Abstract
Background
Expression levels of the cell adhesion molecule syndecan-1 (SDC1) have been shown to be inversely proportional to tumor differentiation and prognosis. However, its role in the development of gallbladder cancer (GBC) remains unclear.
Methods
We knocked down SDC1 in GBC cells by RNA interference and determined its roles in cell proliferation, apoptosis, invasion, and migration by Cell Counting Kit-8, colony-formation, flow cytometry, Hoechst 33342 staining, transwell invasion, and scratch wound assays. Expression levels of epithelial–mesenchymal transition (EMT)-related and extracellular signal-regulated kinase (ERK)/Snail pathway proteins were determined by western blotting and immunofluorescence.
Results
Cell proliferation, invasion, and migration were all increased in GBC cells with SDC1 knockdown, compared with cells in the blank control and negative control groups, but apoptosis was similar in all three groups. E-cadherin and β-catenin expression levels were significantly lower and N-cadherin, vimentin, p-ERK1/2, and Snail expression were significantly higher in the SDC1 knockdown group compared with both controls, while ERK1/2 levels were similar in all groups. Reduced E-cadherin and increased vimentin levels were confirmed by immunofluorescence.
Conclusions
SDC1 knockdown promotes the proliferation, invasion, and migration of GBC cells, possibly by regulating ERK/Snail signaling and inducing EMT and cancer cell invasion.
Keywords: Gallbladder cancer cell, syndecan-1, epithelial–mesenchymal transition, invasion, extracellular signal-regulated kinase/Snail signaling pathway, migration
Introduction
Gallbladder cancer (GBC) is the fifth most common malignant tumor of the biliary system and one of the deadliest human cancers. It accounts for 80% to 95% of malignant bile duct tumors worldwide, with an overall incidence rate of 0.8% to 1.2%, associated with high malignancy and a poor prognosis.1,2 Despite recent advancements in high-throughput technology allowing in-depth molecular biology studies of tumors, there have been no major breakthroughs in the diagnosis and treatment of GBC. There is thus a need to understand the molecular and biological characteristics of GBC fully to improve the diagnosis and treatment of patients with early GBC.
Syndecan-1 (SDC1) is a type-1 transmembrane proteoglycan involved in the regulation of intercellular adhesion and activation of growth factor receptors through its heparan sulfate side chains, which are covalently linked to extracellular ligands such as polypeptide growth factors, cell adhesion molecules, and enzymes. SDC1 has thus become a hot topic of research aimed at understanding tumor invasion and metastasis.3 Some studies showed a reduction in SDC1 expression in several human cancers, including laryngeal carcinoma,4 lung cancer,5 liver cancer,6 mesothelioma,7 and colorectal cancer,8,9 and downregulation of SDC1 has been shown to correlate with high tumor grade, advanced stage, and a poor prognosis in tumors such as hepatocellular carcinoma, gastric cancer, laryngeal cancer, and squamous cell lung carcinoma.10–14 However, despite a large number of published studies, the precise mechanisms explaining the role of SDC1 in these pathologies remain unclear. SDC1 is an important factor in maintaining epithelial stability and integrity, and reduced expression or deletion of SDC1 thus leads to morphological changes in cells and also to decreased intercellular adhesion, potentially accelerating tumor cell invasion into the surrounding tissue.10–12 SDC1 is nonspecific and has been associated with many cancers; however, its role in the invasion and migration of GBC cells and its associated regulatory mechanism have not yet been reported.
Metastasis is the leading cause of mortality in cancer patients, and 90% of patients with solid malignant tumors die as a result of the invasion of primary cancer cells into distant organs. Malignant tumor metastasis is a complex, multi-step, and multi-factor procedure, in which cancer cells first detach from the primary tumor and then migrate and invade the surrounding matrix, including the circulatory or lymphoid system, thereby infiltrating distant sites to form new metastatic foci.13 The dissociation of the tumor from the primary site is the initial step and a prerequisite for the occurrence of tumor metastasis.
The epithelial–mesenchymal transition (EMT) is a critical step in the metastasis of malignant epithelial tumors. It causes polarized epithelial cells to lose their polarity and intercellular cell adhesion and acquire a mesenchymal cell phenotype. The typical features of EMT include the downregulation of epithelial markers (including E-cadherin, β-catenin, desmoplakin, cytokeratins, and laminin), accompanied by the upregulation of interstitial cell markers (including N-cadherin and vimentin), with downregulation of E-cadherin and upregulation of N-cadherin acting as landmarks of epithelial–stromal transformation.14 Snail1, Snail2, ZEB1, and FOXC2 are transcription factors regulating EMT, with the Snail, Twist, and ZEB families forming a core set of transcription factors and E-cadherin-transcription repressors that regulate EMT.15 Snail is highly expressed in gastric cancer, breast cancer, liver cancer, colon cancer, and oral squamous cell carcinoma, and is closely related to tumor histological grade and lymph node metastasis.16 It was also the first transcription factor found to induce EMT and to regulate the epigenetic and transcriptional expression levels of EMT-related marker genes, thereby affecting cell invasion and migration.17 Snail binds to lysine-specific demethylase, multi-comb inhibitory complex, mSin3A, histone deacetylase, and other E-cadherin transcriptional repressors, resulting in the methylation and acetylation of the histones attached to the E-cadherin promoter region, thereby inhibiting transcription of the target gene.18
Although the relationship between EMT and tumor progression has been reported, its molecular mechanism is still unclear. In this study, we used RNA interference technology to knock down SDC1 expression in a GBC cell line, to explore the effects of SDC1 on cell proliferation, apoptosis, invasion, and migration, and EMT in GBC cells. We also explored the molecular mechanism by which SDC1 regulated EMT and GBC cell invasion, including the role of the extracellular signal-regulated kinase (ERK)/Snail signaling pathway. The results of this study provide useful insights into the potential application of SDC1 as a molecular target for the treatment of GBC, as well as a robust theoretical basis for the early diagnosis and targeted therapy of GBC.
Materials and methods
Main reagents and materials
The target plasmid was constructed by GenePharma (Shanghai, China) and short hairpin (sh) RNAs against SDC1 and negative controls were designed and synthesized by GenePharma. Rabbit anti-human phosphorylated (p)-ERK1/2, ERK1/2, and SDC1 antibodies were purchased from ABclonal (Wuhan, China), goat anti-mouse and rabbit IgG secondary antibodies and DyLight 488-conjugated goat anti-mouse IgG secondary antibody were purchased from Abbkine (Wuhan, China). Vimentin, β-catenin, and E-cadherin mouse monoclonal antibodies, and N-cadherin and Snail rabbit polyclonal antibodies were purchased from Beyotime (Shanghai, China).
This study did not include and human or animal studies, and no consent or ethics approval was therefore required.
Cell culture
The GBC-SD human GBC cell line was purchased from the Shanghai Cell Biology Institute of Chinese Academy of Sciences (Shanghai, China), and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 1% penicillin–streptomycin (Beyotime). The cells were incubated at 37°C, 5% CO2, and 90% relative humidity. Cells at 70% to 80% confluency were observed under a light microscope (Olympus, Tokyo, Japan) and sub-cultured in a 1:2 ratio. Cells in good proliferative state and logarithmic growth phase were selected for the following experiments.
Lentivirus transfection and construction of stable transforming strain
SDC1-shRNA (5′-CCGCAAATTGTGGCTACTAAT-3′) targeting the human SDC1 gene was designed and produced by GenePharma. The SDC1 shRNA was confirmed to show no homology with any other gene sequence in the GenBank database. A negative control shRNA (NC-shRNA) was also designed (5′-TCTCCGAACGTTTCACGT-3′). The SDC1-shRNA and NC-shRNA were inserted into the LV3 lentivirus vector (GenePharma), respectively, and lentivirus carrying SDC1-shRNA was prepared. GBC-SD cells were inoculated into 6-well plates (3 × 105 cells/well), cultured for 24 hours, and then infected with SDC1-shRNA at a multiplicity of infection of 50. Green fluorescence intensity was observed after 72 hours by fluorescence microscopy (Olympus). Colonies were selected using 2 μg/mL puromycin for 7 days. GBC-SD cells expressing shSDC1 to inhibit SDC1 (GBC-SD-shSDC1) and cells expressing negative control shRNA (GBC-SD-shNC) were obtained.
Detection of SDC1 mRNA expression by quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the cells using TRIzol (Biotechnology, Shanghai, China) and transcribed to cDNA using a reverse transcription kit (MBI Fermentas, Shenzhen, China). The following primers were used for RT-PCR: SDC1 forward: 5′-CTGGTGGGTTTCATGCTGTA-3′, SDC1 reverse: 5′-CCTGTTTGGTGGGCTTCT-3′; β-actin forward: 5′-GGAAATCGTGCGTGACATTAAG-3′, and β-actin reverse: 5′-GGAAATCGTGCGTGACATTAAG-3′. The PCR amplification conditions were: pre-denaturation at 95°C for 10 minutes; denaturation at 95°C for 10 s; annealing at 60°C for 45 s; and extension at 60°C for 1 minute; for 40 cycles. The relative expression of SDC1 was analyzed by the 2−△△Ct method, with β-actin as an internal control.
Detection of SDC1 protein expression by western blotting
The cells were digested with trypsin (0.25%) at 37°C for 2 minutes, centrifuged at 4°C for 5 minutes at 199 × g, and the supernatant was discarded. Precooled RIPA lysis buffer (Beyotime) was then added to extract total protein. The protein concentration was determined using a BCA protein concentration determination kit (Beyotime). Sample buffer was added to the protein lysates and boiled for 10 minutes to denature the proteins. The samples were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane using a rapid preparation kit (Beyotime). The membrane was blocked with 5% fat-free milk for 2 hours and then incubated with rabbit anti-SDC1 primary antibody (dilution 1:1000) and an anti-β-actin antibody (dilution 1:5000) (Abbkine) overnight at 4°C. The membrane was washed with TBS-Tween and then incubated with goat anti-rabbit secondary antibody (dilution 1:10,000) (Abbkine) for 2 hours at room temperature. Protein signals were visualized using a BeyoECL Plus kit (Beyotime) and observed using a ChemiDoc MP system (Bio-Rad, Hercules, CA, USA). The SDC1 bands were analyzed using Image J 1.44 software (NIH, Bethesda, MA, USA) and normalized to β-actin as a loading control for quantification purposes.
CCK-8 assay
We seeded the cells in 96-well plates (5000 cells/well) and incubated them at 37°C, 5% CO2, and 90% relative humidity for 24, 48, 72, and 96 hours. CCK-8 solution (10 μL; Beyotime) was then added to each well and the plates were incubated at 37°C for another 2 hours. The absorbance was measured at 450 nm using an automatic microplate reader (Bio-Rad).
Colony-formation assay
We seeded the cells in 6-well plates (500 cells/well), incubated them at 37°C, 5% CO2, and 90% relative humidity for 2 weeks, fixed them in 4% paraformaldehyde (PFA) solution (Beyotime), and stained them with crystal violet staining solution (Beyotime) for 10 minutes. The staining solution was then gently washed off with running water, the images were captured, and the rate of cell-clone formation was recorded. The clone-formation rate (%) was calculated as the number of cell clones/number of seeded cells × 100%.
Hoechst 33342 staining assay
The cells were cultured in 10% FBS DMEM for 24 hours, followed by the addition of Hoechst 33342 living cell staining solution (Beyotime) to the medium, and incubation for a further 10 minutes. The cells were then washed twice with phosphate-buffered saline (PBS) and the numbers of apoptotic cells in five random fields of view were counted under a fluorescence microscope, and the apoptotic characteristics were recorded.
Flow cytometry
The cells were washed twice with precooled PBS, suspended in 195 μL Annexin V-fluorescein isothiocyanate (FITC) binding solution (Annexin V-FITC apoptosis detection kit; Beyotime), followed by the addition of 5 μL FITC-tagged Annexin V and 10 μL propidium iodide staining solution. The cells were mixed gently, incubated for 20 minutes at room temperature, and then transferred to a flow cytometry tube and detected by flow cytometry.
Transwell invasion assay
Matrigel (BD Biosciences, MD, USA) was diluted in DMEM (1:8) and used to coat the inner surface of the transwell chamber (Millipore Corporation, MA, USA) to make an artificial basement membrane. Approximately 30,000 cells were then added to the upper chamber and 500 μL DMEM medium containing 10% FBS was added to the lower chamber, and the cells were cultured in an incubator at 37°C, 5% CO2, and 90% relative humidity for 24 hours. The transwell chamber was then removed, the culture medium was discarded, and the upper layer of the filter membrane was gently cleaned with moist cotton swabs and washed with PBS. Finally, the membrane was fixed with 4% PFA and stained with crystal violet staining solution for 20 minutes, and cells were counted in five high-power microscope fields from each fixed part of the membrane.
Scratch wound assay
The cells were digested with trypsin (0.25%) at 37°C for 2 minutes, centrifuged at 4°C for 5 minutes at 199 × g, and the supernatant was discarded. The cells were then suspended in DMEM supplemented with 10% FBS. The cell suspension was then inoculated into 6-well plates (3 × 105 cells/well) with three wells per group. When the cells reached 100% confluence, parallel scratches were made in each well using a 10 µL plastic tip. The old medium was then removed and floating cells were washed off using PBS and the cells were grown in serum-free DMEM medium. The scratch width was observed under a microscope and photographed at 0 and 24 hours. The scratch width was measured using ImageJ software. Each experiment was repeated three times and the cell migration rate (%) was calculated as (width at 0 hours − width at 24 hours)/width at 0 hours × 100%.
Western blot analysis
Cells were digested and centrifuged as above. Precooled RIPA lysis buffer was then added to extract total protein, and the protein concentration was determined using a BCA kit. The protein concentration was adjusted by adding sample buffer and boiled for 10 minutes to denature the protein. The samples were then separated by SDS-PAGE and transferred to PVDF membranes, blocked with 5% skim milk for 2 hours, and incubated with antibodies to ERK1/2 (dilution 1:1000), p-ERK1/2 (dilution 1:1000), Snail (dilution 1:1000), E-cadherin (dilution 1:500), β-catenin (dilution 1:500), N-cadherin (dilution 1:500), vimentin (dilution 1:500), and β-actin (dilution 1:5000) overnight at 4°C. The membranes were then washed three times with PBS, and goat anti-rabbit and goat anti-mouse IgG secondary antibody (dilution 1:10,000) respectively, was added and incubated for 2 hours at room temperature. The membranes were washed three times and the protein signals were visualized using a BeyoECL Plus kit (Beyotime) and analyzed with ImageJ software, using β-actin as a loading control.
Immunofluorescence
We inoculated the cells into 6-well plates (3 × 105 cells/well) on glass slides and cultured them in an incubator at 37°C, 5% CO2, and 90% relative humidity for 24 hours. The cells were then fixed with 4% PFA for 10 minutes, permeabilized with Triton X-100 (Beyotime) for 10 minutes, blocked with 3% bovine serum albumin (Abbkine) for 20 minutes, and then incubated with primary antibodies to E-cadherin (dilution 1:200) and vimentin (dilution 1:200) overnight at 4°C. Nuclei were stained with Hoechst 33342 for 1 hour at room temperature, and images were captured under a fluorescence microscope.
Statistical analysis
The data were analyzed and graphs constructed using SPSS 18.0 (SPSS Inc., Chicago, IL, USA) and Graph Pad Prism 7.0 (Graph Pad Prism Inc., San Diego, CA, USA). Measured data were expressed as mean ± standard deviation and were compared between groups using t-tests or analysis of variance. P < 0.05 was considered statistically significant.
Results
Transfection efficiency
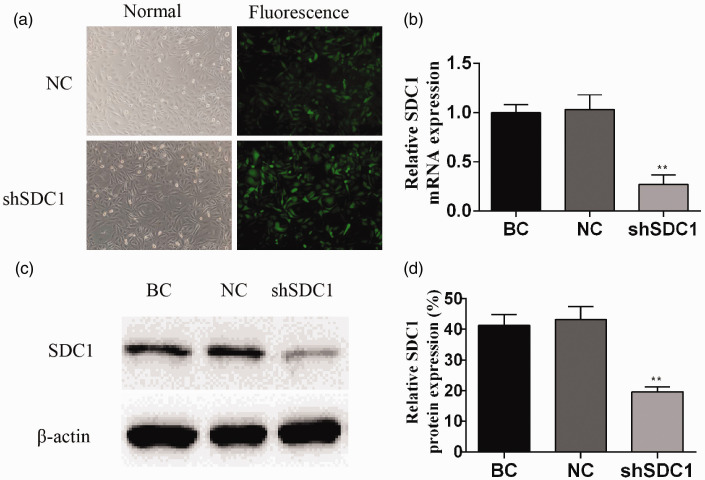
After transfection with the target plasmid expression vector expressing green fluorescent protein for 72 hours, GBC-SD cells with green fluorescence were observed under a fluorescence microscope. The transfection efficiency was >90% in SDC1-knockdown cells compared with the normal controls (Figure 1a).
Figure 1.
Effect of recombinant lentivirus on SDC-1 expression in gallbladder cancer cells. Infection efficiency (a) shown by green fluorescence 72 hours after infection. Relative SDC1 mRNA and protein expression levels shown by quantitative reverse transcription-polymerase chain reaction (b) and western blotting (c,d). Data presented as mean ± standard deviation. NC, negative control; BC, blank control. **P < 0.01.
Recombinant lentivirus downregulated SDC1 protein and mRNA levels in GBC cells
The relative expression levels of SDC1 mRNA were significantly lower in the shSDC1 compared with the BC and NC groups, as shown by qRT-PCR (P<0.01) (Figure 1b). Similarly, SDC1 protein levels were significantly lower in the shSDC1 compared with the BC and NC groups, according to western blotting (P<0.01) (Figure 1c,d). These results confirmed successful construction of the shSDC1 lentivirus vector and effective inhibition of SDC1 transcription in GBC cells.
SDC1 knockdown promoted proliferation of GBC-SD cells
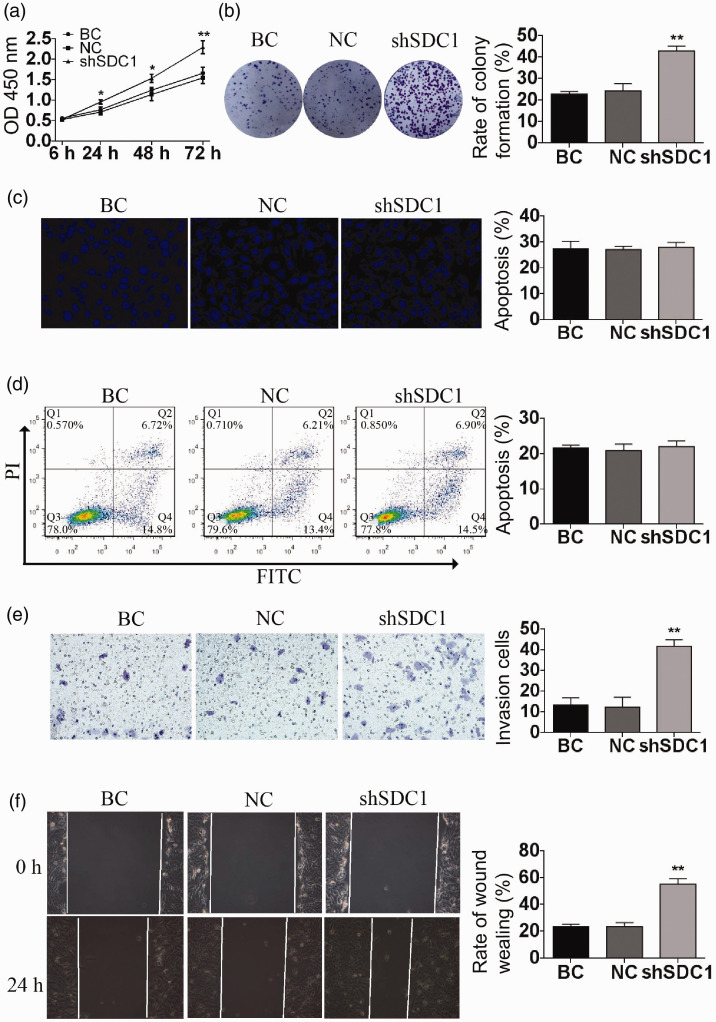
The optical density value of cells in the shSDC1 group was significantly higher compared with the BC and NC groups at 24, 48, 72, and 96 hours, as shown by CCK-8 assay (Figure 2a). The results of the colony-formation experiments showed that the rate of colony formation was significantly higher in the shSDC1 compared with the BC and NC groups (P<0.01) (Figure 2b).
Figure 2.
SDC1 knockdown promoted the proliferation, invasion, and migration of GBC-SD cells. The proliferation of GBC-SD cells was verified by CCK-8 assay (a) and colony-formation assay (b). The apoptosis rate of GBC-SD cells was assessed using Hoechst 33324 staining assay (c) and flow cytometry (d). The invasion and migration of GBC-SD cells were assessed by transwell invasion assay (e) and scratch wound assay (f). Data shown as mean ± standard deviation. OD, optical density; BC, blank control; NC, negative control; FITC, fluorescein isothiocyanate. *P < 0.05, **P < 0.01.
SDC1 knockdown did not affect apoptosis of GBC-SD cells
The percentage of apoptotic cells in the shSDC1 group was similar to those in the BC and NC groups, as demonstrated by Hoechst 33324 staining assay (P>0.05) (Figure 2c). The levels of apoptosis were also similar among the three groups according to flow cytometry (P > 0.05) (Figure 2d).
SDC1 knockdown promoted invasion and migration of GBC-SD cells
The number of invading cells was significantly higher in the shSDC1 group compared with the BC and NC groups after 24 hours, as shown by transwell assay (P<0.01) (Figure 2e). The 24-hour migration rate was also significantly higher in the shSDC1 group (P<0.01) (Figure 2f), according to scratch wound assay.
SDC1 knockdown induced EMT in GBC-SD cells
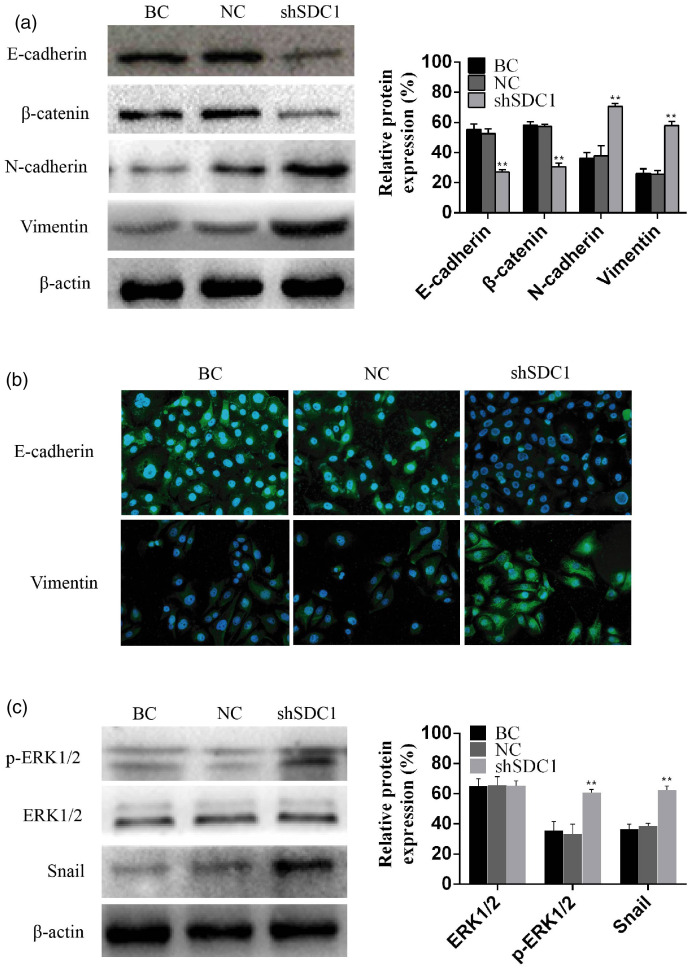
SDC1 knockdown affected the expression of epithelial and interstitial markers. Protein expression levels of the epithelial markers E-cadherin and β-catenin were significantly reduced while the interstitial markers N-cadherin and vimentin were significantly increased by SDC1 knockdown (Figure 3a). We validated the observed changes by determining the subcellular localization of the proteins by immunofluorescence staining. As predicted, E-cadherin immunofluorescence in the cell membrane was weaker and vimentin immunofluorescence was stronger in the shSDC1 group compared with the BC and NC groups (Figure 3b).
Figure 3.
SDC1 knockdown induced epithelial–mesenchymal transition (EMT) via regulating the ERK/Snail signaling pathway. EMT-related proteins were assessed by western blot (a) and by cellular immunofluorescence staining (b). Phosphorylation of ERK was assessed by western blotting (c). Data shown as mean ± standard deviation. BC, blank control; NC, negative control. **P < 0.01.
SDC1 knockdown induced EMT via regulating the ERK/Snail signaling pathway
Levels of phosphorylated ERK and Snail in the shSDC1 group were significantly higher than those in the BC and NC groups (Figure 3c). SDC1 knockdown activated ERK phosphorylation and upregulated the expression of Snail protein, indicating that the ERK/Snail signaling pathway may play a role downstream of SDC1. Thus, downregulation of endogenous SDC1 activated the ERK cascade, upregulated the expression of Snail protein, and inhibited the expression of E-cadherin.
Discussion
There are four syndecan family members, of which SDC1 is the most widely studied. Several studies have reported that SDC1 expression was significantly reduced in a variety of malignant tumor tissues,4–9 leading to speculation regarding its possible correlation with the occurrence and development of GBC. SDC1 is a transmembrane heparan sulfate proteoglycan. The human SDC1 gene is located on chromosome 2 and contains five exons, four introns, and regulatory sequences.19 SDC1 mediates intracellular and extracellular matrix adhesion and regulates the activation of growth factor receptors through its heparan sulfate side chain bound to a series of extracellular ligands.20,21 Numerous studies have confirmed that the expression of SDC1 is downregulated in various cancers, including hepatocellular carcinoma, squamous cell lung cancer, colorectal cancer, and prostate cancer, leading to reduced intercellular adhesion between tumor cells, and increased migration of tumor cells to the surrounding tissues.22–26
Although SDC1 plays a critical role in the progression of various tumors, its mechanism and role in GBC is still unclear. We therefore downregulated SDC1 expression in GBC-SD cells by RNA interference to determine its effect on the progression of GBC. Downregulation of SDC1 promoted the malignant phenotype of GBC cells by promoting their proliferation, invasion, and migration via upregulating the expression of Snail and promoting EMT by activating the ERK signaling pathway.
EMT involves the conversion of some or all epithelial cells to stromal cells under specific physiological or pathological conditions. Tumor cells undergo morphological changes and loss and remodeling of adhesion between cells and the extracellular matrix, promoting the cell movement and invasion abilities via EMT. EMT is thus an essential mechanism for tumor cell infiltration, by allowing them to break through the basement membrane to enter the blood circulation and form metastatic tumors at the implant site.27 In this study, we reduced expression of endogenous SDC1 by RNA interference, to promote the proliferation, invasion, and migration of GBC cells and promote the progression of EMT. Consistent with our results, previous studies have reported a role for SDC1 in EMT. Wang et al.28 found that knockout of endogenous SDC1 activated the ERK pathway, resulting in upregulation of Snail expression and induction of EMT. Sun et al.29 showed that SNC1 and E-cadherin expression were lost in the embryonic palate during EMT. Loussouarn et al.30 found that low SDC1 expression in epithelial tissue had good prognostic value for breast cancer diagnosis, and that decreased SDC1 expression promoted the occurrence of EMT. Kumar-Singh et al.7 showed that SDC1 could be used as an important prognostic indicator of mesothelioma, and its loss may play a crucial role in the epithelial–interstitial transformation of mesothelioma cells.
Studies in prostate cancer have shown that reduced SDC1 expression led to increased Snail expression, thereby inducing the occurrence of EMT.31 Masola et al.32 reported that basic fibroblast growth factor 2 induced EMT based on the interaction between heparanase and SDC1 in renal tubular epithelial cells. In the current study, we used RNA interference to downregulate the expression of SDC1, which resulted in upregulation of p-ERK1/2 and Snail, reduced expression of E-cadherin and β-catenin, and increased expression of N-cadherin and vimentin. SDC1 may thus act by upregulating the expression of Snail by activating the ERK signaling pathway to induce EMT in GBC cells. The results of the present study are in line with those of Wang et al.,28 who showed that low expression of SDC1 might be a useful indicator of the degree of malignancy before invasion of oral squamous cell carcinoma. Activating the ERK pathway can induce Snail expression in breast cancer, gastric cancer, and lung adenocarcinoma.33,34 Luo et al.35 found that toosendanin, a natural product, inhibited transforming growth factor-β1-induced EMT through the ERK/Snail pathway, while Dai et al.36 showed that HNRNPA2B1 also regulated EMT progression of pancreatic cancer through the same signaling pathway. In addition, the expression pattern of SDC1 in prostate cancer suggests that it is involved in EMT and tumor progression.14 The above studies confirmed that expression levels of SDC1 were negatively correlated with EMT progression, consistent with the current results. SDC1 knockdown activated the ERK signaling pathway, upregulated the expression of the Snail transcription factor, inhibited protein expression of E-cadherin, and induced EMT in GBC cells.
Conclusions
The results of the current study showed that knocking down SDC1 expression induced the proliferation, invasion, and migration of GBC cells, upregulated the protein expression of p-ERK1/2, Snail, N-cadherin, and vimentin, downregulated the expression of E-cadherin and β-catenin, and induced the process of EMT. This suggests that SDC1 might induce EMT and cell invasion by regulating the ERK/Snail signaling pathway in GBC cells. These results also provide useful insights into the potential application of SDC1 as a molecular target for the treatment of GBC. This study clarifies the mechanism by which SDC1 affects the aggressiveness of GBC, and provides a basis for further studies aimed at developing a novel therapeutic target for GBC. However, more research is needed to confirm our results and to determine if they provide a true reflection of in vivo changes.
Acknowledgements
The authors thank Dr. Shaobo Zhou for excellent technical assistance with experiments on behalf of the Second Affiliated Hospital of Bengbu Medical College.
Declaration of conflicting interest
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the Natural Science Research Foundation of Anhui province, China [grant number KJ 2018A0240] and Bengbu Medical College Natural Science Key Project, China [grant number BYKY2019129ZD].
References
- 1.Hundal R, Shaffer EA. Gallbladder cancer: epidemiology and outcome. Clin Epidemiol 2014; 6: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Fedewa SA, Miller KD, et al. Cancer statistics for Hispanics/Latinos, 2015. CA Cancer J Clin 2015; 65: 457–480. [DOI] [PubMed] [Google Scholar]
- 3.Teng YH, Aquino RS, Park PW. Molecular functions of syndecan-1 in disease. Matrix Biol 2012; 31: 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pulkkinen JO, Penttinen M, Jalkanen M, et al. Syndecan-1: a new prognostic marker in laryngeal cancer. Acta Otolaryngol 1997; 117: 312–315. [DOI] [PubMed] [Google Scholar]
- 5.Nackaerts K, Verbeken E, Deneffe G, et al. Heparan sulfate proteoglycan expression in human lung-cancer cells. Int J Cancer 1997; 74: 335–345. [DOI] [PubMed] [Google Scholar]
- 6.Matsumoto A, Ono M, Fujimoto Y, et al. Reduced expression of syndecan-1 in human hepatocellular carcinoma with high metastatic potential. Int J Cancer 1997; 74: 482–491. [DOI] [PubMed] [Google Scholar]
- 7.Kumar-Singh S, Jacobs W, Dhaene K, et al. Syndecan-1 expression in malignant mesothelioma: correlation with cell differentiation, WT1 expression, and clinical outcome. J Pathol 1998; 186: 300–305. [DOI] [PubMed] [Google Scholar]
- 8.Day RM, Hao X, Ilyas M, et al. Changes in the expression of syndecan-1 in the colorectal adenoma-carcinoma sequence. Virchows Arch 1999; 434: 121–125. [DOI] [PubMed] [Google Scholar]
- 9.Peretti T, Waisberg J, Mader AM, et al. Heparanase-2, syndecan-1, and extracellular matrix remodeling in colorectal carcinoma. Eur J Gastroenterol Hepatol 2008; 20: 756–765. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S, Qing Q, Wang Q, et al. Syndecan-1 and heparanase: potential markers for activity evaluation and differential diagnosis of Crohn's disease. Inflamm Bowel Dis 2013; 19: 1025–1033. [DOI] [PubMed] [Google Scholar]
- 11.Shepherd TR, Klaus SM, Liu X, et al. The Tiam1 PDZ domain couples to Syndecan1 and promotes cell-matrix adhesion. J Mol Biol 2010; 398: 730–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stepp MA, Pal-Ghosh S, Tadvalkar G, et al. Syndecan-1 and its expanding list of contacts. Adv Wound Care (New Rochelle) 2015; 4: 235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonnomet A, Syne L, Brysse A, et al. A dynamic in vivo model of epithelial-to-mesenchymal transitions in circulating tumor cells and metastases of breast cancer. Oncogene 2012; 31: 3741–3753. [DOI] [PubMed] [Google Scholar]
- 14.Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science 2013; 342: 1234850. [DOI] [PubMed] [Google Scholar]
- 15.Zheng H, Kang Y. Multilayer control of the EMT master regulators. Oncogene 2014; 33: 1755–1763. [DOI] [PubMed] [Google Scholar]
- 16.Zheng M, Jiang Y, Chen W, et al. Snail and Slug collaborate on EMT and tumor metastasis through miR-101-mediated EZH2 axis in oral tongue squamous cell carcinoma. Oncotarget 2015; 6: 6797–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angela NM. A snail tale and the chicken embryo. Int J Dev Biol 2018; 62: 121–126. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Shi J, Chai K, et al. The role of Snail in EMT and tumorigenesis. Curr Cancer Drug Targets 2013; 13: 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunders S, Jalkanen M, O’Farrell S, et al. Molecular cloning of syndecan, an integral membrane proteoglycan. J Cell Biol 1989; 108: 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibrahim SA, Yip GW, Stock C, et al. Targeting of syndecan-1 by microRNA miR-10b promotes breast cancer cell motility and invasiveness via a Rho-GTPase- and E-cadherin-dependent mechanism. Int J Cancer 2012; 131: E884–E896. [DOI] [PubMed] [Google Scholar]
- 21.Kelly T, Suva LJ, Nicks KM, et al. Tumor-derived syndecan-1 mediates distal cross-talk with bone that enhances osteoclastogenesis. J Bone Miner Res 2010; 25: 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nault JC, Guyot E, Laguillier C, et al. Serum proteoglycans as prognostic biomarkers of hepatocellular carcinoma in patients with alcoholic cirrhosis. Cancer Epidemiol Biomarkers Prev 2013; 22: 1343–1352. [DOI] [PubMed] [Google Scholar]
- 23.Anttonen A, Heikkilä P, Kajanti M, et al. High syndecan-1 expression is associated with favourable outcome in squamous cell lung carcinoma treated with radical surgery. Lung Cancer 2001; 32: 297–305. [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto Y, Skacel M, Adams JC. Association of loss of epithelial syndecan-1 with stage and local metastasis of colorectal adenocarcinomas: an immunohistochemical study of clinically annotated tumors. BMC Cancer 2008; 8: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li K, Li L, Wu X, et al. Loss of SDC1 expression is associated with poor prognosis of colorectal cancer patients in Northern China. Dis Markers 2019; 2019: 3768708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farfán N, Ocarez N, Castellón EA, et al. The transcriptional factor ZEB1 represses Syndecan 1 expression in prostate cancer. Sci Rep 2018; 8: 11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science 2011; 331: 1559–1564. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, He J, Zhao X, et al. Syndecan-1 suppresses epithelial-mesenchymal transition and migration in human oral cancer cells. Oncol Rep 2018; 39: 1835–1842. [DOI] [PubMed] [Google Scholar]
- 29.Sun D, Mcalmon KR, Davies JA, et al. Simultaneous loss of expression of syndecan-1 and E-cadherin in the embryonic palate during epithelial-mesenchymal transformation. Int J Dev Biol 1998; 42: 733–736. [PubMed] [Google Scholar]
- 30.Loussouarn D, Campion L, Sagan C, et al. Prognostic impact of syndecan-1 expression in invasive ductal breast carcinomas. Br J Cancer 2008; 98: 1993–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poblete CE, Fulla J, Gallardo M, et al. Increased SNAIL expression and low syndecan levels are associated with high Gleason grade in prostate cancer. Int J Oncol 2014; 44: 647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masola V, Gambaro G, Tibaldi E, et al. Heparanase and syndecan-1 interplay orchestrates fibroblast growth factor-2-induced epithelial-mesenchymal transition in renal tubular cells. J Biol Chem 2012; 287: 1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu YL, Hou MF, Kuo PL, et al. Breast tumor-associated osteoblast-derived CXCL5 increases cancer progression by ERK/MSK1/Elk-1/snail signaling pathway. Oncogene 2013; 32: 4436–4447. [DOI] [PubMed] [Google Scholar]
- 34.Li S, Lu J, Chen Y, et al. MCP-1-induced ERK/GSK-3β/Snail signaling facilitates the epithelial-mesenchymal transition and promotes the migration of MCF-7 human breast carcinoma cells. Cell Mol Immunol 2017; 14: 621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo W, Liu X, Sun W, et al. Toosendanin, a natural product, inhibited TGF-β1-induced epithelial-mesenchymal transition through ERK/Snail pathway. Phytother Res 2018; 32: 2009–2020. [DOI] [PubMed] [Google Scholar]
- 36.Dai S, Zhang J, Huang S, et al. HNRNPA2B1 regulates the epithelial-mesenchymal transition in pancreatic cancer cells through the ERK/snail signalling pathway. Cancer Cell Int 2017; 17: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]