Abstract
In this work, our primary objective was to examine the interrelationship among the serum level of chemokine (C-C motif) ligand 2 (CCL2) and plaque characteristics in coronary culprit lesions. The clinical data of 116 coronary heart disease patients who were hospitalized in the Department of Cardiology of Henan Province People's Hospital from February 2015 to June 2017 were retrospectively analyzed. The study population was subdivided according to the concentration of CCL2 into low CCL2 group and high CCL2 group. The levels of blood lipid, creatinine, and uric acid were measured, and patients underwent coronary angiography. The characteristics of the culprit lesions were detected by intravascular ultrasound, and the correlation between the serum markers and the characteristics of coronary artery plaque was analyzed. Moreover, the coronary artery disease dataset from the Gene Expression Omnibus database was downloaded and the genes regulated were analyzed by CCL2 using gene set enrichment analysis (GSEA). Patients with high CCL2 group had higher LDL-C level and L/H ratio, and lower HDL-C level than the low CCL2 group. Compared with low-level CCL2 group, coronary plaque in the high CCL2 group had higher eccentric plaque and plaque rupture, and thin cap fibroatheromas, fibrofatty and necrotic core and lower fibrous tissue. CCL2 was positively correlated with the percentage of fibrofatty and necrotic core, and negatively correlated with the percentage of fibrous tissue. Furthermore, GSEA analysis showed that samples with high CCL2 expression were enriched for genes involved in different pathways, such as cell adhesion molecules and Nod-like receptor signaling pathway. The CCL2 level was correlated with vulnerable plaques of coronary artery and had certain value in detecting vulnerable plaques. These results indicated that CCL2 could be regarded as a clinical prognostic biomarker for coronary artery disease.
Impact statement
Vulnerable plaques are plaques which are susceptible to rupture or thrombosis and trigger a series of adverse events such as coronary disorders. CCL2 is a soluble basic protein belonging to the CC subfamily. Previous studies have been investigated on the correlation between inflammatory factors and clinical events, but there are few studies on the correlation between CCL2 and plaque characteristics. Our study found that the high expression of CCL2 is involved in multiple processes in the genesis and progression of coronary artery disease, and would be a potential clinical prognostic indicator. In addition, high expression of CCL2 may be related to gene pathways such as Nod-like receptor signaling pathway, suggesting that CCL2 is involved in the inflammatory response and immune process of coronary artery disease.
Keywords: Monocyte chemoattractant protein-1, intravenous ultrasound, vulnerable plaques, coronary artery disease, CCL2
Introduction
Vulnerable plaques are those with a high risk of rupture or thrombosis and can trigger a series of adverse events in cardiovascular diseases such as acute coronary syndrome (ACS),1 which have two major features of inflammation and morphology. Nearly 70% of acute coronary events are caused by acute stenosis caused by sudden plaque rupture and thrombosis.2 The main histopathological features of vulnerable plaques are large lipid cores and thin fibrous caps, accompanied by infiltration of numerous inflammatory factors.3 Inflammation is one of the key mechanisms driving the pathogenesis of atherosclerosis.4 Previous studies showed that serum inflammatory factors are closely related to plaque vulnerability and ACS.5 Intravascular ultrasound (IVUS) is applicable for quantitatively assessing the distribution and severity of coronary plaques. On the basis of gray-scale IVUS, IVUS imaging system (iMAP-IVUS) can improve the accuracy of morphological characteristics of plaque by color coding.6,7
Chemokine (C-C motif) ligand 2 (CCL2) or monocyte chemoattractant protein-1 (MCP-1) is a soluble basic protein belonging to the CC subfamily. Studies have shown that CCL2 stimulates the interaction of endothelial cells with chemokine receptors and increases plaque instability.8 Serum CCL2 levels are related to coronary artery calcification in patients with atherosclerosis.9 CCL2 is abundantly expressed in atherosclerotic plaques and induces aggregation of macrophages in atherosclerotic lesions.10 Furthermore, CCL2 is implicated in the pathological development of various clinical disorders such as acute myeloid leukemia, cancers, and osteoarthritis.11–13 Previous studies indicated that the overexpression of CCL2 exacerbates atherosclerosis in vivo.14 However, previous studies have been based on the relationship between inflammatory factors and clinical events, but there are few studies on the correlation between plaque characteristics.
Gene set enrichment analysis (GSEA) is an important tool recently developed to study the mapping of core genes and is now widely used to analyze gene expression profile data. GSEA is able to analyze the synergistic differences in gene expression in pathways at the level of gene sets. Compared with traditional single-gene studies, GSEA detects weak interference in gene expression by increasing the signal-to-noise ratio to reduce the interference of related genes.15 Meanwhile, GSEA allows us to focus on gene set research rather than traditional high-score genes, making our findings more convincing and helping us interpret the results.15
In this study, we aimed to analyze the vascular plaque properties of coronary sinus based on iMAP-IVUS technique and to explore the correlation between serum CCL2, uric acid (UA), creatinine (CR), blood lipids, and plaque characteristics. In addition, the GSEA method was used to analyze the public database of gene expression in coronary artery disease (GSE40595), and to explore the regulation of CCL2 on coronary plaque vulnerability-related pathways and key genes. These key genes were then subjected to functional enrichment analysis.
Materials and methods
Study subjects
This study was performed in conformity with the ethical standards of Henan Provincial People's Hospital Heart Center (Zhengzhou, China) review committee and followed the 1975 Helsinki declaration and its later amendments. Informed consent was given by enrolled participants. We retrospectively reviewed 263 patients with coronary heart disease that visited our hospital for percutaneous coronary intervention (PCI) from February 2015 to June 2017 in Henan Provincial People's Hospital. We excluded individuals with acute cardiac insufficiency or cardiogenic shock complications (13 patients), acute myocardial infarction (AMI) <72 h (9 patients), refractory angina (25 patients), hemodynamic instability (3 patients), complete occlusion (11 patients), thrombotic lesions in interventional therapy (based on coronary angiography) (6 patients), trauma or a history of surgery in the last six months (4 patients), blood disease (2 patients), malignant tumor (1 patient), severe kidney (3 patients) or liver disease (16 patients), inflammatory state or using glucocorticoids (34 patients), and patients who did not give consent to participate (21 patients). The patients who underwent coronary angiography and IVUS examination with available clinical and imaging features, and who gave consent to participate were finally incorporated in the study population (116 patients). PCI indications were estimated according to the Chinese Percutaneous Coronary Intervention Therapy Guidelines (2016). The included patients were aged 18–75 years old, male or non-pregnant women. All patients were enrolled in the morning on an empty stomach to collect 3 ml of venous blood. The level of CCL2 was detected by a human-specific CCL2 enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems, Inc., Minneapolis, MN, USA). The patients were distributed into two groups on the basis of their overall cohort median CCL2 level (70 pg/mL): low CCL2 group (52 cases, CCL2 ≤70 pg/mL) and high CCL2 group (64 cases, CCL2 >70 pg/mL).
Collection of clinical data and serological examination
The basic clinical data such as age, gender, body mass index (BMI), hypertension, diabetes, hyperlipidemia, smoking history, and medication were collected. Three milliliters of fasting venous blood from all subjects were collected in the early morning and sent to our laboratory for examination. High-density lipoprotein cholesterol (HDL-C), total cholesterol (TC), and triacylglycerol (TG) were detected with peroxidase method. The level of low-density lipoprotein cholesterol (LDL-C) was deduced from the Friedewald's formula,16 and the LDL-C/HDL-C ratio (L/H) was calculated. UA and CR were determined by enzymatic method.
Coronary angiography
Philips PHILIPSUNIQ FD20 and Siemens Artis zee III ceiling angiography machine were used. Through the radial or femoral approach, multiple position projections were performed to determine the location and severity of the coronary lesion. Coronary diameter and lesion stenosis were then examined using quantitative coronary angiography (QCA) at the end of diastole.
IVUS examination and analysis
The IVUS test was performed before the intervention of all enrolled patients. An iLab Ultrasound system and the coronary ultrasound imaging catheter (OptiCross Boston Scientific) with a diameter of 3.0 F and a frequency of 40 MHz were used. The intracoronary ultrasound catheter probe was automatically retracted at a speed of 0.5 mm/s at a distance of 5 mm from the distal end of the lesion. The IVUS image was recorded in real time, and the tissue image was constructed using the Qlvus iMap Basic Viewer 2.1 software (Medis Medical Imaging Systems, Leiden, the Netherlands). Gray-scale IVUS measurements were the minimum lumen area (MLA), the external elastic membrane cross-sectional area (EEMCSA), the plaque area (plaque area = EEMCSA-MLA), the plaque load (plaque load (%) = plaque area/EEMCSA × 100%), and remodeling index (RI). High-risk plaque thin cap fibroatheromas (TCFAs) were detected in culprit plaque.17 The iMAP-IVUS analysis is as follows18: different plaque components are represented by different colors: fibrous tissue (FT, green), necrotic core (NC, red), fibrofatty (FF, yellow), dense calcium (DC, blue); record the percentage of various ingredients.
GSEA analysis
The series matrix data for the coronary artery disease sample data set GSE20681 were downloaded from NCBI's Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). According to the relative expression level of CCL2 in CCL2 expression profile data (GSE20681), it was divided into two categories: high expression group (49 patients) and low expression group (50 patients). The GESA analysis was performed using GSEA v4.0.0 software (https://www.broadinstitute.org/gsea/). The c2.cp.kegg.v7.0.symbols.gmt data set from the GSEA website was used as the reference gene set for analyzing the impact of CCL2 expression on each reference gene set. Gene enrichment analysis was based on the default parameters of the weighted enrichment statistic method, and the number of random combinations was set to 1000 times. Pathways with P <0.05 and false discovery rates (FDR) of <0.25 were those significantly enriched in the GSEA analysis.
Analysis of GO and KEGG enrichment of enriched genes in Nod-like receptor signaling pathway
Functional enrichment analysis based on GO and KEGG pathway were performed using R library clusterProfiler,19 and the enriched genes with corrected P <0.05 were regarded as significantly enriched in each GO category (biological process(BP), cellular component (CC), and molecular function (MF)) and KEGG pathways.
Statistical analysis
Data were processed with SPSS 19.0 statistical software (SPSS, Chicago, IL). Data were expressed as means ± standard deviation. Comparison of means between the CCL2 high and CCL2 low groups was performed with the t test. The correlation between serum CCL2, blood lipid, UA, and CR levels and plaque components was analyzed using the univariate regression analysis. Logistic multivariate regression analysis was done for adjustment. The statistically significant P value cutoff was 0.05.
Results
General information
No statistically significant discrepancy in gender, age, BMI, hypertension, diabetes, smoking history, family history, TG, CR, UA, and oral medication was observed between the low CCL2 group (52 patients) and high CCL2 group (64 patients) (Table 1). The high CCL2 level group had higher LDL-C level ((2.92 ± 0.65) vs. (2.23 ± 0.53), P = 0.008) and L/H ratio ((3.64 ± 0.92) vs. (2.14 ± 0.68), P = 0.003), and lower HDL-C level ((0.83 ± 0.21) vs. (1.06 ± 0.23), P = 0.005) compared to the low CCL2 group (Table 1).
Table 1.
Comparison of basic clinical data between the two groups of patients.
| Item | Low CCL2 group (n = 52) | High CCL2 group (n = 64) | P |
|---|---|---|---|
| Age (years) | 58.2 ± 9.3 | 59.4 ± 9.7 | 0.972 |
| Male (n (%)) | 32 (61.54) | 36 (56.52) | 0.565 |
| BMI (kg/m2) | 25.1 ± 5.2 | 25.4 ± 5.1 | 0.476 |
| Hypertension (n (%)) | 19 (36.54) | 27 (42.19) | 0.536 |
| Diabetes (n (%)) | 17 (32.70) | 28 (43.75) | 0.224 |
| History of smoking (n (%)) | 28 (53.85) | 31 (48.44) | 0.562 |
| Family history (n (%)) | 8 (15.38) | 14 (21.88) | 0.375 |
| Biochemical indicators | |||
| LDL-C (mmol/L) | 2.23 ± 0.53 | 2.92 ± 0.65 | 0.008 |
| HDL-C (mmol/L) | 1.06 ± 0.23 | 0.83 ± 0.21 | 0.005 |
| L/H | 2.14 ± 0.68 | 3.64 ± 0.92 | 0.003 |
| TG (mmol/L) | 1.56 ± 0.46 | 1.67 ± 0.73 | 0.222 |
| TC (mmol/L) | 5.1 ± 0.4 | 5.84 ± 0.25 | 0.055 |
| UA (μmol/L) | 294.56 ± 30.28 | 312.49 ± 33.57 | 0.086 |
| CR (μmol/L) | 73.43 ± 18.47 | 77.58 ± 20.36 | 0.325 |
| Oral medical | |||
| Aspirin (n (%)) | 52 (100%) | 64 (100) | 1 |
| Clopidogrel (n (%)) | 52 (100%) | 64 (100) | 1 |
| β-receptor blocker (n (%)) | 35 (67.31) | 45 (70.31) | 0.728 |
| ACE inhibitor/ARB (n (%)) | 21 (40.38) | 30 (46.88) | 0.484 |
| Statins (n (%)) | 49 (94.23) | 61 (95.31) | 0.794 |
CCL2: CC chemokine ligand 2; BMI: body mass index; LDL-C: low-density lipoprotein cholesterol; HDL-C: high-density lipoprotein cholesterol; L/H: LDL-C/HDL-C ratio; TG: triglyceride; TC: total cholesterol; UA: uric acid; CR: creatinine.
Comparison of plaque indexes under IVUS in two groups of patients
The gray-scale IVUS results of culprit vessel in the two groups are shown in Table 2. The results showed that the high CCL2 group had higher eccentric plaque (51.56% (33/64) vs. 21.15% (11/52), P = 0.001), plaque rupture (23.44% (15/64) vs. 7.69% (4/52), P = 0.023), and reconstruction index (RI) ((1.02 ± 0.15) vs. (0.84 ± 0.11), P = 0.002) than the low CCL2 group. In addition, no significant discrepancy among the high CCL2 group and low CCL2 group in terms of EEMCSA, MLA, plaque area, plaque load, and thrombus formation was recorded (P >0.05, Table 2).
Table 2.
Comparison of IVUS plaque properties between the two groups.
| Items | Low CCL2 group (n = 52) | High CCL2 group (n = 64) | P |
|---|---|---|---|
| Gray scale IVUS | |||
| EEMCSA (mm2) | 11.89±2.39 | 12.15±2.57 | 0.456 |
| MLA (mm2) | 4.13±1.25 | 4.37±1.38 | 0.324 |
| Plaque area (mm2) | 7.13±1.57 | 7.42±1.73 | 0.232 |
| Plaque load (%) | 66.41±10.32 | 68.73±10.67 | 0.187 |
| Eccentric plaque (n (%)) | 11 (21.15) | 33 (51.56) | 0.001 |
| Plaque rupture (n (%)) | 4 (7.69) | 15 (23.44) | 0.023 |
| Thrombus formation (n (%)) | 1 (1.92) | 3 (4.68) | 0.417 |
| RI | 0.84±0.11 | 1.02±0.15 | 0.002 |
| iMAP—IVUS | |||
| TCFAs (n (%)) | 4 (7.69) | 17 (26.56) | 0.009 |
| FT% | 70.54±9.40 | 63.40±10.12 | 0.017 |
| FF% | 9.43±5.32 | 13.56±6.26 | 0.024 |
| DC% | 12.15±6.19 | 11.04±7.35 | 0.151 |
| NC% | 8.31±4.78 | 12.67±6.40 | 0.008 |
CCL2: CC chemokine ligand 2; IVUS: intravascular ultrasound; EEMCSA: external elastic membrane cross-sectional area; MLA: minimum lumen area; RI: remodeling index; TCFAs: thin cap fibroatheromas; FT: fibrous tissue; FF: fibrofatty; DC: dense calcium; NC: necrotic core.
In addition, we further compared the iMAP-IVUS results of culprit vessel in the two groups (Table 2). The detection rate of TCFAs in the high CCL2 level group was notably greater compared to the low CCL2 level group (26.56% vs. 7.69%, P = 0.009). Plaque composition analysis showed that compared to the low CCl2 level group, the high CCL2 level group had a higher FF% ((13.56 ± 6.26)% vs. (9.43 ± 5.32)%, P = 0.024) and NC% ((12.67 ± 6.40)% vs. (8.31 ± 4.78)%, P = 0.008), as well as lower FT% ((63.40 ± 10.12)% vs. (70.54 ± 9.40)%, P = 0.017). No significant difference in DC% was found among high CCL2 group and low CCL2 group (P >0.05).
Univariate regression analysis of CCL2 and plaque characteristics
Univariate regression analysis of serum lipids, CR, UA and CCL2 levels, and plaque characteristics were performed, and the results are shown in Table 3. The results suggested that the levels of CCL2, LDL-C, and L/H were positively and considerably correlated with FF% (r = 0.182, P = 0.006; r = 6.118, P = 0.006; r = 3.280, P = 0.007) and NC% (r = 0.176, P = 0.003; r = 5.776, P = 0.014; r = 3.171, P = 0.01), but negatively and significantly correlated with FT% (r = –0.283, P = 0.008; r=–8.202, P = 0.008; r = –4.663, P = 0.006). However, HDL-C was negatively correlated with FF% (r=–26.119, P = 0.023) and NC% (r=–25.288, P = 0.027), but showed a positive correlation with FT% (r = 41.024, P = 0.013). No significant correlation among UA, CR and TG and plaque characteristics was recorded (P >0.05, Table 3).
Table 3.
Univariate regression analysis of factors associated with CCL2 and plaque components.
| Variable |
Regression coefficients (P) |
|||||
|---|---|---|---|---|---|---|
| EEMCSA | MLA | FT% | FF% | NC% | DC% | |
| LDL-C | 2.961 (0.171) | –0.729 (0.382) | –8.202 (0.008) | 6.118 (0.006) | 5.776 (0.014) | –3.692 (0.237) |
| HDL-C | –13.305 (0.094) | 2.765 (0.408) | 41.024 (0.013) | –26.119 (0.023) | –25.288 (0.027) | 10.384 (0.342) |
| L/H | 1.630 (0.215) | –0.396 (0.176) | –4.663 (0.006) | 3.280 (0.007) | 3.171 (0.010) | –1.787 (0.145) |
| TG | 4.247 (0.435) | 1.232 (0.378) | 14.534 (0.091) | 9.476 (0.149) | 7.621 (0.537) | 5.432 (0.319) |
| UA | 0.016 (0.132) | 0.005 (0.086) | 0.064 (0.332) | –0.042 (0.317) | –0.037 (0.581) | 0.013 (0.216) |
| CR | 0.035 (0.654) | 0.016 (0.468) | 0.168 (0.517) | 0.125 (0.629) | 0.097 (0.571) | 0.035 (0.389) |
| CCL2 | 0.092 (0.098) | –0.018 (0.345) | –0.283 (0.008) | 0.182 (0.006) | 0.176 (0.003) | –0.07 (0.181) |
CCL2: CC chemokine ligand 2; LDL-C: low-density lipoprotein cholesterol; HDL-C: high-density lipoprotein cholesterol; L/H: LDL-C/HDL-C ratio; TG: triacylglycerol; UA: uric acid; CR: creatinine; EEMCSA: external elastic membrane cross-sectional area; MLA: minimum lumen area; FT: fibrous tissue; FF: fibrofatty; DC: dense calcium; NC: necrotic core.
Multivariate regression analysis of CCL2 and plaque components
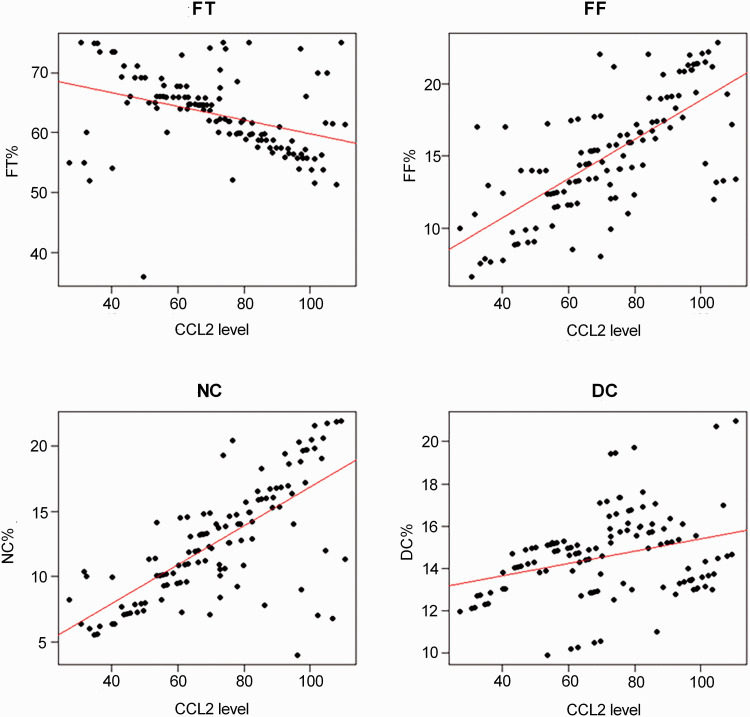
Multivariate analysis based on the factors related to plaque components was performed. The results showed that CCL2 was positively correlated with FF% (β = 0.036, P = 0.005) and NC% (β = 0.045, P = 0.003), but showed a negative correlation with FT% (β = –0.024, P = 0.006) (Table 4, Figure 1). Furthermore, LDL-C and L/H were positively correlated with FF% (β = 0.345, P = 0.006; β = 0.617, P = 0.009) and NC% (β = 0.449, P = 0.012; β = 0.576, P = 0.010), and negatively correlated with FT% (β = –0.254, P = 0.008; β = –0.374, P = 0.007) (Table 4). The opposite results were obtained in HDL-C.
Table 4.
Multivariate regression analysis of different factors.
| Variables |
Β (P) |
|||
|---|---|---|---|---|
| FT% | FF% | NC% | DC% | |
| LDL-C | –0.254 (0.008) | 0.345 (0.006) | 0.449 (0.012) | –0.617 (0.152) |
| HDL-C | 1.045 (0.015) | –0.724 (0.021) | –0.824 (0.021) | 1.325 (0.221) |
| L/H | –0.374 (0.007) | 0.617 (0.009) | 0.576 (0.010) | –0.446 (0.517) |
| CCL2 | –0.024 (0.006) | 0.036 (0.005) | 0.045 (0.003) | 0.041 (0.237) |
CCL2: CC chemokine ligand 2; LDL-C: low-density lipoprotein cholesterol; HDL-C: high-density lipoprotein cholesterol; L/H: LDL-C/HDL-C ratio; FT: fibrous tissue; FF: fibrofatty; DC: dense calcium; NC: necrotic core.
Figure 1.
Correlation of CCL2 with plaque characteristics. (A color version of this figure is available in the online journal.)
FT: fibrous tissue; FF: fibrofatty; DC: dense calcium; NC: necrotic core; CCL2: CC chemokine ligand 2.
Enrichment analysis of functional gene set with high expression of CCL2
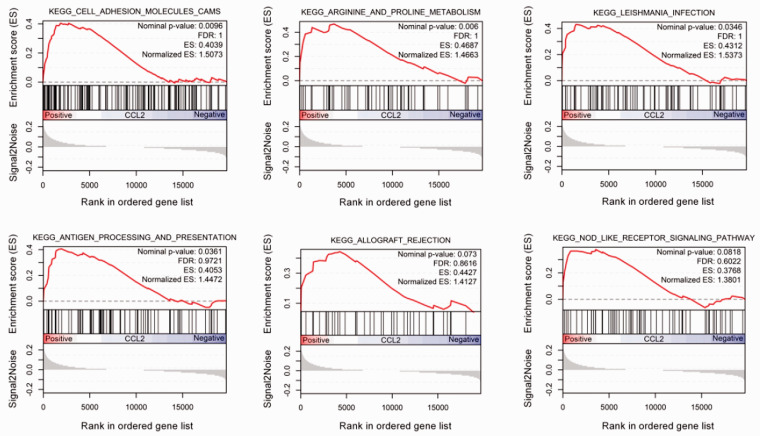
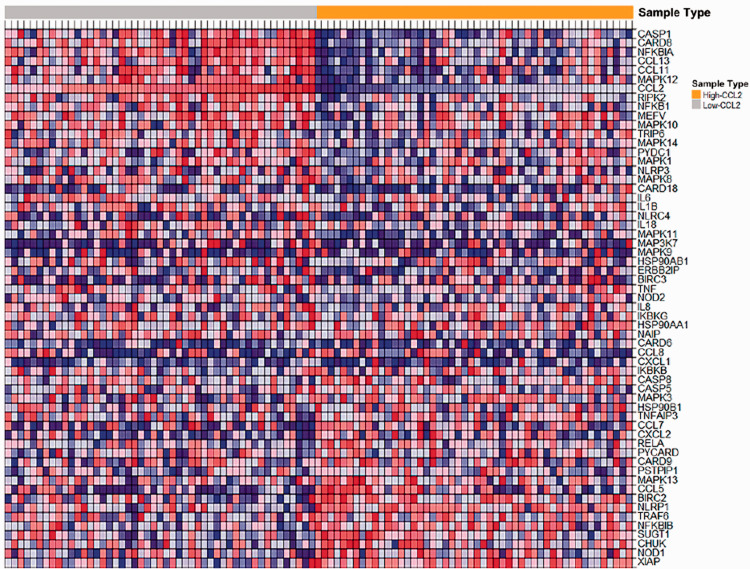
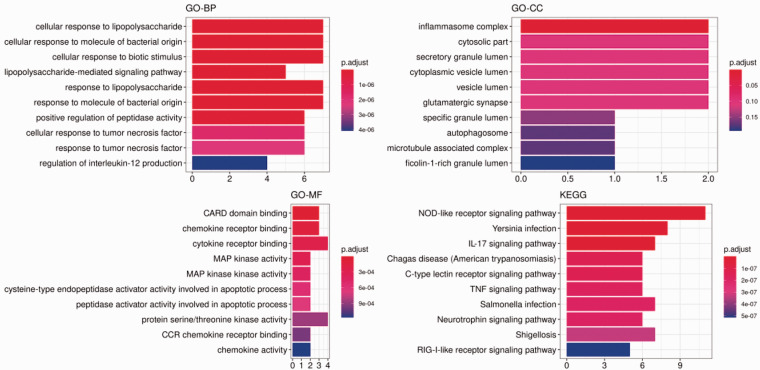
CCL2 high expression samples were enriched in varieties of pathways, such as cell adhesion molecules (CAMs), arginine and proline metabolism, Leishmania infection, antigen processing and presentation, allograft rejection, and Nod-like receptor signaling pathway (Figure 2). Further analysis of the enriched genes of these pathways revealed that the CCL2 gene was only found in the Nod-like receptor signaling pathway. Heatmap showed that CASP1, CARD8, NFKBIA, CCL13, CCL11, and CCL2 genes were highly expressed in the high-level CCL2 group in the Nod-like receptor signaling pathway, while XIAP, NOD1, CHUK, SUGT1, and TRAF6 genes were highly expressed in the low-level CCL2 group (Figure 3). In addition, functional analysis of genes enriched in the Nod-like receptor signaling pathway was found to be involved in cellular response to tumor necrosis factor, response to tumor necrosis factor, and cellular response to lipopolysaccharide in the category of BP, cellular component (such as inflammasome complex), and molecular function (such as MAP kinase activity, CARD domain binding, CCR chemokine receptor binding and chemokine activity) (Figure 4). KEGG analysis hinted that the signal pathways involved in these enriched genes were mainly Nod-like receptor signaling pathway, Yersinia infection, IL-17 signaling pathway, and Shigellosis (Figure 4).
Figure 2.
Results of GSEA analysis between CCL2 high and CCL2 low expression samples. (A color version of this figure is available in the online journal.)
Figure 3.
Heatmap of enriched gene sets in Nod-like receptor signaling pathway obtained from GSEA analysis. (A color version of this figure is available in the online journal.)
Figure 4.
GO function and KEGG pathway enrichment analysis of enriched gene sets in Nod-like receptor signaling pathway. (A color version of this figure is available in the online journal.)
Discussion
Vulnerable plaque features include a slight fibrous cap, multiple macrophages, large necrotic nuclei, spotted calcification, and positive remodeling.20 This plaque is also known as inflammatory TCFA. iMAP-IVUS provides a more intuitive and accurate analysis of coronary plaques to precisely identify the vulnerable plaques.
Inflammation is closely related to the characteristics of vulnerable plaques.21 CCL2 is a CC-like cell chemokine, which has specific chemotactic activation and promotes inflammatory response to mononuclear and macrophages. Research findings have reported that CCL2 plays a key role in multiple stages of atherosclerosis, including initial fat streaks and remodeling after myocardial infarction.22 Studies suggest that CCL2 level is closely related to events such as heart failure and death encountered in coronary heart disease. Previous studies showed that elevated CCL2 baseline level is associated with both traditional atherosclerosis risk factors and increased risk of heart failure or death.23,24 Ding et al.’s25 study suggested that CCL2 level is related to all-cause and increased mortality in coronary heart disease patients. However, previous studies have mostly studied CCL2 level and clinical events, while studies on CCL2 level and coronary plaque characteristics (based on virtual histology) are relatively rare. Studying the correlation between CCL2 level and plaque properties may be helpful in identifying high-risk plaques and high-risk patients.
This study found that patients with high-level CCL2 group had higher eccentric plaque, plaque rupture, and TCFA detection rates than the low-level CCL2 group. The regression analysis showed that CCL2 was positively associated with FF and NC, and negatively associated with FT. The nature and composition of coronary plaques are closely related to plaque stability and coronary events. A VH-IVUS study of triple vessel disease showed a significant proportion of ruptured plaques and TCFA plaques in the coronary arteries of individuals with ACS.26 Stone et al.27 suggested that plaques in acute coronary events are mostly characterized by thin cap fibroids or large plaque loads, small lumen areas, or some combination of these features. Plaque eccentricity, plaque rupture, TCFA, high lipid volume, and high NC are closely related to plaque instability. This study found that coronary plaques in the high CCL2 group had more unstable plaque characteristics and a higher probability of TCFA detection, suggesting that CCL2 may be a serological indicator for predicting high-risk coronary plaques. Early diagnosis of patients with high-risk plaques, intensive medication or interventional therapy may help lessen the occurrence of acute coronary events. In addition, previous studies in mice have shown that downregulated expression of CCL2 can transform vulnerable plaques into a more stable plaque and prevents plaque rupture.28 In the genetic study of CCL2 gene polymorphism and atherosclerotic disease, Angeles-Martinez et al.29 pointed out that four CCL2 gene polymorphisms were highly linked unbalanced, and a haplotype was markedly correlated with the risk of developing early onset coronary heart disease. Future large-scale studies on CCL2 gene polymorphisms and gene loci for CCL2 expression may lead to possible treatment of CCL2 gene therapy, or the stability of coronary plaques can be converted from the genetic level to lessen the prevalence of acute coronary events. In addition, we also found that LDL-C level and L/H ratio have a good positive correlation with plaque instability. Previous studies pointed out that the L/H ratio is the best pertinent index for predicting the lipid component in coronary plaque and is related to plaque vulnerability,30,31 which are similar to the results of this study. Combining multiple serological markers may reduce bias in the diagnosis of vulnerable plaque.
Furthermore, based on the GSEA method, we found that CCL2 high expression samples were enriched for gene pathways such as Nod-like receptor signaling pathway. Nod-like receptors are pattern recognition receptors (PRRs) of intracellular pathogens that act as sensors for “danger signals″ in cells and are involved in inflammation and innate immune responses in mammals.32 NOD1 and NOD2 protein receptors activate nuclear factor-kappa B (NF-κB) phosphorylation and mitogen-activated protein kinase (MAPK) signaling pathways, thereby promoting cytokine production and apoptosis.33 NF-κB, as an important transcription factor, regulates the development of cardiovascular diseases such as ACS and atherosclerosis,34 and is vital in mediating chronic inflammation of blood vessel walls and promoting the release of inflammatory factors.35 Consistent with our results, multiple pathways such as NOD-like receptors were incriminated in the formation of coronary artery disease, suggesting that the activation of multiple signaling pathways mediates chronic inflammation of the vessel wall, leading to persistent shifts in the organization and function of the vessel wall, which ultimately promotes the pathogenesis of coronary artery disease. Functional analysis of genes enriched in the Nod-like receptor signaling pathway was found to be associated with cellular response to tumor necrosis factor, and cellular response to lipopolysaccharide, inflammasome complex, MAP kinase activity, CARD domain binding, CCR chemokine receptor binding, and chemokine activity. Inflammasome formation in the heart of mice during acute myocardial infarction leads to loss of functional myocardial, ultimately resulting in heart failure.36 These results indicated that CCL2 was involved in the inflammatory response and immune process of coronary artery disease.
In summary, this study uses clinical studies and the GEO database to speculate that the high expression of CCL2 is involved in multiple processes in the pathogenesis and development of coronary artery disease and could be regarded as a clinical prognostic indicator for the progression of coronary diseases. However, further studies are needed to definitely validate this conclusion. In addition, the high expression of CCL2 may be related to gene pathways such as Nod-like receptor signaling pathway, suggesting that CCL2 was involved in the inflammatory response and immune process of coronary artery disease.
Authors’ contributions
All authors participated in the interpretation of the studies and analysis of the data; YC contributed to conception and revised the article. ML wrote the first draft of article. YZ, DL, and JL performed the experiments and analyzed the data. All of the authors had read and approved the final version of the article.
Ethical approval
This study was performed in conformity with the ethical standards of Henan Provincial People's Hospital Heart Center (Zhengzhou, China) review committee and followed the 1975 Helsinki declaration and its later amendments. Informed consent was given by enrolled participants.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
This work was supported by the Henan Scientific and Technological Research Projects (162102310034) and the Basic Research Projects of Henan Science and Technology Department (142300410264).
ORCID iD
References
- 1.Muller JE, Tofler GH, Stone PH. Circadian variation and triggers of onset of acute cardiovascular disease. Circulation 1989; 79:733–43 [DOI] [PubMed] [Google Scholar]
- 2.Frobert O, van't Veer M, Aarnoudse W, Simonsen U, Koolen JJ, Pijls NH. Acute myocardial infarction and underlying stenosis severity. Catheter Cardiovasc Interv 2007; 70:958–65 [DOI] [PubMed] [Google Scholar]
- 3.Naghavi M, Libby P, Falk E, Casscells SW, Litovsky S, Rumberger J, Badimon JJ, Stefanadis C, Moreno P, Pasterkamp G, Fayad Z, Stone PH, Waxman S, Raggi P, Madjid M, Zarrabi A, Burke A, Yuan C, Fitzgerald PJ, Siscovick DS, de Korte CL, Aikawa M, Airaksinen KE, Assmann G, Becker CR, Chesebro JH, Farb A, Galis ZS, Jackson C, Jang IK, Koenig W, Lodder RA, March K, Demirovic J, Navab M, Priori SG, Rekhter MD, Bahr R, Grundy SM, Mehran R, Colombo A, Boerwinkle E, Ballantyne C, Insull W, Jr, Schwartz RS, Vogel R, Serruys PW, Hansson GK, Faxon DP, Kaul S, Drexler H, Greenland P, Muller JE, Virmani R, Ridker PM, Zipes DP, Shah PK, Willerson JT. From vulnerable plaque to vulnerable patient: a call for new definitions and risk assessment strategies: part II. Circulation 2003; 108:1772–8 [DOI] [PubMed] [Google Scholar]
- 4.Libby P. Mechanisms of acute coronary syndromes and their implications for therapy. N Engl J Med 2013; 368:2004–13 [DOI] [PubMed] [Google Scholar]
- 5.Lodh M, Goswami B, Parida A, Patra S, Saxena A. Assessment of serum leptin, pregnancy-associated plasma protein a and CRP levels as indicators of plaque vulnerability in patients with acute coronary syndrome. Cardiovasc J Afr 2012; 23:330–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawasaki M, Takatsu H, Noda T, Sano K, Ito Y, Hayakawa K, Tsuchiya K, Arai M, Nishigaki K, Takemura G, Minatoguchi S, Fujiwara T, Fujiwara H. In vivo quantitative tissue characterization of human coronary arterial plaques by use of integrated backscatter intravascular ultrasound and comparison with angioscopic findings. Circulation 2002; 105:2487–92 [DOI] [PubMed] [Google Scholar]
- 7.Okubo M, Kawasaki M, Ishihara Y, Takeyama U, Kubota T, Yamaki T, Ojio S, Nishigaki K, Takemura G, Saio M, Takami T, Minatoguchi S, Fujiwara H. Development of integrated backscatter intravascular ultrasound for tissue characterization of coronary plaques. Ultrasound Med Biol 2008; 34:655–63 [DOI] [PubMed] [Google Scholar]
- 8.Gilbert J, Lekstrom-Himes J, Donaldson D, Lee Y, Hu M, Xu J, Wyant T, Davidson M. Effect of CC chemokine receptor 2 CCR2 blockade on serum C-reactive protein in individuals at atherosclerotic risk and with a single nucleotide polymorphism of the monocyte chemoattractant protein-1 promoter region. Am J Cardiol 2011; 107:906–11 [DOI] [PubMed] [Google Scholar]
- 9.Deo R, Khera A, McGuire DK, Murphy SA, M, Neto Jde P, Morrow DA, de Lemos JA. Association among plasma levels of monocyte chemoattractant protein-1, traditional cardiovascular risk factors, and subclinical atherosclerosis. J Am Coll Cardiol 2004; 44:1812–8 [DOI] [PubMed] [Google Scholar]
- 10.Koenig W, Khuseyinova N. Biomarkers of atherosclerotic plaque instability and rupture. Arterioscler Thromb Vasc Biol 2007; 27:15–26 [DOI] [PubMed] [Google Scholar]
- 11.Macanas-Pirard P, Quezada T, Navarrete L, Broekhuizen R, Leisewitz A, Nervi B, Ramirez PA. The CCL2/CCR2 axis affects transmigration and proliferation but not resistance to chemotherapy of acute myeloid leukemia cells. PLoS One 2017; 12:e0168888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conti I, Rollins BJ. CCL2 (monocyte chemoattractant protein-1) and cancer. Semin Cancer Biol 2004; 14:149–54 [DOI] [PubMed] [Google Scholar]
- 13.Raghu H, Lepus CM, Wang Q, Wong HH, Lingampalli N, Oliviero F, Punzi L, Giori NJ, Goodman SB, Chu CR, Sokolove JB, Robinson WH. CCL2/CCR2, but not CCL5/CCR5, mediates monocyte recruitment, inflammation and cartilage destruction in osteoarthritis. Ann Rheum Dis 2017; 76:914–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aiello RJ, Bourassa PA, Lindsey S, Weng W, Natoli E, Rollins BJ, Milos PM. Monocyte chemoattractant protein-1 accelerates atherosclerosis in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol 1999; 19:1518–25 [DOI] [PubMed] [Google Scholar]
- 15.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102:15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vujovic A, Kotur-Stevuljevic J, Spasic S, Bujisic N, Martinovic J, Vujovic M, Spasojevic-Kalimanovska V, Zeljkovic A, Pajic D. Evaluation of different formulas for LDL-C calculation. Lipids Health Dis 2010; 9:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Granillo GA, Garcia-Garcia HM, McFadden EP, Valgimigli M, Aoki J, de Feyter P, Serruys PW. In vivo intravascular ultrasound-derived thin-cap fibroatheroma detection using ultrasound radiofrequency data analysis. J Am Coll Cardiol 2005; 46:2038–42 [DOI] [PubMed] [Google Scholar]
- 18.Nair A, Kuban BD, Tuzcu EM, Schoenhagen P, Nissen SE, Vince DG. Coronary plaque classification with intravascular ultrasound radiofrequency data analysis. Circulation 2002; 106:2200–6 [DOI] [PubMed] [Google Scholar]
- 19.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 2012; 16:284–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Virmani R, Kolodgie FD, Burke AP, Farb A, Schwartz SM. Lessons from sudden coronary death: a comprehensive morphological classification scheme for atherosclerotic lesions. Arterioscler Thromb Vasc Biol 2000; 20:1262–75 [DOI] [PubMed] [Google Scholar]
- 21.Libby P, Tabas I, Fredman G, Fisher EA. Inflammation and its resolution as determinants of acute coronary syndromes. Circ Res 2014; 114:1867–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birdsall HH, Green DM, Trial J, Youker KA, Burns AR, MacKay CR, LaRosa GJ, Hawkins HK, Smith CW, Michael LH, Entman ML, Rossen RD. Complement C5a, TGF-beta 1, and MCP-1, in sequence, induce migration of monocytes into ischemic canine myocardium within the first one to five hours after reperfusion. Circulation 1997; 95:684–92 [DOI] [PubMed] [Google Scholar]
- 23.de Lemos JA, Morrow DA, Sabatine MS, Murphy SA, Gibson CM, Antman EM, McCabe CH, Cannon CP, Braunwald E. Association between plasma levels of monocyte chemoattractant protein-1 and long-term clinical outcomes in patients with acute coronary syndromes. Circulation 2003; 107:690–5 [DOI] [PubMed] [Google Scholar]
- 24.de Lemos JA, Morrow DA, Blazing MA, Jarolim P, Wiviott SD, Sabatine MS, Califf RM, Braunwald E. Serial measurement of monocyte chemoattractant protein-1 after acute coronary syndromes: results from the a to Z trial. J Am Coll Cardiol 2007; 50:2117–24 [DOI] [PubMed] [Google Scholar]
- 25.Ding D, Su D, Li X, Li Z, Wang Y, Qiu J, Lin P, Zhang Y, Guo P, Xia M, Li D, Yang Y, Hu G, Ling W. Serum levels of monocyte chemoattractant protein-1 and all-cause and cardiovascular mortality among patients with coronary artery disease. PLoS One 2015; 10:e0120633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong MK, Mintz GS, Lee CW, Lee JW, Park JH, Park DW, Lee SW, Kim YH, Cheong SS, Kim JJ, Park SW, Park SJ. A three-vessel virtual histology intravascular ultrasound analysis of frequency and distribution of thin-cap fibroatheromas in patients with acute coronary syndrome or stable angina pectoris. Am J Cardiol 2008; 101:568–72 [DOI] [PubMed] [Google Scholar]
- 27.Stone GW, Maehara A, Lansky AJ, de Bruyne B, Cristea E, Mintz GS, Mehran R, McPherson J, Farhat N, Marso SP, Parise H, Templin B, White R, Zhang Z, Serruys PW. A prospective natural-history study of coronary atherosclerosis. N Engl J Med 2011; 364:226–35 [DOI] [PubMed] [Google Scholar]
- 28.Liu XL, Zhang PF, Ding SF, Wang Y, Zhang M, Zhao YX, Ni M, Zhang Y. Local gene silencing of monocyte chemoattractant protein-1 prevents vulnerable plaque disruption in apolipoprotein E-knockout mice. PLoS One 2012; 7:e33497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Angeles-Martinez J, Posadas-Sanchez R, Alvarez-Leon E, Villarreal-Molina T, Cardoso-Saldana G, Fragoso JM, Juarez-Rojas JG, Medina-Urrutia A, Posadas-Romero C, Vargas AG. Monocyte chemoattractant protein-1 gene (MCP-1) polymorphisms are associated with risk of premature coronary artery disease in Mexican patients from the genetics of atherosclerotic disease (GEA) study. Immunol Lett 2015; 167:125–30 [DOI] [PubMed] [Google Scholar]
- 30.Kawakami R, Matsumoto I, Shiomi M, Kurozumi M, Miyake Y, Ishizawa M, Ishikawa K, Murakami K, Noma T, Takagi Y, Nishimoto N, Minamino T. Role of the low-density lipoprotein-cholesterol/high-density lipoprotein-cholesterol ratio in predicting serial changes in the lipid component of coronary plaque. Circ J 2017; 81:1439–46 [DOI] [PubMed] [Google Scholar]
- 31.Hu X, Zhang W, Zhao N, Zhao R, Li S. Low- to high-density lipoprotein cholesterol ratio followed by coronary computed tomography angiography improves coronary plaque classification accuracy. Oncotarget 2018; 9:7727–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol 2006; 7:1250–7 [DOI] [PubMed] [Google Scholar]
- 33.Caruso R, Warner N, Inohara N, Nunez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity 2014; 41:898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valen G, Yan ZQ, Hansson GK. Nuclear factor kappa-B and the heart. J Am Coll Cardiol 2001; 38:307–14 [DOI] [PubMed] [Google Scholar]
- 35.Brasier AR. The nuclear factor-kappaB-interleukin-6 signalling pathway mediating vascular inflammation. Cardiovasc Res 2010; 86:211–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mezzaroma E, Toldo S, Farkas D, Seropian IM, Van Tassell BW, Salloum FN, Kannan HR, Menna AC, Voelkel NF, Abbate A. The inflammasome promotes adverse cardiac remodeling following acute myocardial infarction in the mouse. Proc Natl Acad Sci U S A 2011; 108:19725–30 [DOI] [PMC free article] [PubMed] [Google Scholar]