Abstract
NRF2 is the master regulator for the cellular oxidative stress response and regulates γ-globin gene expression in human erythroid progenitors and sickle cell disease mice. To explore NRF2 function, we established a human β-globin locus yeast artificial chromosome transgenic/NRF2 knockout (β-YAC/NRF2−/−) mouse model. NRF2 loss reduced γ-globin gene expression during erythropoiesis and abolished the ability of dimethyl fumarate, an NRF2 activator, to enhance γ-globin transcription. We observed decreased H3K4Me1 and H3K4Me3 chromatin marks and association of TATA-binding protein and RNA polymerase II at the β-locus control region (LCR) and γ-globin gene promoters in β-YAC/NRF2−/− mice. As a result, long-range chromatin interaction between the LCR DNase I hypersensitive sites and γ-globin gene was decreased, while interaction with the β-globin was not affected. Further, NRF2 loss silenced the expression of DNA methylcytosine dioxygenases TET1, TET2, and TET3 and inhibited γ-globin gene DNA hydroxymethylation. Subsequently, protein-protein interaction between NRF2 and TET3 was demonstrated. These data support the ability of NRF2 to mediate γ-globin gene regulation through epigenetic DNA and histone modifications.
Impact statement
Sickle cell disease is an inherited hemoglobin disorder that affects over 100,000 people in the United States causing high morbidity and early mortality. Although new treatments were recently approved by the FDA, only one drug Hydroxyurea induces fetal hemoglobin expression to inhibit sickle hemoglobin polymerization in red blood cells. Our laboratory previously demonstrated the ability of the NRF2 activator, dimethyl fumarate to induce fetal hemoglobin in the sickle cell mouse model. In this study, we investigated molecular mechanisms of γ-globin gene activation by NRF2. We observed the ability of NRF2 to modulate chromatin structure in the human β-like globin gene locus of β-YAC transgenic mice during development. Furthermore, an NRF2/TET3 interaction regulates γ-globin gene DNA methylation. These findings provide potential new molecular targets for small molecule drug developed for treating sickle cell disease.
Keywords: NRF2, β-YAC mouse, fetal hemoglobin, epigenetic modification, chromatin
Introduction
The human β-like globin gene locus (HBB) on chromosome 11 contains five developmentally expressed globin genes including ε-Gγ-Aγ-δ-β, which are regulated by a distant locus control region (LCR) enhancer through chromatin loop formation.1 This process is modulated by ubiquitous and stage-specific erythroid transcription factors such as KLF1, BCL11A, FOG1, GATA1, GATA2, TR2/TR4, TAL1, and LDB1/NL1 among others.2 These factors bind the LCR and proximal globin gene promoters in concert with chromatin regulatory molecules such as BRG1, LSD1, G9a, MBD2, HDAC1, and DNMT1 to alter chromatin structure and facilitate hemoglobin switching during development. Understanding the mechanisms involved in this process has been investigated extensively to develop treatments for sickle cell disease (SCD) and β-Thalassemia.3,4
Significant advances contributed to developing strategies of fetal hemoglobin (HbF) induction to inhibit hemoglobin S polymerization, the main pathophysiology of SCD; compounds such as cancer chemotherapy agents and inhibitors of DNA methyltransferases or histone deacetylases have proven effective in people.5 However, these agents are cytotoxic, damage DNA, or suppress erythropoiesis, which limits their clinical utilization. Recently, hematopoietic stem cell transplantation6,7 and gene editing approaches using zinc finger nucleases3,8 or Crispr/Cas9 technology9–11 show great promise for treatment of SCD and β-thalassemia, but safety, efficacy, and health-care cost remain major concerns. Therefore, discovery of additional cellular proteins regulated by small molecules to activate HbF is desirable.
NRF2 as the master regulator of cellular oxidative stress response controls the expression of a wide variety of antioxidant genes.12 Oxidative stress is generated by multiple mechanisms including inflammation, hypoxia/reperfusion injury, and chronic hemolysis leading to stress erythropoiesis.13–16 Previous studies demonstrated NRF2 knockout increased susceptibility to a broad range of chemical toxicants and was associated with disease conditions involving oxidative pathology.17 Genome-wide chromatin immunoprecipitation (ChIP)-seq demonstrated that NRF2 binds a consensus motif, TGACnnnGC, known as the antioxidant response element (ARE) to regulate downstream antioxidant genes.18,19
In addition to controlling the antioxidant stress response, NRF2 regulates human γ-globin gene expression. Drugs that activate NRF2, such as tert-butylhydroquione20 and simvastatin,21 induce HbF expression in human primary erythroid progenitors through NRF2 binding to γ-globin gene promoter ARE. We also demonstrated that dimethyl fumarate (DMF), an FDA approved drug for the treatment of multiple sclerosis mediated HbF induction through NRF2 activation in human erythroid progenitors22 and SCD mice.23 Subsequently, we established a SCD/NRF2 knockout mouse where exacerbated SCD phenotype with reduced γ-globin gene expression and increased expression of inflammatory factors was observed.24 In addition, microRNA-144, which silences NRF2 expression, has been shown to be involved in erythroid differentiation25,26 and associated with the severity of anemia in SCD.27 Recently, we completed genome-wide miRNA analysis in SCD patients, demonstrating higher microRNA-144 levels are associated with lower HbF expression.28 These studies support an important role for NRF2 in erythropoiesis and globin gene regulation.
To investigate the function of NRF2 during in vivo erythropoiesis and hemoglobin switching, we crossbred NRF2 knockout mice12 with β-globin locus-yeast artificial chromosome (β-YAC) transgenic mouse29 to produce β-YAC/NRF2 knockout mouse (β-YAC/NRF2−/−). Using this novel model, we demonstrated that NRF2 is required for normal γ-globin gene expression during erythropoiesis. Furthermore, NRF2 mediated HBB locus DNA hydroxymethylation to facilitate long-range chromatin interactions between LCR and γ-globin genes as part of its mechanism on gene activation. These findings provide the rationale to develop small chemical drugs to activate NRF2 and induce HbF for the treatment of β-hemoglobinopathies.
Materials and methods
Animal models and drug treatment
The C57BL/6J NRF2 knockout (NRF2−/−)12 and β-YAC29 mice were crossbred to generate β-YAC/NRF2−/− transgenic/knockout mice (Supplemental Figure S1A). Mouse genotyping and copy-number determination for the HBB locus were determined by quantitative polymerase chain reaction (qPCR) with gene specific primers (Supplementary Table S1). The 2- to 3-month-old mouse blood samples were analyzed for hematological indices using Micros 60 CS/CT from HORIBA Medical/ABX Diagnostics (Irvine, CA) and reticulocyte counting was performed using Retic-Count reagent (BD Biosciences) and analyzed by flow cytometry. For drug treatments, 2- to 3-month-old male and female wild-type β-YAC and β-YAC/NRF2−/− mice were treated by intraperitoneal injection 5 days a week, for 4 weeks with vehicle 0.08% hydroxypropyl methyl cellulose or 100 mg/kg DMF (Sigma, St. Louis, MO). All animal experiments were approved by the Institutional Animal Care and Use Committee at Augusta University.
RNA and protein analysis
Reverse transcription-quantitative PCR (RT-qPCR) and protein immunoblot analyses were performed as previously published.22
DNA dot-blot, DNA immunoprecipitation and epimark assays
DNA dot blot analysis of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) levels using mouse embryonic day E13.5 fetal liver genomic DNA was performed with a Bio-Dot microfiltration apparatus (Bio-Rad, Hercules, CA). Methylated DNA (MeDIP) and hydroxymethylated DNA (hMeDIP) immunoprecipitations were performed with kits from Active Motif (Carlsbad, CA) according to the manufacturer’s instruction. Signals for 5mC and 5hmC in HBB locus were determined by qPCR with gene specific primers (Supplemental Table S1) after normalization to input DNA. EpiMark 5hmC and 5mC Analysis Kit (New England Biolabs) was used for the detection of 5hmC, 5mC, and cytosine at the γ-globin gene promoter -53 CCGG site.
ChIP and sequential ChIP
ChIP and sequential ChIP were performed with anti-NRF2, anti-TET3, anti-TATA binding protein (TBP), anti-RNA polymerase II (PolII), and anti-histone antibodies (Supplemental Table S2); pulldown DNA was quantified by qPCR with gene specific primers (Supplemental Table S1).22
Chromosome conformation capture (3C) assay
Long-range chromatin interaction was studied using 3C assay with EcoRI restriction digestion as previously published.22,30 The relative interaction frequency was determined by qPCR.
Protein co-immunoprecipitation analysis
HEK293 cells were transient transfected with NRF2, DNA methyltransferase (DNMT1, DNMT3A, DNMT3B, and DNMT3L), and DNA methylcytosine dioxygenase (TET1, TET2, and TET3) expression constructs, followed by immunoprecipitation.31 For endogenous protein immunoprecipitation, nuclear extracts from mouse E13.5 fetal livers were prepared using NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher, Waltham, MA) and immunoprecipitation conducted with various antibodies, followed by immunoblot analysis.
Flow cytometry analysis
To measure the percentage of γ-globin expressing positive cells (F-cells), mouse peripheral blood and fetal liver cells were fixed and permeabilized before staining with following antibodies: isotype control, FITC conjugated anti-γ-globin antibody or PE conjugated anti-CD71 antibody, and FITC conjugated anti-Ter119 antibody for globin expression and erythropoiesis analysis by flow cytometry on a LSR II analyzer (BD Biosciences).22,24
Statistical analysis
Data from at least three different samples (fetal liver or blood) were reported as the mean ± standard deviation (SD). The unpaired Student’s t-test was performed to determine significance and P < 0.05 was considered statistically significant. For chronic drug treatments, at least five age-matched mice were used in each treatment group. The paired t-test was performed to compare before and after drug treatment effects and for comparison among groups, one-way analysis of variance was performed.
See Supplemental Document for detailed methods.
Results
NRF2 loss produced mild changes in erythropoiesis in β-YAC mice
The β-YAC/NRF2−/− mice were generated by crossbreeding β-YAC mice with NRF2−/− knockout mice (Supplemental Figure S1A). This novel mouse model consisting the entire 213 kb HBB locus in normal chromatin configuration allowed us to test physiologic changes produced by NRF2 knockout. Initially, we determined copy number using qPCR (Supplemental Figure S1B) and only mice with a single copy were used for analyses. In contrast to our previous NRF2 knockout SCD mouse model where high mortality was observed,24 β-YAC/NRF2−/− mice appeared healthy and showed no change in fertilization or survival rates (Supplemental Figure S1C and D).
Recent findings showed that NRF2 deficiency caused an expansion of hematopoietic stem and progenitor cell compartments due to cell-intrinsic hyperproliferation, which was accomplished at the expense of hematopoietic stem cell quiescence and self-renewal/survival.33 To determine whether NRF2 affected adult erythropoiesis, we compared the hematologic parameters of β-YAC/NRF2−/− mice with NRF2 wild-type β-YAC (β-YAC/NRF2+/+) mice at 2 to 3 months of age. Automated complete blood counts including red blood cell count, hemoglobin, hematocrit, and platelet count were not changed in β-YAC/NRF2−/− mice compared to β-YAC/NRF2+/+ mice (Supplemental Figure S2A). Interestingly, the levels of lymphocytes and monocytes were decreased significantly in β-YAC/NRF2−/− mice (P < 0.05), but the total white blood cell count was not affected.
Next, we performed flow cytometry to analyze erythroid maturation in β-YAC/NRF2−/− mice. The transferrin receptor (CD71) is expressed by erythroid precursors committed to differentiation and decreases during terminal maturation; therefore, we quantified CD71 expression in peripheral blood (Supplemental Figure S2B). We observed an increased CD71 expression of 2.6% ± 0.7% in β-YAC/NRF2−/− mice compared with 1.7% ± 0.2% in β-YAC/NRF2+/+ mice (P < 0.05). Furthermore, the percentage of reticulocytes was increased to 4.4% in β-YAC/NRF2−/− mice compared to 2.3% in β-YAC/NRF2+/+ (P < 0.05), an observation supported by the mild increase in hemoglobin and hematocrit (Supplemental Figure S2A). Together, these data suggest that NRF2 played a minor role in erythroid differentiation under non-oxidative stressed physiological conditions.
NRF2 modulates γ-globin gene expression during erythroid development
To investigate the role of NRF2 in regulating gene expression in normal human HBB locus, we determined globin mRNA levels during development. During erythropoiesis in β-YAC mice, in E13.5 and E18.5 fetal livers and peripheral red blood cells, NRF2 loss reduced γ-globin gene transcription by 42.7%, 44.0%, and 30.5%, respectively (P < 0.05), whereas no effect on β-globin gene transcription was detected (Figure 1(a)). Moreover, the transcription of mouse endogenous εy and β-major globin genes were not changed by NRF2 loss (Supplemental Figure S3).
Figure 1.
NRF2 loss affected γ-globin expression during erythropoiesis. (a) mRNA levels of human fetal γ-globin and adult β-globin genes were monitored by RT-qPCR in E13.5 and E18.5 fetal livers and peripheral blood of adult (2–3 months old) β-YAC/NRF2+/+ and β-YAC/NRF2−/− mice (n = 8–9); the γ/(γ + β) mRNA ratios were calculated with the mRNA level of mouse GAPDH as an internal control. (b) Protein expression levels of NRF2, HbF and HbA were determined for the whole cell extract of β-YAC/NRF2+/+ and β-YAC/NRF2−/− mice E13.5 fetal livers (n = 3). β-actin was used as a protein loading control and relative protein levels quantitated (below) for β-YAC/NRF2+/+ (white bars) and β-YAC/NRF2−/− (black bars) mice. (c and d) β-YAC/NRF2+/+ and β-YAC/NRF2−/− mice (2–3 months old) were treated by intraperitoneal injection with 100 mg/kg DMF in 0.08% hydroxypropyl methyl cellulose or vehicle control for 4 weeks. At week 0 and week 4, mouse blood samples were analyzed for human globin gene expression by RT-qPCR (c), and the percentage of the HbF expressing cells (F-cell) was determined by flow cytometry (d). Shown are representative flow cytometry dot plots used for quantifications. Data are presented as mean ± SD (n = 5). *P< 0.05. Symbols: blue circle (○), β-YAC/NRF2+/+ mice; red triangle (▵), β-YAC/NRF2−/− mice. DMF: dimethyl fumarate. (A color version of this figure is available in the online journal.)
To ascertain the level of protein expression, we determined HbF expression in E13.5 fetal livers by Western blot analysis. As shown in Figure 1(b), we observed a 52.8% decrease of HbF in β-YAC/NRF2−/− mice compared to β-YAC/NRF2+/+ mice (P < 0.05), with no differences in HbA expression. We also quantified the percentage of erythrocytes containing HbF (F-cells) in peripheral blood samples. In agreement with the γ-globin mRNA levels, the number of F-cells in β-YAC/NRF2−/− mice was decreased by 40% compared to that in β-YAC/NRF2+/+ mice (Supplemental Figure S4).
DMF is a known NRF2 activator that induces γ-globin gene transcription in human erythroid progenitors22 in vitro and SCD mice.23 To demonstrate the presence of NRF2 is required for γ-globin induction by DMF, we treated wild type and NRF2 knockout β-YAC mice, with DMF (100 mg/kg, M-F) by intraperitoneal injections for 4 weeks. In β-YAC/NRF2+/+ mice, DMF increased the γ/γ + β mRNA ratio by 1.6-fold (P < 0.05); however, in β-YAC/NRF2−/− mice, the levels of γ-globin gene transcription were not changed significantly (Figure 1(c)). Moreover, F-cell levels increased from 0.6% to 1.5% in β-YAC/NRF2+/+ mice, whereas no significant change in F-cells occurred in β-YAC/NRF2−/− mice after DMF treatment (Figure 1(d)). These data demonstrated NRF2 modulated γ-globin expression in β-YAC mice and is essential for γ-globin gene induction by DMF.
NRF2 modulated HBB locus chromatin structure and long-range interaction
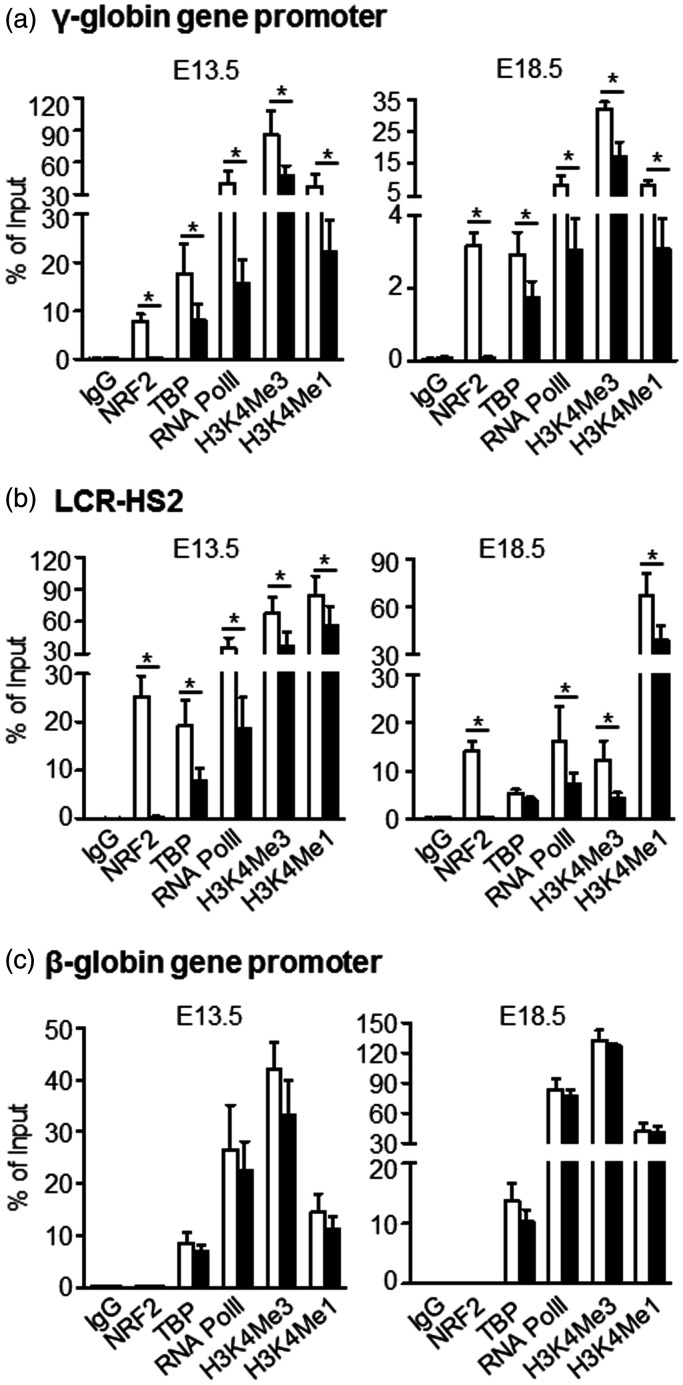
To investigate whether NRF2 mediated chromatin modifications in the HBB locus, we determined the level of active histone marks H3K4Me1 and H3K4Me3 in the LCR and globin gene promoters by ChIP assay. In E13.5 and E18.5 fetal livers of β-YAC/NRF2−/− mice, no detectable binding of NRF2 to either γ-globin gene promoters or LCR-HS2 was observed (Figure 2(a) and (b)), while H3K4Me1 and H3K4Me3 levels were low. In late erythroid developmental stage E18.5 fetal livers, TBP and RNA PolII binding to the γ-globin promoter was lower than E13.5 fetal livers (Figure 2(a)). In contrast, in the β-globin gene promoter, where no NRF2 binding site was identified, NRF2 loss had no effect on in vivo binding (Figure 2(c)). In the mouse NQO1 gene promoter regulated by NRF2, the associations of protein binding and active histone marks were decreased after NRF2 loss (Supplemental Figure S5). Collectively, these data support HBB locus histone modifications in the mechanism of NRF2-mediated γ-globin gene regulation.
Figure 2.
NRF2 loss alters chromatin structure in HBB locus. Chromatin immunoprecipitation (ChIP) assay was performed to determine the association of NRF2, TATA binding protein (TBP), RNA polymerase II (RNA PolII), histone marks H3K4Me1 and H3K4Me3 in HBB locus γ-globin gene promoter (a), LCR-HS2 (b), and β-globin gene promoter (c) for E13.5 and E18.5 fetal livers of β-YAC/NRF2+/+ and β-YAC/NRF2−/− mice. Normal rabbit IgG was used as an antibody control. Primers target NRF2 consensus binding sites in individual gene loci (Supplemental Table S1). Symbols: β-YAC/NRF2+/+, white bars; β-YAC/NRF2−/− mice, black bars. Data after normalized to the input are presented as mean ± SD (n = 5–6); *P< 0.05.
Long-range chromatin interactions between the LCR and globin gene promoters are required to accomplish developmentally regulated hemoglobin switching.1 To investigate whether NRF2 mediates long-range chromatin interaction, chromosome conformation capture (3C) assay was performed, since the entire 213 kb HBB locus and the surrounding regions remained in normal chromatin configuration. Using LCR containing 5’HS4/3/2 as the anchor, 3C assay in E13.5 fetal livers showed interactions between the LCR and γ-globin genes were significantly decreased in the absence of NRF2 (Figure 3(a)). In fetal definitive erythroid day E18.5 liver, where the γ-globin genes are expressed at low levels, the interaction between LCR and γ-globin gene was decreased further (Figure 3(b)). However, the interaction between β-globin gene and LCR was not affected at either erythroid stage (Figure 3(a) and (b)). We also performed 3C assay with the Aγ-globin gene promoter as an anchor. Similarly, we detected decreased interaction between γ-globin and LCR in the absence of NRF2 (Figure 3(c) and (d)) supporting NRF2-mediated HBB locus long-range chromatin interactions are part of its mechanism on γ-globin gene regulation.
Figure 3.
NRF2 enhances HBB locus chromatin interactions with the
γ-globin genes. Chromatin looping in the HBB locus was
analyzed by the chromosome conformation capture (3C) assay using
HBB LCR encompassing 5’HS4, HS3, and HS2 (a and b)
or γ-globin gene promoter as anchors (c and d) for E13.5 (a and c) and
E18.5 (b and d) fetal livers of β-YAC/NRF2+/+ and
β-YAC/NRF2−/− mice (n = 3);
β-YAC/NRF2+/+ mice ( );
β-YAC/NRF2−/− mice (
);
β-YAC/NRF2−/− mice ( ). The positions of
the EcoRI restriction sites are shown (
). The positions of
the EcoRI restriction sites are shown ( ) above the
X-axis and the relative position of the five
HBB locus globin genes by the red arrows
(
) above the
X-axis and the relative position of the five
HBB locus globin genes by the red arrows
( ).
The position of the β-globin locus transcript 3 (BGLT3, B3), a long
non-coding RNA is indicated by the black box (▪); HS5-1, DNase I
hypersensitive sites. The relative distance of individual globin genes
to the anchor encompassing HS4, HS3 and HS2 (black line) is indicated in
kb. The relative cross-linking frequency between the anchor and other
EcoRI fragments was determined by qPCR analysis and shown in a
logarithmic scale. The data are shown as the mean ± SD of three
different fetuses. (A color version of this figure is available in the
online journal.)
).
The position of the β-globin locus transcript 3 (BGLT3, B3), a long
non-coding RNA is indicated by the black box (▪); HS5-1, DNase I
hypersensitive sites. The relative distance of individual globin genes
to the anchor encompassing HS4, HS3 and HS2 (black line) is indicated in
kb. The relative cross-linking frequency between the anchor and other
EcoRI fragments was determined by qPCR analysis and shown in a
logarithmic scale. The data are shown as the mean ± SD of three
different fetuses. (A color version of this figure is available in the
online journal.)
NRF2 loss altered global DNA modifications
DNA epigenetic modifications, such as 5mC and 5hmC, are typically associated with inactive and active gene transcription, respectively. Recent findings support an emerging role for NRF2 in rewiring the metabolism of α-ketoglutarate, a Krebs cycle metabolite in normal and pathological conditions such as cancer. α-Ketoglutarate is a rate limiting substrate for dioxygenases such as histone demethylases, prolyl hydroxylases, and ten-eleven translocation (TET) enzymes that modulate epigenetic histone and DNA modifications.34,35 Thus, to evaluate an effect of NRF2 loss on 5mC and 5hmC levels in β-YAC mice, we performed DNA dot-blot assay with genomic DNA isolated from E13.5 fetal livers. Interestingly, global DNA 5hmC levels were significantly decreased in fetal livers of β-YAC/NRF2−/− mice; however, DNA 5mC levels were not changed (Figure 4(a)). To further determine whether 5hmC DNA modifications occurred locally in the HBB locus to affect globin gene expression, we performed MeDIP and hMeDIP assays. MeDIP results showed there was no significant change in 5mC levels in the HBB locus in E13.5 fetal liver. However, the levels of 5hmC were decreased for LCR-HS2 and γ-globin gene promoters, but not for the β-globin gene promoter (Figure 4(b)).
Figure 4.
NRF2 loss alters epigenetic DNA modifications in E13.5 mouse fetal livers. (a) Genomic DNA samples extracted from E13.5 fetal livers of β-YAC/NRF2+/+ and β-YAC/NRF2−/− mice (n = 4) after two-fold serial dilutions, were measured anti-5mC (top) and anti-5hmC (bottom) levels by DNA dot-blot assay. Methylene blue staining monitored total DNA loading control. The relative genome 5mC and 5hmC signals after NRF2 loss were presented; β-YAC/NRF2+/+ (white bars); β-YAC/NRF2−/− (black bars). (B) DNA 5mC and 5hmC levels in the LCR-HS2, γ-globin gene promoter and β-globin gene promoter regions were determined by methylated DNA and hydroxymethylated DNA immunoprecipitations. Data are presented as mean ± SD (n = 3) after normalized to input DNA. (c) Protein expression levels of DNA methylcytosine dioxygenases TET1, TET2, and TET3 were determined for the whole cell extract of both transgenic lines using E13.5 fetal livers (n = 3). β-actin was used as a protein loading control and the relative protein levels were quantitated (below). *P< 0.05; ns, not significant.
The effect of NRF2 loss on γ-globin gene expression was further analyzed for the levels of 5hmC, 5mC, and unmodified cytosine at a CCGG MspI site located at -53 within the 5’ γ-globin promoter region with Epimark assays. This CCGG site has been demonstrated to regulate γ-globin promoter activity36 and undergoes demethylation rapidly at early stage of erythroid differentiation in fetal liver compared to adult bone marrow in baboon.37 Significantly, levels of 5hmC in the E13.5 fetal livers of β-YAC/NRF2−/− mice (3.9 ± 1.3%) were lower than compared to β-YAC/NRF2+/+ (5.5 ± 1.1%), while 5mC levels were comparable for both models (Supplemental Figure S6).
Subsequently, we tested whether NRF2 loss in E13.5 fetal livers affected the expression of DNA 5hmC modifier methylcytosine dioxygenases, TET1, TET2, and TET3. We found that the expression of all three TET enzymes at mRNA (Supplemental Figure S7) and protein levels in NRF2 knockout mice were decreased by 55.0% ± 16.4%, 46.1% ± 15.0%, and 39.3% ± 12.5%, respectively (P < 0.05) (Figure 4(c)). Thus, NRF2 might influence global and HBB locus DNA 5hmC modifications for gene regulation.
Interaction between NRF2 and TET3 modulated HBB locus DNA methylation status
TET methylcytosine dioxygenases associate with site-specific transcription factors to bind methylated CpG regions to oxidize 5mC, reverse DNA methylation and activate gene expression. We asked whether NRF2 recruits TET proteins to gene targets, HA-tagged NRF2 and FLAG-tagged TET1, TET2 or TET3 were transiently expressed in HEK293 cells and subjected to immunoprecipitation with anti-FLAG antibody followed by immunoblot with anti-HA antibody. We observed FLAG-tagged TET3 in the HA immunoprecipitation assay, indicating the presence of both NRF2 and TET3 in the same protein complex (Figure 5(a)). To detect whether NRF2 also interacts with DNA methyltransferases in regulating DNA 5mC modifications, we determined the interaction between NRF2 and known DNA methyltransferases DNMT3A, DNMT3B or DNMT3L after overexpressed in HEK293 cells similarly. However, there was no interaction between NRF2 and DNMT3A, DNMT3B or DNMT3L (Figure 5(b) and (c)). To further determine whether endogenous interactions between NRF2 and TET3 exist, we performed immunoprecipitation assays with nuclear extracts of E13.5 fetal liver and observed an interaction between NRF2 and TET3, but not with TET1 or TET2 (Figure 5(d), Supplemental Figure S8).
Figure 5.
Interaction between NRF2 and TET3. (a) HEK293 cells were co-transfected with HA-NRF2 and FLAG tagged TET1, TET2, or TET3. The NRF2-TET complexes in HEK293 WCE were immunoprecipitated with FLAG antibody and analyzed by immunoblot with HA antibody. <, indicated the NRF2 band; *, indicated the IgG heavy chain. (b and c) HEK293 cells were co-transfected with HA-NRF2 and FLAG tagged DNMT3A, DNMT3B (b) or YFP tagged DNMT3L (c). The NRF2-DNMT complexes in HEK293 whole cell lysate were immunoprecipitated with either FLAG (b) or YFP (c) antibody and analyzed by immunoblot with HA antibody. (d) Endogenous NRF2 was immunoprecipitated from nuclear extract of β-YAC/NRF2+/+ and β-YAC/NRF2−/− mice E13.5 fetal livers with TET3 antibody and analyzed by immunoblot with NRF2 antibody. (e) Sequential ChIP was conducted to determine co-localization of NRF2 and TET3 on the γ-globin gene promoter, LCR-HS2 and β-globin gene promoter in E13.5 fetal livers of β-YAC/NRF2+/+ mice. The G-CRE was used as a negative control region. Sequential immunoprecipitations with the first antibody (First IP) and second antibody (Second IP) were conducted with indicated antibody and rabbit IgG was used as ChIP pulldown antibody control. G-CRE: Gγ-globin cAMP response element; YFP: yellow fluorescent protein; WCE: whole cell extract.
Next, we asked whether NRF2 interacts with TET3 at the HBB locus in mediating gene regulation. Sequential ChIP assay in E13.5 fetal liver was performed with anti-NRF2 antibody followed by anti-TET3 antibody. As shown in Figure 5(e), an enrichment signal for TET3 was observed in γ-globin gene promoter and LCR-HS2 without significant change in the β-globin promoter. As a negative control, in the Gγ-globin cAMP response element, where no NRF2 binding site exists, we did not detect chromatin enrichment for either NRF2 or TET3. Together, these data support the ability of NRF2 to modulate HBB locus chromatin structure in preferentially regulating γ-globin gene expression through interactions with TET3.
Discussion
Currently, there are four FDA-approved drugs for the treatment of SCD including Hydroxyurea, Endari (L-glutamine), Adakveo® (crizanlizumab), and Oxbryta®. Endari improves clinical symptoms of SCD by suppressing oxidative stress and reducing the adhesion of sickle erythrocytes to endothelial cells.38 Oxbryta® is the first FDA-approved drug that directly inhibits hemoglobin S polymerization and improves total hemoglobin levels.39 Adakveo® (crizanlizumab) is a targeted monoclonal antibody that binds to P-selectin on the surface of activated endothelial cells and platelets40; blocking interactions among endothelial cells, platelets, red blood cells, and white blood cells decreased vaso-occlusive episodes. By contrast, Hydroxyurea ameliorates SCD complications mainly by HbF induction; moreover, this agent restores nitric oxide levels, suppresses the inflammatory response, and inhibits expression of adhesion molecules. Therefore, discovery of additional molecular targets for drug-mediated HbF induction to develop novel effective therapies for β-hemoglobinopathies is desirable.
Recently, we established an NRF2 knockout SCD mouse model to study globin gene expression under chronic hemolysis and oxidant stress conditions.24 Here, we established an NRF2 knockout β-YAC transgenic mouse model with normal ROS levels to investigate γ-globin regulation during normal erythropoiesis. The human β-like globin transgene in SCD mouse is a mini-HBB construct, comprised of 22 kb extracted from the normal 81 kb HBB locus consisting the LCR linked to the 5.5 kb Aγ-globin and 4.1 kb β-globin genes. Accordingly, the SCD mouse model lacks multiple binding sites for critical globin gene regulatory transcription factors including BCL11A, NuRD, Sox6, and LRF.41 Moreover, our SCD/NRF2 knockout model produced more severe phenotypes with lower survival and fertility which limited its application.24 Therefore, as an alternative, we chose the β-YAC mouse model to ascertain the role of NRF2 in globin gene regulation through long-range chromatin interactions that exist in the full-length HBB locus under more physiologic oxidative stress conditions.
In this report, we identified a mild effect of NRF2 loss on erythropoiesis based on CD71 expression in adult mouse peripheral blood. Importantly, NRF2 was previously shown to affect erythropoiesis by modulating ROS stress through glutathione peroxidase family proteins since accumulation of ROS is particularly deleterious to red blood cells leading to hemolysis.42,43 By contrast, activation of glutathione peroxidase and other selenoprotein genes as efficient ROS scavengers44,45 is capable of affecting both iron homeostasis and erythrocyte health46,47 thus may prevent hemolytic anemia.48 Moreover, other than regulating redox stress during hematopoietic cell development, NRF2 also plays a role in hematopoietic stem cell (HPSC) maintenance.49 In fact, NRF2 loss causes increased apoptosis and alters to balance HPSC proliferation, self-renewal, and bone marrow localization.33,50 Moreover, using KEAP1-deficient mice, Murakami et al. demonstrated NRF2 mediated HPSC fate with enhanced granulocyte–monocyte differentiation and a compensatory decrease in erythroid and lymphoid differentiation. On the other hand, combined NRF2/KEAP1 knockout restored lineage commitment,51 while NRF2 loss produces immune-mediated hemolytic anemia in mice.52 Other lines of evidence support a role of NRF2 in the regulation of erythropoiesis. For example, NRF2 binding is enriched in regions with hypomethylated CpG dinucleotides in human adult erythroid cells.53 NRF2 also regulates genes involved in iron homeostasis and heme metabolism, such as ferritin light and heavy chain and ferrochelatase, among others.53 Moreover, NRF2 silencing was recently shown to interrupt the pentose phosphate pathway and alter cellular levels of α-ketoglutarate,34 a metabolic intermediate of the Krebs cycle and an essential cofactor for α-ketoglutarate-dependent dioxygenases containing proteins such as hypoxia inducible factor 1α. Thus, NRF2 might mediate genome-wide DNA and histone epigenetic modifications and hypoxia sensing,54 in addition to its major role in regulating cellular oxidative stress response.
Our data demonstrate that NRF2-mediated global and localized HBB locus DNA hydroxymethylation affects target gene expression by regulating the expression of TET1, TET2, and TET3. We further demonstrated the ability of NRF2 to recruit TET3 to γ-globin promoter and LCR regions in modulating DNA methylation. These data suggest a role of NRF2 in gene regulation through direct DNA binding and indirect DNA epigenetic modifications. Using Epimark assay, we can only demonstrated the methylation status for -53 CCGG, one of six CpG dinucleotides in the γ-globin proximal promoter critical for gene transcription. Whether DNA 5mC and 5hmC modifications on numerous other CpG dinucleotides along the HBB locus are affected by NRF2 and/or TET3 loss is not known.
Previously, NRF2 loss did not affect the expression of mouse α- or β-globin genes at embryonic E13.5 and E15.5 stages.55 In transgenic mouse models carrying the human HBB in a mini construct or entire 213 kb locus, neither NRF2 activation nor knockout affected human β-globin gene expression.22,23 However, in both transgenic mouse models, the expression of γ-globin gene was increased after NRF2 induction and NRF2 loss abolished activation. Moreover, in human normal and sickle erythroid progenitors, NRF2 activators such as tert-butylhydroquione,20 Simvastatin,21 and DMF22 induced γ-globin expression, but not β-globin gene expression. In contrast, small interference RNA,20 lentiviral shRNA,22 or microRNA mimics28 mediated NRF2 silencing and reduced γ-globin expression without an effect on β-globin expression. These studies demonstrated that NRF2 does not regulate β-globin gene expression but preferentially activates γ-globin.
Collectively, our data from SCD24 and β-YAC NRF2 knockout mice suggest a regulatory role of NRF2 on γ-globin gene. Therefore, pharmacologic activators of NRF2 expression might improve the clinical phenotype of SCD patients. Recently, NRF2 activator sulforaphane in the form of broccoli sprouts was tested for SCD treatment. HbF induction was not observed in SCD patients; however, the lack of response might be due to low concentrations of sulforaphane administered.56 Small chemicals that directly inhibit KEAP1 to activate NRF2 such as DMF, a FDA-approved agent for multiple sclerosis, might be more efficacious. Testing small chemical compounds that activate NRF2 and γ-globin expression in β-YAC and SCD mice will identify potential novel agents for treating β-hemoglobinopathies.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_1535370220945305 for NRF2 mediates γ-globin gene regulation through epigenetic modifications in a β-YAC transgenic mouse model by Xingguo Zhu, Caixia Xi, Alexander Ward, Mayuko Takezaki, Huidong Shi, Kenneth R Peterson and Betty S Pace in Experimental Biology and Medicine
ACKNOWLEDGMENTS
The authors thank Drs. Bobby Thomas, Dorothy Tuan, and Hongyan Xu for their insights and helpful discussions of data. NRF2−/− mouse was kindly provided by Dr. Masayuki Yamamoto (Tohoku University, Japan). The authors also want to thank Dr. Yue Xiong (University of North Carolina at Chapel Hill, NC) and Dr. Scott M. Plafker (Oklahoma Medical Research Foundation, Oklahoma City, OK) for providing NRF2 plasmid constructs; Dr. Xiaochun Yu (City of Hope, Duarte, CA) for TET1, TET2, and TET3 plasmid constructs; Dr. Keith D. Robertson (Mayo Clinic, Rochester, MN) for DNA methylation methyltransferases DNMT3A, DNMT3B, and DNMT3L plasmid constructs; and Dr. Tianxiang Hu (Augusta University, GA) for the lentiviral vector pLKO.EF1.GTP.
AUTHORS’ CONTRIBUTIONS
XZ designed and performed research, analyzed data, performed statistical analysis, and wrote the manuscript. CX performed research and analyzed data. AW performed research and analyzed data. MT performed research and analyzed data. HS analyzed data and reviewed manuscript. KRP analyzed data and critically reviewed manuscript. BSP supervised the project, designed research, and wrote the manuscript.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by funding from the National Institutes of Health to XZ (HL117684), KRP (HL111264, DK100595), and BSP (HL069234).
ORCID iD
Xingguo Zhu https://orcid.org/0000-0001-7039-4541
Supplemental Material
Supplemental material for this article is available online.
References
- 1.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell 2002; 10:1453–65 [DOI] [PubMed] [Google Scholar]
- 2.Bauer DE, Kamran SC, Orkin SH. Reawakening fetal hemoglobin: prospects for new therapies for the β-globin disorders. Blood 2012; 120:2945–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breda L, Motta I, Lourenco S, Gemmo C, Deng W, Rupon JW, Abdulmalik OY, Manwani D, Blobel GA, Rivella S. Forced chromatin looping raises fetal hemoglobin in adult sickle cells to higher levels than pharmacologic inducers. Blood 2016; 128:1139–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, Reik A, Gregory PD, Rivella S, Dean A, Blobel GA. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 2014; 158:849–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mabaera R, West RJ, Conine SJ, Macari ER, Boyd CD, Engman CA, Lowrey CH. A cell stress signaling model of fetal hemoglobin induction: what doesn’t kill red blood cells may make them stronger. Exp Hematol 2008; 36:1057–72 [DOI] [PubMed] [Google Scholar]
- 6.Sebastiano V, Maeder ML, Angstman JF, Haddad B, Khayter C, Yeo DT, Goodwin MJ, Hawkins JS, Ramirez CL, Batista LF, Artandi SE, Wernig M, Joung JK. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells 2011; 29:1717–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou J, Mali P, Huang X, Dowey SN, Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood 2011; 118:4599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilber A, Tschulena U, Hargrove PW, Kim YS, Persons DA, Barbas CF, 3rd, Nienhuis AW. A zinc-finger transcriptional activator designed to interact with the gamma-globin gene promoters enhances fetal hemoglobin production in primary human adult erythroblasts. Blood 2010; 115:3033–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, Luc S, Kurita R, Nakamura Y, Fujiwara Y, Maeda T, Yuan GC, Zhang F, Orkin SH, Bauer DE. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 2015; 527:192–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X, Wang Y, Yan W, Smith C, Ye Z, Wang J, Gao Y, Mendelsohn L, Cheng L. Production of Gene-Corrected adult beta globin protein in human erythrocytes differentiated from patient iPSCs after genome editing of the sickle point mutation. Stem Cells 2015; 33:1470–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traxler EA, Yao Y, Wang YD, Woodard KJ, Kurita R, Nakamura Y, Hughes JR, Hardison RC, Blobel GA, Li C, Weiss MJ. A genome-editing strategy to treat β-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat Med 2016; 22:987–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 1997; 236:313–22 [DOI] [PubMed] [Google Scholar]
- 13.Mohanty JG, Nagababu E, Rifkind JM. Red blood cell oxidative stress impairs oxygen delivery and induces red blood cell aging. Front Physiol 2014; 5:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chirico EN, Pialoux V. Role of oxidative stress in the pathogenesis of sickle cell disease. IUBMB Life 2012; 64:72–80 [DOI] [PubMed] [Google Scholar]
- 15.Silva DGH, Belini Junior E, de Almeida EA, Bonini-Domingos CR. Oxidative stress in sickle cell disease: an overview of erythrocyte redox metabolism and current antioxidant therapeutic strategies. Free Radic Biol Med 2013; 65:1101–9 [DOI] [PubMed] [Google Scholar]
- 16.Nur E, Biemond BJ, Otten HM, Brandjes DP, Schnog JJ, CURAMA Study Group. Oxidative stress in sickle cell disease; pathophysiology and potential implications for disease management. Am J Hematol 2011; 86:484–9 [DOI] [PubMed] [Google Scholar]
- 17.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol 2013; 53:401–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y, Bannai S, Yamamoto M. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem 2000; 275:16023–9 [DOI] [PubMed] [Google Scholar]
- 19.Chorley BN, Campbell MR, Wang X, Karaca M, Sambandan D, Bangura F, Xue P, Pi J, Kleeberger SR, Bell DA. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res 2012; 40:7416–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macari ER, Lowrey CH. Induction of human fetal hemoglobin via the NRF2 antioxidant response signaling pathway. Blood 2011; 117:5987–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macari ER, Schaeffer EK, West RJ, Lowrey CH. Simvastatin and t-butylhydroquinone suppress KLF1 and BCL11A gene expression and additively increase fetal hemoglobin in primary human erythroid cell. Blood 2013; 121:830–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu X, Li B, Pace BS. NRF2 mediates γ-globin gene regulation and fetal hemoglobin induction in human erythroid progenitors. Haematologica 2017; 102:e285–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnamoorthy S, Pace B, Gupta D, Sturtevant S, Li B, Makala L, Brittain J, Moore N, Vieira BF, Thullen T, Stone I, Li H, Hobbs WE, Light DR. Dimethyl fumarate increases fetal hemoglobin, provides vascular protection and heme detoxification and corrects anemia in sickle cell disease. JCI Insight 2017; 2:96409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu X, Xi C, Thomas B, Pace BS. Loss of NRF2 function exacerbates the pathophysiology of sickle cell disease in a transgenic mouse model. Blood 2018; 131:558–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim M, Tan YS, Cheng WC, Kingsbury TJ, Heimfeld S, Civin CI. MIR144 and MIR451 regulate human erythropoiesis via RAB14. Br J Haematol 2015; 168:583–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmussen KD, O'Carroll D. The miR-144/451eGFP allele, a novel tool for resolving the erythroid potential of hematopoietic precursors. Blood 2011; 118:2988–92 [DOI] [PubMed] [Google Scholar]
- 27.Sangokoya C, Telen MJ, Chi JT. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood 2010; 116:4338–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, Zhu X, Ward CM, Starlard-Davenport A, Takezaki M, Berry A, Ward A, Wilder C, Neunert C, Kutlar A, Pace BS. MIR-144-mediated NRF2 gene silencing inhibits fetal hemoglobin expression in sickle cell disease. Exp Hematol 2019; 70:85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterson KR, Clegg CH, Huxley C, Josephson BM, Haugen HS, Furukawa T, Stamatoyannopoulos G. Transgenic mice containing a 248-kb yeast artificial chromosome carrying the human beta-globin locus display proper developmental control of human globin genes. Proc Natl Acad Sci Usa USA 1993; 90:7593–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiefer CM, Lee J, Hou C, Dale RK, Lee YT, Meier ER, Miller JL, Dean A. Distinct Ldb1/NLI complexes orchestrate γ-globin repression and reactivation through ETO2 in human adult erythroid cells. Blood 2011; 118:6200–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu X, Wang Y, Pi W, Liu H, Wickrema A, Tuan D. NF-Y recruits both transcription activator and repressor to modulate tissue- and developmental stage-specific expression of human γ-globin gene. PLoS One 2012; 7:e47175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu T, Pi W, Zhu X, Yu M, Ha H, Shi H, Choi JH, Tuan D. Long non-coding RNAs transcribed by ERV-9 LTR retrotransposon act in cis to modulate long-range LTR enhancer function. Nucleic Acids Res 2017; 45:4479–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merchant AA, Singh A, Matsui W, Biswal S. The redox-sensitive transcription factor Nrf2 regulates murine hematopoietic stem cell survival independently of ROS levels. Blood 2011; 118:6572–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 2012; 22:66–79 [DOI] [PubMed] [Google Scholar]
- 35.Sayin VI, LeBoeuf SE, Singh SX, Davidson SM, Biancur D, Guzelhan BS, Alvarez SW, Wu WL, Karakousi TR, Zavitsanou AM, Ubriaco J, Muir A, Karagiannis D, Morris PJ, Thomas CJ, Possemato R, Vander Heiden MG, Papagiannakopoulos T. Activation of the NRF2 antioxidant program generates an imbalance in Central carbon metabolism in cancer. Elife 2017; 6:e28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Busslinger M, Hurst J, Flavell RA. DNA methylation and the regulation of globin gene expression. Cell 1983; 34:197–206 [DOI] [PubMed] [Google Scholar]
- 37.Ruiz MA, Rivers A, Ibanez V, Vaitkus K, Mahmud N, DeSimone J, Lavelle D. Hydroxymethylcytosine and demethylation of the γ-globin gene promoter during erythroid differentiation. Epigenetics 2015; 10:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niihara Y, Zerez CR, Akiyama DS, Tanaka KR. Oral L-glutamine therapy for sickle cell anemia: I. Subjective clinical improvement and favorable change in red cell NAD redox potential. Am J Hematol 1998; 58:117–21 [DOI] [PubMed] [Google Scholar]
- 39.U.S. Food and Drug Administration. FDA approves novel treatment to target abnormality in sickle cell disease, www.fda.gov/news-events/press-announcements/fda-approves-novel-treatment-target-abnormality-sickle-cell-disease (accessed 14 July 2020).
- 40.U.S. Food and Drug Administration. FDA approves first targeted therapy to treat patients with painful complication of sickle cell disease, www.fda.gov/news-events/press-announcements/fda-approves-first-targeted-therapy-treat-patients-painful-complication-sickle-cell-disease (accessed 14 July 2020).
- 41.Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev 2010; 24:783–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fibach E, Rachmilewitz E. The role of oxidative stress in hemolytic anemia. Curr Mol Med 2008; 8:609–19 [DOI] [PubMed] [Google Scholar]
- 43.Ghaffari S. Oxidative stress in the regulation of normal and neoplastic hematopoiesis. Antioxid Redox Signal 2008; 10:1923–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benfeitas R, Selvaggio G, Antunes F, Coelho PM, Salvador A. Hydrogen peroxide metabolism and sensing in human erythrocytes: a validated kinetic model and reappraisal of the role of peroxiredoxin II. Free Radic Biol Med 2014; 74:35–49 [DOI] [PubMed] [Google Scholar]
- 45.Johnson RM, Ho Y-S, Yu D-Y, Kuypers FA, Ravindranath Y, Goyette GW. The effects of disruption of genes for peroxiredoxin-2, glutathione peroxidase-1, and catalase on erythrocyte oxidative metabolism. Free Radic Biol Med 2010; 48:519–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattè A, De Falco L, Federti E, Levi S, Iolascon A, Federico G, Mohandas N, Janin A, Bruno M, Leboeuf C. Peroxiredoxin-2: a novel factor involved in iron homeostasis. Am Soc Hematol 2015; 126:406 [Google Scholar]
- 47.Neumann CA, Krause DS, Carman CV, Das S, Dubey DP, Abraham JL, Bronson RT, Fujiwara Y, Orkin SH, Van Etten RA. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature 2003; 424:561–5 [DOI] [PubMed] [Google Scholar]
- 48.Kawatani Y, Suzuki T, Shimizu R, Kelly VP, Yamamoto M. Nrf2 and selenoproteins are essential for maintaining oxidative homeostasis in erythrocytes and protecting against hemolytic anemia. Blood 2011; 117:986–96 [DOI] [PubMed] [Google Scholar]
- 49.Tsai JJ, Dudakov JA, Takahashi K, Shieh JH, Velardi E, Holland AM, Singer NV, West ML, Smith OM, Young LF, Shono Y, Ghosh A, Hanash AM, Tran HT, Moore MA, van den Brink MR. Nrf2 regulates haematopoietic stem cell function. Nat Cell Biol 2013; 15:309–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murakami S, Motohashi H. Roles of NRF2 in cell proliferation and differentiation. Free Radic Biol Med 2015; 88:168–78 [DOI] [PubMed] [Google Scholar]
- 51.Murakami S, Shimizu R, Romeo PH, Yamamoto M, Motohashi H. Keap1-Nrf2 system regulates cell fate determination of hematopoietic stem cells. Genes Cells 2014; 19:239–53 [DOI] [PubMed] [Google Scholar]
- 52.Lee JM, Chan K, Kan YW, Johnson JA. Targeted disruption of Nrf2 causes regenerative immune-mediated hemolytic anemia. Proc Natl Acad Sci Usa USA 2004; 101:9751–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lessard S, Beaudoin M, Benkirane K, Lettre G. Comparison of DNA methylation profiles in human fetal and adult red blood cell progenitors. Genome Med 2015; 7:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh A, Happel C, Manna SK, Acquaah-Mensah G, Carrerero J, Kumar S, Nasipuri P, Krausz KW, Wakabayashi N, Dewi R, Boros LG, Gonzalez FJ, Gabrielson E, Wong KK, Girnun G, Biswal S. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J Clin Invest 2013; 123:2921–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin F, van Deursen JM, Shivdasani RA, Jackson CW, Troutman AG, Ney PA. Erythroid maturation and globin gene expression in mice with combined deficiency of NF-E2 and nrf-2. Blood 1998; 91:3459–66 [PubMed] [Google Scholar]
- 56.Doss JF, Jonassaint JC, Garrett ME, Ashley-Koch AE, Telen MJ, Chi JT. Phase 1 study of a Sulforaphane-Containing broccoli sprout homogenate for sickle cell disease. PLoS One 2016; 11:e0152895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_1535370220945305 for NRF2 mediates γ-globin gene regulation through epigenetic modifications in a β-YAC transgenic mouse model by Xingguo Zhu, Caixia Xi, Alexander Ward, Mayuko Takezaki, Huidong Shi, Kenneth R Peterson and Betty S Pace in Experimental Biology and Medicine