Abstract
In the present study, a total of 35 S. aureus isolates collected from two different geographical locations viz., Germany and Hungary were tested for their methicillin-resistant phenotype which revealed a high incidence of methicillin-resistant S. aureus. The quantitative test for biofilm production revealed that 73.3% of isolates were biofilm producers. The isolates were further characterized using a set of biochemical and genotypic methods such as amplification and analysis of S. aureus species-specific sequence and mecA gene. The 33 mecA positive isolates were then characterized by the amplification of SCCmec and pvl toxin genes. Further, based on the biofilm-forming phenotype, 15 isolates were selected and characterized through PCR–RFLP of coa gene, polymorphism of spa gene and amplification of biofilm-associated genes. The dendrogram prepared from the results of both biochemical and genotypic analyses of the 15 isolates showed that except for the isolates SA G5 and SA H29, the rest of the isolates grouped themselves according to their locations. Thus, the two isolates were selected for further characterization through whole-genome sequencing. Comparative genome analysis revealed that SA G5 and SA H29 have 97.20% ANI values with 2344 gene clusters (core-genome) of which 16 genes were related to antibiotic resistance genes and 57 genes encode virulence factors. The highest numbers of singleton genes were found in SA H29 that encodes proteins for virulence, resistance, mobile elements, and lanthionine biosynthesis. The high-resolution phylogenetic trees generated based on shared proteins and SNPs revealed a clear difference between the two strains and can be useful in distinguishing closely related genomes. The present study demonstrated that the whole-genome sequence analysis technique is required to get a better insight into the MRSA strains which would be helpful in improving diagnostic investigations in real-time to improve patient care.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02387-y) contains supplementary material, which is available to authorized users.
Keywords: MRSA, Molecular typing, Whole genome sequencing, Comparative genome analysis
Introduction
Staphylococcus aureus is one of the leading causes in both communities- and nosocomial-acquired infections. It acquires an arsenal of antibiotic resistance genes (ARGs) and virulence factors-encoding genes (VFGs) that are subjected to horizontal gene transfer (HGT) and recombination (Hughes and Friedman 2005; Chan et al. 2011). It can cause a diverse range of infections including chronic skin and soft tissue infections to life-threatening illnesses (Stefani and Varaldo 2003; Yamamoto et al. 2013; Mottola et al. 2016). The genomic plasticity of S. aureus has enabled the emergence of hypervirulent and drug-resistant strains and led to challenging issues in antibiotic therapy. Consequently, the morbidity and mortality rates caused by S. aureus infections have a substantial impact on health concern (Denis 2017). Methicillin-resistant S. aureus (MRSA) acquired methicillin resistance gene (mecA) which is present within Staphylococcal Chromosomal Cassette mec (SCCmec) (Zhang et al. 2012) and reduced affinity of penicillin and β-lactam antibiotics (Jansen et al. 2006; Mistry et al. 2016). S. aureus anchors epithelial surfaces and produces biofilm (Strandén et al. 2003; Goudarzi et al. 2017). Biofilms act as a barrier against many antibiotics and other stressors and prevent their entry into the cells. Biofilms also defend the bacterial cells from host-immune evasion (Mah and O’Toole 2001; Donlan and Costerton 2002; Le et al. 2018; Vestby et al. 2020). Biofilm is a complex 3D structure of sessile microbial community covered by an exopolysaccharide glycocalyx (Otto 2008; Deka et al. 2019). The biofilm-forming ability depends on several physicals, chemical, and biological factors (Garrett et al. 2008). The biofilm-forming processes of S. aureus are determined by the icaADBC gene cluster, responsible for the synthesis of polysaccharide intracellular adhesin (PIA), and capsular polysaccharide/adhesion (PS/A) (Chaieb et al. 2005; Arciola et al. 2015; Hoang et al. 2019). The PIA is composed of β-1,6-linked N-acetylglucosamine with partially deacetylated residues, a major component of the exopolysaccharide matrix that surrounds bacterial cells in the biofilm (Mack et al. 1996; Vuong et al. 2004). It was reported that the co-existence of icaA and icaD increase N-acetylglucosaminyltransferase activity and slime production (Arciola et al. 2001, 2006). Additionally, S. aureus possesses microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), such as elastin (ebps), laminin (eno), collagen (cna), fibronectin A and B (fnbA and fnbB), fibrinogen (fib), bone sialoprotein (bbp) and clumping factors A and B (clfA and clfB) and these molecules are present on the bacterial surface to enable adherence to host tissues, thus playing a pivotal role in pathogenesis (Lindsay et al. 2006; Foster et al. 2014; Ghasemian et al. 2015; Dufrêne and Viljoen 2020).
Methods for molecular typing of MRSA that depend on gene-specific polymerase chain reaction such as SCCmec, pvl (Panton-Valentine leukocidin), coa (coagulase), and spa (S. aureus protein A) genes and followed by restriction enzyme digestion (PCR–RFLP) has proven its good discriminatory power (DP) and is used routinely for typing MRSA strains (Faria et al. 2008; Omar et al. 2014; Al-Obaidi et al. 2018; Tenover et al. 2019; Alkharsah et al. 2019). SCCmec typing classifies the MRSA into hospital-associated (HA-MRSA) and community-associated (CA-MRSA) strains (Appelbaum 2007). HA-MRSA isolates carried SCCmec types I to III, but CA-MRSA isolates has a novel, small variant of SCCmec IV and V, but also has the locus for pvl gene (Baba et al. 2002; Shukla et al. 2012). The coagulase enzyme, a virulence factor encoded by the coa gene contains several tandem repeats suitable to generate polymorphic RFLP patterns among different isolates. These molecular typing methods may be helpful in determining the relatedness among geographically diverse MRSA (Singh et al. 2006; Grundmann et al. 2010).
Phenotypic analysis including the antibiotic resistance patterns and molecular typing methods are beneficial for identifying the risk factors associated with MRSA infections which support the establishment of adequate infection control programs (Zhang et al. 2012; Mistry et al. 2016). Epidemiological studies of MRSA apply various molecular typing techniques such as Pulsed-Field Gel Electrophoresis (PFGE), SCCmec, spa genes typing, Multi-Locus Sequence Typing (MLST), and detection of pvl gene as well as PCR–RFLP of coa gene (Zhang et al. 2012; Al-Obaidi et al. 2018). Many of these established techniques are costly and time-consuming, and the discriminatory abilities of these techniques are also different (Du et al. 2011). However, the spa typing method has been considered as a rapid and inexpensive method for genotyping and it provides high discriminating power than other typing methods (Shittu et al. 2011; O’Hara et al. 2016; Goudarzi et al. 2017; Ali et al. 2019; Rezai et al. 2019; Kareem et al. 2020). Both phenotypic and molecular typing methods have been used widely to detect and differentiate several MRSA strains, but these techniques have certain limitations in infection control and investigating the nosocomial transmission as these techniques provide low resolution and more time-consuming. Because of that in recent times whole genome-based typing has been used as it offers an excellent resolution in global and local epidemiological investigations of pathogen outbreaks and offers further data mining activities essentially for ARGs and VFGs profiling (Köser et al. 2012). So, the Next Generation Sequencer (NGS) based-genome sequencing technique has become a vital tool in the clinical microbiology arenas for comparative genomic analysis of several other species of the Staphylococcus genus in terms of the niche adaptation, combat antibiotics, and emergence of new virulent strains in real-time (Al-Obaidi et al. 2018; McClure et al. 2018; Tenover et al. 2019; Maljkovic Berry et al. 2019; Raven et al. 2020). In the current study, we performed characterization through molecular typing methods and their integrated polyphasic approach to determine the S. aureus lineage strains. The lineage strains were compared deeply into a genomic level based on Average Nucleotide Identity (ANI), genome distance, orthologue gene/ clusters, and the evolutionary relationship. Further, ARGs and VFGs were characterized to understand their crucial role in pathogenesis and defense. The information on genomic characteristics and comparative genomics of S. aureus will facilitate investigations into the molecular basis of pathogenesis and improve diagnostic investigations of infectious diseases in real-time and improve patient care.
Materials and methods
Collection and preliminary identification of the isolates
In this study, 35 S. aureus strains were collected from the Department of Medical Microbiology and Immunology Laboratory, Medical School, University of Pecs, Hungary. The Hungarian S. aureus strains (60%) were previously recovered during February to July 2016 from wounds (31.42%), blood (8.57%), tracheas (5.71%), ears (2.85%), lungs (2.85%), nostrils (2.85%), skins (2.85%) and throats (2.85%) while the German S. aureus strains (40%) were recovered from body sites without documentation. The isolates were identified as Staphylococcal strain based on colony morphology on Nutrient agar, Blood Agar, Mannitol Salt Agar, Gram staining, and different biochemical tests (Bergey and Holt 1994). The isolates were tested for catalase, coagulase, urease, DNase production, and mannitol fermentation test (Collee et al. 1996).
Antibiotic susceptibility test
The 35 S. aureus clinical strains were screened for MRSA using BBL™ CHROMagar™ MRSA II media (BD, USA). Susceptibility of S. aureus strains to oxacillin (1 μg), cefoxitin (30 μg), erythromycin (15 μg) and vancomycin (30 μg) were determined using disk diffusion method according to Clinical and Laboratory Standards Institute (CLSI) guidance (CLSI 2014). The entire antimicrobial susceptibility test (AST) was repeated three times using the S. aureus ATCC25923 and ATCC700698 as MRSA negative and positive controls, respectively. The diameter zone of inhibition was measured in millimetre (mm).
Biofilm formation assay
Biofilm formation was performed as previously described (Rahimi et al. 2016) with some modifications. Briefly, S. aureus strains were cultured overnight at 37 °C in tryptic soy broth (TSB) (BD, Germany) containing 0.25% (w/v) glucose. The cell density was adjusted to a final concentration of 106 CFU/ml in TSB supplemented with 0.25% (w/v) glucose. Cell suspensions (200 µl) were loaded into 96-well round-bottomed microtiter plate (Sarstedt, Germany), and incubated at 37 °C for 18 h without shaking. Cells were washed three times with 200 µl sterile PBS (pH 7.2), dried at room temperature and fixed with methanol (99% v/v). The dried biofilm was stained with 200 µl of 0.16% (w/v) crystal violet for 15 min. To remove the unbound dye, biofilms were washed three times with PBS and air dry. Finally, the biofilm-bound dye was solubilized with 200 µl of 95% (v/v) ethanol, and absorbance was measured at 540 nm wavelength using a Multiskan Ex microtiter plate reader (Thermo Electron Corporation, USA) in a flat-bottom 96-well plate (Costar 3599; Corning; USA). Experiments were performed in triplicates with S. aureus ATCC25923 as a biofilm-positive control strain.
Molecular identification and genotyping
The DNA of the 35 strains was extracted from the overnight culture of S. aureus using QIAamp DNA Mini Kit (Qiagen GmbH, Hilden, Germany). The extracted DNA concentration was assayed by the Nanodrop-2000 spectrophotometer. Genomic DNA was used for the detection of S. aureus species-specific sequence (Martineau et al. 1996), mecA (Strommenger et al. 2003), and the pvl toxin (Hisata et al. 2005; Karahan and Çetinkaya 2007) genes. S. aureus ATCC25923, ATCC700698, and ATCC700699 strains were used as reference strains for mecA negative and positive controls respectively. Multiplex PCR typing of SCCmec gene was performed on mecA-positive S. aureus strains using primers as described previously (Zhang et al. 2005). The MRSA isolates that showed unexpected amplified fragments or no amplification were defined as non-typeable (NT). For PCR–RFLP, the coa gene amplicons were digested with HaeIII (Fermentas, USA) restriction enzyme (Khoshkharam-Roodmajani et al. 2014) and a heat-map with dendrogram was generated from the restriction banding pattern using Morpheus web-based program (https://software.broadinstitute.org/morpheus/) using the Euclidean distance feature. Polymorphism of the spa gene was detected based on a previously described primer set (Harmsen et al. 2003). The PCR products were purified using the ZR-96 DNA Clean-up Kit (Zymo Research, USA). Concentration was determined by Qubit 3.0 and sequencing reactions were performed using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). Sequencing reactions were run on ABI PRISM 310 Genetic Analyzer (Applied Biosystems, USA). The spa sequence types were assigned using spaTyper (https://spatyper.fortinbras.us/) and confirmed by a spa database (https://spa.ridom.de/) in DNAGear software (Al-Tam et al. 2012). The spa sequences were aligned, and a phylogenetic tree was constructed by the UPGMA method in MEGA X software (Kumar et al. 2018). The DPs of coa and spa typing were calculated based on Simpson's index using the DP calculator, available online (https://insilico.ehu.es/mini_tools/discriminatory_power/).
Biofilm-encoding genes were amplified using the primer sets listed in Supplementary Table 2. The PCR amplification was performed using DreamTaq PCR Master Mix according to the manufacturer's recommendation (Thermo Fisher Scientific, USA) in a Veriti™ 96-Well Thermal Cycler (Applied Biosystem, USA) as follows: 96 °C for 3 min; 96 °C for 30 s, 54 °C for 30 s, 72 °C for 1 min repeated for 35 cycles; final extension was performed at 72 °C for 7 min. The amplified products were electrophorized on 2% (w/v) agarose gel, stained using 0.5 µg/ml ethidium bromide solution, and captured using the FluroChem Q system (ProteinSimple™, USA).
Data setting for a polyphasic approach
Individual result of the applied techniques was converted into the unweighted binary code (0, 1), Jacquard’s similarity index was generated and visualized according to the Neighbour-Joining (NJ) clustering method using Past 3.x (Hammer et al. 2001). Besides the binary data (Supplementary file 2) was used to perform a logistic Principal Component Analysis (PCA) in R software.
Whole-genome sequencing
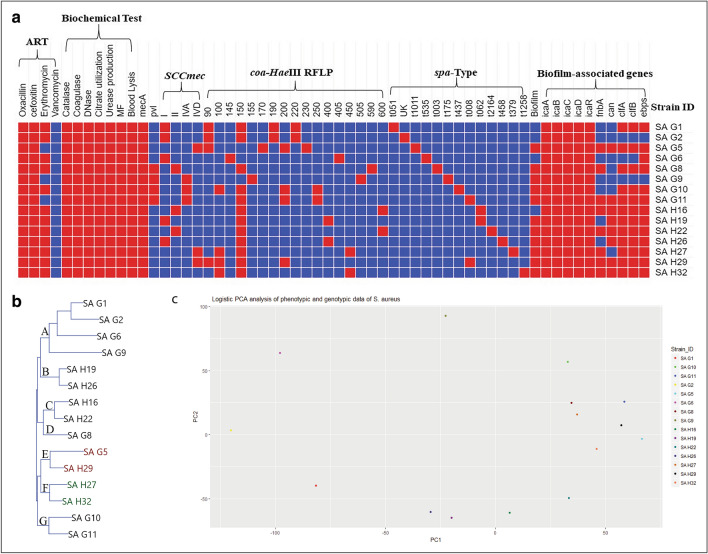
Based on the dendrogram and PCA plot generated from combined results of the phenotypic and genotypic analysis (binary data) of the S. aureus strains it was observed that two strains viz., SA G5 (collected from Germany) and SA H29 (collected from Hungary) were found in the same group (Fig. 1a–c). Therefore, these two strains (SA G5 and SA H29) were selected for further analysis through whole-genome sequencing to get a better insight into their genomic background.
Fig. 1.
Polyphasic comparison of S. aureus clinical isolates. a Colored similarity matrix categories representing resistant test against different antibiotics (ART), biochemical test, PCR based mecA and pvl genes detection, SCCmec typing, coa-HaeIII RFLP, spa type, biofilm-forming assay and detection of biofilm-associated genes. Red and blue colors indicate the presence ( +) and absence (−) of particular properties respectively. b NJ-dendrogram prepared from the results of biochemical and molecular analysis of the 15 S. aureus clinical isolates (SA). The isolates were labeled according to their geographical origin where G and H indicate Germany and Hungary. Clusters are labeled as A-F. c Principal component analysis of the results of biochemical and molecular analysis of the 15 S. aureus clinical isolates
Genomic DNA extraction, library preparation, and sequencing
Whole-genome sequencing was performed at Microbial Biotechnology Research Group, Szentágothai Research Centre, University of Pecs, Hungary with the following procedures: the genomic DNA was extracted using the GenElute™ Bacterial Genomic DNA Kit (Sigma, USA) following the manufacturer’s recommendation. The extracted DNA samples were quantified in a Qubit 3.0 fluorometer (Invitrogen, USA) using dsDNA High Sensitivity (HS) Assay Kit (Thermo Fisher Scientific Inc. USA) and subsequently, DNA quality was visualized by agarose gel electrophoresis. Genomic libraries were prepared using the NEB Next Fast DNA Fragmentation and Library Preparation Kit, developed for Ion Torrent (New England Biolabs, USA) and used according to 200 bp protocol. After chemical fragmentation, DNA size selection was performed on precast 2% E-Gel Size Select Gel (Thermo Fisher Scientific Inc. USA). The quality of the libraries was verified using Agilent high sensitivity DNA assay kit (Agilent Technologies Inc. USA) in Agilent 2100 Bioanalyzer System (Agilent Technologies Inc. USA). For the template preparation, Ion PGM Hi-Q View OT2 Kit was used (Thermo Fisher Scientific Inc. USA). The template positive beads were loaded on Ion 316v2 Chip and sequenced using Ion PGM Hi-Q View Sequencing Kit on Ion Torrent Personal Genome Machine (PGM) (Thermo Fisher Scientific Inc. USA).
Genome assembly, annotation, and comparison
Quality trimming of the reads was performed with the Ion Torrent Suite 5.4.0 (Thermo Fisher Scientific Inc. USA) with default settings. De novo genome assembly was performed by SPAdes 3.7.1 software with default parameters for Ion Torrent data and k-mer settings of 21, 33, 55, 77, 99, and 127 (Nurk et al. 2013). Closely related reference genomes were identified using kmerFinder 3.1 and NCBI microbial genome blast web-platforms. Identified high-quality reference genomes, CP006044.1 (S. aureus CA-347), and CP023390 (S. aureus subsp. aureus str. Newman) were applied to orientate and order contigs of SA G5 and SA H29 respectively using the ‘Move Contigs’ algorithm in Mauve 2.4.0 (Darling et al. 2010). Scaffolds were generated using the reference-based scaffolder MeDuSa (Bosi et al. 2015). Gene annotation was performed by two independent automated pipelines based on Rapid Annotation using Subsystem Technology (RAST) (Aziz et al. 2008) and NCBI Prokaryotic Genome Annotation Pipeline (Tatusova et al. 2016).
The values for ANI were calculated using the OrthoANIu algorithm (Yoon et al. 2017). Also, dDDH (digital DNA-DNA Hybridization) estimates were obtained from Genome to Genome Distance Calculator 2.1 (Meier-Kolthoff et al. 2013). RAST server-based SEED viewer was used for the subsystem functional categorization (Overbeek et al. 2014). The protein-coding sequence identified by RAST was analyzed for orthologous genes/clusters and subsequent functional annotation using web platform OrthoVenn2 (Xu et al. 2019). Graphical map of sequence features embedded circular genomes of SA G5, CP006044.1, SA H29, and CP023390.1 were generated using CGView Server (Grant and Stothard 2008) with default parameters against the reference genome Staphylococcus aureus subsp. aureus NCTC 8325 (NC_007795.1). ARGs and VFGs were identified by the Comprehensive Antibiotic Resistance Database (CARD) and the virulence factor database (VFDB) in PATRIC 3.5.43 (Chen et al. 2016; Wattam et al. 2017). The genome assemblies were screened for plasmid replicon (rep) genes in nonaligned contigs or scaffold regions using PlasmidFinder version 2.1 (Carattoli et al. 2014) with default parameters. The identified nonaligned contigs or scaffolds associated with plasmid sequences were extracted and used for the identification of full-length plasmid regions using PLSDB (Plasmid Database) version-2020-03-04 with search strategy Mash screen, and the default values were a maximum P value of 0.1 and a minimum identity of 0.99 (Galata et al. 2019).
Phylogenetic analysis
The 16S rRNA sequences obtained from the whole genome data of the two strains were aligned with the other closely related 16S rRNA sequences of S. strains and Bacillus cereus ATCC14579 (AE016877.1) using MUSCLE algorithm in MEGA X (Kumar et al. 2018). The phylogenetic tree was then constructed by the NJ method with 500 replicates. Phylogenetic trees were generated based on the concatenated alignment of all shared proteins (core) of S. aureus genome sequences with B. cereus ATCC14579 (AE016877.1) as outgroup by RAxML in PATRIC 3.5.43 (Wattam et al. 2017). SNPs were filtered from the genome sequences and generate phylogeny based on the concatenated alignment of the high-quality SNPs using CSI phylogeny 1.4 servers (https://cge.cbs.dtu.dk/services/CSIPhylogeny/) and the tree was visualized in MEGA X (Kumar et al. 2018).
Sequence supporting data
The genomic data of this study were deposited in the NCBI genome database under the GenBank accession numbers: CP032160 and CP032468-CP032470.
Results
Phenotypic characterization
The colony characteristics of the strain are yellow colored, moist, round, glistening opaque colonies with β or weak hemolysis on blood agar. The strains are Gram-positive cocci showing typical staphylococcal bunch.
Antibiotic susceptibility test
The 35 S. aureus clinical strains were screened for MRSA using BBL™ CHROMagar™ MRSA II media which revealed that 94.28% of the strains were MRSA. The tested strains were highly resistant to beta-lactam antibiotics viz. oxacillin (94.28%) and cefoxitin (94.28%), but less resistant to non-beta lactam antibiotics, erythromycin (71.43%), and none of the strain displayed resistant to vancomycin (Table 1).
Table 1.
Antibiotic susceptibility patterns of S. aureus isolates
| Strain ID | Origin | Diameter of inhibition zone (mm) | |||
|---|---|---|---|---|---|
| Ox-1 µg | Cfox-30 μg | Ery-15 µg | Van-30 μg | ||
| SA G1 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 19.5 ± 0.7 (S) |
| SA G2 | HA | 11.5 ± 0.7 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 20 ± 0.7 (S) |
| SA G3 | HA | 28 ± 0 (S) | 18.5 ± 0.7 (S) | 27 ± 0 (S) | 21 ± 1.4 (S) |
| SA G4 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 24.5 ± 7.7 (S) | 20.5 ± 0.7 (S) |
| SA G5 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 24.5 ± 7.7 (S) | 19.5 ± 0.7 (S) |
| SA G6 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 20 ± 0 (S) |
| SA G7 | HA | 24.5 ± 0.7 (S) | 18 ± 0 (S) | 30 ± 0 (S) | 18.5 ± 0.7 (S) |
| SA G8 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 18.5 ± 0.7 (S) |
| SA G9 | CA | 6 ± 0 (R) | 6 ± 0 (R) | 24 ± 8 (S) | 18 ± 0 (S) |
| SA G10 | CA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 20 ± 1.4 (S) |
| SA G11 | CA | 6 ± 0 (R) | 6 ± 0 (R) | 10 ± 0 (S) | 19 ± 1.4 (S) |
| SA G12 | CA | 6 ± 0 (R) | 7.5 ± 2.1 (R) | 30 ± 0 (S) | 19.5 ± 07 (S) |
| SA G13 | CA | 6 ± 0 (R) | 6 ± 0 (R) | 30 ± 0 (S) | 19 ± 0 (S) |
| SA G14 | CA | 6 ± 0 (R) | 6 ± 0 (R) | 30 ± 0 (S) | 19 ± 0 (S) |
| SA H15 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 19 ± 0 (S) |
| SA H16 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 20 ± 0 (S) |
| SA H17 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 18 ± 0 (S) |
| SA H18 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 18.5 ± 0.7 (S) |
| SA H19 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 19.5 ± 0.7 (S) |
| SA H20 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 19 ± 1.4 (S) |
| SA H21 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 11 ± 0 (R) | 18.5 ± 0.7 (S) |
| SA H22 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 11 ± 0 (R) | 18.5 ± 0.7 (S) |
| SA H23 | HA | 6 ± 0 (R) | 7.5 ± 2.1 (R) | 6 ± 0 (R) | 20 ± 0 (S) |
| SA H24 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 19 ± 0 (S) |
| SA H25 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 20 ± 0 (S) |
| SA H26 | HA | 8 ± 2.8 (R) | 14.5 ± 0.7 (R) | 6 ± 0 (R) | 19 ± 1.4 (S) |
| SA H27 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 16.14 ± 0 (R) | 19.5 ± 0.7 (S) |
| SA H28 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 24.5 ± 0.7 (R) | 19 ± 1.4 (S) |
| SA H29 | HA | 10.5 ± 0.7 (R) | 8 ± 2.8 (R) | 6 ± 0 (R) | 19 ± 1.4 (S) |
| SA H30 | HA | 6 ± 0 (R) | 3 ± 0 (R) | 6 ± 0 (R) | 18.5 ± 2.1 (S) |
| SA H31 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 18.5 ± 0.7 (S) |
| SA H32 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 19 ± 1.4 (S) |
| SA H33 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 19 ± 1.4 (S) |
| SA H34 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 19.5 ± 0.7 (S) |
| SA H35 | HA | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 19 ± 1.4 (S) |
| ATCC700698 | – | 6 ± 0 (R) | 6 ± 0 (R) | 6 ± 0 (R) | 19.5 ± 0.7 (S) |
| ATCC25923 | – | 26.5 ± 0.7 (S) | 25 ± 0 (S) | 33.5 ± 0.7 (S) | 20.5 ± 0.7 (S) |
Zone of inhibition in mm is given as Mean ± SD; R and S denote the resistant and susceptible, Ox oxacillin (≤ 10 mm = Resistant), Cfox cefoxitin (≤ 14 mm = Resistant), Ery erythromycin (≤ 13 mm = Resistant), Van vancomycin (≤ 9 mm = Resistant)
aSA G and SA H represent strains isolated from Germany and Hungary respectively. HA and CA represent the Hospital and Community-associated S. aureus strains respectively
Biofilm production test
The quantitative test for biofilm production revealed that among the 33 methicillin-resistant strains, 87.87% of strains produced biofilm. Fifteen strains were selected among 33 methicillin-resistant strains based on non- (26.7%), moderate- (40%), and strong- (33.3%) biofilm-forming abilities for molecular typing and discrimination. (Supplementary Table 1).
Molecular characterization and genotyping
The PCR amplified products showed that 100% strains of S. aureus were positive to S. aureus species-specific sequence represented by a distinct band of 107 base pairs (bp) (Supplementary Fig. 1). The mecA and pvl genes were detected in 94.28% with an amplicon size of 532 bp and 22.85% with the amplicon size 1918 bp respectively (Supplementary Figs. 2, 3). The AST and mecA gene detection revealed concordant results, thus 94.28% of S. aureus isolates were confirmed as MRSA.
SCCmec typing revealed that SCCmec type II (33.33%) was the most predominant followed by type I (21.21%), type IVa (15.5%), type IVd (9.09%), IVb (3.03%) and the rest of the strains were non-typeable (12.12%). Among the 35 strains, eight strains were positive for pvl of which four strains belonged to SCCmec II, three strains to SCCmec IV and, one to SCCmec V (Supplementary Fig.4). The distribution of SCCmec types showed that 36.36% of strains were CA-MRSA and 54.54% strains were HA-MRSA (Table 2).
Table 2.
Biochemical tests and PCR based molecular detection of mecA, pvl and SCCmec genes of S. aureus isolates
| aStrain ID | Biochemical tests | Molecular detection | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Catalase | Coagulase | DNase | Citrate utilization | Urease production | MF | Blood Lysis | Biofilm | mecA | pvl | SCCmec type | |
| SA G1 | + | + | + | + | + | + | + | − | + | − | I |
| SA G2 | + | + | + | + | + | + | + | − | + | − | I |
| SA G3 | + | + | + | + | + | + | + | + | − | − | NT |
| SA G4 | + | + | + | + | + | + | + | + | + | − | NT |
| SA G5 | + | + | + | + | + | + | + | + | + | − | IVd |
| SA G6 | + | + | + | + | + | + | + | − | + | − | I |
| SA G7 | + | + | + | + | + | + | + | + | − | − | NT |
| SA G8 | + | + | + | + | + | + | + | + | + | + | II |
| SA G9 | + | + | + | + | + | + | + | + | + | + | IVa |
| SA G10 | + | + | + | + | + | + | + | + | + | + | IVa |
| SA G11 | + | + | + | + | + | + | + | + | + | + | IVa |
| SA G12 | + | + | + | + | + | + | + | + | + | − | IVa |
| SA G13 | + | + | + | + | + | + | + | + | + | − | IVa |
| SA G14 | + | + | + | + | + | + | + | + | + | + | V |
| SA H15 | + | + | + | + | + | + | + | + | + | − | II |
| SA H16 | + | + | + | + | + | + | + | − | + | + | II |
| SA H17 | + | + | + | + | + | + | + | + | + | − | II |
| SA H18 | + | + | + | + | + | + | + | + | + | − | I |
| SA H19 | + | + | + | + | + | + | + | + | + | − | I |
| SA H20 | + | + | + | + | + | + | + | + | + | − | II |
| SA H21 | + | + | + | + | + | + | + | + | + | − | IVb |
| SA H22 | + | + | + | + | + | + | + | + | + | − | II |
| SA H23 | + | + | + | + | + | + | + | + | + | − | IVa |
| SA H24 | + | + | + | + | + | + | + | + | + | − | II |
| SA H25 | + | + | + | + | + | + | + | + | + | − | II |
| SA H26 | + | + | + | + | + | + | + | + | + | − | I |
| SA H27 | + | + | + | + | + | + | + | + | + | − | IVd |
| SA H28 | + | + | + | + | + | + | + | + | + | − | IVa |
| SA H29 | + | + | + | + | + | + | + | + | + | − | IVd |
| SA H30 | + | + | + | + | + | + | + | + | + | + | II |
| SA H31 | + | + | + | + | + | + | + | + | + | − | I |
| SA H32 | + | + | + | + | + | + | + | + | + | − | NT |
| SA H33 | + | + | + | + | + | + | + | + | + | − | II |
| SA H34 | + | + | + | + | + | + | + | + | + | + | II |
| SA H35 | + | + | + | + | + | + | + | + | + | − | NT |
| ATCC700698 | + | + | + | + | + | + | + | + | + | − | II |
| ATCC25923 | + | + | + | + | + | + | + | + | − | − | − |
aMF represents mannitol production, Blood lysis represents hemolysis on blood agar, Biofilm represents biofilm production, mecA represents gene encode for penicillin-binding protein 2A (PBP2A); pvl represents gene encode for Panton-Valentine leukocidin toxin. + and – represent present and absent; NT denotes SCCmec cassette non-typeable
The coa gene amplification produced amplicons size in the range of 550 to 800 bp, the amplicons of 700 bp showed the highest frequency (33.33%) followed by amplicon sizes of 550 bp (26.66%), 650 bp (13.33%), 740 bp (0.66%), 660 bp (0.66%), 800 bp (0.66%) and 600 bp (0.66%) (Supplementary Fig. 5). The result of the coa gene PCR–RFLP is summarized in supplementary Table 3 and supplementary Fig. 6. To get more insight into the similarity and difference of complex RFLP banding pattern, presence/absence heat-map and dendrogram was generated. Visualisation of banding patterns revealed six distinct clusters, namely A-F with calculated prevalence of 6.6, 13.3, 13.3, 20, 33.3 and 13.3%, respectively. (Supplementary Fig. 7). Typing of coa gene and HaeIII RFLP, as well as DIs, were presented in supplementary Table 3.
Typing of the spa gene revealed 10 amplicons, ranging in size from 355 to 560 bp (Supplementary Fig. 8). The DI of spa-PCR typing was 0.9429. Analysis of the spa gene revealed twelve known spa types. SA G11 and SA H29 strains possessing SCCmec-IV gene (CA-MRSA) and both were in the same t008 spa-type. SA H16 and SA H19 strains clustered together into t062 spa-type and both were found resistant to erythromycin, also classified into HA-MRSA group (Supplementary Table 4). The phylogenetic tree based on spa sequences revealed four distinct clusters, designated as A, B, C, and D with a prevalence of 33.33%, 46.66%, 13.33%, and 6.66% respectively (Supplementary Fig. 9).
Although, all the selected strains did not produce biofilm each of them harbors genes for intracellular adhesion (icaADBC) and regulation (icaR). The presence of fnbA and fnbB genes were detected in 73.3% and 66.6% strains, respectively. Genes also associated with biofilm-forming ability viz. cna, clfA, clfB, and ebps were found present in 53.33%, 80%, 73.3%, and 86.6% strains, respectively. Agarose gel electrophoresis pictures of biofilm-associated genes are presented in supplementary figs. 10–20.
Cluster analysis based on phenotypic and genotypic data
Although, the individual test, for example, performed AST, catalase, coagulase, DNase, citrate utilization, urease production, mannitol fermentation, blood lysis, and biofilm production assays has the advantage of being cost-effective, but often cannot differentiate among the strains. PCR-based detection of mecA gene and genes responsible for PIA (icaADBC and icaR) could not differentiate the clinical isolates in the present study. PCR-based detection of pvl gene and genes encoding for MSCRAMMs (fnaA, fnaB, clfA, clfB, cna, and ebps) also showed poor DP, while SCCmec, coa-HaeIII RFLP and spa typing revealed moderate DP. To avoid the misleading conclusion, the data from all applied methods were coupled to perform cluster and PCA analysis. The generated dendrogram suggested that strains collected from the same geographical region shared the same cluster F. Except for cluster E in which strains SA G5 and SA H29 from different geographical locations clustered together (Fig. 1b). The PCA analysis also revealed that SA G5 and SA H29 shared less variance than the other strains (Fig. 1c).
General features of sequenced genomes
Ion-Torrent PGM sequencing of SA G5 and SA H29 genomes generated reads range of 18.55 and 25.98 million bases (Mb) per sample covering more than 98% of the reference genome (ASM1342v1) with an average depth of 152.4X. SPAdes assembled sequences were further analyzed by QUAST producing genomes with 43 and 47 contigs and N50 ranging from 85 and 185 kb after filtering contigs size < 200 bp. Mauve contigs ordering and MeDuSa scaffolder produced SA G5 and SA H29 final genomes sequence lengths of 2,760,385 and 2,834,624 bp respectively. The total GC contents were 32.77 and 32.65%. The SA G5 and SA H29 genomes contain 2689 and 2843 coding sequences (CDS) with 9 and 6 rRNAs, and 59 and 51 tRNAs, respectively. The summarized genomic features were shown in Table 2.
Comparative genome analysis
OrthoANIu identity found ANI of 97.20% among the SA G5 and SA H29 genome sequences. Similarly, dDDH was calculated using the GGDC tool revealed that the probability of DDH (DNA-DNA hybridization) is 84.5% with a genetic distance of 0.0308, suggesting that SA G5 and SA H29 occupy identical taxonomic status which is also supported by the different genotypic data.
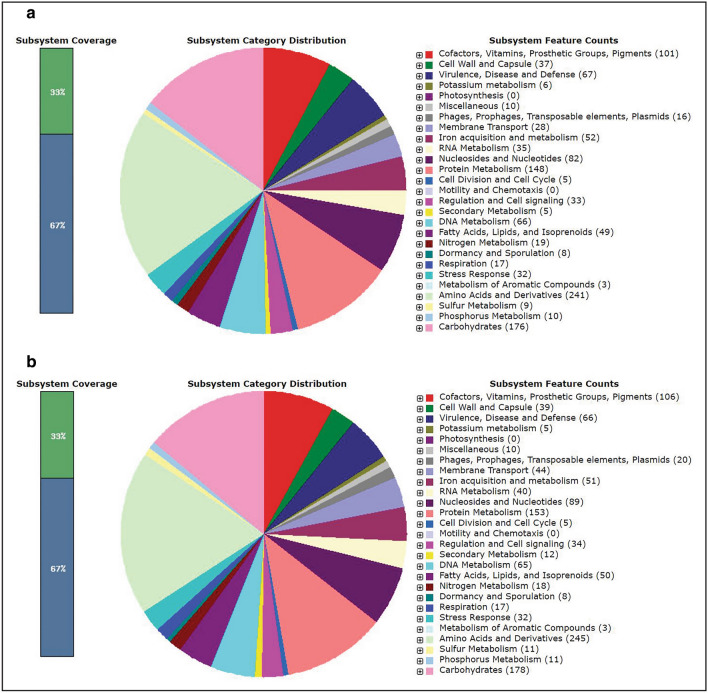
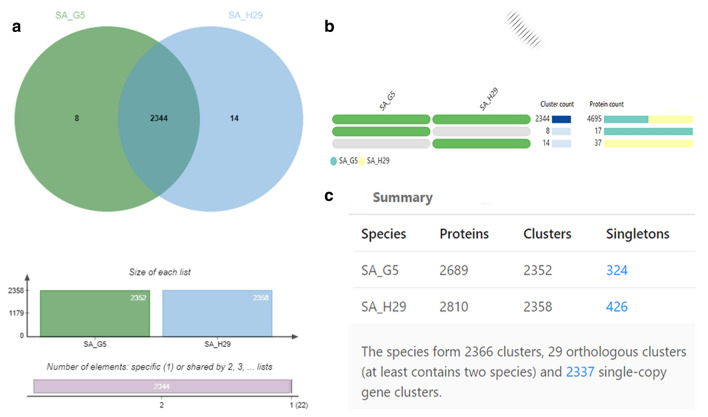
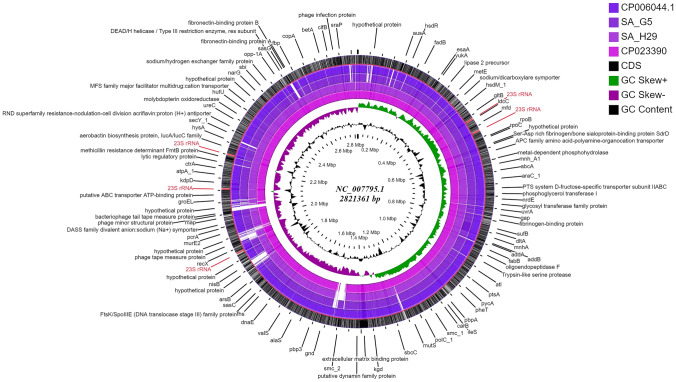
According to SEED subsystems, SA H29 genome contains amino acids and derivatives encoding genes (245 CDSs), virulence, disease and defense genes (67 CDSs), carbohydrate utilization genes (178 CDSs) and genes derived from phages, prophages, transposable elements and plasmid (20 CDSs) as presented in the Fig. 2a, b. In addition, the relationship between SA G5 and SA H29 genomes were analyzed using web platform OrthoVenn2, which identified 2366 gene clusters (pan-genome), of which 2344 orthologous clusters were shared between SA G5 and SA H29, and 2337 were single-copy gene clusters (Fig. 3c). These two strains shared 2344 gene clusters (core-genome) comprising 4695 proteins (Fig. 3a, b). SA H29 genome possesses the highest singleton genes covering 37 numbers of proteins that are responsible for virulence, resistance, mobile genetic elements, and lanthionine biosynthesis. Whole-genome circular comparative map of two genomes (SA G5 and SA H29) and their close genomes against Staphylococcus aureus subsp. aureus NCTC 8325 (NC_007795.1) was generated using CGView server based on BLAST sequence similarities (Fig. 4). Each genome was represented by a different color and the darker areas in the circular genome showed a 100% sequence similarity with the reference genome, whereas the lighter areas showed a 70% sequence similarity.
Fig. 2.
Subsystem category distribution of (a) SA G5 and (b) SA H29 genomes. The genomes of SA G5 and SA H29 annotated using the RAST Server classified subsystems into 270 and 276 respectively. The green part in the bar-chart at the leftmost position corresponds to the percentage of proteins included and the pie-chart in the right panel demonstrates the subsystem category distribution
Fig. 3.
Comprehensive comparative protein analysis of SA G5 and SA H29. a Venn diagram showing the distribution of shared orthologous clusters (core-genome) among the genomes. b Occurrence pattern of shared orthologous groups among SA G5 and SA H29. The pattern to the left indicates SA G5 and SA H29 genomes are in the clusters, the number of clusters shared between genomes (cluster count), and the number of protein members in the shared clusters (protein count). c Showing the numbers of proteins, cluster genes, and singletons with respect to species
Fig. 4.
Circular genome comparison map showing homologous chromosome of S. aureus genomes against S. aureus subsp. aureus NCTC 8325 (NC_007795.1) genome using CGviewer. The inner scales designate the coordinates in mega-base pairs (Mbp). White spaces indicate regions with no identity to the reference genome and the genome features were indicated by the outermost (black) ring
Epidemiological characterizations of SCCmec type, spa type, and multilocus sequence type (MLST) are presented in Table 2. In silico analysis of ARGs using CARD database revealed 18 genes related to antibiotic resistance and among them, 16 genes were shared which are responsible for the resistance against methicillin (mecA), beta-lactams (blaZ), fluoroquinolone (norA, gyrA, and gyrB), tetracycline (tet-38), glycylcycline (mepA), multidrug and toxic compound (mepR), fluoroquinolone and acridine dye (arlS and arlR), tetracycline, penam, cephalosporin, glycylcycline, rifamycin, phenicol, triclosan and fluoroquinolone (mgrA), daptomycin (clsA), diaminopyrimidine (dfrC), nitroimidazole (msbA), rifamycin (rpoB32) and defensin (mprF). In addition, genes that provide resistance against macrolide, lincosamide, streptogramin (emrA) were detected in the SA H29 genome. In-silico identification of ARGs and the results of in-vitro AST (for erythromycin, vancomycin, and beta-lactam antibiotics) are in good agreement.
The VFGs were predicted against the VFDB in the PATRIC annotation system. Genomic comparison of the two strains identified 78 genes that encode virulence factors of which 57 are common. These virulence factors are responsible for adherence, toxins production, antiphagocytosis, immune evasion, exoenzyme activity, iron uptake, and secretion system. Genomes annotation revealed common genes encoding MSCRAMMs i.e. adhesion (sdrC, sdrD, and sdrE) elastin binding protein (ebps), polysaccharide intercellular adhesion proteins (icaA, icaB, icaC, icaD, and icaR), clumping factor (clfB) and cell wall anchored protein (sasH). The genome of SA H29 isolate harbored all these genes extended with additional genes encoding clumping factor A and B (clfA and clfB), fibronectin-binding protein A (fnbA), serine-aspartate repeat-containing proteins (sdrC and sdrD), and fibronectin-binding protein (fnbp), while the genome of SA G5 contained additional genes responsible for collagen adhesion (cna), and extracellular adherence protein (eap/map).
Several hemolysin toxin genes such as alpha (hla), beta (hlb), delta (hld) and gamma A, B, and C (hlgA, hlgC, and hlgB) were identified in both the strains. Bicomponent leukotoxins, leukocidin lukE, and lukD genes were identified in the SA H29 genome. In the sequenced genomes 10 Staphylococcal enterotoxins (SEs) types were detected. The most prevalent enterotoxin genes, sea, and seb were presented in SA H29. The genome of SA G5 possessed 8 enterotoxin gene types such as sec, sec3, seg, sei, sel, sem, seo, and seu. Toxin genes involved in immune evasions such as IgG-binding proteins (sbi and spa), staphylococcal complement inhibitor (scn), and staphylokinase (sak) were present in both genomes. Chemotaxis-inhibiting protein (CHIPS) encoded by chp was identified in the SA G5 genome. The antiphagocytosis capsular stereotype encoding genes such as cap5C, cap5D, cap5F, cap5G, cap5O, cap5P, cap8M, and cap8N were present in both genomes, however, cap5E gene was detected in SA H29 genome. Moreover, capsular stereotype 8 genes (cap8E and cap8H) were detected in SA G5 genomes. S. aureus secret ESAT-6-like proteins consist of eight genes cluster namely esxA, esxB, esaA, esaB, esaC, essA, essB, and essC. These eight secretary system genes were identified in both genomes. Several exoenzyme encoding genes namely staphylocoagulase (coa), catalase (katA), hyaluronidase (hysA), von Willebrand factor binding protein (vwb), zinc metalloproteinase aureolysin (aur), V8 protease/glutamyl endopeptidase (sspA), staphopain B (sspB), staphostatin B (sspC) and lipase (geh) were present in both genomes. Genes involved in iron uptake mechanism such as isdA, isdB, isdC, isdD, isdE, isdF, srtB, and isdG that encodes for cell surface protein, cell surface receptor, cell wall anchor protein, heme-transporter component, high-affinity heme-uptake system protein, heme–iron transport system permease protein, sortase B and heme-degrading monooxygenase/staphylobilin-producing respectively, were identified in both genomes.
The putative plasmids were identified in nonaligned contigs that displayed an unexpected high coverage level after the genome assemblies. Two putative plasmids, p1H29 and p2H29 of 17,165 bp and 9020 bp lengths, respectively were identified in nonaligned contigs (scaffolds 2 and 3) of SA H29 genome, and these two plasmids constitute rep20 and rep7C type genes. Plasmid p1H29 has and showed 44.20% and 57.34% sequence coverage with plasmids pBU108b (KF831356.1) and pPS00089.1A.1 (NZ_CP022911.1) respectively. Plasmid p2H29 has a length of and showed 23.42% and 23.45% sequence coverage with plasmid: II (LT671860.1), and UP_1395 plasmid (NZ_CP047821.1) respectively. Identified plasmid p1H29 carried genes encoding cadmium resistance (CadD) and transportation (CadX) proteins, and enterotoxins (EntA, EntD and EntG). Identified plasmid, p2H29 has blaZ (beta-lactamase), blaR, and mecI genes that conferred resistance to penicillin. The identified plasmids of S. aureus encode no transfer factors; thus, these plasmids may transfer via bacteriophage generalized transduction (McCarthy and Lindsay 2012).
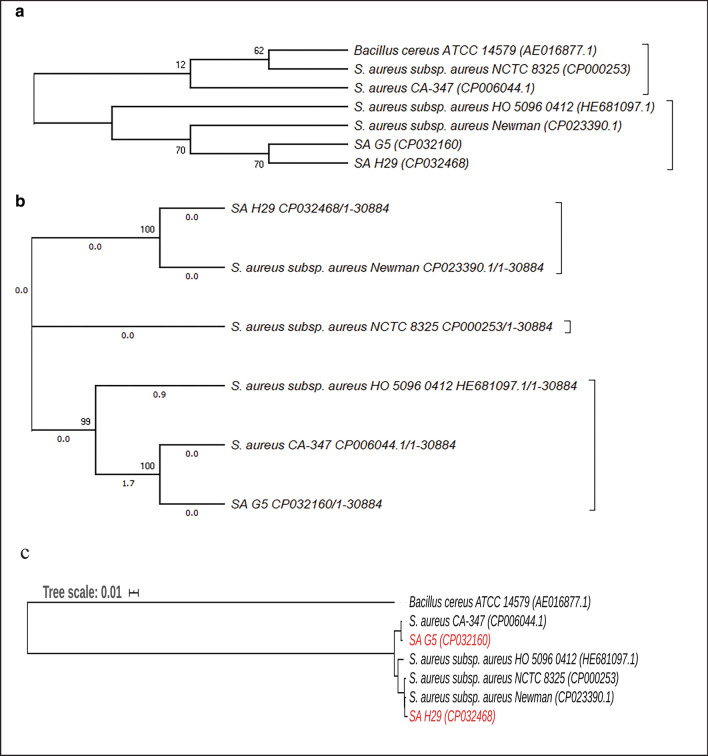
A comparative phylogenetic relationship was analyzed using three different gene sequences (viz., 16S rRNA genes, SNPs, and genes encoding core protein) to compare at the strain level. The phylogenetic tree generated using 16S rRNA gene sequences showed that SA G5 and SA H29 strains are highly similar and grouped them in the same group (Fig. 5b), however, the phylogenetic tree prepared from the SNP sequences and protein-coding gene sequences grouped them differently suggesting that 16S rRNA gene sequences are not sufficient to discriminate them on the strain level, possibly due to the low resolution of their evolutionary relationships (Konstantinidis and Tiedje 2007). It is interesting that both these methods and ANI values support each other and can be useful in distinguishing the genomes even in the strain level (Fig. 5b, c).
Fig. 5.
Comparative phylogenetic analysis of SA G5 and SA H29 strains with their closely related S. aureus strains. a Neighbor-joining tree prepared from the 16S rRNA gene sequences of SA G5, SA H29, and other related strains. Bacillus cereus ATCC 14,579 was used as an out-group and branch values are calculated from 500 bootstrap replicates. b Maximum likelihood (ML) tree based on all shared proteins (core) obtained from the S. aureus genome sequences, where B. cereus ATCC14579 was used as an out-group. The ML tree was prepared with the help of RAxML program. c Phylogeny tree based on the concatenated alignment of the high-quality SNPs using CSI phylogeny tool
Discussion
In the present study thirty-five European S. aureus clinical isolates were characterized by several different typing methods. The data generated from typing results along with virulence genes detection were evaluated through cluster analysis. Antibiotic susceptibility test and detection of mecA gene result showed 33 (94.28%) strains were MRSA while 2 (5.72%) strains were MSSA. Such a validation process was reported by other researchers (Skov et al. 2006; Kumurya 2015). Among the tested strains, 26 strains (71.43%) were found to be resistant to erythromycin. Prevalence of erythromycin-resistant strains collected from Hungary representing 90.47% in a good agreement with previous data (Szabó et al. 2009). It was also reported that the frequency of erythromycin-resistance during 2010–2015 was 72% in Germany (Walter et al. 2017) and 60% in Greece (Stefanaki et al. 2017). However, in the present study, no vancomycin-resistant strains were found which is supported by the observations of previous studies (Szabó et al. 2009; Chaudhari et al. 2014). Vancomycin kills Gram-positive bacteria by interfering with peptidoglycan synthesis and peptidoglycan assembly (McGuinness et al. 2017). The vancomycin susceptibility can be helpful in the treatment of MRSA infections. The resistance of S. aureus to vancomycin is governed by many factors. Previous studies have shown that point mutation in different regulatory loci associated with cell wall metabolism including two-component regulatory systems (guanylate kinase gene (gmk); walRK, graSR, and vraSR) resulted in vancomycin-resistant S. aureus strains (McAleese et al. 2006; Howden et al. 2008; Cui et al. 2009, 2010; Cameron et al. 2012; Shekarabi et al. 2017). In addition to cell wall thickening, altered surface protein profile, enhanced capsule, and agr gene dysfunction are also found to be the cause of the generation of vancomycin-resistant S. aureus. The presence of the vanA operon is also found to confer vancomycin-resistant (Périchon and Courvalin 2009; McGuinness et al. 2017). Also, it was observed that vancomycin-susceptibility in MRSA due to the activation of the WalRK two-component regulatory system (Cameron et al. 2016). The stains of the present study may neither have these mutations or cell wall-related modifications nor do they carry the genes required for vancomycin resistance and thus showed susceptible phenotype. However, a detailed study will be required to confirm the presence or absence of these factors.
Some of the MRSA strains produce PVL, encoded by two genes, lukS-PV, and lukF-PV which have been shown to play a role in the pathogenicity of S. aureus by provoking necrosis, accelerating apoptosis and destruction of polymorphonuclear and mononuclear-cells, thereby contributing to morbidity and mortality (Lina et al. 1999). It was reported that the low prevalence of pvl has found 5% and 4.9% in MRSA strains isolated from France and the UK respectively (Holmes et al. 2005). In the present study, 24.24% (8/33) strains were found positive to the pvl gene. Among these strains, three carried SCCmec type IV and one classified as SCCmec type V, however, four strains carried SCCmec II (Table 2). According to previous reports, MRSA strains belong to SCCmec types I, II, and III are dominant among the HA-MRSA, while SCCmec types IV and V characteristic of CA-MRSA (Monecke et al. 2011; Chua et al. 2014). In the present study, we found that SCCmec type II prevalence (12.12%) was higher in comparison with other SCCmec types. It was also reported that SCCmec type II usually presents in multidrug-resistant MRSA strains (Ito et al. 2001; Hiramatsu et al. 2001) and were dominant outside European countries (Kilic et al. 2006; Makgotlho et al. 2009; de Oliveira et al. 2015). Our data related to SCCmec type IV showed a higher prevalence (27.6%). The reason behind this observation is probably due to the easy acquisition of short size SCCmec type IV cassette (Robinson and Enright 2004). Even though the representation of non-typeable SCCmec in our case complies with the previous finding (Makgotlho et al. 2009), a few non-typeable SCCmec can be reduced by applying the new SCCmec cassette detection (Kaya et al. 2018). Some of the MRSA strains harbored SCCmec IVa showed signs of pvl gene negative (Table 2), which is similar to the finding, reported earlier (Moroney et al. 2007). Our data related to pvl gene detection revealed that MRSA strains harboring SCCmec IVb and SCCmec IVd were found negative. Our finding also supports the idea that the harboring bacteriophage pvl gene by MRSA strains may not be a promising marker for CA-MRSA (Rossney et al. 2007). This conclusion is supported by other studies about SCCmec typing for the classification of HA-MRSA and CA-MRSA (Monecke et al. 2011; Chua et al. 2014). Taken together, our finding suggested that the SCCmec typing method is more informative in problem-solving approaches (control and prevent infections caused by MRSA strains) for the clinicians and epidemiologists.
S. aureus secretes the coagulase enzyme, a polypeptide that helps promote the clotting of plasma or blood (Cheng et al. 2010). The coa gene shows heterogenicity in the 81 base-pair long tandem repeats region differing in number and location of restriction sites among the S. aureus isolates (Goh et al. 1992; Schwarzkopf and Karch 1994). The assay based on PCR amplification of the coa gene followed by RFLP was used to differentiate among the geographically diverse MRSA strains. This technique is simple, rapid, specific, inexpensive, and reproducible; allowing early recognition of an epidemic strain in a hospital setting (Martineau et al. 1996; Hookey et al. 1998; Shopsin et al. 2000; Mahmoudi et al. 2017). In this study, coa-PCR typing yielded seven different amplicons in size range from 550 to 800 bp. Among 15 S. aureus strains, the highest occurrence size is 700 bp (33.33%) as shown in Table 4, however, it was earlier reported that 600 bp amplicon is the most predominant (Mahmoudi et al. 2017). Discrimination of coa gene-specific amplicon pattern was further improved by HaeIII restriction enzyme digestion, which yielded 11 types of patterns with DI of 0.9619. Our data were in good agreement with the previous result in which DI was improved by digestion (Janwithayanuchit et al. 2006). S. aureus produces protein A, an antiphagocytic protein that is coded by the spa gene (Shakeri et al. 2010). The analysis of spa-sequence revealed high diversity, however, two strains belonged to spa-type t008, and another two were classified to spa-type t062 (Supplementary Table 3). In a previous study conducted on German, S. aureus isolates reported that t003 and t008 were predominant spa types (Strommenger et al. 2008). Also, a recently published article stated that t008 was the most prevalent spa type in Europe and America (Asadollahi et al. 2018). The spa-PCR typing method produced eleven different genotypes with variable amplicons size ranges from 335 to 560 bp and revealed 0.9429 DI, which provides similar DI with coa-HaeIII RFLP method which is supported by a previous study (Omar et al. 2014). This study suggested that spa-typing has performed better than other molecular typing methods and showed better DP. This typing method is useful for studying the genetic diversity of S. aureus for the epidemiological tracking of the source of infections (Koreen et al. 2004) and offering several advantages in comparison with alternatives methods, such as a publicly available comprehensive and curated database for analyzing spa sequence with standard nomenclature (Strommenger et al. 2008).
In a good agreement with previous data, we found that not all ica-positive isolates produce biofilms (Møretrø et al. 2003; Fitzpatrick et al. 2005), however, the clinical origin of the used S. aureus isolates was supported by the presence of icaADBC and icaR, indicating the ability to discriminate between the normal floras and virulence strains representatives (Galdbart et al. 2000). In this study, we observed that 66% of the isolates harbor two fnb genes almost similar to the results reported by Peacock et al. (2000) for European S. aureus strains. However, the presence of fnb genes in an isolate does not guarantee the biofilm-forming ability of the isolate.
In this study, the cna gene was found to be present in 53.3% isolates. Earlier, the prevalence of the cna gene was reported in a range from 22 to 56.5% (Peacock et al. 1999; Rohde et al. 2007; Zmantar et al. 2008), The clfA, clfB, and epbs genes play an initial role for biofilm development (Ghasemian et al. 2015), however, our data showed that the presence or absence of these genes does not represent a clear discriminative marker for differentiating strains in terms of biofilm-forming ability.
The grouping of the isolates based on the results of coa-HaeIII-RFLP and spa gene sequences produced different groups with different strains. Therefore, the results of all the biochemical and genotypic tests were used to prepare a dendrogram followed by PCA analysis. The dendrogram generated from the combined data of phenotypic and genotypic methods showed that the strain SA G5 and SA H29 collected from different geographical regions clustered together in the same group (Fig. 1b). Similar results were also obtained from the PCA analysis (Fig. 1c). To gain a better idea about their genetic background and closeness between the two strains, both strains were subjected to whole-genome sequencing and analysis. Comparative genome analysis of these two strains revealed 97.03% OrthoANI identity with a genetic distance of 0.0308, sharing 2344 gene clusters. The genes for virulence, resistance, plasmids, pathogenicity islands, prophage-like elements, and mobile elements are encoded by singleton genes of the SA H29 genome, suggesting that these singleton genes might be inherited through horizontal gene transfer (HGT) events (Lindsay 2014). Besides, the genes (5 genes) encoding lanthionine biosynthesis proteins were also found as singleton genes in the SA H29 genome. The presence of such genes influences the niche adaptation, pathogenesis, and contributes to evolution (Hacker and Carniel 2001). The map revealed a small gap with colorless region against the reference genome, which is due to the change in GC % content, this change in GC % content is due to the acquisition of gene through HGT (Hayek 2013) and the GC skewed regions indicated the regions where HGT occurred.
The 16S rRNA gene sequence-based phylogenetic analysis has been used widely to study the evolutionary relationships of microbes (Janda and Abbott 2002, 2007; Goswami et al. 2017). The phylogenetic tree generated using 16S rRNA gene sequences of the two isolates viz., SA G5, and SA H29 also clustered them in the same group whereas the other publicly available isolates were found in other groups (Fig. 5a) supporting the observations of the dendrogram and PCA analysis. However, the other two phylogenetic trees generated based on the SNP and core protein-coding gene sequences grouped the strain SA G5 and SA H29 in different groups. This indicated that these novel approaches of phylogenetic analysis using core protein-coding gene sequences and SNPs (Fig. 5b, c) are more powerful than the16S rRNA gene sequence-based phylogeny and are generally acceptable to distinguish the genomes even in the genus or species level, higher strain-level resolution. It has been reported that 16S rRNA gene sequence analysis is not powerful enough to discern clearly among the closely related species (Fox et al. 1992). Overall, the results of the present study indicate that whole-genome sequence analysis is more powerful than the individual genotypic test and provide better insight into the taxonomic and genotypic background of the test isolates.
Conclusion
Methicillin-resistant S. aureus is the leading cause of nosocomial and community infections and the emergence of hypervirulent strains and becoming a greater threat to the public. The high-frequency emergence of antibiotic-resistant could be due to the acquisition of resistance determinants such as plasmids, integrons, and transposons through horizontal or vertical gene transfer and partly by improper administration of antibiotics. The phenotypic and genotypic characterizations are important for identifying the risk factors associated with S. aureus infections and useful to monitor and control the circulation or transmission of these strains. But a comparative analysis of the pathogens based on the NGS-based genome sequencing technique could extend our understanding of pathogenesis and evolution at the molecular level and has the potential to a breakthrough in diagnosis, treatment, and infection control. In this study, the comparative genomic analysis revealed that niche-specific differences between the S. aureus strains in terms of genes and genes clusters that are related to amino acid metabolism, carbohydrate metabolism, cell envelope biogenesis, defense mechanisms, secondary metabolism, and phage-like elements. The difference in the presence of resistance genes (penicillin, methicillin, erythromycin, aminoglycoside, streptothricin acetyltransferase, and cadmium), VFGs (hlb, chp, scn, ear, qsa, sea, seb, sel, seg, lukD, lukE, sasH, clfA, and eap), plasmids and phage-related genes between the strains were also observed which may be due to the events like HGT and homologous recombination. Also, the comparative genome analysis provides high resolution to distinguish between the closely related sequenced strains which are indistinguishable by SCCmec and spa typing. The whole-genome analysis technique is a feasible tool to improve clinical diagnostic investigations of clinical infectious diseases in real-time and provides the goal of improving patient care.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was partially supported by the European Regional Development Fund (ERDF) under the grant numbers GINOP-2.3.2-15-2016-00021 and GINOP-2.3.2-15-2016-00047. The authors acknowledge the anonymous reviewers for their valuable suggestions that helped improve the quality of the manuscript.
Funding
Open access funding provided by University of Pécs.
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest.
References
- Al-Obaidi MMJ, Suhaili Z, Desa MNM (2018) Genotyping approaches for identification and characterization of Staphylococcus aureus. IntechOpen: London, UK, 2018
- Al-Tam F, Brunel A-S, Bouzinbi N, et al. DNAGear—a free software for spa type identification in Staphylococcus aureus. BMC Res Notes. 2012;5:642. doi: 10.1186/1756-0500-5-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MS, Isa NM, Abedelrhman FM, et al. Genomic analysis of methicillin-resistant Staphylococcus aureus strain SO-1977 from Sudan. BMC Microbiol. 2019;19:1–9. doi: 10.1186/s12866-019-1470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkharsah KR, Rehman S, Alnimr A, et al. Molecular typing of MRSA isolates by spa and PFGE. J King Saud Univ Sci. 2019;31:999–1004. doi: 10.1016/j.jksus.2018.07.018. [DOI] [Google Scholar]
- Appelbaum PC. Microbiology of antibiotic resistance in Staphylococcus aureus. Clin Infect Dis. 2007;45:S165–S170. doi: 10.1086/519474. [DOI] [PubMed] [Google Scholar]
- Arciola CR, Baldassarri L, Montanaro L. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J Clin Microbiol. 2001;39:2151–2156. doi: 10.1128/JCM.39.6.2151-2156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciola CR, Campoccia D, Baldassarri L, et al. Detection of biofilm formation in Staphylococcus epidermidis from implant infections. Comparison of a PCR-method that recognizes the presence ofica genes with two classic phenotypic methods. J Biomed Mater Res Part A. 2006;76A:425–430. doi: 10.1002/jbm.a.30552. [DOI] [PubMed] [Google Scholar]
- Arciola CR, Campoccia D, Ravaioli S, Montanaro L. Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects. Front Cell Infect Microbiol. 2015;5:7. doi: 10.3389/fcimb.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadollahi P, Farahani NN, Mirzaii M, et al. Distribution of the most prevalent spa types among clinical isolates of methicillin-resistant and -susceptible Staphylococcus aureus around the world: a review. Front Microbiol. 2018;9:163. doi: 10.3389/fmicb.2018.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Takeuchi F, Kuroda M, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- Bergey DH, Holt JG. Bergey’s manual of determinative bacteriology, Ninth edit. Baltimore: Williams & Wilkins, Philadelphia; 1994. [Google Scholar]
- Bosi E, Donati B, Galardini M, et al. MeDuSa: a multi-draft based scaffolder. Bioinformatics. 2015;31:2443–2451. doi: 10.1093/bioinformatics/btv171. [DOI] [PubMed] [Google Scholar]
- Cameron DR, Jiang JH, Kostoulias X, et al. Vancomycin susceptibility in methicillin-resistant Staphylococcus aureusis mediated by YycHI activation of the WalRK essential two-component regulatory system. Sci Rep. 2016;6:1–11. doi: 10.1038/srep30823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DR, Ward DV, Kostoulias X, et al. Serine/Threonine phosphatase Stp1 contributes to reduced susceptibility to vancomycin and virulence in Staphylococcus aureus. J Infect Dis. 2012;205:1677–1687. doi: 10.1093/infdis/jis252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A, Zankari E, García-Fernández A, et al. In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaieb K, Mahdouani K, Bakhrouf A. Detection of icaA and icaD loci by polymerase chain reaction and biofilm formation by Staphylococcus epidermidis isolated from dialysate and needles in a dialysis unit. J Hosp Infect. 2005;61:225–230. doi: 10.1016/j.jhin.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Chan CX, Beiko RG, Ragan MA. Lateral transfer of genes and gene fragments in staphylococcus extends beyond mobile elements. J Bacteriol. 2011;193:3964–3977. doi: 10.1128/JB.01524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari CN, Tandel K, Grover N, et al. In vitro vancomycin susceptibility amongst methicillin resistant Staphylococcus aureus. Med J Armed Forces India. 2014;70:215–219. doi: 10.1016/j.mjafi.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zheng D, Liu B, et al. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016;44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AG, McAdow M, Kim HK, et al. Contribution of Coagulases towards Staphylococcus aureus disease and protective immunity. PLoS Pathog. 2010;6:e1001036. doi: 10.1371/journal.ppat.1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua KYL, Howden BP, Jiang J-H, et al. Population genetics and the evolution of virulence in Staphylococcus aureu. Infect Genet Evol. 2014;21:554–562. doi: 10.1016/j.meegid.2013.04.026. [DOI] [PubMed] [Google Scholar]
- CLSI (2014) Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. CLSI document M100-S24. Clinical and laboratory standards institute, Wayne, PA, USA
- Collee JG, Fraser AG, Marmion BP. Mackie and McCartney Practical Medical Microbiology. 14. India: Elsevier Health Sciences; 1996. [Google Scholar]
- Cui L, Isii T, Fukuda M, et al. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob Agents Chemother. 2010;54:5222–5233. doi: 10.1128/AAC.00437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Neoh HM, Shoji M, Hiramatsu K. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53:1231–1234. doi: 10.1128/AAC.01173-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. ProgressiveMauve: multiple genome alignment with gene gain loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira LM, van der Heijden IM, Golding GR, et al. Staphylococcus aureus isolates colonizing and infecting cirrhotic and liver-transplantation patients: comparison of molecular typing and virulence factors. BMC Microbiol. 2015;15:264. doi: 10.1186/s12866-015-0598-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deka P, Goswami G, Das P, et al. Bacterial exopolysaccharide promotes acid tolerance in Bacillus amyloliquefaciens and improves soil aggregation. Mol Biol Rep. 2019;46:1079–1091. doi: 10.1007/s11033-018-4566-0. [DOI] [PubMed] [Google Scholar]
- Denis O. Route of transmission of Staphylococcus aureus. Lancet Infect Dis. 2017;17:124–125. doi: 10.1016/S1473-3099(16)30512-6. [DOI] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Chen C, Ding B, et al. Molecular characterization and antimicrobial susceptibility of nasal Staphylococcus aureus isolates from a Chinese Medical College Campus. PLoS ONE. 2011;6:e27328. doi: 10.1371/journal.pone.0027328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne YF, Viljoen A. Binding strength of gram-positive bacterial adhesins. Front Microbiol. 2020;11:1457. doi: 10.3389/fmicb.2020.01457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria NA, Carrico JA, Oliveira DC, et al. Analysis of typing methods for epidemiological surveillance of both methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Clin Microbiol. 2008;46:136–144. doi: 10.1128/JCM.01684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick F, Humphreys H, O’Gara JP. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J Clin Microbiol. 2005;43:1973–1976. doi: 10.1128/JCM.43.4.1973-1976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TJ, Geoghegan JA, Ganesh VK, Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox GE, Wisotzkey JD, Jurtshuk P. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- Galata V, Fehlmann T, Backes C, Keller A. PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res. 2019;47:D195–D202. doi: 10.1093/nar/gky1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdbart J, Allignet J, Tung H, et al. Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J Infect Dis. 2000;182:351–355. doi: 10.1086/315660. [DOI] [PubMed] [Google Scholar]
- Garrett TR, Bhakoo M, Zhang Z. Bacterial adhesion and biofilms on surfaces. Prog Nat Sci. 2008;18:1049–1056. doi: 10.1016/j.pnsc.2008.04.001. [DOI] [Google Scholar]
- Ghasemian A, Peerayeh SN, Bakhshi B, Mirzaee M. The microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) genes among clinical isolates of Staphylococcus aureus from hospitalized children. Iran J Pathol. 2015;10:258–264. doi: 10.7508/ijp.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh SH, Byrne SK, Zhang JL, Chow AW. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J Clin Microbiol. 1992;30:1642–1645. doi: 10.1128/JCM.30.7.1642-1645.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami G, Deka P, Das P, et al. Diversity and functional properties of acid-tolerant bacteria isolated from tea plantation soil of Assam. 3 Biotech. 2017;7(3):229. doi: 10.1007/s13205-017-0864-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi M, Seyedjavadi SS, Nasiri MJ, et al. Molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from patients with bacteremia based on MLST, SCCmec, spa, and agr locus types analysis. Microb Pathog. 2017;104:328–335. doi: 10.1016/j.micpath.2017.01.055. [DOI] [PubMed] [Google Scholar]
- Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann H, Aanensen DM, van den Wijngaard CC, et al. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 2010;7:e1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. EMBO Rep. 2001;2:376–381. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:3–9. [Google Scholar]
- Harmsen D, Claus H, Witte W, et al. Typing of methicillin-resistant Staphylococcus aureus in a University Hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayek N. Lateral transfer and GC content of bacterial resistant genes. Front Microbiol. 2013;4:41. doi: 10.3389/fmicb.2013.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu K, Cui L, Kuroda M, Ito T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2001;9:486–493. doi: 10.1016/S0966-842X(01)02175-8. [DOI] [PubMed] [Google Scholar]
- Hisata K, Kuwahara-Arai K, Yamanoto M, et al. Dissemination of methicillin-resistant staphylococci among healthy Japanese children. J Clin Microbiol. 2005;43:3364–3372. doi: 10.1128/JCM.43.7.3364-3372.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TM, Zhou C, Lindgren JK, et al. Transcriptional regulation of icaADBC by both IcaR and TcaR in Staphylococcus epidermidis. J Bacteriol. 2019;201:e00524–e618. doi: 10.1128/JB.00524-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A, Ganner M, McGuane S, et al. Staphylococcus aureus isolates carrying panton-valentine Leucocidin genes in England and Wales: frequency, characterization, and association with clinical disease. J Clin Microbiol. 2005;43:2384–2390. doi: 10.1128/JCM.43.5.2384-2390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hookey JV, Richardson JF, Cookson BD. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J Clin Microbiol. 1998;36:1083–1089. doi: 10.1128/JCM.36.4.1083-1089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden BP, Stinear TP, Allen DL, et al. Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob Agents Chemother. 2008;52:3755–3762. doi: 10.1128/AAC.01613-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://insilico.ehu.es/mini_tools/discriminatory_power/ Discriminatory power Calculator. Accessed 8 Feb 2019
- https://spa.ridom.de/ Ridom SpaServer
- https://spatyper.fortinbras.us/ spa Type Finder/Identifier. https://spatyper.fortinbras.us/. Accessed 8 Feb 2019
- https://cge.cbs.dtu.dk/services/CSIPhylogeny/ CSI Phylogeny 1.4 (Call SNPs & Infer Phylogeny). Accessed 2 Mar 2019
- Hughes AL, Friedman R. Nucleotide substitution and recombination at orthologous loci in Staphylococcus aureus. J Bacteriol. 2005;187:2698–2704. doi: 10.1128/JB.187.8.2698-2704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Katayama Y, Asada K, et al. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda JM, Abbott SL. 16S rRNA Gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007;45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda JM, Abbott SL. Bacterial identification for publication: when is enough enough? J Clin Microbiol. 2002;40:1887–1891. doi: 10.1128/JCM.40.6.1887-1891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen WTM, van der Bruggen JT, Verhoef J, Fluit AC. Bacterial resistance: a sensitive issue. complexity of the challenge and containment strategy in Europe. Drug Resist Updat. 2006;9:123–133. doi: 10.1016/j.drup.2006.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janwithayanuchit I, Ngam-Ululert S, Paungmoung P, Rangsipanuratn W. Epidemiologic study of methicillin-resistant Staphylococcus aureus by coagulase gene polymorphism. ScienceAsia. 2006;32:127–132. doi: 10.2306/scienceasia1513-1874.2006.32.127. [DOI] [Google Scholar]
- Karahan M, Çetinkaya B. Coagulase gene polymorphisms detected by PCR in Staphylococcus aureus isolated from subclinical bovine mastitis in Turkey. Vet J. 2007;174:428–431. doi: 10.1016/j.tvjl.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Kareem SM, Aljubori SS, Ali MR. Novel determination of spa gene diversity and its molecular typing among Staphylococcus aureus Iraqi isolates obtained from different clinical samples. New Microbes New Infect. 2020;34:100653. doi: 10.1016/j.nmni.2020.100653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H, Hasman H, Larsen J, et al. SCCmecFinder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere. 2018;3(1):e00612. doi: 10.1128/mSphere.00612-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshkharam-Roodmajani H, Sarvari J, Bazargani A, et al. Molecular typing of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from Shiraz teaching hospitals by PCR-RFLP of coagulase gene. Iran J Microbiol. 2014;6:246–252. [PMC free article] [PubMed] [Google Scholar]
- Kilic A, Li H, Stratton CW, Tang Y-W. Antimicrobial susceptibility patterns and staphylococcal cassette chromosome mec types of, as well as panton-valentine leukocidin occurrence among, methicillin-resistant Staphylococcus aureus isolates from children and adults in middle Tennessee. J Clin Microbiol. 2006;44:4436–4440. doi: 10.1128/JCM.01546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol. 2007;10:504–509. doi: 10.1016/j.mib.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Koreen L, Ramaswamy SV, Graviss EA, et al. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macro variation. J Clin Microbiol. 2004;42:792–799. doi: 10.1128/jcm.42.2.792-799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köser CU, Ellington MJ, Cartwright EJP, et al. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog. 2012;8:e1002824. doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumurya A. One step PCR for detection of Staphylococcus aureus specific sequence gene and mecA gene in Northwestern Nigerian hospitals. Antimicrob Resist Infect Control. 2015;4:P196. doi: 10.1186/2047-2994-4-S1-P196. [DOI] [Google Scholar]
- Le KY, Park MD, Otto M. Immune evasion mechanisms of Staphylococcus epidermidis biofilm infection. Front Microbiol. 2018;9:359. doi: 10.3389/fmicb.2018.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina G, Piémont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- Lindsay JA. Staphylococcus aureus genomics and the impact of horizontal gene transfer. Int J Med Microbiol. 2014;304:103–109. doi: 10.1016/j.ijmm.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Lindsay JA, Moore CE, Day NP, et al. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J Bacteriol. 2006;188:669–676. doi: 10.1128/JB.188.2.669-676.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D, Fischer W, Krokotsch A, et al. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178:175–183. doi: 10.1128/JB.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah T-FC, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Mahmoudi H, Arabestani MR, Mousavi SF, Alikhani MY. Molecular analysis of the coagulase gene in clinical and nasal carrier isolates of methicillin-resistant Staphylococcus aureus by restriction fragment length polymorphism. J Glob Antimicrob Resist. 2017;8:41–45. doi: 10.1016/j.jgar.2016.10.007. [DOI] [PubMed] [Google Scholar]
- Makgotlho PE, Kock MM, Hoosen A, et al. Molecular identification and genotyping of MRSA isolates. FEMS Immunol Med Microbiol. 2009;57:104–115. doi: 10.1111/j.1574-695X.2009.00585.x. [DOI] [PubMed] [Google Scholar]
- Maljkovic Berry I, Melendrez MC, Bishop-Lilly KA, et al. Next generation sequencing and bioinformatics methodologies for infectious disease research and public health: approaches, applications, and considerations for development of laboratory capacity. J Infect Dis. 2019;221:S292–S307. doi: 10.1093/infdis/jiz286. [DOI] [PubMed] [Google Scholar]
- Martineau F, Picard FJ, Roy PH, et al. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus epidermidis. J Clin Microbiol. 1996;34:2888–2893. doi: 10.1128/JCM.34.12.2888-2893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAleese F, Wu SW, Sieradzki K, et al. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S.aureus-type resistance to vancomycin. J Bacteriol. 2006;188:1120–1133. doi: 10.1128/JB.188.3.1120-1133.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy AJ, Lindsay JA. The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol. 2012;12:104. doi: 10.1186/1471-2180-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure J-AM, Lakhundi S, Kashif A, et al. Genomic comparison of highly virulent, moderately virulent, and avirulent strains from a genetically closely-related MRSA ST239 sub-lineage provides insights into pathogenesis. Front Microbiol. 2018;9:1531. doi: 10.3389/fmicb.2018.01531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness WA, Malachowa N, DeLeo FR. Vancomycin resistance in Staphylococcus aureus. Yale J Biol Med. 2017;90:269–281. [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry H, Sharma P, Mahato S, et al. Prevalence and characterization of oxacillin susceptible mecA-positive clinical isolates of Staphylococcus aureus causing bovine mastitis in India. PLoS ONE. 2016;11:e0162256. doi: 10.1371/journal.pone.0162256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke S, Coombs G, Shore AC, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE. 2011;6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møretrø T, Hermansen L, Holck AL, et al. Biofilm formation and the presence of the intercellular adhesion locus ica among staphylococci from food and food processing environments. Appl Environ Microbiol. 2003;69:5648–5655. doi: 10.1128/AEM.69.9.5648-5655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney SM, Heller LC, Arbuckle J, et al. Staphylococcal cassette chromosome mec and panton-valentine leukocidin characterization of methicillin-resistant Staphylococcus aureus clones. J Clin Microbiol. 2007;45:1019–1021. doi: 10.1128/JCM.01706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottola C, Semedo-Lemsaddek T, Mendes JJ, et al. Molecular typing, virulence traits and antimicrobial resistance of diabetic foot staphylococci. J Biomed Sci. 2016;23:33. doi: 10.1186/s12929-016-0250-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Bankevich A, Antipov D, et al. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) Berlin, Heidelberg: Springer; 2013. Assembling genomes and mini-metagenomes from highly chimeric reads; pp. 158–170. [Google Scholar]
- O’Hara FP, Suaya JA, Ray GT, et al. Spa typing and multilocus sequence typing show comparable performance in a macro epidemiologic study of Staphylococcus aureus in the United States. Microb Drug Resist. 2016;22:88–96. doi: 10.1089/mdr.2014.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar NY, Ali HAS, Harfoush RAH, El Khayat EH. Molecular typing of methicillin resistant Staphylococcus aureus clinical isolates on the basis of protein A and coagulase gene polymorphisms. Int J Microbiol. 2014;2014:650328. doi: 10.1155/2014/650328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Olson R, Pusch GD, et al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock SJ, Day NP, Thomas MG, et al. Clinical isolates of Staphylococcus aureus exhibit diversity in fnb genes and adhesion to human fibronectin. J Infect. 2000;41:23–31. doi: 10.1053/jinf.2000.0657. [DOI] [PubMed] [Google Scholar]
- Peacock SJ, Foster TJ, Cameron BJ, Berendt AR. Bacterial fibronectin-binding proteins and endothelial cell surface fibronectin mediate adherence of Staphylococcus aureus to resting human endothelial cells. Microbiology. 1999;145:3477–3486. doi: 10.1099/00221287-145-12-3477. [DOI] [PubMed] [Google Scholar]
- Périchon B, Courvalin P. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53:4580–4587. doi: 10.1128/AAC.00346-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi F, Katouli M, Karimi S. Biofilm production among methicillin resistant Staphylococcus aureus strains isolated from catheterized patients with urinary tract infection. Microb Pathog. 2016;98:69–76. doi: 10.1016/j.micpath.2016.06.031. [DOI] [PubMed] [Google Scholar]
- Raven KE, Blane B, Kumar N, et al. Defining metrics for whole-genome sequence analysis of MRSA in clinical practice. Microb Genomics. 2020;6:e000354. doi: 10.1099/mgen.0.000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezai S, Valadan R, Ahangarkani F, Rezai MS. The spa typing and characterization of nasal carriage methicillin-resistant Staphylococcus aureus isolates from healthy children. J Pediatr Rev. 2019;8:59–64. doi: 10.32598/jpr.8.1.59. [DOI] [Google Scholar]
- Robinson DA, Enright MC. Multilocus sequence typing and the evolution of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2004;10:92–97. doi: 10.1111/j.1469-0691.2004.00768.x. [DOI] [PubMed] [Google Scholar]
- Rohde H, Burandt EC, Siemssen N, et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28:1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Rossney AS, Shore AC, Morgan PM, et al. The emergence and importation of diverse genotypes of methicillin-resistant Staphylococcus aureus (MRSA) harboring the panton-valentine leukocidin gene (pvl) reveal that pvl Is a poor marker for community-acquired MRSA strains in Ireland. J Clin Microbiol. 2007;45:2554–2563. doi: 10.1128/JCM.00245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzkopf A, Karch H. Genetic variation in Staphylococcus aureus coagulase genes: potential and limits for use as epidemiological marker. J Clin Microbiol. 1994;32:2407–2412. doi: 10.1128/JCM.32.10.2407-2412.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakeri F, Shojai A, Golalipour M, et al. Spa diversity among MRSA and MSSA strains of Staphylococcus aureus in North of Iran. Int J Microbiol. 2010;2010:351397. doi: 10.1155/2010/351397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekarabi M, Hajikhani B, Salimi Chirani A, et al. Molecular characterization of vancomycin-resistant Staphylococcus aureus strains isolated from clinical samples: a three year study in Tehran, Iran. PLoS ONE. 2017;12:1–10. doi: 10.1371/journal.pone.0183607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shittu AO, Okon K, Adesida S, et al. Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol. 2011;11:92. doi: 10.1186/1471-2180-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopsin B, Gomez M, Waddington M, et al. Use of Coagulase Gene Repeat Region Nucleotide Sequences for Typing of Methicillin-Resistant Staphylococcus aureus; Strains. J Clin Microbiol. 2000;38:3453–3456. doi: 10.1128/JCM.38.9.3453-3456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SK, Pantrang M, Stahl B, et al. Comparative whole-genome mapping to determine Staphylococcus aureus genome size, virulence motifs, and clonality. J Clin Microbiol. 2012;50:3526–3533. doi: 10.1128/JCM.01168-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Goering RV, Simjee S, et al. Application of molecular techniques to the study of hospital infection. Clin Microbiol Rev. 2006;19:512–530. doi: 10.1128/CMR.00025-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skov R, Smyth R, Larsen AR, et al. Phenotypic detection of methicillin resistance in Staphylococcus aureus by disk diffusion testing and Etest on Mueller-Hinton agar. J Clin Microbiol. 2006;44:4395–4399. doi: 10.1128/JCM.01411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanaki C, Ieronymaki A, Matoula T, et al. Six-year retrospective review of hospital data on antimicrobial resistance profile of Staphylococcus aureus isolated from skin infections from a single institution in Greece. Antibiotics. 2017;6:39. doi: 10.3390/antibiotics6040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani S, Varaldo PE. Epidemiology of methicillin-resistant staphylococci in Europe. Clin Microbiol Infect. 2003;9:1179–1186. doi: 10.1111/j.1469-0691.2003.00698.x. [DOI] [PubMed] [Google Scholar]
- Strandén A, Frei R, Widmer AF. Molecular typing of methicillin-resistant Staphylococcus aureus: can PCR replace pulsed-field gel electrophoresis? J Clin Microbiol. 2003;41:3181–3186. doi: 10.1128/jcm.41.7.3181-3186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strommenger B, Braulke C, Heuck D, et al. spa Typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J Clin Microbiol. 2008;46:574–581. doi: 10.1128/JCM.01599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strommenger B, Kettlitz C, Werner G, Witte W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J Clin Microbiol. 2003;41:4089–4094. doi: 10.1128/JCM.41.9.4089-4094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó J, Dombrádi Z, Dobay O, et al. Phenotypic and genetic characterisation of methicillin-resistant Staphylococcus aureus strains isolated from the university hospitals of Debrecen. Eur J Clin Microbiol Infect Dis. 2009;28:129–136. doi: 10.1007/s10096-008-0588-1. [DOI] [PubMed] [Google Scholar]
- Tatusova T, DiCuccio M, Badretdin A, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, Tickler IA, Le VM, et al. Updating molecular diagnostics for detecting methicillin-susceptible and methicillin-resistant Staphylococcus aureus isolates in blood culture bottles. J Clin Microbiol. 2019;57:e01195–e1219. doi: 10.1128/JCM.01195-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestby LK, Grønseth T, Simm R, Nesse LL. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics. 2020;9:59. doi: 10.3390/antibiotics9020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong C, Kocianova S, Voyich JM, et al. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- Walter J, Noll I, Feig M, et al. Decline in the proportion of methicillin resistance among Staphylococcus aureus isolates from non-invasive samples and in outpatient settings, and changes in the co-resistance profiles: an analysis of data collected within the Antimicrobial Resistance Surveillance Network, Germany 2010 to 2015. BMC Infect Dis. 2017;17:169. doi: 10.1186/s12879-017-2271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam AR, Davis JJ, Assaf R, et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017;45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Dong Z, Fang L, et al. OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019;47:W52–W58. doi: 10.1093/nar/gkz333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Hung W-C, Takano T, Nishiyama A. Genetic nature and virulence of community-associated methicillin-resistant Staphylococcus aureus. BioMedicine. 2013;3:2–18. doi: 10.1016/j.biomed.2012.12.001. [DOI] [Google Scholar]
- Yoon S-H, Ha S, Lim J, et al. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- Zhang K, McClure J-A, Conly JM. Enhanced multiplex PCR assay for typing of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. Mol Cell Probes. 2012;26:218–221. doi: 10.1016/j.mcp.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Zhang K, McClure J-A, Elsayed S, et al. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec Types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmantar T, Chaieb K, Makni H, et al. Detection by PCR of adhesins genes and slime production in clinical Staphylococcus aureus. J Basic Microbiol. 2008;48:308–314. doi: 10.1002/jobm.200700289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.