muc5ac+/+ mice at baseline and during DSS colitis (Fig. 3). Acute DSS colitis is characterized by significant increases in colonic levels of chemokines and cytokines, along with substantial infiltration of leukocytes into the colon, in particular an early recruitment of neutrophils.28, 30, 42, 43 Analysis of colonic levels of the neutrophil chemoattractant keratinocyte-derived chemokine (KC) demonstrated a significant increase in the concentration of KC in the colon of Muc5ac-/- mice compared with that of Muc5ac+/+ mice during DSS but not at baseline (Fig. 3A). Flow cytometric analysis of the colonic lamina propria demonstrated an increased frequency of neutrophils in the colon following DSS compared with water control mice, as previously observed (Fig. 3B).42, 44 Note that neutrophil frequency in the colon was not different between Muc5ac-/- and Muc5ac+/+ mice at baseline but was significantly elevated in Muc5ac-/- mice compared with wildtype mice during DSS (Fig. 3B). Furthermore, flow cytometry demonstrated no significant differences in the frequency of eosinophils, dendritic cells, resident macrophages, monocytes, and differentiating monocytes in the colon of Muc5ac-/- mice compared with that of Muc5ac+/+ mice at baseline or during DSS (Supplementary Fig. 5). This result suggests a specific increase in neutrophil recruitment to the colon in Muc5ac-/- mice during DSS. Further analysis of whole colonic tissue revealed a robust increase in the concentration of proinflammatory cytokines known to be elevated in DSS42 in the colon of Muc5ac-/- mice compared to control mice following DSS (TNFα, interleukin [IL]-12p70, IL-10, IL-1β, and IL-6; Fig. 3C). However, no difference in cytokine concentration in the colon was observed between the 2 strains at baseline (Supplementary Fig. 6). Taken together, these data suggest that the loss of Muc5ac results in exacerbated colonic inflammation during DSS colitis.
FIGURE 3.
Loss of Muc5ac results in increased neutrophil frequency and cytokine concentration in the colonic lamina propria during experimental colitis. A and C, Administered DSS (3%) or water to Muc5ac-deficient mice (Muc5ac-/-) and their wildtype control mice (Muc5ac+/+). After sacrifice on day 7, post-DSS whole colon digested for cytokine analysis by Meso Scale. Data displayed relative to protein concentration as determined by BCA. Results represent 1 individual experiment with n = 9 to 13 mice/group. Statistical differences determined by unpaired Student t test. B, Administered DSS (3%) or water to Muc5ac-/- and Muc5ac+/+. Following sacrifice on day 7 or 9, post-DSS whole colon digested for flow cytometric analysis to identify neutrophils (% live CD45.2+ MHCIINeg SiglecFNeg Ly6G+ cells). Results represent 2 independent experiments with n = 3 mice/water group and n = 8 mice/DSS group. Two-way ANOVA with posthoc Bonferroni t test used to determine statistical differences. In all cases, data displayed as mean ± SEM. ANOVA indicates analysis of variance; SEM, standard error of the mean. *P< 0.05.
Host-Bacteria Contact Is Enhanced in Muc5ac-Deficient Mice During Experimental Colitis
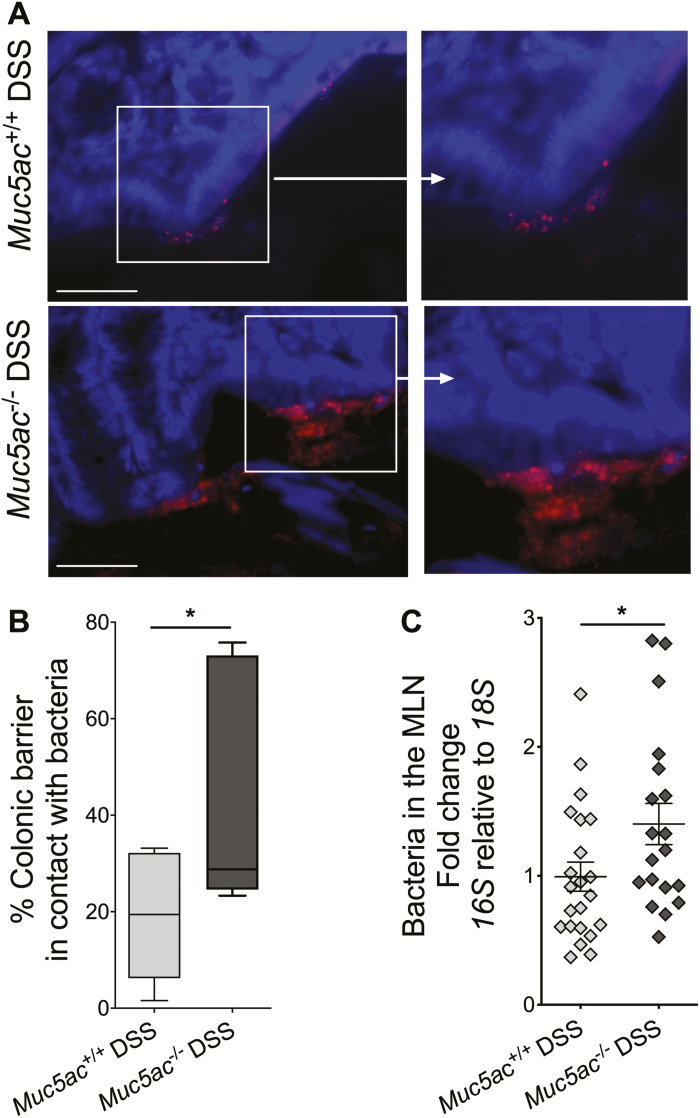
The primary purpose of neutrophil recruitment during inflammation is to aid in bacterial killing and clearance.45 Inhibition of neutrophil recruitment or genetic deletion of neutrophil bacterial killing mechanisms are associated with enhanced disease severity and failure to clear bacteria in murine colitis.46, 47 We hypothesized that greater neutrophil frequency in the colon of Muc5ac-/- mice compared with that in wildtype mice during DSS (Fig. 3B) was because of the increased need for bacterial clearance in Muc5ac-/- mice. Fluorescence in situ hybridization quantification of bacteria in contact with the colonic barrier during DSS showed an approximately 50% increase in the surface area of the colonic barrier in direct contact with bacteria in Muc5ac-/- mice compared with that observed in wildtype control mice (Figs. 4A, B). PCR detection of the 16S rRNA gene showed a robust elevation in bacterial content in the MLN of Muc5ac-/- mice compared with that of Muc5ac+/+ mice during DSS (Fig. 4C). In addition, the 16S rRNA gene was below the level of detection in the MLN of mice on normal drinking water, so we were unable to measure if bacterial translocation was altered in Muc5ac-/- mice at baseline using this technique. These data suggest that loss of Muc5ac is associated with greater bacterial contact with the colonic barrier and translocation of bacteria to the draining lymph nodes during colitis.
FIGURE 4.
Loss of Muc5ac increases bacterial contact with epithelium and translocation to MLN during experimental colitis. Administered DSS (3%) to Muc5ac-deficient mice (Muc5ac-/-) and their wildtype control mice (Muc5ac+/+) for 6 days, followed by water for 3 days. Whole colon and MLN were harvested. A, Performed 16S FISH to identify bacteria (red) in contact with the epithelium. Used 4′,6-diamidino-2-phenylindole as counterstain (blue). Bar represents 50 µm, images acquired at 40x. Images represent n = 7 to 9 mice/group. B, Length of epithelium in contact with bacteria displayed as percentage of total epithelial surface measured in 3 random images acquired from each mouse following 16S FISH as in Fig. 4A; n = 7 to 9 mice/group. C, Used 16S RT-PCR to detect bacterial load in MLN. Normalized PCR data to 18S; n = 19 to 22 mice/group. All results represent 3 independent studies and displayed as mean ± SEM. Two-tailed unpaired Student t test used to determine statistical differences. FISH indicates fluorescence in situ hybridization; SEM, standard error of the mean. *P< 0.05.
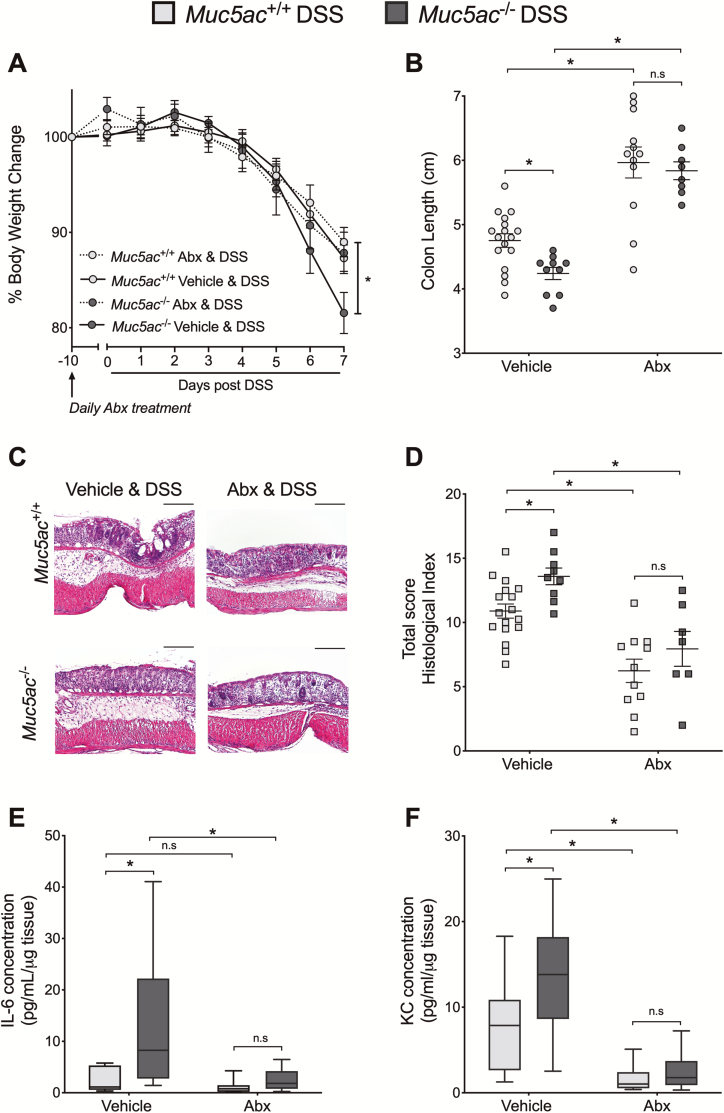
Antibiotic Administration Abrogates Differences Between Muc5ac-Deficient and Wildtype Mice During Colitis
To determine if Muc5ac expression during colitis protects the colon from microbial-mediated inflammation and injury, we depleted the host microbiome by daily antibiotic administration for 10 days before (day -10) and during DSS or water treatment (Fig. 5, Supplementary Fig. 7). Antibiotic administration did not result in any significant differences in body weight change or colon length between Muc5ac-/- and wildtype mice during water treatment (Supplementary Fig. 7). Previous studies have shown that effective antibiotic treatment results in an enlarged cecum.48-50 Cecum weight was measured in a representative group of mice and showed robust enlargement in antibiotic-treated mice compared with vehicle-treated mice during water or DSS exposure (Supplementary Fig. 8), suggesting that antibiotic treatment was effective. A limitation of this study is that we did not attempt to culture live fecal bacteria or determine bacterial load by PCR to confirm bacterial load depletion following antibiotic treatment. However, 16S PCR measurement may be difficult to interpret given that DSS is a known inhibitor of the PCR reaction.29
FIGURE 5.
Antibiotic treatment abrogates differences between Muc5ac-deficient (Muc5ac-/-) and wildtype (Muc5ac+/+) mice during experimental colitis. Muc5ac-/- mice and Muc5ac+/+ mice were gavaged daily with antibiotic cocktail (ampicillin 1 mg/mL, gentamicin 1 mg/mL, metronidazole 1 mg/mL, neomycin 1 mg/mL, and vancomycin 0.5 mg/mL) or vehicle for 10 days before (day -10) administration of DSS (3%), with continued daily delivery of antibiotics or vehicle during DSS. A, Weight measurements obtained for each group of mice and displayed as percentage of body weight at day 0 of antibiotic administration. B, Following sacrifice on day 7, post-DSS colons harvested and measured. C, Representative histological sections from colon harvested at day 7 post-DSS (bar represents 200 µm; images acquired at 10x). D, Histological analysis of whole colon (proximal and distal combined scores) harvested at day 7 post-DSS provided by pathologist blinded to the groups and the study. E and F, Following sacrifice on day 7, post-DSS whole colon digested for cytokine analysis by Meso Scale. Data displayed relative to protein concentration as determined by BCA. Results represent 2 independently performed experiments and displayed as mean ± SEM; n = 7 to 17 mice/group. Two-way ANOVA with posthoc Bonferroni t test used to determine statistical differences. ANOVA indicates analysis of variance; SEM, standard error of the mean. *P< 0.05.
Vehicle-treated Muc5ac-/- mice showed increased severity of DSS colitis compared with wildtype mice, as already observed (Figs. 2, 3C). As previously shown, antibiotic administration to wildtype mice reduced the severity of DSS compared with vehicle treatment as measured by colon length (Fig. 5B), histological inflammation (Fig. 5C, D) and tissue cytokine production (Fig. 5E, F). However, antibiotic administration abolished the differences observed in body weight loss (Fig. 5A), colon length (Fig. 5B), colon histological scores (Fig. 5D) and tissue cytokine production (Fig. 5E, F) following DSS in Muc5ac-/- and Muc5ac+/+ mice. These data reinforce our findings that the loss of Muc5ac is detrimental in DSS colitis (Figs. 2, 3C) and supports a causative role for host bacteria in exacerbated colitis observed in Muc5ac-/- mice.
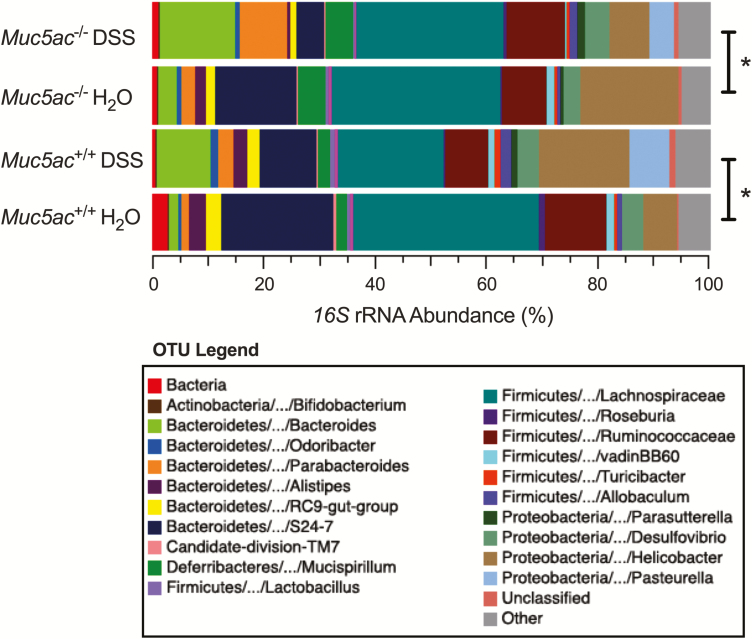
Significant Alteration in 16S rRNA Abundance in the Colonic Mucosa During Colitis in Muc5ac-/- and Muc5ac+/+ Mice
Antibiotic treatment studies (Fig. 5) suggested a causative role for colonic bacteria in exacerbated colitis in Muc5ac-/- mice. Given that bacteria reside in the colonic mucus gel layer5, 12 and that the mucins contained within this layer can support bacterial growth based on their glycan repertoire (reviewed in Schroeder51), we tested the hypothesis that loss of Muc5ac may lead to an altered colonic microbiome that contributes to increased severity of experimental colitis in Muc5ac-/- mice. Experiments were performed using littermate control mice from multiple different breeding pairs to reduce the influence of microbiome heritability as a confounding factor (reviewed in Spor et al33). In addition, 4 males and 4 females were used in each group to account for gender differences.52 We performed 16S rRNA gene sequencing on samples taken from the colonic mucosal surface to determine the composition and diversity of the mucosally associated microbiota. We chose to examine microbiota at the colonic luminal surface because this is where mucins are expressed in the colon. Furthermore, the colonic mucosal microbiota are distinct from fecal microbiota,52, 53 and bacterial dysbiosis at the colonic luminal surface is linked to colitis severity in mice.52
We found that DSS colitis studies revealed increased body weight loss and colon shortening in Muc5ac-/- mice compared with their littermate control mice (Supplementary Fig. 9), confirming earlier studies (Figs. 2, 5). In addition, 16S rRNA gene sequencing revealed a significant difference in the overall bacterial community composition (ie, beta diversity) between water-treated or DSS-treated mice of both genotypes (Fig. 6). An increase in the abundance of Bacteroidetes taxa and a decrease in the abundance of Bacteroidales S24-7 and Firmicutes such as Lachnospiraceae were observed in DSS-exposed Muc5ac-/- and wildtype littermates when compared to their respective water control mice, mirroring previous colitis studies in mice.54-57 These common findings in each genotype suggest that similar alterations to the mucosally resident microbial community occur in both genotypes during DSS. There was no significant difference evident in beta diversity between Muc5ac-/- water-treated mice and their water-treated wildtype littermate control mice. Similarly, no significant difference in beta diversity was observed between Muc5ac-/- DSS-treated mice and their DSS-treated wildtype littermate control mice. These findings suggest that alterations in the composition of the microbial communities resident in the colonic mucosa are not responsible for the increased severity of experimental colitis in Muc5ac-deficient mice.
FIGURE 6.
Colonic mucosal microbiome is altered in Muc5ac-deficient (Muc5ac-/-) and wildtype (Muc5ac+/+) mice during experimental colitis. Administered DSS (3%) or water to Muc5ac-/- and Muc5ac+/+ littermates generated by a heterozygous breeding strategy. On day 6, DSS replaced with normal drinking water. Mice sacrificed after 3 days of normal drinking water (day 9) and whole colon excised. Extracted DNA from the colonic mucosal lining and performed 16S rRNA gene sequencing to profile bacterial taxa in each group. Colored bars represent relative abundance (%) of particular taxa averaged in each group. Data represent >3 independent experiments; n = 8 mice/group. Statistical differences calculated using nonparametric permutation-based multiple analysis of variance tests. *P< 0.05.
DISCUSSION
Depletion and/or disruption of the MGL has long been observed in patients with active UC.8-11 The secreted gel-forming mucin, MUC2/Muc2, is the major constituent of the MGL in the healthy small and large intestine.4, 5 Mice with a genetic deletion of Muc2 demonstrate a reduction in MGL thickness and develop spontaneous colitis that progresses to colon cancer.6, 7 These studies suggest a predominant role for Muc2 in maintaining intestinal homeostasis.4-7 However, during inflammation as observed in active UC an alternative secreted gel-forming mucin, MUC5AC, is expressed in the colonic mucosa. Given the known role of Muc2 in controlling intestinal homeostasis, the reason for the induction of MUC5AC in active UC has not yet been explained. Our study was designed to assess the possible functional role of MUC5AC during intestinal inflammation as observed in UC by using mice with genetic deletion of Muc5ac in a model of experimental colitis. We demonstrate for the first time that Muc5ac expression during active colitis reduces intestinal inflammation and injury to protect the colon.
Initial studies were designed to measure the colonic expression of MUC5AC/Muc5ac in active UC and in experimental colitis. Consistent with previous reports,16, 18, 20, 24 we observed a significant increase in MUC5AC mRNA expression in visibly inflamed tissue from patients with active UC compared with normal control patients (Fig. 1A). These findings were mirrored by studies in experimental colitis (DSS) where we observed a marked increase in Muc5ac mRNA expression in mucosal scrapings of the distal colon during DSS colitis compared with noncolitic (water) controls (Fig. 1C). Based on previous observations,58-60 it is likely that induction of MUC5AC/Muc5ac in the actively inflamed colon during UC or experimental colitis is mediated via proinflammatory cytokines known to be increased in the actively inflamed colon. We conclude from our expression analysis that MUC5AC/Muc5ac colonic mucosal expression is increased during active colitis. In this study, there was no significant correlation between MUC5AC expression levels and disease severity in UC (not shown). In addition, we measured MUC5AC expression in UC biopsies that were not visibly inflamed (not shown). However, MUC5AC was beyond the level of detection in greater than 50% of these noninflamed samples (6 out of 11). These results could suggest that MUC5AC expression is absent or even decreased in noninflamed tissue in UC. In the future, it would be interesting to measure MUC5AC expression in a greater number of samples to determine a potential relationship between MUC5AC expression and disease activity in UC.
Although our expression data confirmed previous observations, the potential functional role for MUC5AC/Muc5ac expression in active colitis is not known. To begin to address a possible functional role for Muc5ac in colitis, we used mice deficient in Muc5ac (Muc5ac-/-) in the DSS experimental model of colitis. Muc5ac-/- mice were observed to have exacerbated intestinal injury and inflammation compared with control mice in DSS colitis. These findings are consistent with previous reports where Muc5ac was induced during intestinal nematode infection and was determined to be critical for expulsion of the nematode.26, 61 We did not note any differences in intestinal inflammation or injury in Muc5ac-/- mice at baseline, which aligns with previous observations.26 Therefore, our data support the previous suggestion that MUC5AC/Muc5ac is not critical for intestinal homeostasis26, 61 but that it is induced as a protective mechanism during intestinal injury, infection, or inflammation.
In determining the mechanisms by which Muc5ac protects the inflamed colon, we considered the mechanism by which experimental colitis is induced and the possible functional role of mucins in the MGL. Studies in DSS colitis,12, 62 the Muc2-deficient12, 63 and IL-10-deficient12 spontaneous models of colitis, and UC biopsies12, 63 showed early and sustained contact of host bacteria with the colonic surface, which is associated with greater severity of tissue inflammation. Neutrophil recruitment is an early event in inflammation in acute experimental colitis28, 42, 43 and is a hallmark of UC.64 The primary purpose of tissue neutrophil recruitment is to aid in bacterial killing and clearance.45 Functional studies have shown that inhibiting steps in this neutrophil cascade can reduce the severity of colitis.46, 47
Overall, the neutrophil recruitment in DSS colitis is consistent with the potentiated activation of innate proinflammatory responses by gut bacteria. Indeed, antibiotic treatment decreases the severity of tissue inflammation in models of colitis,65-67 suggesting that bacteria interaction with the host causes greater tissue injury and inflammation in these models. Furthermore, blocking excessive neutrophil recruitment suppresses colonic inflammation in models of colitis.28, 68-70 Taking these findings into consideration, it could be proposed that increased early and continued interaction of bacteria with the host epithelium and/or mucosal immune system results in excessive neutrophil recruitment, which leads to enhanced tissue cytokine production along with greater tissue inflammation and injury as observed in colitis. Enhancing colonic MGL function could be a mechanism for controlling bacterial-induced neutrophil recruitment to the colon.
Our demonstration that Muc5ac-/- mice have increased bacterial contact with the colonic barrier, a greater concentration of the neutrophil chemoattractant KC, and increased frequency of neutrophils in colonic tissue compared with wildtype control mice during DSS colitis supports the concept that adaptation of an MGL mucin component is a protective mechanism. In addition, because antibiotic treatment normalizes the differences in the outcome of DSS between Muc5ac-/- and control mice, our findings suggest that Muc5ac-mediated modulation of MGL function is driven in part by suppression of bacterial interaction with the host.
The effectiveness of antibiotic treatment in normalizing the colitis phenotype in Muc5ac-/- and control mice prompted us to consider that the loss of Muc5ac may alter microbial communities residing in the gut to promote a colitogenic environment. Given the known importance of environment and genetics on the microbial composition of the gut, we decided to investigate the bacterial taxa resident in the colon of Muc5ac-/- and control mice using a littermate control strategy.33 A limitation of our study was that the majority of experiments were performed with cohoused nonlittermate control mice. Therefore, repetition of our study with littermate control mice addressed this limitation while using the most appropriate strategy to assess microbial abundance at the colonic mucosal surface. Note that Muc5ac-/- mice showed greater body weight loss and colon shortening than their littermate control mice during DSS colitis, supporting our earlier studies. In addition, 16S rRNA gene sequencing did not reveal significant changes in bacterial community composition in the colonic mucosa between Muc5ac-/- and littermate control mice at baseline or during experimental colitis.
These initial findings suggest that differences in the resident bacterial population of the colon between Muc5ac-/- and wildtype control mice are not responsible for increased colitis severity in Muc5ac-/- mice. However, on close inspection these studies present some possibly interesting findings. Indeed, a number of taxa approached significance, with a lower abundance of Roseburia observed in Muc5ac-/- mice than in littermate control mice at baseline and during DSS. Roseburia spp. specifically colonize mucins,71 and a decrease in Roseburia is observed in inflammatory bowel disease.72, 73 In addition, a greater abundance of Mucispirillium was observed in Muc5ac-/- mice compared with littermate control mice at baseline and during DSS. Previous studies linked the presence of Mucispirillium to colitis in genetically susceptible hosts.74, 75 These data did not reach significance, perhaps because of underpowering in the study. These preliminary findings could suggest that the absence of Muc5ac can alter the colonic mucosal microbiome to a more colitogenic environment. Therefore, further studies with greater power may be necessary to address this question.
While our study shows that Muc5ac plays a previously unrecognized role in regulating host-bacteria interactions during colonic inflammation, the question of the exact mechanism by which Muc5ac performs this function remains. Helminth infection studies have shown that Muc5ac expression is increased during infection and that Muc5ac-/- mice fail to effectively clear worms.26, 61 This circumstance was associated with a more porous MGL in the cecum of Muc5ac-/- mice, but it is not yet known if changes in MGL porosity during helminth infection may result from loss of mucin content or a change in the organization of the MGL in Muc5ac-/- mice. Future studies would seek to define the porosity of the MGL in Muc5ac-/- colitic mice. Recent studies have revealed that intestinal mucins have immunomodulatory capabilities through direct signaling on dendritic cells.76, 77 Depending on the context, Muc2 has been suggested to regulate tolerogenic77 or proinflammatory76 responses in dendritic cells. These controversies leave open the question that Muc5ac may induce immunomodulatory responses in leukocytes that are trafficking to the inflamed intestine. While these intriguing questions remain, the fact that an alternative mucin to Muc2 can be induced to protect the colonic barrier supports the importance of maintaining a mucin-mediated functional barrier in the colon.
Taken together, it is clear that additional studies are needed to determine how the biophysical properties of the mucus barrier in the colon are affected by MUC2/Muc2 and MUC5AC/Muc5ac mixtures. At present, there are no specific therapeutic interventions designed to support MGL function in inflammatory bowel disease. The importance of an intact MGL in intestinal homeostasis has been shown in mice with the genetic deletion of Muc2 that develop spontaneous colitis.6, 7 More recently, studies on the impact of a Western-style78 or fiber-free79 diet showed that loss of complex plant fibers leads to bacterial-mediated degradation of the MGL, resulting in enhanced MGL permeability and pathogen invasion. Replacing fiber has been shown to be beneficial to MGL function in these studies; however, they conflicted as to whether MGL penetrability or thickness are affected by fiber supplementation.78, 79 Both of these studies suggested that the presence of fiber in the diet can alter the colonic microbiome to limit MGL depletion. While these 2 studies did not link the expression of MUC5AC/Muc5ac to the beneficial effects of fiber, they provided evidence that supporting MGL function is important for intestinal health.
Our findings suggest that the loss of Muc5ac leads to enhanced bacterial contact with the host. Therefore, we predict that enhancing MUC5AC/Muc5ac expression would be protective during colonic inflammation by supporting MGL function. Future studies will focus on the exact mechanisms to drive MUC5AC/Muc5ac expression as a means to support MGL function in the inflamed intestine.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge technical assistance from Dan Koyanagi, Loni Perrenoud, Melissa Ledezma, Rachael Dran and Tracy Young.
Author contributions: K.O. designed, performed, and analyzed the experiments and cowrote the manuscript. C.R. designed, performed, and analyzed experiments and reviewed the manuscript. L.O.C. cowrote the manuscript. C.B.C. designed, performed, and analyzed experiments and reviewed the manuscript. E.N.M. designed, performed, and analyzed experiments and reviewed the manuscript. O.J. designed, performed, and analyzed experiments and reviewed the manuscript. P.J. provided blinded histological analysis of all tissues and reviewed the manuscript. K.C.A. designed, performed, and analyzed experiments and reviewed the manuscript. M.S.G. designed, performed, and analyzed experiments and reviewed the manuscript. M.E.G. collected patient biopsies, designed experiments, and reviewed the manuscript. D.N.F. designed and analyzed experiments and reviewed the manuscript. D.I. designed, performed, and analyzed experiments and reviewed the manuscript. C.E.R. designed experimental analysis tools and reviewed the manuscript. C.M.E. provided the Muc5ac-deficient mice, advised on experimental design and analysis, and reviewed the manuscript. C.M.A. conceptualized the study; designed, performed, and analyzed the experiments; and cowrote the manuscript.
Supported by: This work was supported by National Institutes of Health (K01-DK099485, R03-DK114545, R01-DK111856 to C.M.A.), (R01-HL130938 and R01-HL080396 to C.M.E.), the Crohn’s and Colitis Foundation (#276536 and #601121 to C.M.A.), and the GI and Liver Innate Immune Program Pilot Award to C.M.A. D.N.F., D.I., and C.E.R. were supported by the GI and Liver Innate Immune Program.
Conflicts of interest: The authors have declared that no conflict of interest exists.
REFERENCES
- 1. Johansson ME, Sjövall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol. 2013;10:352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wagner CE, Wheeler KM, Ribbeck K. Mucins and their role in shaping the functions of mucus barriers. Annu Rev Cell Dev Biol. 2018;34:189–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pelaseyed T, Bergström JH, Gustafsson JK, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260:8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tytgat KM, Büller HA, Opdam FJ, et al. Biosynthesis of human colonic mucin: Muc2 is the prominent secretory mucin. Gastroenterology. 1994;107:1352–1363. [DOI] [PubMed] [Google Scholar]
- 5. Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4659–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Velcich A, Yang W, Heyer J, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–1729. [DOI] [PubMed] [Google Scholar]
- 7. Van der Sluis M, De Koning BA, De Bruijn AC, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. [DOI] [PubMed] [Google Scholar]
- 8. Swidsinski A, Loening-Baucke V, Theissig F, et al. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut. 2007;56:343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fraser GM, Clamp JR. Proceedings: changes in human colonic mucus in ulcerative colitis. Gut. 1975;16:832–833. [PubMed] [Google Scholar]
- 10. Pullan RD, Thomas GA, Rhodes M, et al. Thickness of adherent mucus gel on colonic mucosa in humans and its relevance to colitis. Gut. 1994;35:353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Strugala V, Dettmar PW, Pearson JP. Thickness and continuity of the adherent colonic mucus barrier in active and quiescent ulcerative colitis and Crohn’s disease. Int J Clin Pract. 2008;62:762–769. [DOI] [PubMed] [Google Scholar]
- 12. Johansson ME, Gustafsson JK, Holmén-Larsson J, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tytgat KM, Opdam FJ, Einerhand AW, et al. MUC2 is the prominent colonic mucin expressed in ulcerative colitis. Gut. 1996;38:554–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weiss AA, Babyatsky MW, Ogata S, et al. Expression of MUC2 and MUC3 mRNA in human normal, malignant, and inflammatory intestinal tissues. J Histochem Cytochem. 1996;44:1161–1166. [DOI] [PubMed] [Google Scholar]
- 15. Hanski C, Born M, Foss HD, et al. Defective post-transcriptional processing of MUC2 mucin in ulcerative colitis and in Crohn’s disease increases detectability of the MUC2 protein core. J Pathol. 1999;188:304–311. [DOI] [PubMed] [Google Scholar]
- 16. Taman H, Fenton CG, Hensel IV, et al. Transcriptomic landscape of treatment-naïve ulcerative colitis. J Crohns Colitis. 2018;12:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tytgat KM, van der Wal JW, Einerhand AW, et al. Quantitative analysis of MUC2 synthesis in ulcerative colitis. Biochem Biophys Res Commun. 1996;224:397–405. [DOI] [PubMed] [Google Scholar]
- 18. Vancamelbeke M, Vanuytsel T, Farré R, et al. Genetic and transcriptomic bases of intestinal epithelial barrier dysfunction in inflammatory bowel disease. Inflamm Bowel Dis. 2017;23:1718–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hensel KO, Boland V, Postberg J, et al. Differential expression of mucosal trefoil factors and mucins in pediatric inflammatory bowel diseases. Sci Rep. 2014;4:7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shaoul R, Okada Y, Cutz E, et al. Colonic expression of MUC2, MUC5AC, and TFF1 in inflammatory bowel disease in children. J Pediatr Gastroenterol Nutr. 2004;38:488–493. [DOI] [PubMed] [Google Scholar]
- 21. Van Klinken BJ, Van der Wal JW, Einerhand AW, et al. Sulphation and secretion of the predominant secretory human colonic mucin MUC2 in ulcerative colitis. Gut. 1999;44:387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Corfield AP, Myerscough N, Bradfield N, et al. Colonic mucins in ulcerative colitis: evidence for loss of sulfation. Glycoconj J. 1996;13:809–822. [DOI] [PubMed] [Google Scholar]
- 23. van der Post S, Jabbar KS, Birchenough G, et al. Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut. 2019:gutjnl–2018–317571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Forgue-Lafitte ME, Fabiani B, Levy PP, et al. Abnormal expression of M1/MUC5AC mucin in distal colon of patients with diverticulitis, ulcerative colitis and cancer. Int J Cancer. 2007;121:1543–1549. [DOI] [PubMed] [Google Scholar]
- 25. Borralho P, Vieira A, Freitas J, et al. Aberrant gastric apomucin expression in ulcerative colitis and associated neoplasia. J Crohns Colitis. 2007;1:35–40. [DOI] [PubMed] [Google Scholar]
- 26. Hasnain SZ, Evans CM, Roy M, et al. Muc5ac: a critical component mediating the rejection of enteric nematodes. J Exp Med. 2011;208:893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aherne CM, Collins CB, Rapp CR, et al. Coordination of ENT2-dependent adenosine transport and signaling dampens mucosal inflammation. JCI Insight. 2018;3. doi: 10.1172/jci.insight.121521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aherne CM, Collins CB, Masterson JC, et al. Neuronal guidance molecule netrin-1 attenuates inflammatory cell trafficking during acute experimental colitis. Gut. 2012;61:695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kerr TA, Ciorba MA, Matsumoto H, et al. Dextran sodium sulfate inhibition of real-time polymerase chain reaction amplification: a poly-A purification solution. Inflamm Bowel Dis. 2012;18:344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aherne CM, Saeedi B, Collins CB, et al. Epithelial-specific A2B adenosine receptor signaling protects the colonic epithelial barrier during acute colitis. Mucosal Immunol. 2015;8:1324–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Collins CB, Aherne CM, Yeckes A, et al. Inhibition of N-terminal ATPase on HSP90 attenuates colitis through enhanced Treg function. Mucosal Immunol. 2013;6:960–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hill DA, Hoffmann C, Abt MC, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3:148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. [DOI] [PubMed] [Google Scholar]
- 34. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 36. Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Robertson CE, Harris JK, Wagner BD, et al. Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics. 2013;29:3100–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Analytical Methods Committee, AMCTB No. 69 Using the Grubbs and Cochran tests to identify outliers. Anal Methods. 2015;7:7948–7950. [DOI] [PubMed] [Google Scholar]
- 41. Araki Y, Mukaisyo K, Sugihara H, et al. Increased apoptosis and decreased proliferation of colonic epithelium in dextran sulfate sodium-induced colitis in mice. Oncol Rep. 2010;24:869–874. [DOI] [PubMed] [Google Scholar]
- 42. Yan Y, Kolachala V, Dalmasso G, et al. Temporal and spatial analysis of clinical and molecular parameters in dextran sodium sulfate induced colitis. PLoS One. 2009;4:e6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alex P, Zachos NC, Nguyen T, et al. Distinct cytokine patterns identified from multiplex profiles of murine DSS and TNBS-induced colitis. Inflamm Bowel Dis. 2009;15:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Neudecker V, Haneklaus M, Jensen O, et al. Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasome. J Exp Med. 2017;214:1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou GX, Liu ZJ. Potential roles of neutrophils in regulating intestinal mucosal inflammation of inflammatory bowel disease. J Dig Dis. 2017;18:495–503. [DOI] [PubMed] [Google Scholar]
- 46. Conway KL, Goel G, Sokol H, et al. p40phox expression regulates neutrophil recruitment and function during the resolution phase of intestinal inflammation. J Immunol. 2012;189:3631–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Spehlmann ME, Dann SM, Hruz P, et al. CXCR2-dependent mucosal neutrophil influx protects against colitis-associated diarrhea caused by an attaching/effacing lesion-forming bacterial pathogen. J Immunol. 2009;183:3332–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grasa L, Abecia L, Forcén R, et al. Antibiotic-induced depletion of murine microbiota induces mild inflammation and changes in toll-like receptor patterns and intestinal motility. Microb Ecol. 2015;70:835–848. [DOI] [PubMed] [Google Scholar]
- 49. Zarrinpar A, Chaix A, Xu ZZ, et al. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat Commun. 2018;9:2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reikvam DH, Erofeev A, Sandvik A, et al. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One. 2011;6:e17996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schroeder BO. Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep (Oxf). 2019;7:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kozik AJ, Nakatsu CH, Chun H, et al. Comparison of the fecal, cecal, and mucus microbiome in male and female mice after TNBS-induced colitis. PLoS One. 2019;14:e0225079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ericsson AC, Gagliardi J, Bouhan D, et al. The influence of caging, bedding, and diet on the composition of the microbiota in different regions of the mouse gut. Sci Rep. 2018;8:4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bloom SM, Bijanki VN, Nava GM, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Berry D, Schwab C, Milinovich G, et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. Isme J. 2012;6:2091–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Maharshak N, Packey CD, Ellermann M, et al. Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes. 2013;4:316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Osaka T, Moriyama E, Arai S, et al. Meta-analysis of fecal microbiota and metabolites in experimental colitic mice during the inflammatory and healing phases. Nutrients. 2017;9:1329. doi: 10.3390/nu9121329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Song KS, Lee WJ, Chung KC, et al. Interleukin-1 beta and tumor necrosis factor-alpha induce MUC5AC overexpression through a mechanism involving ERK/p38 mitogen-activated protein kinases-MSK1-CREB activation in human airway epithelial cells. J Biol Chem. 2003;278:23243–23250. [DOI] [PubMed] [Google Scholar]
- 59. Raja SB, Murali MR, Devaraj H, et al. Differential expression of gastric MUC5AC in colonic epithelial cells: TFF3-wired IL1 β/Akt crosstalk-induced mucosal immune response against Shigella dysenteriae infection. J Cell Sci. 2012;125:703–713. [DOI] [PubMed] [Google Scholar]
- 60. Enss ML, Cornberg M, Wagner S, et al. Proinflammatory cytokines trigger MUC gene expression and mucin release in the intestinal cancer cell line LS180. Inflamm Res. 2000;49:162–169. [DOI] [PubMed] [Google Scholar]
- 61. Hasnain SZ, Wang H, Ghia JE, et al. Mucin gene deficiency in mice impairs host resistance to an enteric parasitic infection. Gastroenterology. 2010;138:1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Johansson ME, Gustafsson JK, Sjöberg KE, et al. Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLoS One. 2010;5:e12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wenzel UA, Magnusson MK, Rydström A, et al. Spontaneous colitis in Muc2-deficient mice reflects clinical and cellular features of active ulcerative colitis. PLoS One. 2014;9:e100217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. DeRoche TC, Xiao SY, Liu X. Histological evaluation in ulcerative colitis. Gastroenterol Rep (Oxf). 2014;2:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Madsen KL, Doyle JS, Tavernini MM, et al. Antibiotic therapy attenuates colitis in interleukin 10 gene-deficient mice. Gastroenterology. 2000;118:1094–1105. [DOI] [PubMed] [Google Scholar]
- 66. Hans W, Schölmerich J, Gross V, et al. The role of the resident intestinal flora in acute and chronic dextran sulfate sodium-induced colitis in mice. Eur J Gastroenterol Hepatol. 2000;12:267–273. [DOI] [PubMed] [Google Scholar]
- 67. Nakanishi Y, Sato T, Ohteki T. Commensal gram-positive bacteria initiates colitis by inducing monocyte/macrophage mobilization. Mucosal Immunol. 2015;8:152–160. [DOI] [PubMed] [Google Scholar]
- 68. Farooq SM, Stillie R, Svensson M, et al. Therapeutic effect of blocking CXCR2 on neutrophil recruitment and dextran sodium sulfate-induced colitis. J Pharmacol Exp Ther. 2009;329:123–129. [DOI] [PubMed] [Google Scholar]
- 69. Kwon JH, Keates AC, Anton PM, et al. Topical antisense oligonucleotide therapy against LIX, an enterocyte-expressed CXC chemokine, reduces murine colitis. Am J Physiol Gastrointest Liver Physiol. 2005;289:G1075–G1083. [DOI] [PubMed] [Google Scholar]
- 70. Bento AF, Leite DF, Claudino RF, et al. The selective nonpeptide CXCR2 antagonist SB225002 ameliorates acute experimental colitis in mice. J Leukoc Biol. 2008;84:1213–1221. [DOI] [PubMed] [Google Scholar]
- 71. Van den Abbeele P, Belzer C, Goossens M, et al. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. Isme J. 2013;7:949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Herp S, Brugiroux S, Garzetti D, et al. Mucispirillum schaedleri antagonizes salmonella virulence to protect mice against colitis. Cell Host Microbe. 2019;25:681–694.e8. [DOI] [PubMed] [Google Scholar]
- 75. Caruso R, Mathes T, Martens EC, et al. A specific gene-microbe interaction drives the development of Crohn’s disease-like colitis in mice. Sci Immunol. 2019;4:eaaw4341. doi: 10.1126/sciimmunol.aaw4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Melo-Gonzalez F, Fenton TM, Forss C, et al. Intestinal mucin activates human dendritic cells and IL-8 production in a glycan-specific manner. J Biol Chem. 2018;293:8543–8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shan M, Gentile M, Yeiser JR, et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342:447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schroeder BO, Birchenough GMH, Ståhlman M, et al. Bifidobacteria or fiber protects against diet-induced microbiota-mediated colonic mucus deterioration. Cell Host Microbe. 2018;23:27–40.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Desai MS, Seekatz AM, Koropatkin NM, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167:1339–1353.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.