Abstract
We have previously shown that the incomplete splicing of exon 1 to exon 2 of the HTT gene results in the production of a small polyadenylated transcript (Httexon1) that encodes the highly pathogenic exon 1 HTT protein. There is evidence to suggest that the splicing factor SRSF6 is involved in the mechanism that underlies this aberrant splicing event. Therefore, we set out to test this hypothesis, by manipulating SRSF6 levels in Huntington’s disease models in which an expanded CAG repeat had been knocked in to the endogenous Htt gene. We began by generating mice that were knocked out for Srsf6, and demonstrated that reduction of SRSF6 to 50% of wild type levels had no effect on incomplete splicing in zQ175 knockin mice. We found that nullizygosity for Srsf6 was embryonic lethal, and therefore, to decrease SRSF6 levels further, we established mouse embryonic fibroblasts (MEFs) from wild type, zQ175, and zQ175::Srsf6+/− mice and transfected them with an Srsf6 siRNA. The incomplete splicing of Htt was recapitulated in the MEFs and we demonstrated that ablation of SRSF6 did not modulate the levels of the Httexon1 transcript. We conclude that SRSF6 is not required for the incomplete splicing of HTT in Huntington’s disease.
Subject terms: Molecular biology, Neuroscience, Diseases, Neurology
Introduction
Huntington’s disease is a monogenic neurodegenerative disorder that manifests with psychiatric, motor and cognitive symptoms1. It is caused by a CAG repeat expansion mutation in the first exon of the huntingtin (HTT) gene2, which encodes an expanded polyglutamine tract in the HTT protein. There are currently no effective treatments to delay the onset or slow the progression of the disease and current efforts are largely focused on knocking down HTT mRNA3–6. This is a particularly attractive target as it is upstream of the pathogenic HTT protein, and therefore, understanding how the HTT mRNA is processed in the context of Huntington’s disease could inform future therapeutic strategies.
Both human HTT and mouse Htt are known to be transcribed into five mature protein-coding mRNAs: three endogenous 67 exon full-length transcripts that differ by the length of their 3′ UTRs and encode a 350 kDa protein7,8 and, in the context of an expanded CAG repeat, two transcripts that contain exon 1 and intron 1 sequences (HTTexon1), and are translated to produce the exon 1 HTT protein9. The HTTexon1 mRNA is generated by the incomplete splicing of exon 1 to exon 2 of the HTT gene and the aberrant activation of one of two cryptic polyadenylation (polyA) signals in intron 1 resulting in premature termination. These cryptic polyA signals are located at 680 and 1,145 bp into intron 1 for mouse Htt9,10 and 2,710 and 7,327 bp into intron 1 for human HTT11. We have previously shown that HTTexon1 is present in Huntington’s disease patient post-mortem brains and fibroblast lines11 as well as all Huntington’s disease mouse models that contain a mutant version of the complete mouse or human gene12,13. The exon 1 HTT protein, that this small transcript encodes, is highly toxic14,15 and aggregation-prone16.
The RNA processing mechanisms that result in the production of Httexon1 have yet to be elucidated17. Expanded CAG repeats within RNA form hairpin loops18 and are known to sequester a number of RNA binding proteins in a CAG repeat-dependent fashion18,19. In other microsatellite repeat disorders such as amyotrophic lateral sclerosis—frontal temporal dementia20 and myotonic dystrophy21, functional sequestration of RNA binding proteins by expanded polynucleotide repeat RNA is known to impact cellular RNA processing events and lead to pathogenesis. It is therefore plausible that the incomplete splicing of HTT is caused by sequestration of RNA-binding proteins in a CAG repeat-dependent fashion. Bioinformatic analysis indicated that the predicted recognition sequence for serine/arginine-rich splicing factor 6 (SRSF6) included a CAG repeat, and an SRSF6 antibody immunoprecipitated greater levels of 5′ Htt RNA sequences from the zQ175 knockin brain lysates than from wild type mice9. That SRSF6 has a greater binding affinity to HTTexon1 with an expanded CAG repeat than to HTTexon1 with a CAG repeat in the normal range was supported by an independent study19. Finally, we showed that incomplete splicing of a mouse Htt minigene decreased following SRSF6 knockdown by RNA interference and increased following SRSF6 overexpression in cell culture10. SRSF6 is a member of the serine/arginine-rich (SR) family of proteins, an evolutionary conserved group defined by one or two N-terminal RNA-binding domains and a C-terminal SR domain, which mediates protein–protein interactions22,23. SR proteins mainly localise to the nucleus but some also shuttle between the nucleus and cytoplasm24. They all function as constitutive and alternative splicing factors but have also been shown to operate in a number of co-transcriptional and co-translational processes.
In this study, we set out to investigate whether SRSF6 reduction modulated Htt splicing in vivo. To do this, a constitutive Srsf6 knockout mouse model was generated and nullizygosity for Srsf6 was found to be embryonic lethal. Therefore, heterozygous Srsf6+/− mice were bred to the zQ175 knockin mouse model of Huntington’s disease to examine the effect of decreasing SRSF6 to 50% of wild type levels. This was found to have no effect on the levels of Httexon1 in various zQ175 brain regions. We then derived mouse embryonic fibroblasts (MEFs) from the progeny of the Srsf6+/− x zQ175 cross to investigate the effects of further lowering SRSF6 levels by RNA interference. We found that the incomplete splicing of Htt was recapitulated in the MEFs and that SRSF6 ablation had no effect on the incomplete splicing of Htt. We conclude that SRSF6 levels do not play a role in the mechanism that underlies this aberrant splicing event.
Results
Characterisation of the Srsf6 knockout mouse model
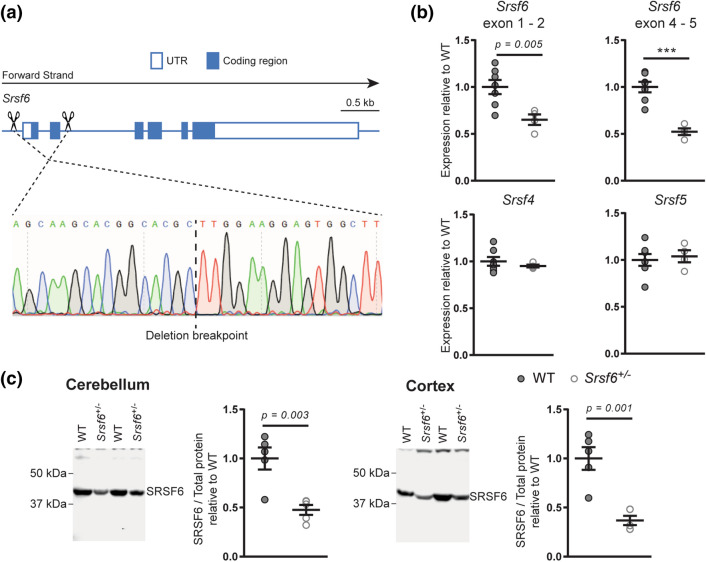
In order to study the effect of SRSF6 reduction on the incomplete splicing of Htt, Srsf6 knockout mice were generated by the Jackson Laboratory using CRISPR/Cas9. Guide RNAs targeting sequences upstream of the 5′ untranslated region and within intron 2 were used to generate the Srsf6 knockout alleles (Fig. 1a), giving rise to four founder lines. We used TaqMan real-time quantitative PCR (qPCR) assays to measure cortical transcripts encoding SR proteins in two of the founder lines with deletion sizes of 990 bp and 956 bp. mRNA levels were measured for Srsf6 and also Srsf4 and Srsf5, the two paralogs with the greatest RNA and protein sequence homology22. To determine whether transcriptional inactivation of the Srsf6 knockout allele had occurred, we used two Srsf6 assays: one at the boundary of exon 1–2 (corresponding to part of the deleted sequence of the knockout allele) and the other at the boundary of exon 4–5 (which remained intact on the knockout allele). The transcripts for the SR proteins were normalised to Canx, Ubc and Atp5b as reference genes. We found that Srsf6 levels were 50% of wild type (WT) in both heterozygous Srsf6 strains (Srsf6+/−(Δ990bp) and Srsf6+/−(Δ956bp)) using both the exon 1–2 and exon 4–5 assays (Fig. 1b, Supplementary Fig. S1). This implies that transcription of the knockout allele had been inactivated. There were no significant differences in Srsf4 and Srsf5 levels in either of the heterozygous Srsf6 founder strains compared to WT indicating that the Srsf6 paralogs are not upregulated to compensate for the deleted gene (Fig. 1b, Supplementary Fig. S1). Since we saw no differences between the two founder lines at the mRNA level, we performed all succeeding experiments with the Srsf6+/−(Δ990bp) mice, which shall be denoted Srsf6+/− from this point forward. The deletion breakpoint was confirmed by DNA sequencing (Fig. 1a) and we verified that the promoter of Srsf6, as predicted by the eukaryotic promoter database25, had been removed. We observed a concurrent two-fold reduction in SRSF6 protein levels in cortex and cerebellum of Srsf6+/− compared to WT (Fig. 1c).
Figure 1.
Characterisation of the Srsf6 knockout mouse line. (a) Schematic of mouse Srsf6 with scissors denoting approximate Cas9 cleavage sites used to generate the knockout allele. Sanger sequencing was used to confirm the deletion breakpoint for the Δ990 bp line. UTR untranslated region. (b) qPCR analysis showed Srsf6 mRNA levels to be 50% of WT whereas, Srsf4 or Srsf5 levels were unchanged in cortex from 2 month old Srsf6+/− Δ990 mice. n = 7 WT and 4 Srsf6+/− mice. (c) Western blot analysis showed a 50% reduction in SRSF6 in cerebellum and cortex from 2 month old Srsf6+/− mice compared to WT littermates. See Supplementary Fig. S7 for uncropped blots and total protein loading controls. n = 5/genotype. Statistical analyses were by unpaired Student’s t-tests. Test statistics can be found in Supplementary Table S4. WT wild type.
To assess the viability of homozygous Srsf6 knockout mice (Srsf6−/−), we intercrossed Srsf6+/− heterozygous mice. From the twenty-eight progeny, we observed that nine (32.1%) were WT, nineteen (67.9%) were Srsf6+/− and none were Srsf6−/− homozygotes (Table 1). We confirmed that our observed distribution was significantly different from the expected distribution assuming that Srsf6−/− mice were viable (χ2(2) = 9.357, p = 0.009) but not significantly different from the expected distribution assuming that Srsf6−/− mice were inviable (χ2(1) = 0.018, p = 0.895) (Table 1). Therefore, we conclude that homozygosity for Srsf6 knockout is embryonically lethal in mice and all in vivo experiments going forward utilised Srsf6+/− heterozygous mice.
Table 1.
Genotypes of the progeny from the Srsf6+/− intercross.
| Genotype | Progeny observed | Percentage | Hypothesis 1 expected values (Srsf6−/− viable) | Hypothesis 2 expected values (Srsf6−/− inviable) |
|---|---|---|---|---|
| WT | 9 (4 m, 5f.) | 32.1% | 7 (25%) | 9.33 (33.3%) |
| Srsf6+/− | 19 (9 m, 10f.) | 67.9% | 14 (50%) | 18.66 (66.7%) |
| Srsf6−/− | 0 | 0% | 7 (25%) | 0 (0%) |
| Statistics | ||||
| Χ2 | 9.357 | 0.018 | ||
| Degrees of freedom | 2 | 1 | ||
| p-value | 0.009 | 0.895 | ||
| Statistical significance? | Yes—distributions different | No—distributions the same | ||
Heterozygosity for Srsf6 knockout does not change Htt splicing patterns in zQ175 mouse brain
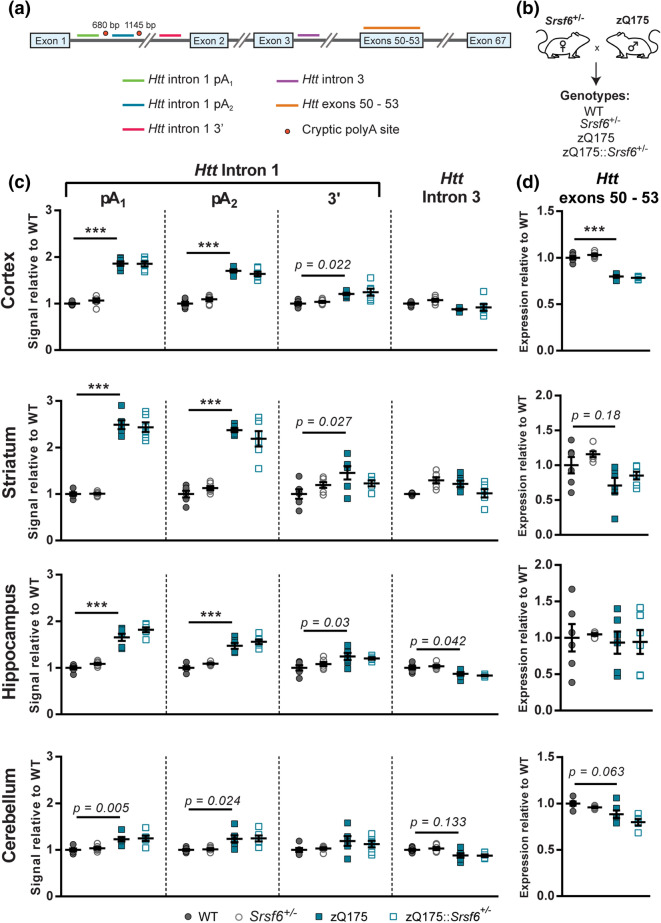
We used the zQ175 knockin model of Huntington’s disease to investigate the effect of heterozygous Srsf6 knockout on incomplete splicing. The zQ175 mice were generated by replacing mouse Htt exon 1 with human HTT exon 1 with an expanded CAG repeat26–28, and the Httexon1 transcript can be readily detected in this model using a previously designed high-throughput QuantiGene plex assay29. The QuantiGene plex included the following probe sets: five Htt, Srsf4, Srsf5, two Srsf6 and a number of housekeeping reference genes (Supplementary Table S1). Four of the five Htt targets were intronic sequences and the fifth spanned exons 50–53, which measured the level of the processed full-length Htt transcript (Fig. 2a). Three intronic Htt targets were in intron 1: before the first cryptic polyA signal (Htt intron 1 pA1), between the two cryptic polyA signals (Htt intron 1 pA2) and at the 3′ end of intron 1 (Htt intron 1 3′) (Fig. 2a). Htt intron 1 pA1 detected the Httexon1 mRNA which terminated at the first or second polyA signal and Htt intron 1 Htt pA2 targets Httexon1 terminated at the second polyA signal. Htt intron 1 3′ and Htt intron 3 detected unprocessed mRNAs that contained intron 1 or intron 3 sequences (Fig. 2a).
Figure 2.
QuantiGene analysis of Htt transcripts in brain regions from the progeny of the zQ175 and Srsf6+/− mouse cross. (a) Schematic of the location of the QuantiGene plex probe sets on the mouse Htt transcript. (b) Srsf6+/− female mice were bred to zQ175 knockin male mice to generate progeny with four genotypes: WT, Srsf6 heterozygous knockout (Srsf6+/−), zQ175 knockin and double mutants (zQ175::Srsf6+/−). (c) Httexon1 was detected in the cortex, striatum, hippocampus and cerebellum of 2 month old zQ175 mice but was not altered by heterozygosity for Srsf6 knockout. (d) Full-length Htt was measured in the cortex, striatum, hippocampus and cerebellum using the Htt exon 50–53 assay. Cortical full-length Htt was significantly lower in zQ175 mice compared to WT and this was not changed by heterozygosity for Srsf6 knockout. n = 6/genotype. Statistical analysis was by one-way ANOVA with Bonferroni correction for multiple pairwise comparisons, ***p < 0.001, p < 0.2 values are indicated. Test statistics can be found in Supplementary Table S5. WT wild type.
To investigate the effect of SRSF6 reduction on incomplete splicing, we bred Srsf6+/− mice to zQ175 knockins, generating four genotypes: WT, zQ175, Srsf6+/− and zQ175::Srsf6+/− (Fig. 2b). The QuantiGene plex was used to measure the levels of the SR protein transcripts and determine the effect of SRSF6 reduction on Htt and Httexon1 transcripts in cortex, striatum, hippocampus and cerebellum from 2 month-old mice. As expected, Srsf6 was reduced to 50% of WT levels in both the Srsf6+/− and zQ175::Srsf6+/− mice whereas Srsf4 and Srsf5 were unchanged (Supplementary Fig. S2). The Httexon1 transcript was present in all brain regions in zQ175 mice, with the highest levels detected in striatum and the lowest levels detected in cerebellum (Fig. 2c), consistent with previous data29. The level of the Httexon1 transcript detected by the Htt intron 1 pA2 probe set was comparable to that detected by Htt intron 1 pA1, suggesting that the majority of Httexon1 terminated at the second cryptic polyA site (Fig. 2c). The reduction of SRSF6 in zQ175 mice had no effect on the level of Httexon1. Comparison of the levels of the 3′ end of intron 1 and intron 3 in the four genotypes, revealed small changes, with an increase in intron 1 and a decrease in intron 3 in some brain regions, possibly reflecting effects of the mutation on Htt mRNA processing (Fig. 2c).
The level of the full-length Htt transcript was decreased in cortex, which may reflect a combination of incomplete splicing and a reduction in transcription as a consequence of the expanded CAG repeat30 (Fig. 2d). In contrast to our previous data, we did not detect a reduction in full-length Htt in the striatum and hippocampus29. The exon 50–53 probe set produced more variable data than probe sets for full-length Htt that we have designed to the 3′UTR29, which we would recommend using for future studies. Heterozygosity for Srsf6 in WT mice did not affect the levels of full-length Htt in any of these brain regions (Fig. 2d).
Httexon1 can be detected in zQ175 mouse embryonic fibroblasts (MEFs)
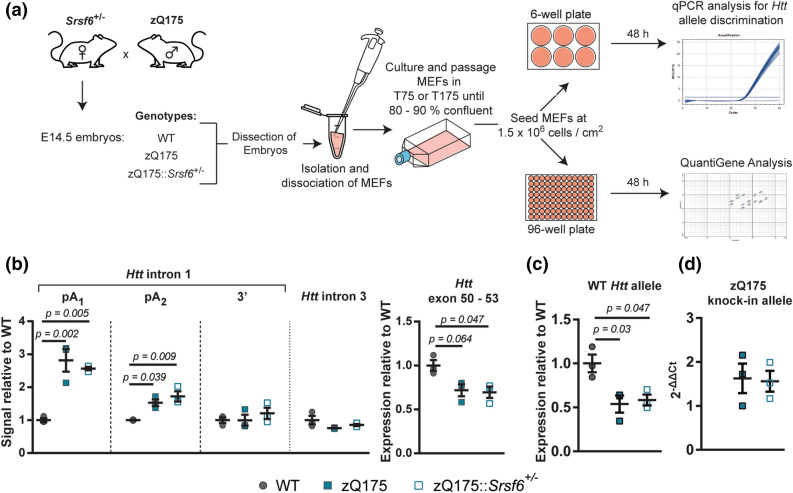
We sought to generate a cell model in which Httexon1 mRNA could be detected. zQ175 males were bred to Srsf6+/− females, E14.5 embryos were harvested and mouse embryonic fibroblasts (MEFs) from these embryos were isolated and cultured (Fig. 3a). QuantiGene analysis demonstrated that Httexon1 mRNA could be detected in the zQ175 and zQ175::Srsf6+/− MEFs at comparable levels (Fig. 3b), consistent with our in vivo data. Most of the Httexon1 transcript terminated at the first cryptic polyA signal, with much lower levels extending to the second cryptic polyA site (Fig. 3b). Full-length Htt levels were reduced by approximately 30% in both zQ175 and zQ175::Srsf6+/− MEFs compared to WT (Fig. 3b). To investigate whether this reduction in Htt had occurred for both the WT and knockin alleles, we employed a qPCR assay that specifically detected WT full-length Htt. We found that the level of WT Htt in the zQ175 and zQ175::Srsf6+/− MEFs, was 50% of that in the WT MEFs (Fig. 3c). Therefore, the observed decrease in full-length Htt in the zQ175 and zQ175::Srsf6+/− MEFs represented a reduction in the levels of the knockin allele by approximately 60% of WT Htt levels. We next used a qPCR assay that specifically targeted the full-length knockin Htt allele, and demonstrated that this is present at equivalent levels in the zQ175 and zQ175::Srsf6+/− MEFs (Fig. 3d). For the SR protein genes: Srsf6 levels were 50% lower in zQ175::Srsf6+/− compared to WT or zQ175 MEFs, Srsf4 was increased by about 1.2-fold, contrary to the situation in all brain regions except striatum (Supplementary Fig. S2), and there was no change in Srsf5 levels (Supplementary Fig. S3). Taken together, we show that zQ175 MEFs express detectable and quantifiable levels of Httexon1 and that, as we observed in brain, Httexon1 and full-length Htt mRNA levels are unaffected by heterozygosity for Srsf6 knockout.
Figure 3.
Generation and characterisation of zQ175 mouse embryonic fibroblasts (MEFs). (a) Schematic shows workflow for derivation and characterisation of MEF cell cultures. Srsf6+/− mice were bred to zQ175 mice. The female was sacrificed at approximately E14.5 and embryos were dissected. Mouse embryonic fibroblasts (MEFs) were isolated, cultured, passaged and expanded as required. MEFs were seeded for qPCR or QuantiGene assays as required. (b) QuantiGene analysis showed that Httexon1 was present in both the zQ175 and zQ175::Srsf6+/− MEFs at comparable levels, and that full-length Htt was decreased in both the zQ175 and zQ175::Srsf6+/− cells to a similar degree. n = 3 biological replicates/genotype. Statistical analysis was by one-way ANOVA with Bonferroni correction for multiple pairwise comparisons, ***p < 0.001. Test statistics can be found in Supplementary Table S6. (c,d) Htt allele discrimination qPCRs were used to measure the (c) WT and (d) zQ175 knockin Htt alleles in the MEF lines. n = 3 biological replicates/genotype. Statistical analysis was by unpaired Student’s t-test or one-way ANOVA with Bonferroni correction for multiple pairwise comparisons, ***p < 0.001, p < 0.2 values are indicated. Test statistics can be found in Supplementary Table S7.
Silencing SRSF6 does not alter incomplete Htt splicing in zQ175 MEFs
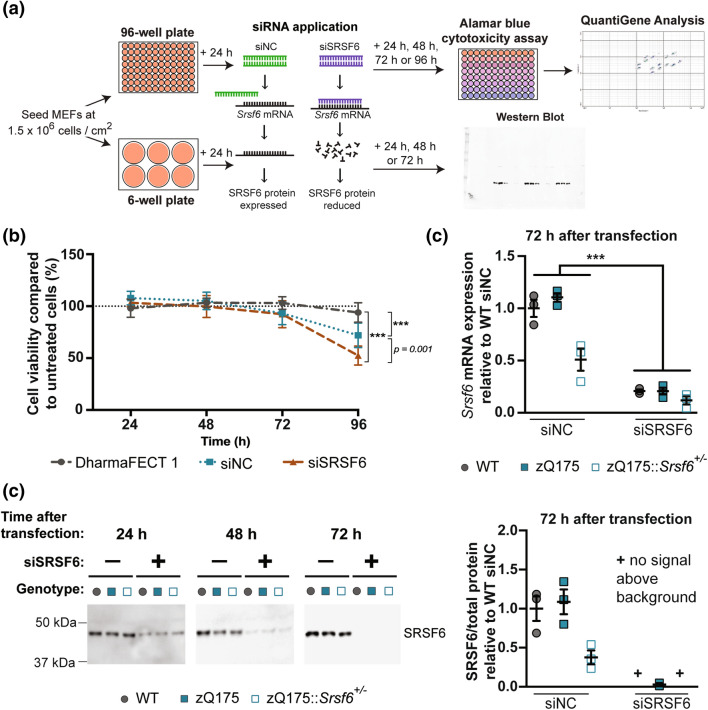
As knocking out a single allele of Srsf6 did not affect Htt mRNA processing in zQ175 mouse brain or MEFs, we used RNA interference to investigate whether further SRSF6 reduction in the zQ175 MEFs might alter Httexon1 mRNA levels (Fig. 4a). An siRNA targeting Srsf6 (siSRSF6) and a non-targeting negative control (siNC) were selected, transfected into WT, zQ175 and zQ175::Srsf6+/− MEFs using DharmaFECT 1, and cell viability was assessed using the alamarBlue assay after 24, 48, 72 and 96 h post-transfection. Neither siSRSF6 nor siNC were cytotoxic as compared to vehicle control (DharmaFect 1 transfection reagent) up to 72 h but both were cytotoxic to the MEFs by 96 h (Fig. 4b, Supplementary Fig. S4). Srsf6 levels were reduced to around 80–90% 72 h after transfection with siSRSF6 as compared to siNC (Fig. 4c, Supplementary Fig. S5). Western blotting showed that although Srsf6 mRNA levels were not completely silenced, SRSF6 protein was ablated by 72 h (Fig. 4d). This is likely because siRNAs degrade mRNA via cytoplasmic P-bodies31 and therefore nuclear Srsf6 mRNA may be preserved. This experiment confirmed that Srsf4 mRNA was increased in MEFs heterozygous for Srsf6 knockout at 24 h post-transfection (Supplementary Fig. S5), but this was not apparent at later time points. At 48 h post-transfection, there was a modest increase in Srsf5 levels (1.1-fold) in MEFs heterozygous for Srsf6 knockout, and reduction of the Srsf6 transcripts by 80–90% resulted in a modest increase in Srsf5 mRNA in wild type and zQ175 MEFs (Supplementary Fig. S5).
Figure 4.
RNA interference experimental plan, cytotoxicity of MEFs, Srsf6 transcript and SRSF6 protein quantification. (a) Schematic showing experimental workflow for RNA interference experiments in WT, zQ175 and zQ175::Srsf6+/− MEFs. MEFs were seeded and transfected with either an Srsf6-targeting (siSRSF6) siRNA or a non-targeting negative control (siNC) using DharmaFECT 1 transfection reagents and harvested after 24, 48, 72 or 96 h. Cells were used in alamarBlue, QuantiGene or western blot experiments. (b) Cytotoxicity was measured using the alamarBlue assay. The viability was calculated as the signal from siSRSF6 or siNC transfected cells as a percentage of the signal from DharmaFect 1 vehicle-treated MEFs. n = 9 cell cultures/treatment group. Data were analysed by one-way ANOVAs, ***p < 0.001. Test statistics can be found in Supplementary Table S8. Data stratified by genotype can be found in Supplementary Fig. S4. (c) QuantiGene analysis showed that Srsf6 mRNA levels decreased to 80–90% of that in the siNC-treated cells by 72 h post transfection. n = 3 biological replicates/genotype. Statistical analysis was by two-way ANOVA, ***p < 0.001. Test statistics can be found in Supplementary Table S9. (d) Western blots of the SRSF6 protein in MEFs 72 h after siSRSF6 or siNC transfection. To quantify SRSF6 levels, the SRSF6 signal was normalised to a total protein loading control. SRSF6 protein levels are plotted relative to WT. n = 3 biological replicates/genotype. Uncropped blots and total protein loading controls in Supplementary Figs. S8, S9 and S10.
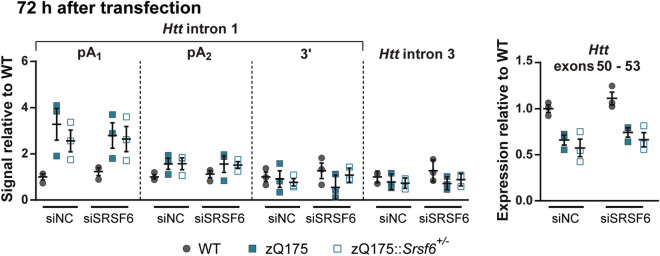
Despite the dramatic reduction in SRSF6 protein levels, culminating in its ablation after 72 h post-transfection with siSRSF6, we saw no change in the levels of Httexon1 or full-length Htt in the zQ175 or zQ175::Srsf6+/− MEFs as compared to those that had been transfected with siNC (Fig. 5, Supplementary Fig S6).
Figure 5.
Measurement of Httexon1 and Htt transcripts in WT, zQ175 and zQ175::Srsf6+/− MEFs after transfection with an siRNA targeting Srsf6 (siSRSF6). QuantiGene analysis was used to measure Httexon1 and Htt mRNA levels 72 h after siSRSF6 or siNC transfection. Neither Httexon1 nor full-length Htt levels were changed in MEFs treated with siSRSF6 compared to siNC. n = 3 biological replicates/genotype. Statistical analysis was by two-way ANOVA, ***p < 0.001. Test statistics can be found in Supplementary Table S10.
Discussion
Our previous data suggested that the splicing factor SRSF6 might play a critical role in the mechanism that, in the context of expanded CAG repeats, underlies the incomplete splicing of the HTT gene resulting in the production of the highly pathogenic exon 1 HTT protein9,10. In order to validate this hypothesis in vivo, we generated Srsf6 knockout mice and demonstrated that a 50% reduction in the levels of SRSF6 did not alter the production of the Httexon1 transcript in brain regions from the zQ175 knockin mouse model of Huntington’s disease. We found that MEFs derived from the zQ175 mice provided a very useful cell culture system in which methods to modulate the splicing of exon 1 to exon 2 of Htt can be tested. Using an siRNA to Srsf6, to ablate SRSF6 in the MEFs, we were able to show that the absence of SRSF6 has no effect on the incomplete splicing of Htt.
We generated constitutive Srsf6 knockout mice to investigate the role of SRSF6 in the incomplete splicing of Htt in the context of the endogenous gene in vivo. We found that heterozygosity for Srsf6 resulted in a 50% reduction in Srsf6 mRNA and SRSF6 protein levels throughout the brain. Srsf4 and Srsf5 are the two paralogs with the greatest gene and protein sequence alignment to Srsf622. Srsf4 and Srsf5 brain levels were comparable between the wild type and Srsf6+/− knock-out mice, indicating that there was no biological compensatory upregulation. This is also consistent with the report showing that the removal of the promoter in the knockout design ensures that compensatory expression of paralogous genes or homologous alleles, due to genetic manipulation, does not occur at the RNA level32. We were unable to generate any viable homozygous Srsf6 knockout mice, implying that SRSF6 plays a critical role in mammalian embryonic development. Similarly, genetic ablation of Srsf1, Srsf2 and Srsf3 are all embryonic lethal in mice33–35.
We previously designed a QuantiGene assay to measure the level of Httexon1 and full-length Htt transcripts in brain regions from zQ175 knockin mice29, and used this to compare the levels of these Htt transcripts in the brains of wild type, zQ175 and zQ175::Srsf6+/− mice. Consistent with our previous data, the Httexon1 transcript predominantly terminated at the second cryptic polyA site and the highest levels were detected in striatum, followed by cortex, hippocampus and the lowest levels were in cerebellum. We observed 20% lower level of full-length Htt in zQ175 cortex compared to wild type, which could be a consequence of incomplete splicing and a decrease in transcription through the expanded CAG repeat. The QuantiGene probe set exons 50–53 used here for full-length Htt gave quite variable data for the other three brain regions, and we would instead recommend using probe sets to the 3′UTR that we published in Papadopoulou et al.29 for this purpose. From this analysis, we were able to conclude that heterozygosity for Srsf6 knockout had no effect on Httexon1 or full-length Htt levels in the zQ175 brain.
We established MEFs from wild type, zQ175 and zQ175::Srsf6+/− E14.5 embryos to see whether these might provide a cell culture system that could be used to explore the effects of further reducing Srsf6 levels on the levels of the Httexon1 and full-length Htt transcripts in the context of the knockin allele. The QuantiGene assay was optimised for cells grown in 96-well plates and we found that the Httexon1 transcript could be readily detected. In contrast to the zQ175 brain regions, Httexon1 mostly terminated at the first cryptic polyA site. We saw a concurrent reduction in full-length Htt to approximately 70% of wild type, and using allele specific qPCR assays, showed that this corresponds to a reduction in the mutant and not wild type transcript. As in brain, we found that heterozygosity for Srsf6 knockout did not alter the levels of Httexon1 or full-length Htt. Having validated the MEFs as a useful model, we used an siRNA targeting the Srsf6 transcript (siSRSF6) to further reduce SRSF6 in zQ175 MEFs. We found that the Srsf6 transcript was reduced by up to 90% and the SRSF6 protein was completely ablated 72 h after transfection. Unlike the situation in zQ175 brain, heterozygosity for Srsf6 knockout resulted in modest increases in Srsf4 at 24 h, and Srsf5 at 48 h post-transfection. Ablation of SRSF6 had no effect on the incomplete splicing of Htt in the zQ175 MEFs.
Our interest in SRSF6 originated from the bioinformatic prediction that it binds to CAG and CAGCAA repeats, and therefore we proposed a model in which its ectopic location might sequester the U1 spliceosome complex, locally decreasing the probability of exon 1 splicing to exon 2 at the same time exposing cryptic polyA sites within intron 110. In this study, we have conclusively shown that SRSF6 is not required for the incomplete splicing of Htt in models in which the CAG expansion occurs in the context of the endogenous gene. Although, in the MEFs, we saw a slight increase in the levels of Srsf4 and Srsf5, these splicing factors are not predicted to bind CAGs, and therefore, any compensatory roles that they are playing are likely occurring at the transcriptome-level, and not in a context relevant to Htt splicing. The factors that bind to Htt mRNA and influence either the efficiency of splicing and/or the probability of premature termination are currently unknown. A number of RNA-binding proteins have been shown to bind Htt mRNA19 and the zQ175 MEFs will provide a powerful resource for screening candidates that may influence incomplete splicing of Htt, thereby increasing our understanding of this mechanism. The MEFs will also be invaluable for screening therapeutics that specifically target the HTTexon1 transcript that encodes the highly pathogenic exon 1 HTT protein.
Methods
Generation of the Srsf6 knockout mouse lines
Srsf6 knockout mice were generated by the Jackson Laboratory (Bar Harbor, Maine, USA) using CRIPSR/Cas9. Guide RNAs targeted the 5′UTR (AGCAAGCACGGCACGCGCCG and GGGCCGCTCCGGATGTGTTG) and intron 2 (CCCCGCGCTCCGGTGTCCCA and CCGTGGGACACCGGAGCGCG). Four founders with different deletions were generated on a C57BL/6J background and preliminary experiments were conducted on two of the four strains (Δ990 bp and Δ956 bp).
Animal colony maintenance and breeding
All animal procedures were undertaken in compliance with the Animals (Scientific Procedures) Act 1986 with the authorisation of the University College London Ethical Review Process Committee. zQ175 and Srsf6+/− colonies were maintained by backcrossing males to C57BL6/J females (Charles River). To generate the zQ175 x Srsf6+/− cross, zQ175 males were mated with Srsf6+/− females. Mice had unrestricted access to food and water, were provided with environmental enrichment which included chew sticks and play tubes, and were maintained under a 12 h light/dark cycle. The animal facility was barrier-maintained and quarterly non-sacrificial FELASA screens found no evidence of pathogens. Two month-old mice were sacrificed, dissected, and their tissues were snap frozen in liquid nitrogen and stored at − 80 °C.
DNA extractions, genotyping and CAG repeat sizing
To isolate genomic DNA, ear notches and tails from 10 day-old mice and E14.5 embryos respectively were lysed overnight in 500 μL of lysis solution (0.05 M TRIS–HCl pH 8.0, 0.1 M EDTA, 0.5% SDS and 0.5 mg/mL proteinase K). 300 μL of saturated 36% (w/v) NaCl was thoroughly mixed with lysates and centrifuged at 1.7 × 105 g for 30 min. The supernatant was thoroughly mixed with 650 μL of 100% ethanol in a fresh Eppendorf tube and centrifuged at 1.7 × 105 g for 20 min. The supernatant was removed and the pellet was washed by addition of 200 μL 70% ethanol followed by centrifugation at 1.7 × 105 g for 5 min. The supernatant was removed and pellets were air dried at RT for 2 h. Pellets were re-suspended in 100 μL of 5 mM TRIS–HCl pH 8. Genomic DNA was quantified using a Nanodrop (Thermo Fisher Scientific).
Mice from the zQ175 colony were genotyped using a forward primer (AGGAGCCGCTGCACCGA) and a reverse primer (CTCTTCACAACAGTCATGTGCG). The PCR thermal cycling program was as follows: 98 °C for 30 s followed by 35 cycles of 98 °C for 15 s, 64 °C for 15 s and 72 °C for 30 s, and lastly 72 °C for 5 min. Mice from the Srsf6 knockout colony were genotyped using a forward primer (GCGTGTACTCAACGAAACCA) and two reverse primers (GCTGTCAGTCTAGGCCATCT for the WT allele and AGCCTCCCAGCTCCTAAGAC for the knockout allele). The PCR thermal cycling program was as follows: 94 °C for 3 min followed by 35 cycles of 96 °C for 30 s, 64 °C for 30 s and 72 °C for 30 s and lastly 72 °C for 5 min. Genotyping PCRs were performed using the GoTaq system (Promega). PCR amplicons and 100 bp DNA size standard ladder (New England BioLabs) were electrophoresed in 2% agarose gels containing 0.003% SYBR Safe (Thermo Fisher Scientific) and imaged using a Gel Doc XR (BioRad). The zQ175 PCR yielded a WT band of 324 bp and a knockin band of 240 bp. The Srsf6 PCR yielded a WT band of 104 bp and a knockout band of 516 bp.
CAG repeat sizing was performed using a 6-FAM-labelled forward primer (ATGAAGGCCTTCGAGTCCCTCAAGTCCTTC) and a reverse primer (GGCGGCTGAGGAAGCTGAGGA). The PCR thermal cycling program was as follows: 94 °C for 90 s, then 35 cycles of 94 °C for 30 s, 65 °C for 30 s and 72 °C for 90 s, and lastly 72 °C for 10 min. The CAG repeat sizing PCR was performed using the AmpliTaq system (Thermo Fisher Scientific). 2 μL of DNA amplicon was mixed with 8 μL of HiDi Formamide (Thermo Fisher Scientific) and 0.05 μL of MapMarker Rox 1000 (Bioventures) in a 96-well plate. DNA was denatured at 95 °C for 10 min then rapidly cooled on ice for 10 min protected from light. Data were acquired by capillary electrophoresis on a 3,730 DNA analyser (Applied Biosystems) and repeat sizing was analysed using GeneMarker software (SoftGenetics). The CAG repeat size for the zQ175 mice was 203.39 ± 4.97 (SD), for zQ175::Srsf6+/− mice was 204.28 ± 4.89 (SD), for the zQ175 MEFs was 202 ± 3.61 (SD) and for the zQ175::Srsf6+/− MEFs was 205.33 ± 3.12 (SD).
DNA sequencing for Srsf6 knockout deletion breakpoint
The Srsf6 knockout allele was amplified by the genotypic PCR protocol and the band was excised. The amplicon was extracted using a Qiagen gel extraction kit (Qiagen) as per the manufacturer’s instructions. DNA was quantified using a Qubit fluorometer (Thermo Fisher Scientific). To generate fragments for sequencing, 200 ng of the extracted amplicon was added to a BigDye Terminator V Mastermix (Thermo Fisher Scientific) as follows: 0.5 μL of Big Dye v3.1 terminator (Thermo Fisher Scientific), 2 μL of 5 × Big Dye Sequencing Buffer (Thermo Fisher Scientific), 3.2 μM of a primer in a final volume of 10 μL. Forward primers were GCTGAAGGGAAAGAGCAACC, CCTGGGCACAAGAACAGTTT and GCGAAATCAACTCCCAGCAA and reverse primers were ACCCCAGCCTTCCTAGAAAC, CCTCGCTTTCAATGGCAGAA and CCCAAGTCAGTGCCAAAGAG. PCR reactions were prepared in a 96-well plate and were run using the following thermal cycling program: 96 °C for 2 min, then 30 cycles of 96 °C for 30 s, 50 °C for 15 s and 60 °C for 2 min. Following the thermal cycling, 30 μL of 100% ethanol and 2.5 μL of 125 mM EDTA pH 8.0 was added to each sample mixed by vortexing and incubated at RT protected from light. The plate was centrifuged at 1 × 104 g for 40 min. The supernatant was removed by inverting the plate and replaced with 50 μL of 70% ethanol. The plate was vortexed and centrifuged at 1 × 104 g for 30 min and the supernatant removed. The plate was then centrifuged upside down on a paper towel at 6.5 × 103 g for 1 min. The pellets were re-suspended in 10 µL of HiDi formamide and denatured at 95 °C for 10 min then rapidly cooled on ice for 10 min protected from light. Data were acquired by capillary electrophoresis by a 3,730 DNA analyser (Applied Biosystems) and analysed using SnapGene software (SoftGenetics).
Cell culture, RNA interference (RNAi) and alamarBlue cytotoxicity assay
Mouse embryonic fibroblasts (MEFs) were isolated from E14.5 embryos from a cross between zQ175 and Srsf6+/− mice. MEFs were maintained in DMEM/10% FBS/1% penicillin–streptomycin at 37 °C in 5% CO2 and passaged roughly once a week when MEFs were around 90% confluent.
MEFs were seeded in 6-well plates (for western blot or qPCR analysis) or 96-well plates (for AlamarBlue or QuantiGene analysis) at a density of 1.5 × 106 cells/cm2 in DMEM/6% FBS. RNAi experiments were performed one day post-seeding when the medium was changed to DMEM/OptiMEM (in a 1:1 ratio)/3% FBS before siRNA transfection. 10 nM of Silencer Select siRNAs (Thermo Fisher Scientific) targeting Srsf6 (assay ID: s86053) and a non-targeting negative control siRNA (assay ID: 4390843) were transfected into MEFs using 2.5 nL of DhramaFECT 1 transfection reagent (Horizon Discovery) per μL of total media in each well as per the manufacturer’s instructions.
Cytotoxicity of siRNAs was assessed using the AlamarBlue cytotoxicity assay (Thermo Fisher Scientific). The media was aspirated and replaced with 90 μL of fresh DMEM/3% FBS and 10 μL of the AlamarBlue reagent was added to each well followed by an incubation at 37 °C in 5% CO2 for 3 h. Absorbance was measured using a SPECTROstar Nano Microplate Reader (BMG Labtech). Media containing AlamarBlue reagent was aspirated, replaced with 100 μL of DMEM/3% FBS and prepared for QuantiGene analysis (see following section).
For MEFs in 6-well plates, the media was aspirated and the cells were washed once with PBS. For western blot analysis: 250 μL Trypsin–EDTA 0.25% (Thermo Fisher scientific) was applied and incubated at 37 °C in 5% CO2 for 3 min to dislodge the MEFs from the bottom of the well. 500 μL of DMEM/3% FBS was added to each well to neutralise the trypsin and the cell suspension was added to a fresh 1.5 mL centrifuge tube. MEFs were pelleted by centrifugation at 500g at RT. The supernatant aspirated and the pellet washed in ice-cold PBS by centrifugation at 500g at 4 °C. The supernatant was aspirated and the pellets were snap-frozen on dry ice and processed for western blot analysis (see following section). For qPCR experiments: Qiazol (Qiagen) was added to each well, the MEFs were scraped from the bottom of the well and the lysate was transferred to a fresh 1.5 mL centrifuge tube. Cells were processed for qPCR (see section below).
RNA isolation and cDNA synthesis
Cortex was homogenised in Qiazol (Qiagen) using a 1 mL syringe with a 23G followed by an 18G needle. Total RNA was isolated from cortex and MEFs using an RNeasy mini-kit (Qiagen) according to the manufacturer’s instructions. This included a DNase step using an RNase-Free DNase Set (Qiagen) as per the manufacturer’s instructions. RNA was eluted in 30 μL of nuclease-free water (Sigma) and quantified using a Nanodrop (Thermo Fisher Scientific). cDNA was synthesised by reverse transcription from 1 μg of RNA using M-MLV Reverse Transcriptase (Invitrogen) and an oligo-dT(18) primer (Invitrogen) as per the manufacturer’s instructions. A reverse transcription negative control was included.
Real-time quantitative PCR (qPCR) assays
To quantify specific transcripts, commercially available Taqman real-time quantitative PCR (qPCR) assays (Thermo Fisher Scientific) were used to measure genes of interest and housekeeping genes (Supplementary Table S2). We also used proprietary Taqman qPCR assays targeting the WT29 and zQ175 knockin Htt alleles separately. For the knockin allele, the forward primer was GCCCGGCTGTGGCTGA, the reverse primer was TTCACACGGTCTTTCTTGGTGG and the ZEN probe was TGCACCGACCAAAGAAGGAACTCT (Integrated DNA Technologies). cDNA was diluted 1:50 nuclease-free water (Sigma) and plated in 96-well thin wall Hard-Shell PCR plates (BioRad). Each 15 μL reaction per sample contained 1 × Taqman Fast Advanced Mastermix (Thermo Fisher Scientific), 1 × Taqman Gene expression assays and 3 μL of diluted cDNA (1:50 in nuclease-free water) and was aliquoted into the 96-well plates and sealed. Plates were centrifuged at 8 × 103 g for 30 s and then analysed using a BioRad CFX96 thermal cycler with the following program: 95 °C for 40 s, followed by 40 cycles of 95 °C for 7 s, 60 °C for 20 s. All biological replicates were run in triplicate. Cq values deviating by ± 0.25 from the mean of the triplicate were removed from the analysis. Data for genes of interest were normalised to reference genes (CanX, Ubc and Atp5b) as per the 2-ΔΔCt2 method36.
Tissue lysis and QuantiGene assays
Brain samples were prepared by lysis in 60 μL of QuantiGene homogenising solution (Thermo Fisher Scientific) and 30 μg proteinase K (Thermo Fisher Scientific) per mg of tissue using a 1 mL syringe with a 23G followed by a 18G needle in a 1.5 mL microfuge tube. Lysates were incubated at 50 °C on a dry heat block for 18 h and then centrifuged at 1.7 × 105 g for 10 min. The supernatant was transferred to a fresh 1.5 mL microfuge tube, snap frozen on dry ice and stored at − 80 °C until required. Lysates were thawed on a heat block at 50 °C for 30 min and diluted (for tissue specific dilutions, refer to Supplementary Table S3). Cell lysates were prepared by adding 50 μL of QuantiGene Lysis Mixture containing 25 μg of proteinase K to each 96-well of a cell culture plate (which contained 100 μL of cell culture medium per well) and pipetting up and down three to four times. 96-well plates containing lysates were snap frozen on dry ice and stored at − 80 °C until required. Prior to analysis, QuantiGene cell lysates were incubated at 50 °C for 1 h and lysis was confirmed under a light microscope. Cell lysates were transferred to 1.5 mL centrifuge tubes and centrifuged at RT at 1.7 × 105 g for 10 min to pellet any debris. The QuantiGene plex assay was run in duplicate as per the manufacturer’s instructions except that the Streptavidin R-Phycoerythrin conjugate (SAPE) was incubated at 51 °C. After subtraction of the background, the median fluorescence intensity (MFI) for the genes of interest was normalised to the geometric mean of the MFI for the reference genes (See Supplementary Table S1 for QuantiGene probe sets).
Western Blot
Tissue or cell lysates were prepared in ice-cold KCL buffer (50 mM Tris–HCl pH8, 10% Glycerol, 5 mM EDTA, 150 mM KCl, 1 mM DTT, 1 mM PMSF) containing cOmplete proteinase inhibitor cocktail (Sigma-Aldrich) and kept on ice throughout the entire lysis process. Tissues were sheared in KCl buffer using a 1 mL syringe with a 23G and then an 18G needle. Cell pellets were re-suspended and samples were sonicated using a Q125 Sonicator (Qsonica) six times for 10 s with a 10 s break between each sonication at an amplitude setting of 40%. Debris was removed by centrifugation at 4 °C at 1.7 × 105 g and lysates were stored at − 80 °C until required. Protein was quantified using Pierce BCA Protein Assay (Thermo Fisher Scientific). 40 μg or 15 μg of protein in Laemmli loading buffer from brain tissue or cells respectively was denatured at 90 °C for 5 min and resolved on Criterion TGX Stain-Free precast gels (BioRad) by electrophoresis at 13.5 V/cm2. Total protein was measured directly from the Criterion TGX Stain-Free precast gels by UV transillumination of the blot for 2.5 min using a Gel Doc XR + transilluminator (BioRad). Protein was transferred to a 0.45 μm nitrocellulose membrane (Biorad) by submerged transfer in transfer buffer (25 mM Tris base, 192 mM glycine, 20% v/v methanol). The membrane was blocked in PBS/0.1% TWEEN 20 (PBST)/5% milk at RT for 1 h and then incubated with anti-SRSF6 (Ab140623; Abcam) at a dilution of 1:1,000 at 4 °C overnight in PBS/0.1% TWEEN 20 (PBST)/1% milk. Blots were washed four times for 5 min in PBS/0.1% TWEEN 20 (PBST) and then incubated with 1:5,000 HRP-conjugated goat anti-rabbit (A31460; Invitrogen) at RT for 1 h. Blots were washed four times for 5 min in PBS/0.1% TWEEN 20. Clarity Western ECL Substrate (BioRad) was applied to the membranes and images were acquired using a ChemiDoc (BioRad). Blots were analysed using Image Lab software (BioRad).
Statistical analysis
Data were screened for outliers using ROUT test (GraphPad Software, California, USA) and any outliers were removed before between-group comparisons. Statistical analysis was performed with SPSS v26 (IBM, Portsmouth, UK) using two-tailed independent samples Student’s t-test, one-way ANOVA or two-way ANOVA with Bonferroni post-hoc tests as indicated. Graphs were prepared using Prism v7 (GraphPad Software, California, USA). A significance threshold of α = 0.05 was set for all statistical tests.
Supplementary information
Acknowledgements
We thank David Howland for the zQ175 mutant Htt specific qPCR sequences. This work was supported by Grants from the CHDI foundation, Medical Research Council (MR/L003627/1) and the UK Dementia Research Institute, which receives its funding from DRI Ltd, funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK.
Author contributions
G.P.B acquired funding for this study. G.P.B, A.N. and M.A.M designed the study. A.S.P., K.S, G.P.B and M.A.M developed the methodology. C.G.P and M.A.M. provided study reagents and performed the experiments. M.A.M analysed and visualised the data. M.A.M. and G.P.B. wrote the manuscript and all authors read and approved the manuscript.
Data availability
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-71111-w.
References
- 1.Bates GP, et al. Huntington disease. Nat. Rev. Dis. Prim. 2015;1:15005. doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 2.HD Colloabrative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 3.Kordasiewicz HB, et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of Huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tabrizi SJ, et al. Targeting Huntingtin expression in patients with Huntington’s disease. N. Engl. J. Med. 2019;380:2307–2316. doi: 10.1056/NEJMoa1900907. [DOI] [PubMed] [Google Scholar]
- 5.El-Andaloussi S, et al. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat. Protoc. 2012;7:2112. doi: 10.1038/nprot.2012.131. [DOI] [PubMed] [Google Scholar]
- 6.DiFiglia M, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin B, et al. Differential 3′ polyadenylation of the Huntington disease gene results in two mRNA species with variable tissue expression. Hum. Mol. Genet. 1993;2:1541–1545. doi: 10.1093/hmg/2.10.1541. [DOI] [PubMed] [Google Scholar]
- 8.Romo L, Ashar-Patel A, Pfister E, Aronin N. Alterations in mRNA 3′ UTR isoform abundance accompany gene expression changes in human Huntington’s disease brains. Cell Rep. 2017;20:3057–3070. doi: 10.1016/j.celrep.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sathasivam K, et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. 2013;110:2366–2370. doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neueder A, Dumas AA, Benjamin AC, Bates GP. Regulatory mechanisms of incomplete huntingtin mRNA splicing. Nat. Commun. 2018;9:1–3. doi: 10.1038/s41467-018-06281-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neueder A, et al. The pathogenic exon 1 HTT protein is produced by incomplete splicing in Huntington’s disease patients. Sci. Rep. 2017;7:1307. doi: 10.1038/s41598-017-01510-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gipson TA, Neueder A, Wexler NS, Bates GP, Housman D. Aberrantly spliced HTT, a new player in Huntington’s disease pathogenesis. RNA Biol. 1947;1011:1647–1652. doi: 10.4161/rna.26706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franich NR, et al. Phenotype onset in Huntington’s disease knock-in mice is correlated with the incomplete splicing of the mutant huntingtin gene. J. Neurosci. Res. 2019;97:1590–1605. doi: 10.1002/jnr.24493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/S0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 15.Barbaro BA, et al. Comparative study of naturally occurring huntingtin fragments in Drosophila points to exon 1 as the most pathogenic species in Huntington’s disease. Hum. Mol. Genet. 2015;24:913–925. doi: 10.1093/hmg/ddu504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scherzinger E, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/S0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 17.Gipson TA, Neueder A, Wexler NS, Bates GP, Housman D. Aberrantly spliced HTT, a new player in Huntington’s disease pathogenesis. RNA Biol. 2013;10:1647–1652. doi: 10.4161/rna.26706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Mezer M, Wojciechowska M, Napierala M, Sobczak K, Krzyzosiak WJ. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011;39:3852–3863. doi: 10.1093/nar/gkq1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schilling J, et al. Deregulated splicing is a major mechanism of RNA-induced toxicity in Huntington’s disease. J. Mol. Biol. 2019;431:1869–1877. doi: 10.1016/j.jmb.2019.01.034. [DOI] [PubMed] [Google Scholar]
- 20.Balendra R, Isaacs AM. C9orf72-mediated ALS and FTD: Multiple pathways to disease. Nat. Rev. Neurol. 2018;14:544–558. doi: 10.1038/s41582-018-0047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodwin M, et al. MBNL sequestration by toxic RNAs and RNA misprocessing in the myotonic dystrophy brain. Cell Rep. 2015;12:1159–1168. doi: 10.1016/j.celrep.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong S. SR proteins: Binders, regulators, and connectors of RNA. Mol. Cells. 2017;40:1. doi: 10.14348/molcells.2017.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Twyffels L, Gueydan C, Kruys V. Shuttling SR proteins: More than splicing factors. FEBS J. 2011;278:3246–3255. doi: 10.1111/j.1742-4658.2011.08274.x. [DOI] [PubMed] [Google Scholar]
- 25.Dreos R, Ambrosini G, Groux R, Cavin Périer R, Bucher P. The eukaryotic promoter database in its 30th year: Focus on non-vertebrate organisms. Nucleic Acids Res. 2017;45:D51–D55. doi: 10.1093/nar/gkw1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet M. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J. Comp. Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 27.Menalled LB, et al. Comprehensive behavioral and molecular characterization of a new knockin mouse model of Huntington’s disease: zQ175. PLoS ONE. 2012;7:e49838. doi: 10.1371/journal.pone.0049838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heikkinen T, et al. Characterization of neurophysiological and behavioral changes, MRI brain volumetry and 1H MRS in zQ175 knockin mouse model of Huntington’s disease. PLoS ONE. 2012;7:e50717. doi: 10.1371/journal.pone.0050717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papadopoulou AS, et al. Extensive expression analysis of Htt transcripts in brain regions from the zQ175 HD Mouse model using a QuantiGene multiplex assay. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-52411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landles C, et al. Subcellular localisation and formation of huntingtin aggregates determine onset age and rate of disease progression. Brain Commun. 2020;2:fcaa066. doi: 10.1093/braincomms/fcaa066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 32.El-Brolosy MA, et al. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019;568:193–197. doi: 10.1038/s41586-019-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jumaa H, Wei G, Nielsen PJ. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr. Biol. 1999;9:899–902. doi: 10.1016/S0960-9822(99)80394-7. [DOI] [PubMed] [Google Scholar]
- 34.Xu X, et al. ASF/SF2-regulated CaMKIIδ alternative splicing temporally reprograms excitation–contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 35.Wang H-Y, Xu X, Ding J-H, Bermingham JR, Jr, Fu X-D. SC35 plays a role in T cell development and alternative splicing of CD45. Mol. Cell. 2001;7:331–342. doi: 10.1016/S1097-2765(01)00181-2. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.