Abstract
Purpose
Retinoblastoma (RB) is the most common primary intraocular tumor in children. Chemoresistance is the major obstacle for treatment of these tumors. This study aims to determine whether or not downregulating microRNA-222 (miR-222) could serve as a potential therapeutic target for preventing chemoresistance in RB treatment.
Methods
Differentially expressed miR-222 in RB samples and its downstream target genes were predicted using bioinformatics methods. The expression of miR-222 was altered by mimic or inhibitor to examine its role in RB cell in response to the chemotherapeutic agent vincristine (VCR). Further bioinformatic analysis predicted involvement of the stability of hypoxia-inducible factor 1α (HIF1α) protein in regulation of the von Hippel–Lindau (VHL) tumor suppressor, followed by characterization of the effect of VHL on the ubiquitin–proteasome degradation of HIF1α. Next, VHL or HIF1α was overexpressed to determine their effects on RB cell activities after VCR treatment. In vivo assays were performed on nude mice to further verify the in vitro results.
Results
miR-222 is highly expressed in RB tissues and cells and was found to facilitate resistance of RB cells to VCR. Of note, miR-222 specifically bound to and negatively regulated VHL. VHL could inhibit the stability of HIF1α and promote the degradation of ubiquitin–proteasome, thus reducing HIF1α expression to attenuate VCR resistance in RB cells. Moreover, inhibition of miR-222 in combination with VCR suppressed tumor formation in nude mice.
Conclusions
miR-222 promotes the expression of HIF1α by targeting VHL, thus accelerating the resistance of RB cells to the chemotherapeutic agent VCR.
Keywords: retinoblastoma, microRNA-222, VHL, HIF1α, chemoresistance, vincristine
Retinoblastoma (RB) is the most frequent intraocular cancer in children1 and usually occurs before the age of five years.2 RB accounts for 3% of childhood cancer and typically presents with leukocoria and strabismus.3 At present, the disease has a 98% survival rate in developed countries, but the survival rate in developing countries is still low.4 Chemotherapy agents such as vincristine (VCR) and topotecan have been widely applied in the treatment of RB,5,6 but the resistance of tumor cells to chemotherapy limits the therapeutic efficacy.7 The chemoresistance in RB has been linked with weakened apoptosis and augmented proliferative potential in RB cells, which may be associated with dysregulation of carcinogenesis-related molecules.8 Previous studies have suggested a link between the dysregulation of microRNAs (miRNAs) and chemoresistance,9–11 which may serve as a target for attenuating chemoresistance in RB.
miRNAs are a class of small noncoding RNAs that can block the translation and stability of messenger RNA (mRNA) to control genes participating in cellular processes12 capable of affecting almost all biological pathways.13 It has been determined that the differentially expressed miRNAs in RB can be used as therapeutic targets for RB cell resistance to chemotherapeutic agents.14–16 Moreover, findings obtained from previous studies indicate that microRNA-222 (miR-222) plays a role promoting the progression of chemoresistance, such as in non-small cell lung cancer,17 liver cancer,18 and breast cancer.19 Additionally, miR-222 confers radiation resistance in colorectal cancer by targeting the phosphatase and tensin homolog deleted on chromosome 10 (PTEN) gene.20 In our study, we verified the differential expression of miR-222 in RB-based microarray data analysis, and the von Hippel–Lindau (VHL) tumor suppressor could be the downstream target for miR-222. Therefore, we hypothesized that miR-222 affected RB cell resistance to VCR by targeting VHL.
VHL is the substrate binding subunit of the VHL E3 ubiquitin ligase, which can bind protein targets, causing ubiquitination and degradation.21 The proline of hypoxia-inducible factor α (HIFα) is hydroxylated in an oxygen-containing environment and binds to VHL protein to trigger the ubiquitin–proteasome proteolytic pathway, which is degraded by ubiquitination.22,23 Also, VHL has been previously demonstrated to target the hydroxylated α subunit of HIFα for ubiquitination and subsequent proteasomal degradation.24 More importantly, previous research has reported that there exists a correlation between hypoxia-inducible factor 1α (HIF1α) and drug resistance.25,26 Therefore, we speculated that VHL affects the resistance of RB cells to VCR by regulating the stability of HIF1α protein. Additionally, miR-222 was reported to play a role in drug resistance through regulation of the PTEN/Akt/FOXO1 pathway.27 Based on our preliminary data and previous studies, we hypothesized that miR-222 played a role in RB cell resistance to VCR, which may provide a new therapeutic strategy for the treatment of RB.
Methods
Ethics Statement
The study was conducted under the approval of the Ethics Committee of Linyi People's Hospital. All participants or their guardians signed written informed consents. Animal experiments were performed in compliance with recommendations in the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Microarray-Based Gene Expression Analysis
RB-associated miRNA GSE41321 and gene GSE97508 expression profiles were retrieved from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo). There were six samples in the GSE41321 expression profile, including three normal control and three RB samples. There were nine samples in the GSE97508 expression profile, including three normal control and six RB samples. The miR-222 expression data for the GSE41321 expression profile were analyzed using the R language (R Foundation for Statistical Computing, Vienna, Austria), and a box plot was then generated to determine its expression trend. miRWalk (binding P > 0.9, energy < –20, accessibility < 0.01, arbitrary units (AU) > 0.55) (http://mirwalk.umm.uni-heidelberg.de/), miRDIP (integrated score > 0.1, number of sources > 5) (http://ophid.utoronto.ca/mirDIP/), and TargetScan (cumulative weighted context++ score < –0.1) (http://www.targetscan.org/vert_71/) were applied to predict and select the target genes of miR-222. The critical genes were obtained by taking the intersection. The protein–protein interaction (PPI) network of critical genes was constructed using the online tool String (https://string-db.org/) to screen the top three genes, followed by obtaining the core target genes. The online analysis tools String and GeneMANIA (http://genemania.org/) were adopted to construct the PPI networks to predict genes related to the core target genes. Differentially expressed genes were selected in the GSE97508 expression profile using the limma package (http://www.bioconductor.org/packages/release/bioc/html/limma.html) in R language and identified with |log2 fold change| > 1.0 and P < 0.05 as threshold. The intersections among the String, GeneMANIA, and GSE97508 expression profiles were selected to find the possible downstream genes of critical genes.
Study Subjects
RB tissues from 56 RB patients and normal retinal tissues from 17 patients with ruptured eyeballs who were not age matched with RB patients were collected at Linyi People's Hospital from March 2017 to June 2019. Tissues after enucleation were stored at –80°C. All of these patients were newly diagnosed and were treated for the first time without laser photocoagulation, chemotherapy, or lesion treatment.
Cell Culture and Transfection
The normal human retinal epithelial cell line ARPE-19 and the RB cell lines Y79, Weri-Rb1, SO-Rb50, and HXO-RB44 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured with Gibco RPMI-1640 medium (Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and 10% penicillin and streptomycin (Life Technologies) and maintained in a 37°C incubator containing 5% CO2. The cells were treated with 0.25% trypsin and passaged at 1:3. Cells were seeded into six-well plates at 3 × 105 cells per well. When confluency reached about 70% to 80%, cells in the logarithmic growth phase were collected for further study.
Y79 cells in the logarithmic growth phase were seeded into six-well plates at 4 × 105 cells per well. When cells reached 70% to 80% confluence, they were collected for transfection according to the instructions for the Invitrogen Lipofectamine 2000 kit (11668-019; Thermo Fisher Scientific, Waltham, MA, USA). Y79 cells were transfected with VHL overexpression plasmid (oe-VHL), HIF1α overexpression plasmid (oe-HIF1α), miR-222 mimic (15 nM), miR-222 inhibitor (50 nM), or their negative controls (mimic-NC, inhibitor-NC, and oe-NC). The transfection plasmids were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). Each experiment was repeated three times.
pSIH1-H1-copGFP, the lentiviral vector silencing the gene, was constructed in our study. The packaged lentivirus expressing inhibitor-NC or miR-222 inhibitor was transfected into human embryo kidney 293T (HEK293T) cells (American Type Culture Collection, Manassas, VA, USA). Cells were cultured for 48 hours prior to supernatant collection. The virus in the logarithmic growth phase was collected and used for infection. Upon reaching the logarithmic growth phase, cell suspensions of 5 × 104/mL were prepared and seeded into six-well plates at 2 mL per well and cultured in the incubator at 37°C. After 48 hours of infection, efficiency indicated by green fluorescent protein (GFP) expression was monitored under fluorescence microscopy. Reverse-transcription quantitative polymerase chain reaction (RT-qPCR) was applied to detect the expression of related genes in each group of cells.
RT-qPCR
Total RNA from cells was extracted using Trizol reagent (16096020; Thermo Fisher Scientific). Reverse transcription was performed with 5 µg of total RNA according to the instructions for the RevertAid First Strand cDNA Synthesis Kit (K1622; Thermo Fisher Scientific) to generate cDNA. Real-time quantitative PCR assays were conducted using the Thermo Fisher Scientific TaqMan MicroRNA Assay and TaqMan Universal PCR Master Mix (normalized to U6) based on TaqMan Gene Expression Assay protocols (normalized to glyceraldehyde 3-phosphate dehydrogenase [GAPDH]); see Supplementary Table S1. The relative expression of target genes was measured by the 2−ΔΔCt method.
Western Blot Analysis
Total protein from tissues and cells was extracted using radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China). Protein concentrations were assessed using a bicinchoninic acid protein assay kit (Yeasen Biotech Co., Ltd., Shanghai, China). Isolated proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). After separation, the protein was transferred to a polyvinylidene fluoride (PVDF) membrane (IPVH85R, Millipore, Darmstadt, Germany) and then blocked with 5% BSA for 1 hour. After being washed with Tris Buffered Saline with Tween (TBST; Sigma-Aldrich, St. Louis, MO, USA), the PVDF membrane was probed with diluted primary antibodies overnight at 4°C: Rabbit Anti-Von Hippel Lindau/VHL antibody (ab83307, 1:100), Mouse Anti-HIF1 beta antibody (ab2771, 1:4000), and Recombinant Anti-GAPDH antibody (ab128915, 1:10,000), all from Abcam (Cambridge, UK). After being washed with TBST again, the membrane was incubated with the secondary antibody Goat Anti-Rabbit IgG H&L (HRP) (ab205718, 1:10,000) labeled with horseradish peroxidase for 1 hour at room temperature. After the membrane was washed, substrate was applied for developing. Images were subjected to grayscale analysis using ImageJ 1.48u software (National Institutes of Health, Bethesda, MD, USA). The relative ratio was calculated with GAPDH as the internal reference.
MTT Assay for Cell Survival
Cells were plated in 96-well plates at a density of 8 × 103 cells per well and were cultured with complete medium (100 µl per well) for 24 hours. Cells were then treated with VCR serial dilution for 48 hours. The methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay was used to analyze the survival rate of cells; 20 µl of 5-mg/ml MTT solution was added to each well, and all cells were incubated at 37°C for 4 hours before removing the medium. Then, 150 µl of dimethyl sulfoxide was added, and the solution was mixed to ensure that the crystals of formazan in the cells were dissolved. The absorbance at 490 nM was determined using a microplate reader (CliniBio 128; ASYS Hitech, Eugendorf, Austria). The maximum half-inhibitory concentration (IC50) of VCR was calculated from three independent experiments.
Flow Cytometry
Cell apoptosis was assessed using the Annexin-V-FITC/PI Apoptosis Assay (C1062M; Beyotime Biotechnology). This was followed by washing 1 × 106 cells with cold PBS, and 5 µl of Annexin-V-FITC and 10 µl of propidium iodide were added to the cell suspension and incubated for 15 minutes in the dark. The cell apoptosis rate was determined by flow cytometry (BD FACSVerse, Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
Dual-Luciferase Reporter Assays
The promoter region of VHL was added to the pmirGLO Dual-Luciferase miRNA Target Expression vector (Promega, Madison, WI, USA) to generate the recombinant vector VHL-wild type (pmirGLO-VHL-WT). Then the promoter of VHL was mutated and added to the pmirGLO vector to be a recombinant vector of VHL mutants (pmirGLO-VHL-MUT). The correctly sequenced luciferase reporter plasmid was co-transfected into HEK293T cells with mimic-NC and miR-222 mimic. Luciferase activity was measured using the Promega Dual-Luciferase Reporter Assay (E1910). Then, 100 µl firefly luciferase solution and 100 µl renilla luciferase were added to each cell sample to determine firefly luciferase and renilla luciferase, respectively. Each experiment was repeated three times.
Co-immunoprecipitation Assay
Transfected cells were lysed in lysis buffer (50-mM Tris-HCl, pH 7.4; 150-mM NaCl; 10% glycerol; 1-mM ethylenediaminetetraacetate; 0.5% 40 ethylene oxide units per molecule [NP-40]; and protease inhibitor cocktail). The supernatant was collected after centrifugation. The supernatant was incubated with anti-HA or anti-FLAG antibody (Sigma-Aldrich) alone by the addition of protein A along with G beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 2 hours. After being washed with the lysis buffer, the beads were boiled at 100°C for 5 minutes. The protein was separated by SDS-PAGE, then transferred to a Millipore nitrocellulose membrane, followed by western blot analysis.
Tumor Xenografts in Nude Mice
Fifty BALB/c mice (ages 4–6 weeks; weighing about 16–20 g) were raised in laminar flow cabinets (specific-pathogen-free grade). Regular indoor ultraviolet irradiation was carried out. The cage, litter, drinking water, and feed were disinfected, and room temperature was maintained at 24°C to 26°C and 40% to 60% relative humidity. Y79 cells were mixed with PBS to prepare single-cell suspensions at a concentration of 1 × 106 cells/ml. Next, 50 µl of the cell suspension was subcutaneously injected into the right side of each mouse. One week later, the mice were randomly divided into four groups (n = 10) and treated accordingly: PBS, VCR (0.5 mg/kg body weight), VCR + lentivirus expressing inhibitor-NC (lv-inhibitor-NC), or VCR + lentivirus expressing miR-222 inhibitor (lv-miR-222 inhibitor). After subcutaneous tumor formation in nude mice, lentiviruses carrying different plasmids were injected into the tumor sites, and then the mice were treated with VCR. The mice in the control group were given PBS, and the mice in the VCR group were given VCR (0.5 mg/kg body weight) once a week for 6 weeks. For mice in the VCR + lv-inhibitor-NC group and mice in the VCR + lv-miR-222 inhibitor group, VCR (0.5 mg/kg body weight) was intraperitoneally injected once a week for 6 weeks, and the lentiviruses carrying corresponding plasmids were injected into the tumor sites. On the 30th day, the nude mice were anesthetized and euthanized with 100 mg/kg of pentobarbital sodium (P3761, Sigma-Aldrich). The short diameter (a) and long diameter (b) of the tumor were measured with a vernier caliper. Tumor volume changes in nude mice were recorded based on the formula tumor volume = π(a2b)/6. The weight of a tumor was measured on a balance.28
Statistical Analysis
The data were processed using SPSS Statistics 21.0 (IBM, Armonk, NY, USA). Measurement data are expressed as mean ± SD. Unpaired data in compliance with normal distributions and homogeneity between two groups were compared using unpaired t-tests. Comparisons among multiple groups were conducted by one-way ANOVA with Tukey's post hoc test. Statistical analysis in relation to time-based measurements within each group was carried out using repeated-measures ANOVA, followed by Bonferroni's post hoc test. Pearson's correlation coefficient was employed to analyze the relationship between two indices. A value of P < 0.05 indicated significant difference.
Results
miR-222 Is Highly Expressed in RB and Promotes the Resistance of RB Cells to VCR
Box plots drawn based on the miR-222 expression data in the GSE41321 expression profile from the GEO database suggested that miR-222 was highly expressed in RB tissues (Supplementary Fig. S1A). To verify the expression of miR-222 in RB, RT-qPCR was adopted to examine the expression of miR-222 in retinal tissues of RB patients and normal retinal tissues. The experimental data confirm that miR-222 was upregulated in the retinal tissue of RB patients compared to normal retinal tissue (Supplementary Fig. S1B). Furthermore, relative to normal human retinal epithelial cell line ARPE-19, miR-222 was highly expressed in the RB cell lines Y79, Weri-Rb1, SO-Rb50, and HXO-RB44, especially in the Y79 cell line (Supplementary Fig. S1C). Thus, the Y79 cell line was selected for subsequent experiments.
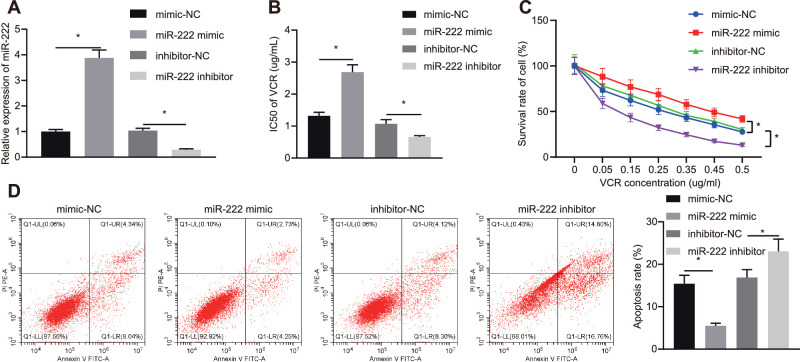
To further investigate the role of miR-222 in the resistance of RB cells to the chemotherapeutic agent VCR, Y79 cells were transfected with miR-222 mimic (15 nM), miR-222 inhibitor (50 nM), or their corresponding NC. RT-qPCR assays revealed that the expression of miR-222 in cells transfected with miR-222 mimic was elevated compared to those transfected with mimic-NC (Fig. 1A). The expression of miR-222 in cells transfected with miR-222 inhibitor was diminished versus that in cells transfected with inhibitor-NC. The IC50 value of transfected Y79 cells was assessed by MTT assay, which revealed that, relative to mimic-NC transfected cells, the IC50 of VCR on miR-222 mimic transfected cells was elevated, whereas the IC50 on miR-222 inhibitor transfected cells was reduced in comparison with the inhibitor-NC transfected cells (Fig. 1B). Moreover, the survival rate and the apoptosis rate of Y79 cells were assessed by MTT assay and flow cytometry (Figs. 1C, 1D). Cell survival was elevated and apoptosis was reduced in response to miR-222 mimic relative to those transfected with mimic-NC. Compared to inhibitor-NC transfected cells, the cell survival rate of miR-222 inhibitor transfected cells was diminished and apoptosis was augmented. These findings indicate that miR-222 enhanced the resistance of RB cells to VCR.
Figure 1.
miR-222 enhances the resistance of RB cells to VCR. (A) Expression of miR-222 in transfected Y79 cells examined by RT-qPCR assay. (B) The IC50 of VCR on Y79 cells detected by MTT assay. (C) Cell viability of Y79 cells assessed by MTT assay. (D) Apoptosis of Y79 cells examined by flow cytometry. *P < 0.05 versus Y79 cells transfected with mimic-NC or inhibitor-NC. Measurement data are expressed as mean ± SD. Data comparisons between two groups were performed by unpaired t-tests. Comparisons among multiple groups were conducted by ANOVA with Tukey's post hoc test. Statistical analysis in relation to time-based measurements within each group was conducted using repeated-measures ANOVA followed by Bonferroni's post hoc test.
miR-222 Targets and Downregulates VHL in RB
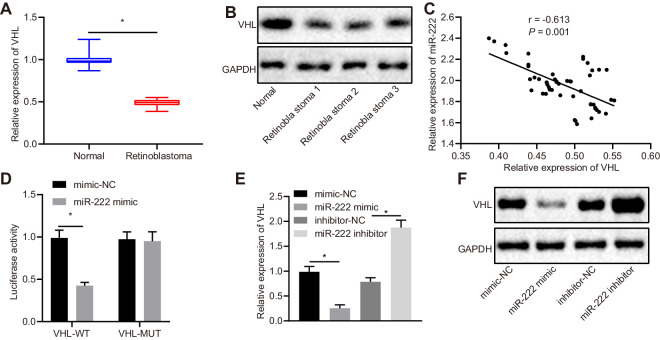
These results indicate that miR-222 accelerated the resistance of RB cells to VCR. In order to explore the downstream regulatory mechanism of miR-222, the downstream target genes of miR-222 were predicted based on the online databases miRWalk, miDIP, and TargetScan, from which we obtained 1442, 5618, and 1432 genes, respectively. A total of 178 genes were obtained via Venn diagrams (Supplementary Fig. S2A). A PPI network was constructed using the online analysis tool String. After analyzing the core degrees and scores of the 178 genes, we identified the top three genes: CYCS, PPP4R2, and VHL (Supplementary Fig. S2B, Table). Prior evidence has shown that the gene VHL is closely related to RB,29 but its specific regulatory mechanism is still unknown; thus, it is worth further research and study. VHL was hypothesized to be the downstream target gene of miR-222 in our study. TargetScan revealed that there were potential binding sites between VHL and miR-222 (Supplementary Fig. S2C). The expression of VHL in the retinal tissue of RB patients and normal retinal tissue was assessed by RT-qPCR and western blot analysis (Figs. 2A, 2B). The expression of VHL in the retinal tissues of RB patients was appreciably lower than that in normal retinal tissues. Pearson's correlation coefficient was employed to analyze the relationship between VHL and miR-222. The experimental data show that the expression of miR-222 was negatively correlated with that of VHL (Fig. 2C). Dual-Luciferase Reporter Assays were performed to verify whether VHL was a downstream target of miR-222. The experimental data revealed that the luciferase activity of miR-222/VHL-WT co-transfected HEK293T cells was diminished in comparison with those transfected with mimic-NC (P < 0.05) (Fig. 2D). However, there was no significant difference in the luciferase activity of the VHL-MUT transfected group, which contained a mutation site in 3′ untranslated region (UTR) (P > 0.05). This suggests that miR-222 could bind to the 3′UTR of VHL. Furthermore, Y79 cells were transfected with miR-222 mimic or miR-222 inhibitor, followed by determination of the expression of VHL in Y79 cells by RT-qPCR and western blot analysis (Figs. 2E, 2F). The experimental data demonstrate that the mRNA and protein expression of VHL was diminished in cells transfected with miR-222 mimic versus those with mimic-NC. Compared to cells transfected with inhibitor-NC, the mRNA and protein expression of VHL in miR-222-inhibitor was elevated (both, P < 0.05). The experimental data further indicate that miR-222 targeted and downregulated VHL expression in RB.
Table.
Top Five Genes Based on Core Degree and Sum Combined Score in PPI Network
| Rank | Gene | Degree | SUM Combined Score |
|---|---|---|---|
| 1 | CYCS | 9 | 5.948 |
| 2 | PPP4R2 | 6 | 3.634 |
| 3 | VHL | 5 | 4.126 |
| 4 | CAND1 | 5 | 4.067 |
| 5 | TCF12 | 5 | 3.453 |
Degree refers to the number of interactions between this gene and other genes. SUM combined score refers to the sum of the combined values between the gene and the other genes.
Figure 2.
VHL is downregulated in RB tissues and it is a target gene of miR-222. (A) Normalized expression of VHL in retinal tissues of RB patients (n = 50) and normal retinal tissues (n = 17) detected by RT-qPCR assay. (B) Expression of VHL in retinal tissues of RB patients (n = 50) and normal retinal tissues (n = 17) examined by western blot analysis. (C) Pearson's correlation analysis between VHL and miR-222. (D) Dual-Luciferase Reporter Assays show the binding between miR-222 and VHL. (E) Normalized expression of VHL in transfected Y79 cells by RT-qPCR assay. (F) Protein expression of VHL in transfected Y79 cells measured by western blot analysis. In A and B, *P < 0.05 versus normal retinal tissues. In D, *P < 0.05 versus mimic-NC group. In E and F, *P < 0.05 versus Y79 cells transfected with mimic-NC or inhibitor-NC. Measurement data are expressed as mean ± SD. Data comparisons between two groups were performed by unpaired t-tests. Pearson's correlation coefficient was used to analyze the relationship between miR-222 and VHL. The cell experiment was repeated three times.
Overexpression of VHL Reduces the Resistance of RB Cells to VCR by Inhibiting the Stability of HIF1α
We obtained 10 VHL-related genes using the online analysis tool String and 20 from GeneMANIA. Also, two sets of PPI networks were constructed (Supplementary Figs. S3A, S3B). In total, 2259 differently expressed genes were selected from GSE97508 expression profiles on GEO databases in R language (Supplementary Fig. S3C). By taking the intersection of results from String, GeneMANIA, and GSE97508 expression profiles, the two most relevant genes were identified as HIF1α and EPAS1 (Supplementary Fig. S3D). It is well known that VHL is a component of the E3 ubiquitin ligase, which is closely related to the stability of HIF1α protein and its ubiquitination and degradation.23,30 The proline of HIF1α is hydroxylated under normoxic conditions and binds to VHL protein to trigger the ubiquitin–proteasome proteolytic pathway, which is degraded by ubiquitination.22
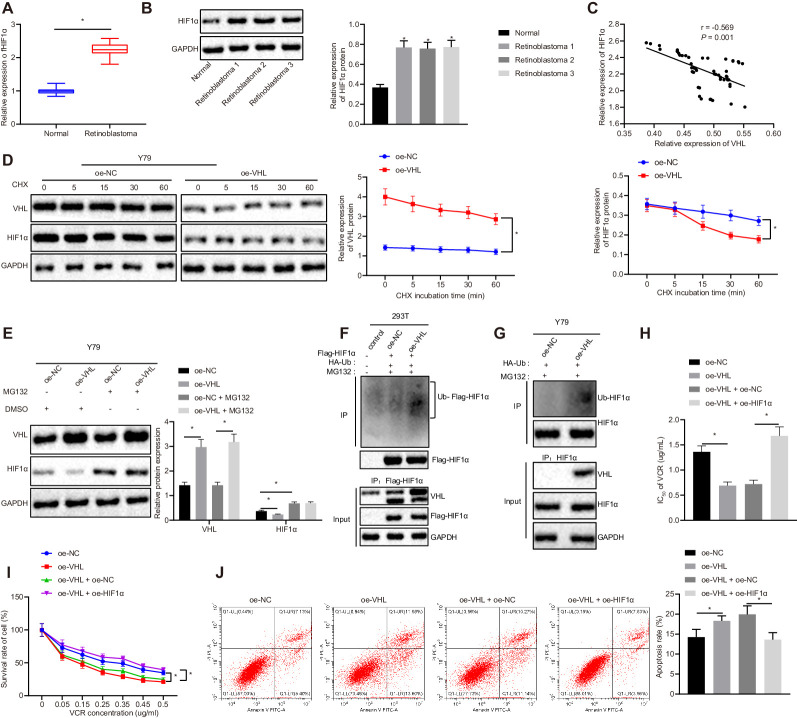
Previous literature has reported that HIF1α is also associated with drug resistance.25,26 Therefore, we speculated that VHL could affect cell resistance to VCR by regulating the stability of HIF1α in RB cells. Western blot analysis was performed to examine the expression of HIF1α in clinical samples of RB patients. The experimental data indicate that HIF1α was highly expressed in RB patients (Figs. 3A, 3B). Pearson correlation analysis revealed that the expression of HIF1α and VHL was negatively correlated (Fig. 3C). To further investigate whether the stability of HIF1α was regulated by VHL, Y79 cells were treated with 100-µM cycloheximide (CHX). The expression of VHL and HIF1α was determined by Western blot analysis. The experimental data indicate that the expression of HIF1α in cells transfected with oe-VHL was diminished relative to cells transfected with oe-NC (Fig. 3D). This result suggests that overexpression of VHL reduced the stability of HIF1α. In order to further explore how VHL reduced the stability of HIF1α, we treated transfected Y79 cells with MG132. Western blot analyses were used to measure the expression of VHL and HIF1α. The experimental data suggest that the expression of VHL was appreciably higher for oe-VHL compared to oe-NC, but the expression of HIF1α was appreciably lower. The expression of VHL was not appreciably changed in the oe-NC transfected cells with MG132 treatment relative to the oe-NC transfected cells without MG132 treatment, but the expression of HIF1α was elevated. After cells were treated with MG132, the expression of VHL in oe-VHL transfected cells was elevated compared to oe-NC transfected cells, but the expression of HIF1α remained the same (Fig. 3E). This result further suggests that VHL contributed to the degradation of HIF1α.
Figure 3.
VHL reduces the resistance of RB cells to VCR by decreasing of the stability of HIF1α. (A) Normalized expression of HIF1α in retinal tissues of RB patients (n = 50) and normal retinal tissues (n = 17) examined by RT-qPCR assay. (B) Expression of HIF1α in retinal tissues of RB patients (n = 50) and normal retinal tissues (n = 17) determined by western blot analysis. (C) Pearson's correlation analysis between HIF1α and VHL. (D) Expressions of VHL and HIF1α in transfected Y79 cells treated with 100-µM CHX measured by western blot analysis. (E) Expression of VHL and HIF1α in transfected Y79 cells treated with 40-µM MG132 determined by western blot analysis. (F) Effect of VHL on HIF1α ubiquitination in 293T cells detected by IP assay. (G) The effect of VHL on HIF1α ubiquitination in Y79 cells assessed by immunoprecipitation assay. (H) The IC50 of VCR on transfected Y79 cells examined by MTT assay. (I) Cell survival rate of transfected Y79 cells measured by MTT assay. (J) Apoptosis rate of transfected Y79 cells analyzed by flow cytometry. In A and B, *P < 0.05 versus normal retinal tissues. In D, *P < 0.05 versus oe-NC transfected Y79 cells. In E, *P < 0.05 versus oe-NC and oe-NC transfected Y79 cells. In F to J, *P < 0.05 versus oe-NC or oe-VHL + oe-NC group. Measurement data are expressed as mean ± SD. Data comparisons between two groups were performed by unpaired t-test. Statistical analysis in relation to time-based measurements within each group was conducted using repeated-measures ANOVA followed by a Bonferroni's post hoc test. Pearson's correlation coefficient was used to analyze the relationship between VHL and HIF1α. The experiment was conducted in triplicate.
Subsequently, we verified whether the VHL facilitated HIF1α degradation via enhancing HIF1α ubiquitination. Ubiquitination of HIF1α was assessed after HEK293T cells overexpressing VHL were treated with HA-Ub and Flag-HIF1α, as well as 40-µM MG132, for 48 hours. We observed that the ubiquitination of HIF1α was appreciably accelerated in the cells overexpressing VHL (Fig. 3F). Y79 cells overexpressing VHL were treated with HA-Ub and Flag-HIF1α, as well as 40-µM MG132, for 48 hours to determine the ubiquitination of HIF1α. The results suggest that ubiquitination of HIF1α was boosted in oe-VHL transfected cells (Fig. 3G). These findings demonstrate that VHL attenuated the stability of HIF1α, promoting its ubiquitin–proteasome degradation and decreasing the expression level.s
After confirming that VHL could promote the degradation of HIF1α, we then investigated the effect of VHL and HIF1α on RB chemoresistance. The IC50 of VCR on transfected Y79 cells was assessed by MTT assay. The experimental data indicate that, relative to oe-NC transfected cells, IC50 was reduced in oe-VHL transfected cells. The IC50 was augmented in oe-VHL/oe-HIF1α co-transfected cells compared to oe-VHL/oe-NC co-transfected cells (Fig. 3H). The survival rate and apoptosis rate of Y79 cells were assessed by MTT assay and flow cytometry (Figs. 3I, 3J). Relative to oe-NC transfected cells, the cell survival rate of oe-VHL transfected cells was diminished, accompanied by elevated apoptosis rate. In comparison with cells transfected with oe-VHL and oe-NC, cells transfected with oe-VHL and oe-HIF1α had elevated cell survival rates and reduced apoptosis rates. These findings indicate that VHL reduced the resistance of RB cells to VCR by attenuating the stability of HIF1α.
miR-222/VHL/HIF1α Axis Regulates RB Cell Resistance to VCR
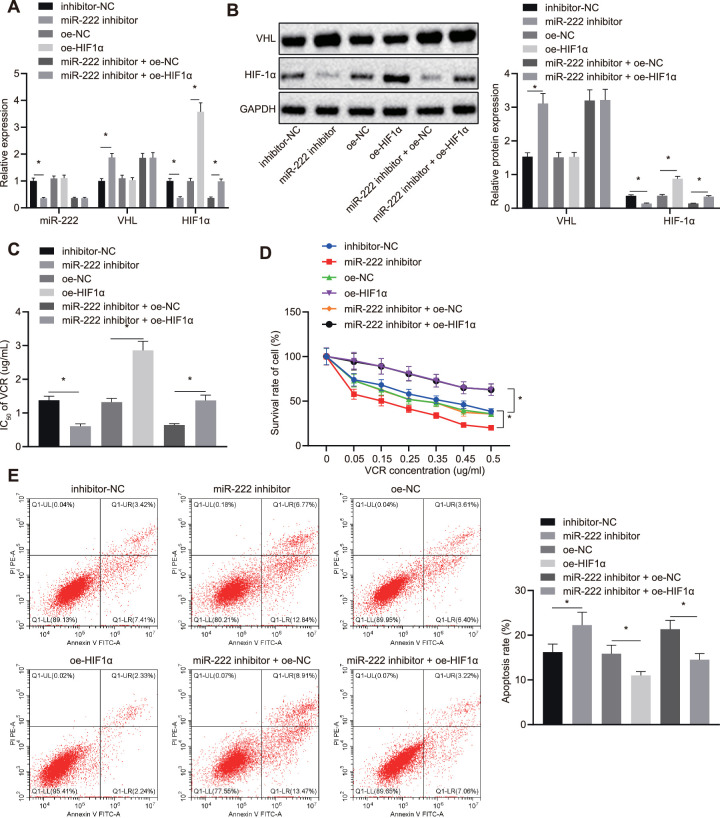
After we determined that miR-222 downregulated VHL, which could promote the ubiquitination of HIF1α, we further explored the effect of the miR-222/VHL/HIF1α axis on RB cell resistance to chemotherapeutic agent VCR. After Y79 cells were transfected with different plasmids and their corresponding controls, the expression of miR-222, VHL, and HIF1α was assessed by RT-qPCR assay and western blot analysis (Figs. 4A, 4B). The experimental data indicate that the expression of miR-222 and HIF1α in cells transfected with miR-222-inhibitor were diminished, accompanied with high expression of VHL versus inhibitor-NC transfected cells. Compared to oe-NC transfected cells, there was no significant difference in the expression of miR-222 and VHL in oe-HIF1α transfected cells, but HIF1α was overexpressed. After cells were exposed to VCR, MTT assays were conducted to examine the IC50 of VCR on Y79 cells. The experimental data show that VCR demonstrated lower IC50 values on the cells transfected with miR-222-inhibitor and higher IC50 values on cells transfected with oe-HIF1α. There was no difference in the miR-222 inhibitor and oe-HIF1α transfected cells (Fig. 4C).
Figure 4.
miR-222/VHL/HIF1α axis regulates RB cells resistance to VCR. (A) Normalized expression of miR-222, VHL, and HIF1α in Y79 cells detected by RT-qPCR assay. (B) Expression of VHL and HIF1α in Y79 cells measured by western blot analysis. (C) The IC50 of VCR on Y79 cells in assessed by MTT assay. (D) The cell survival rate of Y79 cells determined by MTT assay. (E) Apoptosis of Y79 cells examined by flow cytometry. *P < 0.05 versus oe-NC or inhibitor-NC transfected Y79 cells. Measurement data are expressed as mean ± standard SD. Comparisons among multiple groups were conducted by ANOVA with Tukey's post hoc test. Statistical analysis in relation to time-based measurements within each group was conducted using repeated-measures ANOVA followed by a Bonferroni's post hoc test.
MTT assays and flow cytometry (Figs. 4D, 4E) were applied to detect the viability and apoptosis rate of Y79 cells. The experimental data indicate that, relative to inhibitor-NC transfected cells, the cell survival rate of miR-222-inhibitor transfected cells was diminished and apoptosis was elevated. The cell survival rate of the cells was elevated and the apoptosis rate was diminished in response to transfection of cells with oe-HIF1α versus those with oe-NC. Compared to the miR-222 inhibitor and oe-NC transfected cells, the miR-222 inhibitor and oe-HIF1α transfected cells had appreciably higher viability and lower apoptosis rates. These findings confirm that the miR-222/VHL/HIF1α axis mediated RB cells resistance to VCR.
Inhibition of miR-222 Combined with VCR Treatment Suppresses Tumor Formation in Nude Mice
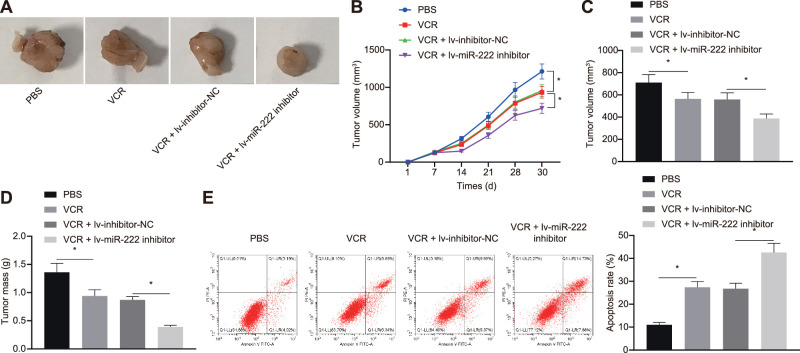
In order to study the effect of inhibition of miR-222 combined with VCR on RB in vivo, xenografts were induced in nude mice. After tumor formation, lentivirus carrying different plasmids were injected into the tumor sites, followed by VCR treatment. The tumor growth curves (Figs. 5A–5D) suggest that, after VCR treatment, the growth rate, weight, and volume of mouse tumors were reduced. Compared to mice injected with cells transfected with inhibitor-NC, the growth rate, weight, and volume of tumors in mice injected with cells transfected with miR-222-inhibitor were diminished. The flow cytometric data show that the apoptosis of cells exposed to VCR was elevated relative to cells treated with PBS. In comparison with mice injected with inhibitor-NC transfected cells, apoptosis was reduced in mice injected with miR-222-inhibitor transfected cells (Fig. 5E). These findings suggest that inhibition of miR-222 combined with VCR treatment restrained the formation of RB in nude mice.
Figure 5.
Inhibition of miR-222 combined with VCR treatment suppresses tumor formation in nude mice. (A) Representative images of formed tumors of RB in nude mice (n = 10). (B) Quantitative analysis of tumor volume. (C) Quantitative analysis of the volume of tumors. (D) Quantitative analysis of the weight of tumors. (E) Apoptosis in tumor tissues of nude mice detected by flow cytometry. *P < 0.05 versus the PBS group or the VCR + lv-inhibitor-NC group. Measurement data are expressed as mean ± SD. Data comparisons between two groups were performed by unpaired t-tests. Statistical analysis in relation to time-based measurements within each group was conducted using repeated-measures ANOVA followed by a Bonferroni's post hoc test (n = 10).
Discussion
VCR is the primary drug utilized in the treatment of many pediatric malignancies,31,32 including RB,33 but it is well known that drug resistance is the main obstacle to cancer treatment. Thus, novel therapies developed with the aid of molecular biomarkers are urgently needed to prevent resistance of RB cells to VCR. A growing number of studies have highlighted that miRNAs function as oncomiRs or tumor suppressors that play important roles in chemotherapy resistance.10,16 Interestingly, miR-222 has proven to be a key regulator in numerous cellular processes of ocular tumors, including uveal melanoma34 and RB.35 Identification of new molecules involved in RB resistance will greatly facilitate the development of novel therapies. A prior study has revealed that miR-222 enhances the resistance of breast cancer cells to doxorubicin (DOX).36
This study aimed to investigate the effects of miR-222 on RB resistance to VCR, as well as the potential downstream molecules and possible signaling pathways. Bioinformatics analysis predicted the putative target gene VHL of miR-222. Collectively, the data of the present study suggest that miR-222 can enhance RB resistance to VCR by downregulating VHL to elevate the expression of HIF1α through inhibition of ubiquitin–proteasome degradation (Fig. 6).
Figure 6.
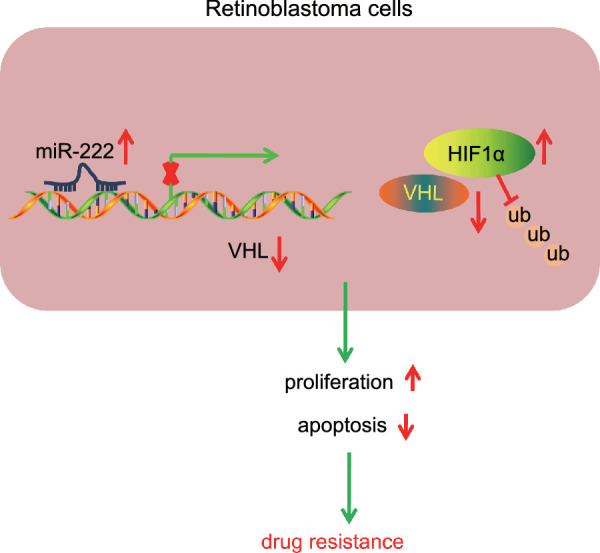
Schematic representation and function of miR-222/VHL/HIF1α axis in chemoresistance of RB cells. miR-222 downregulated VHL to elevate the expression of HIF1α through inhibition of ubiquitin–proteasome degradation, thereby enhancing RB cell proliferation and attenuating apoptosis, which accelerates RB resistance to VCR.
The first finding of the present study was that miR-222 was highly expressed in RB tissues and cells. Moreover, we verified that high expression of miR-222 was associated with the resistance of RB cells to VCR by gain- and loss-of-function experiments. miR-222 has been demonstrated to diminish cell apoptosis induced by cisplatin in bladder cancer.37 These findings collectively suggest that miR-222 accelerates the resistance of RB cells to VCR, which is in agreement with a prior study reporting that miR-222 enhanced the resistance of breast cancer cells to DOX.36 Additionally, miR-222 has been reported to promote the resistance of the breast cancer cell line MDA-MB-231 to carboplatin.38 Although there is apparently no report on the effect of miR-222 on resistance to etoposide, a study did show that miR-222 advanced the resistance of SGC7901 cells to radiotherapy in gastric cancer epithelial cells by targeting PTEN,39 and the PTEN/phosphatidylinositol 3-kinase signaling pathway plays an important role in etoposide resistance.40 We are convinced that a high level of miR-222 will still result in drug resistance with regard to the combination of three drugs used for RB treatment, but it is resistant to different drugs through different signaling pathways.
A previous study documented differentially expressed miRNAs related to RB by analyzing the miRNA expression profiles of GSE7072 and GSE41321.41 However, the GSE7072 dataset in this study contained only three RB patient sequencing samples, whereas the GSE97508 dataset we used contained three infectious RB patient sequencing samples and three non-infectious RB patient sequencing samples. Due to the small number of sequencing samples in the GSE7072 dataset and the failure to distinguish the samples according to infectivity, there may be some differences between the results of their bioinformatics analysis and ours.
The expression profiles used in this study were RB-associated GSE41321 and GSE97508 datasets, and we identified VHL as being the downstream target of miR-222. These results indicate that low expressions of VHL were observed in RB cells. Notably, a series of assays confirmed that miR-222 upregulated the expression of VHL in RB. It has been confirmed that miR-222 confers breast cancer cell resistance to Adriamycin via the inhibition of p27 expression.42 Moreover, miR-222-3p induces DOX resistance in glioblastoma by restraining FOXP2.43
In our study, we identified the interaction between miR-222 and VHL by using the Dual-Luciferase Reporter Assay, and we demonstrated that miR-222 elevated the resistance of RB cells to VCR by downregulating VHL. A previous study showed that depletion of VHL suppressed RB formation in the retina of Rb1/Rbl1-deficient mice but resulted in subretinal vascular growth and retinal capillary hemangioblastoma.44 Our data suggest that the deficiency of VHL in RB leads to enhanced vincristine resistance. Our study revealed abnormally high expression of VHL in the retinal tissues of RB patients. Moreover, downregulation of VHL aggravated RB formation in our work. The two models in the two studies used different species, and additionally, we did not knockdown the expression of the Rb1/Rbl1 gene in the current study, resulting in differences between the observed phenomena. For this reason, the downstream regulation mechanisms were further clarified.
VHL is known to be a crucial substrate recognition subunit of Cullin–RING E3 ubiquitin ligase complexes; it is employed to induce ubiquitination and subsequent proteasomal degradation of target proteins.45 More importantly, VHL is able to regulate the stability of HIF1α.23 HIFα is hydroxylated in an oxygen-containing environment and binds to VHL protein to trigger the ubiquitin–proteasome proteolytic pathway, following degradation by ubiquitination.22 Furthermore, mounting evidence suggests that HIF1α is involved in the development of chemotherapeutic resistance in cancer cells.26,46,47 Targeting HIF1α by either RNAi or siRNA successfully reduces chemotherapeutic resistance; for example, a previous study suggests that suppression of HIF1α expression successfully reverses the resistance of lung cancer cells to curcumin.48 Additionally, inhibition of HIF1α hydroxylation and degradation is conducive to reducing ovarian cancer cell resistance to platinum.49 In parallel with these results, our study showed that VHL diminished the resistance of RB cells to VCR by inhibiting the stability of HIF1α protein.
The evidence provided by the present study supports the proposal that the miR-222/VHL/HIF1α axis may serve as a new target for reducing RB cell resistance to chemotherapy. Our study provides evidence suggesting that miR-222 downregulates VHL and thus increases the expression of HIF1α through the inhibition of ubiquitin–proteasome degradation, thus enhancing RB cell resistance to VCR. However, whether the therapeutic target is applicable to clinical situations remains to be verified.
Supplementary Material
Acknowledgments
The authors thank and appreciate our colleagues for their valuable efforts and comments on this paper.
Disclosure: C. Li, None; J. Zhao, None; W. Sun, None
References
- 1. Kamihara J, Bourdeaut F, Foulkes WD, et al.. Retinoblastoma and neuroblastoma predisposition and surveillance. Clin Cancer Res. 2017; 23: e98–e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lohmann DR, Gallie BL. Retinoblastoma. In: Adam MP Ardinger HH Pagon RA, et al., (Eds.). GeneReviews®. Seattle, WA: University of Washington, Seattle; 2000. [PubMed] [Google Scholar]
- 3. Rao R, Honavar SG. Retinoblastoma. Indian J Pediatr. 2017; 84: 937–944. [DOI] [PubMed] [Google Scholar]
- 4. Lumbroso-Le Rouic L. Retinoblastoma. Rev Prat. 2017; 67: 888–891. [PubMed] [Google Scholar]
- 5. Chawla B, Hasan F, Seth R, et al.. Multimodal therapy for stage III retinoblastoma (International Retinoblastoma Staging System): a prospective comparative study. Ophthalmology. 2016; 123: 1933–1939. [DOI] [PubMed] [Google Scholar]
- 6. Brennan RC, Qaddoumi I, Mao S, et al.. Ocular salvage and vision preservation using a topotecan-based regimen for advanced intraocular retinoblastoma. J Clin Oncol. 2017; 35: 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013; 13: 714–726. [DOI] [PubMed] [Google Scholar]
- 8. Yin X, Liao Y, Xiong W, Zhang Y, Zhou Y, Yang Y. Hypoxia-induced lncRNA ANRIL promotes cisplatin resistance in retinoblastoma cells through regulating ABCG2 expression. Clin Exp Pharmacol Physiol. 2020; 47: 1049–1057. [DOI] [PubMed] [Google Scholar]
- 9. Garofalo M, Croce CM. MicroRNAs as therapeutic targets in chemoresistance. Drug Resist Updat. 2013; 16: 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen W, Wang P, Lu Y, et al.. Decreased expression of mitochondrial miR-5787 contributes to chemoresistance by reprogramming glucose metabolism and inhibiting MT-CO3 translation. Theranostics. 2019; 9: 5739–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wei X, Gao M, Ahmed Y, et al.. MicroRNA-362-5p enhances the cisplatin sensitivity of gastric cancer cells by targeting suppressor of zeste 12 protein. Oncol Lett. 2019; 18: 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014; 9: 287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gam JJ, Babb J, Weiss R. A mixed antagonistic/synergistic miRNA repression model enables accurate predictions of multi-input miRNA sensor activity. Nat Commun. 2018; 9: 2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu K, Huang J, Xie M, et al.. MIR34A regulates autophagy and apoptosis by targeting HMGB1 in the retinoblastoma cell. Autophagy. 2014; 10: 442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jia M, Wei Z, Liu P, Zhao X. Silencing of ABCG2 by MicroRNA-3163 inhibits multidrug resistance in retinoblastoma cancer stem cells. J Korean Med Sci. 2016; 31: 836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang G, Fu Y, Lu X, Wang M, Dong H, Li Q. miR34a regulates the chemosensitivity of retinoblastoma cells via modulation of MAGEA/p53 signaling. Int J Oncol. 2019; 54: 177–187. [DOI] [PubMed] [Google Scholar]
- 17. Wei F, Ma C, Zhou T, et al.. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol Cancer. 2017; 16: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma S, Sun J, Guo Y, et al.. Combination of AAV-TRAIL with miR-221-Zip therapeutic strategy overcomes the resistance to TRAIL induced apoptosis in liver cancer. Theranostics. 2017; 7: 3228–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rao X, Di Leva G, Li M, et al.. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011; 30: 1082–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu L, Wang HJ, Meng T, et al.. lncRNA GAS5 inhibits cell migration and invasion and promotes autophagy by targeting miR-222-3p via the GAS5/PTEN-signaling pathway in CRC. Mol Ther Nucleic Acids. 2019; 17: 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Buckley DL, Van Molle I, Gareiss PC, et al.. Targeting the von Hippel-Lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF-1alpha interaction. J Am Chem Soc. 2012; 134: 4465–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee JW, Bae SH, Jeong JW, Kim SH, Kim KW. Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions. Exp Mol Med. 2004; 36: 1–12. [DOI] [PubMed] [Google Scholar]
- 23. Schonenberger D, Harlander S, Rajski M, et al.. Formation of renal cysts and tumors in Vhl/Trp53-deficient mice requires HIF1alpha and HIF2alpha. Cancer Res. 2016; 76: 2025–2036. [DOI] [PubMed] [Google Scholar]
- 24. Soares P, Gadd MS, Frost J, et al.. Group-based optimization of potent and cell-active inhibitors of the von Hippel-Lindau (VHL) E3 ubiquitin ligase: structure-activity relationships leading to the chemical probe (2S,4R)-1-((S)-2-(1-cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298). J Med Chem. 2018; 61: 599–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schoning JP, Monteiro M, Gu W. Drug resistance and cancer stem cells: the shared but distinct roles of hypoxia-inducible factors HIF1alpha and HIF2alpha. Clin Exp Pharmacol Physiol. 2017; 44: 153–161. [DOI] [PubMed] [Google Scholar]
- 26. Wang P, Wan W, Xiong S, et al.. HIF1alpha regulates glioma chemosensitivity through the transformation between differentiation and dedifferentiation in various oxygen levels. Sci Rep. 2017; 7: 7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shen H, Wang D, Li L, et al.. MiR-222 promotes drug-resistance of breast cancer cells to adriamycin via modulation of PTEN/Akt/FOXO1 pathway. Gene. 2017; 596: 110–118. [DOI] [PubMed] [Google Scholar]
- 28. Abdelfattah N, Rajamanickam S, Panneerdoss S, et al.. MiR-584-5p potentiates vincristine and radiation response by inducing spindle defects and DNA damage in medulloblastoma. Nat Commun. 2018; 9: 4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jain PK, Beberwyck BJ, Fong LK, Polking MJ, Alivisatos AP. Highly luminescent nanocrystals from removal of impurity atoms residual from ion-exchange synthesis. Angew Chem Int Ed Engl. 2012; 51: 2387–2390. [DOI] [PubMed] [Google Scholar]
- 30. Ordonez-Navadijo A, Fuertes-Yebra E, Acosta-Iborra B, et al.. Mutant versions of von Hippel-Lindau (VHL) can protect HIF1alpha from SART1-mediated degradation in clear-cell renal cell carcinoma. Oncogene. 2016; 35: 587–594. [DOI] [PubMed] [Google Scholar]
- 31. Sands S, Ladas EJ, Kelly KM, et al.. Glutamine for the treatment of vincristine-induced neuropathy in children and adolescents with cancer. Support Care Cancer. 2017; 25: 701–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Skiles JL, Chiang C, Li CH, et al.. CYP3A5 genotype and its impact on vincristine pharmacokinetics and development of neuropathy in Kenyan children with cancer. Pediatr Blood Cancer. 2018; 65: e26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qaddoumi I, Billups CA, Tagen M, et al.. Topotecan and vincristine combination is effective against advanced bilateral intraocular retinoblastoma and has manageable toxicity. Cancer. 2012; 118: 5663–5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng Y, Cheng T, Zhao Y, Qu Y. HMGA1 exacerbates tumor progression by activating miR-222 through PI3K/Akt/MMP-9 signaling pathway in uveal melanoma. Cell Signal. 2019; 63: 109386. [DOI] [PubMed] [Google Scholar]
- 35. Liu Y, Zhang X, Zhong X, et al.. miR-222 promotes retinoblastoma cell proliferation and invasion by targeting RB1. China Oncology. 2016; 26: 743–749. [Google Scholar]
- 36. Dai H, Xu LY, Qian Q, Zhu QW, Chen WX. MicroRNA-222 promotes drug resistance to doxorubicin in breast cancer via regulation of miR-222/bim pathway. Biosci Rep. 2019; 39: BSR20190650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zeng LP, Hu ZM, Li K, Xia K. miR-222 attenuates cisplatin-induced cell death by targeting the PPP2R2A/Akt/mTOR Axis in bladder cancer cells. J Cell Mol Med. 2016; 20: 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li S, Li Q, Lu J, et al.. Targeted inhibition of miR-221/222 promotes cell sensitivity to cisplatin in triple-negative breast cancer MDA-MB-231 cells. Front Genet. 2019; 10: 1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chun-Zhi Z, Lei H, An-Ling Z, et al.. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010; 10: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu HG, Ai YW, Yu LL, et al.. Phosphoinositide 3-kinase/Akt pathway plays an important role in chemoresistance of gastric cancer cells against etoposide and doxorubicin induced cell death. Int J Cancer. 2008; 122: 433–443. [DOI] [PubMed] [Google Scholar]
- 41. Yang Y, Mei Q. miRNA signature identification of retinoblastoma and the correlations between differentially expressed miRNAs during retinoblastoma progression. Mol Vis. 2015; 21: 1307–1317. [PMC free article] [PubMed] [Google Scholar]
- 42. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016; 29: 452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang H, Deng Z, Chen X, et al.. Downregulation of miR-222-3p reverses doxorubicin-resistance in LoVo cells through upregulating forkhead box protein P2 (FOXP2) protein. Med Sci Monit. 2019; 25: 2169–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wei R, Ren X, Kong H, et al.. Rb1/Rbl1/Vhl loss induces mouse subretinal angiomatous proliferation and hemangioblastoma. JCI Insight. 2019; 4: e127889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Girardini M, Maniaci C, Hughes SJ, Testa A, Ciulli A. Cereblon versus VHL: hijacking E3 ligases against each other using PROTACs. Bioorg Med Chem. 2019; 27: 2466–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Feng X, Liu N, Deng S, Zhang D, Wang K, Lu M. miR-199a modulates cisplatin resistance in ovarian cancer by targeting Hif1alpha. Onco Targets Ther. 2017; 10: 5899–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu FQ, Fang T, Yu LX, et al.. ADRB2 signaling promotes HCC progression and sorafenib resistance by inhibiting autophagic degradation of HIF1alpha. J Hepatol. 2016; 65: 314–324. [DOI] [PubMed] [Google Scholar]
- 48. Ye MX, Zhao YL, Li Y, et al.. Curcumin reverses cis-platin resistance and promotes human lung adenocarcinoma A549/DDP cell apoptosis through HIF-1alpha and caspase-3 mechanisms. Phytomedicine. 2012; 19: 779–787. [DOI] [PubMed] [Google Scholar]
- 49. Li Z, Zhou W, Zhang Y, et al.. ERK regulates HIF1alpha-mediated platinum resistance by directly targeting PHD2 in ovarian cancer. Clin Cancer Res. 2019; 25: 5947–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.