Bats host diverse coronaviruses, including taxa capable of pandemic spread in humans. We report the genome of an alphacoronavirus from a neotropical bat species (Desmodus rotundus) in Peru, which contributes to our understanding of bat coronaviruses in nature.
ABSTRACT
Bats host diverse coronaviruses, including taxa capable of pandemic spread in humans. We report the genome of an alphacoronavirus from a neotropical bat species (Desmodus rotundus) in Peru, which contributes to our understanding of bat coronaviruses in nature.
ANNOUNCEMENT
Coronaviruses (CoVs) (family Coronaviridae) are positive-sense, single-stranded RNA viruses that naturally circulate in many vertebrates. CoVs are relatively common and genetically diverse in bats and include zoonotic species that cause severe acute respiratory syndrome (SARS-CoV), Middle East respiratory syndrome (MERS-CoV), and CoV disease 2019 (SARS-CoV-2) (1–5). Due to the geographic origin of the most prominent zoonotic CoVs, most knowledge of bat CoVs is derived from Old World species. Nonetheless, comparatively limited sampling has revealed diverse CoVs among bats in North and South America (6–9). Additional knowledge of the diversity and distribution of CoVs in these species will aid efforts to understand virus ecology within bat reservoirs, anticipate zoonotic risk, and accelerate identification of reservoir hosts following emergence.
We report the genome of an alphacoronavirus from common vampire bats (Desmodus rotundus, family Phyllostomidae), termed DesRot/Peru/Amazonas/CoV (isolate AMA_L_F). The genome was derived from shotgun sequencing of 10 pooled samples collected with noninvasive rectal swabs from D. rotundus bats from two colonies in Rio Escondido, Amazonas, Peru (ENA accession number ERR2756788) (10). Briefly, total nucleic acid was extracted using a BioSprint One-for-All veterinary kit (Qiagen) and a Kingfisher 96 Flex system. Following DNase I (Ambion) treatment, rRNA depletion (Ribo-Zero; Illumina), and cDNA synthesis (Maxima H Minus first-strand cDNA synthesis kit; Thermo Fisher Scientific), sequencing libraries were prepared using the KAPA DNA library preparation kit for Illumina (Kapa Biosystems). Sequencing was performed on an Illumina NextSeq 500 system (read length, 150 bp). A total of 17,760,709 raw reads were processed through an in-house pipeline, including quality filtering with Trim Galore v.0.4.0 and prinseq-lite v.0.20.4 (11, 12), assembly with SPAdes v.3.10.1 (13), and classification with DIAMOND blastx v.0.8.20 (14), leading to a genome of 29,097 bp with a mean coverage of 226.7 reads. BLASTn analysis against GenBank showed that the most similar full genome was that of an alphacoronavirus from a microbat (GenBank accession number MK472070.1) (70.7% nucleotide similarity); that virus was used to determine genome termini. The two genomes were of similar initial size (MK472070.1, 28,009 bp; DesRot/Peru/Amazonas/CoV, 29,140 bp), and the new genome aligned over the length of the reference with relatively few gaps (final untrimmed alignment, 29,887 bp). DesRot/Peru/Amazonas/CoV had a GC content (42.9%) and genomic organization of major open reading frames (ORFs) (5′-ORF1a/ORF1ab-S-ORF3-E-M-N-3′) similar to those of other alphacoronaviruses.
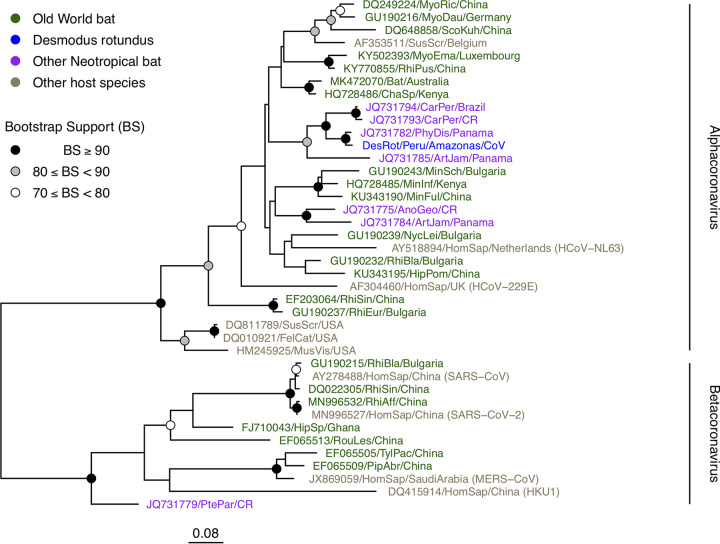
A 272-amino-acid section of the RdRp gene (5) was aligned with other representative CoVs using MAFFT v.7.017 (15). Maximum likelihood phylogenetic analysis performed in RAxML v.8.2.8 (16), using the LG+I+G substitution model identified by ProtTest 3 (17), showed that the vampire bat sequence was more closely related to other neotropical bat CoVs (96.2% nucleotide similarity, over 816 bp, to the most similar sequence [GenBank accession number JQ731782]) and fell within a clade of alphacoronaviruses from other Phyllostomidae bats (Fig. 1). Two Brazilian vampire bat sequences, which were within ORF1b but did not overlap completely with the RdRp section used for phylogenetic analysis, were compared to DesRot/Peru/Amazonas/CoV separately, displaying pairwise nucleotide identities of 69.2% over 52 bp (EU236685.1) and 98.1% over 572 bp (KU552072.1). In summary, DesRot/Peru/Amazonas/CoV is a genomic representative of neotropical bat alphacoronaviruses, providing a new resource for understanding the global diversity of CoVs.
FIG 1.
RdRp phylogeny including the novel vampire bat alphacoronavirus. The maximum likelihood tree was based on a 272-amino-acid alignment of 40 RdRp sequences, including the novel vampire bat CoV. Human-infecting species are indicated by names in parentheses. The scale bar represents the mean expected rate of substitutions per site. Node support is from 1,000 bootstrap replicates.
Data availability.
The complete genome sequence for DesRot/Peru/Amazonas/CoV has been deposited in GenBank under accession number MT663548. Raw data were deposited in the ENA under run accession number ERR2756788, experiment accession number ERX2769781, and study accession number PRJEB28138.
ACKNOWLEDGMENTS
Funding was provided by the Wellcome Trust (Wellcome-Beit Prize 102507/Z/13/A and Wellcome Senior Research Fellowship 217221/Z/19/Z) and the Human Frontier Science Program (grant RGP0013/2018).
REFERENCES
- 1.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang L-F. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 2.Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, Alhakeem R, Durosinloun A, Al Asmari M, Islam A, Kapoor A, Briese T, Daszak P, Al Rabeeah AA, Lipkin WI. 2013. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis 19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poon LLM, Chu DKW, Chan KH, Wong OK, Ellis TM, Leung YHC, Lau SKP, Woo PCY, Suen KY, Yuen KY, Guan Y, Peiris JSM. 2005. Identification of a novel coronavirus in bats. J Virol 79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drexler JF, Gloza-Rausch F, Glende J, Corman VM, Muth D, Goettsche M, Seebens A, Niedrig M, Pfefferle S, Yordanov S, Zhelyazkov L, Hermanns U, Vallo P, Lukashev A, Muller MA, Deng H, Herrler G, Drosten C. 2010. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J Virol 84:11336–11349. doi: 10.1128/JVI.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominguez SR, O'Shea TJ, Oko LM, Holmes KV. 2007. Detection of group 1 coronaviruses in bats in North America. Emerg Infect Dis 13:1295–1300. doi: 10.3201/eid1309.070491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrington CVF, Foster JE, Zhu HC, Zhang JX, Smith GJD, Thompson N, Auguste AJ, Ramkissoon V, Adesiyun AA, Guan Y. 2008. Detection and phylogenetic analysis of group 1 coronaviruses in South American bats. Emerg Infect Dis 14:1890–1893. doi: 10.3201/eid1412.080642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anthony SJ, Ojeda-Flores R, Rico-Chávez O, Navarrete-Macias I, Zambrana-Torrelio CM, Rostal MK, Epstein JH, Tipps T, Liang E, Sanchez-Leon M, Sotomayor-Bonilla J, Aguirre AA, Ávila-Flores R, Medellín RA, Goldstein T, Suzán G, Daszak P, Lipkin WI. 2013. Coronaviruses in bats from Mexico. J Gen Virol 94:1028–1038. doi: 10.1099/vir.0.049759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corman VM, Rasche A, Diallo TD, Cottontail VM, Stocker A, Souza B, Correa JI, Carneiro AJB, Franke CR, Nagy M, Metz M, Knornschild M, Kalko EKV, Ghanem SJ, Morales KDS, Salsamendi E, Spinola M, Herrler G, Voigt CC, Tschapka M, Drosten C, Drexler JF. 2013. Highly diversified coronaviruses in neotropical bats. J Gen Virol 94:1984–1994. doi: 10.1099/vir.0.054841-0. [DOI] [PubMed] [Google Scholar]
- 10.Bergner LM, Orton RJ, da Silva Filipe A, Shaw AE, Becker DJ, Tello C, Biek R, Streicker DG. 2019. Using noninvasive metagenomics to characterize viral communities from wildlife. Mol Ecol Resour 19:128–143. doi: 10.1111/1755-0998.12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 12.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 15.Katoh K, Misawa K, Kuma K-I, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 17.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence for DesRot/Peru/Amazonas/CoV has been deposited in GenBank under accession number MT663548. Raw data were deposited in the ENA under run accession number ERR2756788, experiment accession number ERX2769781, and study accession number PRJEB28138.