Abstract
Purpose
Glycemic control has been recognized as an important modifiable risk factor for diabetic retinopathy (DR). Whether hemoglobin A1c (HbA1c), as an indicator of glycemic control, could modify the genetic susceptibility to severe DR remains to be investigated. This study aimed to investigate whether HbA1c could modulate the genetic susceptibility to severe DR in Chinese patients with type 2 diabetes.
Methods
A total of 3,093 Chinese individuals with type 2 diabetes were included in the cross-sectional case-control study: 1,051 with sight-threatening DR (STDR) and 2,042 without STDR. Sixty-nine top-ranked single nucleotide polymorphisms (SNPs) identified from previous genome-wide association studies were examined for their associations with STDR and proliferative DR as a subgroup analysis. SNPs showing suggestive associations with DR were examined in the stratified analysis by dichotomized HbA1c (<7% vs. ≥7%). An interaction analysis was performed by including an interaction term of SNP × HbA1c in the regression model.
Results
Four SNPs showed suggestive associations with STDR. In the stratified analysis, patients with adequate glycemic control (HbA1c <7%) had a 42% lower risk of STDR for carrying each additional protective C allele of COL5A1 rs59126004 (P = 1.76 × 10−4; odds ratio, 0.58; 95% confidence interval, 0.44–0.77). rs59126004 demonstrated a significant interaction with dichotomized HbA1c on the risk of STDR (Pinteraction = 1.733 × 10−3). In the subgroup analysis for proliferative DR, the protective effect of rs59126004 was even more pronouncedly demonstrated (P = 8.35 × 10−5; odds ratio, 0.37; 95% confidence interval, 0.22–0.60) and it showed similar interactions with dichotomized HbA1c (Pinteraction = 1.729 × 10−3).
Conclusions
Our data provided evidence for possible interactions between HbA1c and COL5A1 rs59126004 on the risk of severe DR. These findings may provide new insight into the pathophysiologic mechanism of DR.
Keywords: diabetic retinopathy, single nucleotide polymorphisms, hemoglobin A1c, gene–environment interactions
As a common microvascular complication of diabetes, diabetic retinopathy (DR) remains the leading cause of irreversible vision loss among the working population around the world.1,2 In 2010, the estimated global prevalences of any DR and sight-threatening DR (STDR), the advanced form of the disease, were 34.6% and 10.2%, respectively, among patients with diabetes.2 DR is a multifactorial disorder arising from the complex interplay of numerous environmental and genetic factors. Over the past few decades, epidemiologic studies have revealed a range of risk factors associated with DR, including long duration of diabetes, hypertension (HTN), hemoglobin A1c (HbA1c), and dyslipidaemia.2 Microvascular complications of diabetes are often exacerbated by prolonged hyperglycemia. Large-scale epidemiologic studies such as the Diabetes Control and Complications Trial and United Kingdom Prospective Diabetes Project revealed the strong relationship between inadequate glycemic control with the development and progression of DR.3,4 HbA1c, as a well-established indicator of glycemic control,5 has been recognized as one of the most important modifiable risk factors for microvascular complications, including DR.2 In view of the growing prevalence of diabetic complications, international organizations, such as the American Diabetes Association, have therefore recommended an HbA1c of <7% (53 mmol/mol) as the general target for glycemic control to reduce complications among patient with diabetes.6
Noteworthy, heterogeneity in the frequency and severity of DR among patients with diabetes can only be partly explained by the conventional risk factors.7 Such observations implicate that other factors, including genetic determinants, are involved in the pathogenesis of DR. Familial aggregation studies have clearly demonstrated the genetic components of DR.8 To date, several genome-wide association studies (GWAS) for DR have been reported in multiple ethnic groups and a number of DR-susceptibility loci have been identified.9–17 However, statistically robust evidence of associations is lacking because the majority of the GWAS-identified association signals have failed to reach genome-wide significance.9–17 Furthermore, inconsistent results have been yielded in subsequent follow-up replication studies.18–22 Such discrepancies could have been due to differences in study design and variation in the degree of exposure to environmental factors. Similar to other complex diseases, the GWAS-identified susceptibility variants to date cannot fully explain the heritability of DR. Gene–environment interactions may partially account for the unexplained heritability. To the best of our knowledge, no previous study has assessed the interactions between the susceptibility variants identified from recent GWAS and HbA1c on the risk of severe DR, including both STDR and the most severe form of DR, proliferative DR (PDR). In this study, we aimed to investigate whether HbA1c could modulate the genetic susceptibility to severe DR in Chinese patient with type 2 diabetes (T2DM).
Methods
Patients
A cross-sectional case-control study on STDR was performed in 3093 Southern Chinese patients with T2DM. This study involved 2042 non-STDR controls and 1051 STDR cases, including 409 cases with PDR, the most severe form of DR. The current study was an extension of our previous study22 that comprised 567 cases and 1490 controls, and included only 38 DR-associated single nucleotide polymorphisms (SNPs) identified from GWAS reported up to that juncture.9–12 To enhance the statistical power of our study, we further expanded the sample size by including additional cases and controls. We also investigated an additional 31 DR-associated SNPs to form a more comprehensive list of SNPs in the current study, including those that were identified in more recently published GWAS for DR.13–15,18 These 31 additional SNPs are indicated in Supplementary Table S1 and Supplementary Table S2. The STDR cases were recruited from the Hong Kong West Diabetes Registry, as well as the ophthalmology clinics at Queen Marry Hospital, Tseung Kwan O Hospital, and the United Christian Hospital, Hong Kong. All non-STDR controls were recruited from the Hong Kong West Diabetes Registry cohort. Details of the study cohorts have been described previously.22 At assessment, detailed family, medical, and drug histories of the participants were recorded using a standardized questionnaire. Anthropometric parameters and clinical data were collected with written informed consent. Body mass index (BMI) was defined as weight in kilogram divided by the square of height in meters. HTN was defined as blood pressure of ≥140/90 mm Hg or on antihypertensive medications. Blood samples were drawn from the patients after an overnight fasting of ≥8 hours for biochemical and genetic analysis. The measurement of HbA1c was performed in whole blood using cation exchange HPLC on Bio-Rad Variant (BioRad Laboratories, Inc., Hercules, CA). The HbA1c test was performed using a method certified by the National Glycohemoglobin Standardization Program and standardized to the Diabetes Control and Complications Trial reference assay.23 The study protocol was approved by the institutional review boards of the University of Hong Kong/Hospital Authority. All study procedures of this research were in accordance with the Declaration of Helsinki.
Phenotype Characterization
The DR status of the participants was determined on the basis of digital, color fundal photographs taken with the fundus cameras (Topcon TRC50-DX Type 1A, Tokyo, Japan) with two photographic fields (45°) for each eye (one centered at the macula and one centered at the optic disc). Visual acuity was determined with the Early Treatment of Diabetic Retinopathy Study chart using the auto-chart projector (Topcon Auto-Chart Projector ACP-7EM). All STDR cases were assessed systematically by specialist ophthalmologists to determine the severity of DR according to the English National Screening Program guideline for DR.24 STDR cases were defined as T2DM patients having either PDR (grading R3), pre-PDR (grading R2), or with clinically significant macular edema.25 Non-STDR controls were defined as T2DM patients without retinopathy (grading R0) or with background DR (grading R1). Participants with ungradable fundus photographs were excluded from this study.
Genetic Analysis
Genomic DNA was extracted using the ReliaPrep Blood gDNA Miniprep System (Promega, Madison, WI) extraction kits according to the manufacturer's instructions. Sixty-nine top-ranked SNPs selected from previous GWAS (P < 5 × 10−4; r2 < 0.9)9–15,18 were investigated in the current study. SNPs reported in two recent GWAS of DR16,17 were not included because SNP selection and the genotyping procedures were completed before these GWAS were published. For the reported SNPs that showed strong linkage disequilibrium (LD; r2 ≥ 0.9) on the 1000 Genome Project for Southern Chinese, only one representative SNP with the most significant association was selected for investigation. SNPs that were monomorphic or with minor allele frequency of <1% in the Southern Chinese population were excluded. The majority of SNPs were genotyped using the Sequenom iPLEX Gold genotyping platform at the Centre of Genomic Sciences, the University of Hong Kong. Six SNPs were incompatible with the Sequenom multiplexing design and were therefore replaced by a proxy SNP (CDC42BPA rs3014267 replaced by rs2953655 [r2 = 0.82], UBE2E2 rs11927173 replaced by rs79941515 [r2 = 0.97], AKAP11-FABP3P2 rs238250 replaced by rs117850847 [r2 = 1], CCDC68-TCF4 rs1970671 replaced by rs12607567 [r2 = 1], COL5A1 rs6537949 replaced by rs59126004 [r2 = 1], rs10910200 replaced by rs6662352 [r2 = 1], KIAA1804-KCNK1 and HS6ST3 rs2038823 replaced by rs16953072 [r2 = 1]). Eight SNPs that failed to be genotyped using the Sequenom platform were then genotyped using the predesigned TaqMan SNP genotyping assays (rs2518344, assay ID: C_1972331_10; rs487083, assay ID: C_2379367_10; rs1224329, assay ID: C_2729878_10; rs10499298, assay ID: C_27436426_10; rs713050, assay ID: C_12026888_10; rs11867934, assay ID: C_31635648_10; rs11867934, assay ID: C_31635648_10) and the custom TaqMan SNP genotyping assay (rs6909083, assay ID: ANZTEFG). Hardy-Weinberg equilibrium for each SNP was examined by the exact test using PLINK version 1.09.26 The average successful genotyping call rate was 99.8%. All SNPs passed quality control and were included in further analysis.
Statistical Analysis
Statistical analyses were performed using PLINK version 1.0926 and IBM SPSS Statistics 24. All continuous variables were reported as mean ± standard deviation or median with interquartile range, as appropriate. Continuous variables that did not follow a normal distribution as suggested by a significant P value in the Kolmogorov-Smirnov test, were natural logarithmically transformed before the analyses. All continuous variables were standardized using the z-score formula before analysis (mean, 0 ± 1). Continuous and categorical parameters were compared between cases and controls by one-way ANOVA and χ2 tests, respectively. Only risk factors that showed a significant association in the univariate analysis or were biologically relevant were included as covariates in the adjustment model. Associations between the SNPs and STDR, or PDR in the subgroup analysis, were examined by multiple logistic regression analyses with adjustments for age, sex, BMI, duration of diabetes, the presence of HTN and HbA1c, under an additive model. Stratified analyses were performed on participants with a HbA1c of <7% and those with a HbA1c of ≥7%. Interaction analyses of SNPs showing suggestive associations with STDR or PDR (P < 0.05) and HbA1c were examined by including an interaction term of the SNP and HbA1c (dichotomized as <7% vs. ≥7%) in the multiple logistic regression model adjusted for the listed covariates. A two-tailed P value of <0.05 was considered as statistically significant. To account for multiple comparisons, the false discovery rate (FDR) correction with a cutoff of 6.25 × 10−3 (= 0.05/8 independent tests) was used. The FDR-adjusted P value, q-value, was calculated using the Benjamini-Hochberg FDR correction method.27
Results
All 69 SNPs identified from previous GWAS for DR (P < 5 × 10−4; r2 > 0.9) were successfully genotyped in 1051 STDR cases and 2042 non-STDR controls. Table 1 shows the clinical characteristics of the participants. As expected, STDR cases had a longer duration of diabetes, higher HbA1c, and with a greater proportion diagnosed with HTN, compared with the non-STDR controls. However, a higher BMI was observed in non-STDR controls. Smoking and the presence of dyslipidemia did not show any significant difference between STDR cases and non-STDR controls.
Table 1.
Clinical Characteristics of Study Participants
| Variables | Non-STDR | STDR | PDR |
|---|---|---|---|
| Number | 2,042 | 1,051 | 409 |
| Age (years) | 62.9 ± 13.0 | 63.9 ± 12.6 | 60.3 ± 10.9* |
| Sex (male %) | 57.9 | 57.0 | 58.6 |
| Diabetes duration (years)† | 7 (3–12) | 14 (6–22)* | 13 (5–22)* |
| HTN (%) | 84.6 | 93.1* | 94.7* |
| SBP (mm Hg)‡ | 143.61 ± 21.14 | 151.08 ± 21.60* | 151.68 ± 21.06* |
| DBP (mm Hg)‡ | 79.13 ± 10.08 | 78.59 ± 11.19 | 80.10 ± 11.40 |
| HbA1c (%)† | 7.0 (6.5–7.7) | 7.5 (6.7–8.5)* | 7.5 (6.8–8.6)* |
| BMI (kg/m2) | 26.17 ± 4.26 | 25.66 ± 4.07* | 25.99 ± 4.15 |
| Dyslipidemia (%) | 70.2 | 68.3 | 70.1 |
| Ever smoke (%) | 33.3 | 31.3 | 31.3 |
DBP, diastolic blood pressure; SBP, systolic blood pressure.
HTN was defined as a BP of ≥140/90 mm Hg or taking any antihypertensive drugs. Dyslipidemia was indicated by a documented history of dyslipidemia in patient's record or taking lipid-lowering drugs.
P < 0.01 when compared with non-STDR controls.
Natural log-transformed before analysis.
SBP + 10 mm Hg and DBP + 5 mm Hg if on antihypertensive drugs.
Associations of SNPs With STDR and PDR
Four SNPs showed nominal associations with STDR after adjustment for age, sex, BMI, duration of diabetes, the presence of HTN and HbA1c (Table 2). These SNPs included COL5A1 rs59126004 (P = 0.034; odds ratio [OR], 0.84; 95% confidence interval [95% CI], 0.71–0.98), IGSF21-KLHDC7A rs3007729 (P = 0.034; OR, 0.87; 95% CI, 0.77–0.99), CREB5 rs11765845 (P = 0.036; OR, 0.87; 95% CI, 0.76–0.99), and LOC728275-LOC72831 rs227455 (P = 0.048; OR, 0.89; 95% CI, 0.79–0.99). With a view to increase power and minimize phenotypic heterogeneity, a subgroup analysis was conducted by comparing the non-STDR controls (grading R0 or R1) with the PDR cases (grading R3), the most severe manifestation of DR. In the subgroup analysis, COL5A1 rs59126004 (P = 0.011; OR, 0.73; 95% CI, 0.57–0.93) and IGSF21-KLHDC7A rs3007729 (P = 0.023; OR, 0.81; 95% CI, 0.67–0.97) also showed suggestive association with PDR. Furthermore, INSR rs2115386 (P = 5.65 × 10−3; OR, 1.25; 95% CI, 1.06–1.47), UBE2E2 rs79941515 (P = 6.75 × 10−3; OR, 0.72; 95% CI, 0.57–0.91), MYT1L-LOC729897 rs10199521 (P = 8.93 × 10−3; OR, 1.24; 95% CI, 1.05–1.47), ZNRF1 rs17684886 (P = 0.022; OR, 0.82; 95% CI, 0.67–0.97), AKAP11-FABP3P2 rs117850847 (P = 0.030; OR, 0.65; 95% CI, 0.44–0.96), and CCDC68-TCF4 rs12607567 (P = 0.030; OR, 0.83; 95% CI, 0.70–0.98) showed nominal associations only with PDR but not with STDR. However, none of these SNPs were able to survive correction for multiple testing at a FDR cut off of 6.25 × 10−3. Supplementary Table S1 and Supplementary Table S2 show the results of association analysis with STDR and PDR, respectively.
Table 2.
SNPs Showing Significant Associations With STDR/PDR
| MAF | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phenotype | Nearest Gene(s) | SNP | Position | A1 | A2 | Non-STDR | STDR | OR (95% CI) | P Value* | q-Value§ |
| STDR | COL5A1 | rs59126004‡ | 9:137674341 | C | T | 0.17 | 0.14 | 0.84 (0.71–0.98) | 0.034 | 0.798 |
| IGSF21-KLHDC7A | rs3007729 | 1:18795255 | T | C | 0.32 | 0.30 | 0.87 (0.77–0.99) | 0.034 | 0.798 | |
| CREB5 | rs11765845 | 7:28391142 | A | G | 0.29 | 0.28 | 0.87 (0.76–0.99) | 0.036 | 0.798 | |
| LOC728275-LOC728316 | rs227455† | 6:165478051 | C | T | 0.48 | 0.47 | 0.89 (0.79–0.99) | 0.048 | 0.798 | |
| PDR | INSR | rs2115386† | 19:7196565 | C | T | 0.47 | 0.52 | 1.25 (1.06–1.47) | 5.65 × 10−3 | 0.190 |
| UBE2E2 | rs79941515‡ | 3:23225738 | T | C | 0.18 | 0.14 | 0.72 (0.57–0.91) | 6.75 × 10−3 | 0.190 | |
| MYT1L-LOC729897 | rs10199521† | 2:2519513 | T | C | 0.35 | 0.39 | 1.24 (1.05–1.47) | 8.93 × 10−3 | 0.190 | |
| COL5A1 | rs59126004‡ | 9:137674341 | C | T | 0.17 | 0.13 | 0.73 (0.57–0.93) | 0.011 | 0.190 | |
| ZNRF1 | rs17684886† | 16:75086875 | A | T | 0.47 | 0.43 | 0.82 (0.69–0.97) | 0.022 | 0.259 | |
| IGSF21-KLHDC7A | rs3007729 | 1:18795255 | T | C | 0.32 | 0.29 | 0.81 (0.67–0.97) | 0.023 | 0.259 | |
| AKAP11-FABP3P2 | rs117850847 | 13:42909215 | A | C | 0.06 | 0.04 | 0.65 (0.44–0.96) | 0.030 | 0.259 | |
| CCDC68-TCF4 | rs12607567† | 18:52858659 | G | A | 0.47 | 0.44 | 0.83 (0.70–0.98) | 0.030 | 0.259 | |
A1, minor allele; A2, major allele; MAF, minor allele frequency.
SNPs are ranked by P value. Chromosomal position corresponds with human reference genome hg19. OR corresponds with the minor allele.
Adjusted for age, sex, BMI, duration of diabetes, the presence of HTN, and HbA1c.
Independent FDR correction for each test.
Direction of effect consistent with original report.
Direction of effect not available in original report.
Stratified Analysis by Dichotomized HbA1c
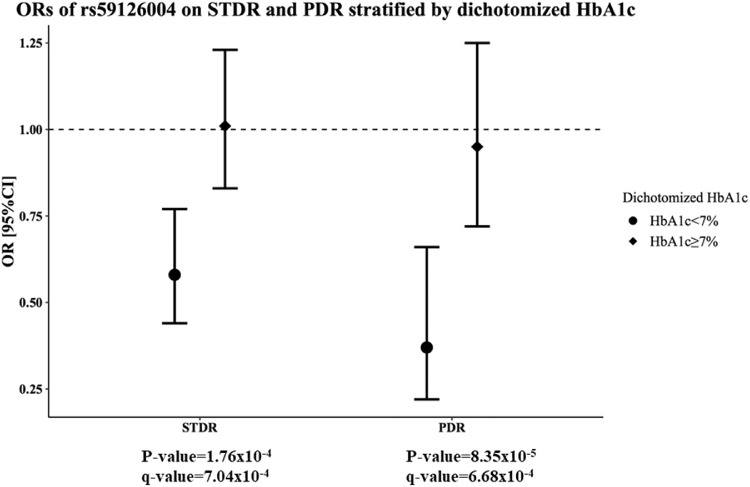
We then performed the stratified analysis in participants with adequate glycemic control (HbA1c <7%) or inadequate glycemic control (HbA1c ≥7%) for the four SNPs, which showed suggestive associations with STDR. The C allele of COL5A1 rs59126004 was only associated with a reduced risk of STDR in patients with HbA1c level of <7% (P = 1.76 × 10−4; OR, 0.58; 95% CI, 0.44–0.77), but not in patients with a HbA1c of ≥7% (Table 3 and Figure). This association was able to survive the correction for multiple testing at FDR ≤6.25 × 10−3 (q-value = 7.04 × 10−4). In the subgroup analysis for PDR, COL5A1 rs59126004 showed an even more evident protective effect in patients with a HbA1c level of <7% (P = 8.35 × 10−5; OR, 0.37; 95% CI, 0.22–0.60), but a significant association in patients with a HbA1c of ≥7% was not observed (Table 3 and Figure). Again, this association was able to survive the correction for multiple testing at an FDR of ≤6.25 × 10−3 (q-value = 6.68 × 10−4). However, none of the other SNPs was able to survive correction for multiple testing at an FDR of ≤6.25 × 10−3.
Table 3.
Significant Findings in Interaction Analysis of SNP × Dichotomized HbA1c for STDR/PDR
| STDR | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HbA1c <7% (case/control = 328/962) | HbA1c ≥7% (case/control = 723/1,080) | ||||||||||
| Nearest Gene(s) | SNP | A1 | A2 | OR (95% CI) | P -Value* | q-Value§ | OR (95% CI) | P -Value* | q-Value§ | P interaction * | q-Value§ |
| COL5A1 | rs59126004 | C | T | 0.58 (0.44–0.77) | 1.76 × 10−4 | 7.04 x10−4 | 1.01 (0.83–1.23) | 0.893 | 0.893 | 1.73 × 10−3 | 6.92 × 10−3 |
| CREB5 | rs11765845 | A | G | 0.82 (0.66–1.01) | 0.069 | 0.092 | 0.90 (0.77–1.06) | 0.219 | 0.292 | 0.433 | 0.633 |
| IGSF21-KLHDC7A | rs3007729 | T | C | 0.81 (0.66–1.00) | 0.056 | 0.092 | 0.89 (0.76–1.04) | 0.143 | 0.286 | 0.523 | 0.633 |
| LOC728275-LOC728316 | rs227455 | C | T | 0.92 (0.76–1.12) | 0.434 | 0.434 | 0.86 (0.75–0.99) | 0.047 | 0.188 | 0.633 | 0.633 |
| PDR | |||||||||||
| HbA1c <7% (case/control = 121/962) | HbA1c ≥7% (case/control = 288/1080) | ||||||||||
| Nearest Gene(s) | SNP | A1 | A2 | OR (95% CI) | P -Value * | q-Value § | OR (95% CI) | P -Value * | q-Value § | Pinteraction * | q-Value § |
| COL5A1 | rs59126004 | C | T | 0.37 (0.22–0.60) | 8.35 × 10−5 | 6.68 × 10−4 | 0.95 (0.72–1.25) | 0.727 | 0.727 | 1.73 × 10−3 | 0.014 |
| MYT1L-LOC729897 | rs10199521 | T | C | 1.13 (0.84–1.51) | 0.397 | 0.454 | 1.32 (1.08–1.62) | 6.51 × 10−3 | 0.036 | 0.392 | 0.843 |
| CCDC68-TCF4 | rs12607567 | G | A | 0.93 (0.70–1.23) | 0.619 | 0.619 | 0.81 (0.67–0.99) | 0.047 | 0.075 | 0.478 | 0.843 |
| INSR | rs2115386 | C | T | 1.17 (0.89–1.55) | 0.241 | 0.386 | 1.3 (1.06–1.58) | 9.03 × 10−3 | 0.036 | 0.519 | 0.843 |
| IGSF21-KLHDC7A | rs3007729 | T | C | 0.85 (0.62–1.15) | 0.297 | 0.396 | 0.77 (0.62–0.96) | 0.023 | 0.046 | 0.527 | 0.843 |
| ZNRF1 | rs17684886 | A | T | 0.81 (0.60–1.09) | 0.172 | 0.386 | 0.84 (0.69–1.02) | 0.086 | 0.098 | 0.764 | 0.979 |
| UBE2E2 | rs79941515 | T | C | 0.71 (0.48–1.05) | 0.094 | 0.376 | 0.71 (0.53–0.94) | 0.020 | 0.046 | 0.967 | 0.979 |
| AKAP11-FABP3P2 | rs117850847 | A | C | 0.64 (0.31–1.32) | 0.233 | 0.386 | 0.67 (0.43–1.05) | 0.085 | 0.098 | 0.979 | 0.979 |
A1, minor allele; A2, major allele; MAF, minor allele frequency.
SNPs are ranked by Pinteraction. Chromosomal position corresponds with human reference genome hg19. OR corresponds with the minor allele.
Adjusted for age, sex, BMI, duration of diabetes and the presence of HTN.
Independent FDR correction for each test.
Figure.
ORs of COL5A1 rs59126004 in the stratified analyses for STDR and PDR.
Interactions Between DR Susceptibility SNPs and Dichotomized HbA1c
We then further conducted the gene–environment interaction analysis. COL5A1 rs59126004 showed a significant interaction with dichotomized HbA1c on the risk of STDR (Pinteraction = 1.733 × 10−3; q-value = 6.92 × 10−3) (Table 3). In the subgroup analysis for PDR, COL5A1 rs59126004 yielded similar interaction with dichotomized HbA1c on PDR risk (Pinteraction = 1.729 × 10−3; q-value = 0.014) (Table 3). However, the interactions between COL5A1 rs59126004 and dichotomized HbA1c on the risk of both STDR and PDR were slightly attenuated after correction for multiple comparisons by the FDR correction. None of the other SNPs showed a significant interaction with dichotomized HbA1c on the risk of STDR or PDR.
Discussion
In this study, we conducted the first evaluation on the possible gene–environment interactions between genetic variants identified from recent GWAS and the effect of glycemic control, as indicated by HbA1c, on the risk of severe DR (STDR and PDR). The current study showed suggestive associations of several susceptibility variants with severe DR and demonstrated that HbA1c could modify the genetic susceptibility to severe DR. Among the SNPs investigated, COL5A1 rs59126004 provided the most convincing results in the current study. In the stratified analysis, the protective effect of COL5A1 rs59126004 against both STDR and PDR was clearly demonstrated in patients with adequate glycemic control (HbA1c <7%) but was not observed in those with inadequate glycemic control (HbA1c ≥7%).
Hyperglycemia plays a prominent role in a cascade of damaging molecular and cellular effects, such as oxidative stress, abnormal glycosylation, and inflammation, that may contribute to the development and severity of DR.28 Glycemic control is a crucial modifiable risk factor for DR. In this study, we showed that COL5A1 rs59126004 was nominally associated with a decreased risk of severe DR, even after adjustment for the traditional risk factors. In the stratified analysis, we showed that individuals with adequate glycemic control had a 42% and 63% decreased risk of STDR and PDR, respectively, for carrying each additional C allele of rs59126004. In contrast, the protective effect of this SNP against STDR or PDR was not observed in individuals who had inadequate glycemic control. Such observation suggested the possible interaction between COL5A1 rs59126004 and HbA1c. Indeed, suggestive interactions between COL5A1 rs59126004 and HbA1c on the risk of STDR and PDR were observed in the current study. Our data showed that the protective effect of this SNP was of a greater magnitude in those who had achieved the recommended target of adequate glycemic control, thereby provided further support to encourage a good glycemic control in patients with T2DM. Understanding the role of gene–environment interactions might have potential implications for the management of T2DM patients.
rs59126004 is located at the intronic region of COL5A1 and is in complete LD with rs6537949, the originally reported SNP that was identified in the subanalysis of a GWAS meta-analysis for severe DR (Pmeta = 4.7 × 10−5) in patients with T1DM.10 However, because the direction of effect was not indicated in the original report, whether rs59126004 (i.e., a proxy for rs6537949) has shown the consistent direction of effect as in the original study could not be determined. A follow-up in silico analysis suggested that the COL5A1 rs6537949 maps to a region with transcriptional binding signals for the CCCTC-binding factor (CTCF) in the ENCODE Consortium ChIP-seq data.29 The binding affinity of CTCF was found to be decreased by the G allele of rs6537949, which show strong linkage with the protective C allele of rs59126004 reported in the current study (r2 = 1). CTCF is a highly conserved zinc finger protein that has a multifunctional effect on gene regulation.30 For instance, it can function as both a transcriptional activator and repressor, a genomic insulator, and a mediator of long-range genomic interactions.30 Gene expression profile suggested that CTCF is highly expressed in multiple eye structures, which implies its essential role in eye development.31 CTCF has been reported to influence retinal cell differentiation through downregulation of Pax6 expression.32 A previous study demonstrated that CTCF restrained retinal angiogenesis by preventing the enhancer-mediated activation of the vascular endothelial growth factor gene.33 The role of COL5A1 in the pathogenesis of DR is yet to be elucidated. COL5A1 encodes an important component of type V collagen, the collagen type V alpha 1 chain. Type V collagen appears to play a critical role in fibrogenesis.34 Type V collagen has also been reported to inhibit angiogenesis via inhibition of endothelial budding in an angiogenesis assay.35 Variants of COL5A1 were shown to be associated with central cornea thickness.36 A patient with classic Ehlers–Danlos syndrome carrying a missense mutation (p.Gly1393Asp) in COL5A1 was reported to show features of T2DM and retinopathy.37 Furthermore, COL5A1 has been suggested to be involved in several pathways, such as the extracellular matrix–receptor interaction pathway, that were upregulated in the active and inactive fibrovascular membranes associated with PDR.38 We speculate that COL5A1 rs6537949 might perturb the binding of CTCF, thereby influencing the expression of COL5A1 or its neighboring genes, and ultimately the risk of DR. Further functional studies to elucidate the role of CTCF and COL5A1 in the pathogenesis of DR are warranted.
It is recognized that the small effect of genetic variants can be masked from detection in direct analysis for gene-to-disease association owing to genotype-specific environmental effects on the disease status.39 Our data have provided evidence that HbA1c might modify the genetic susceptibility to severe DR. These findings may give novel insight into the pathophysiologic mechanism of DR and help to stratify patients into different risk groups for personalized clinical management. Further studies in independent cohorts to validate our findings are required before definitive conclusions can be drawn. Consideration of different environmental exposures, such as high blood pressure and obesity, is recommended in future studies, because some variants may only demonstrate the substantial effect when specific environmental exposure is present.
Several limitations of this study should be acknowledged. First, the current study adopted a cross-sectional rather than a prospective study design and hence causality could not be established. Second, the current study has not included SNPs reported in two recently published GWAS,16,17 including the largest GWAS to date.16 A more comprehensive list of study SNPs would have strengthened the current study. Third, because the majority of the studied SNPs were unable to achieve the genome-wide significant level in the original reports, some of them could have been false-positive findings. Fourth, the measurement of HbA1c was a one-time measurement at the time of blood sample collection. We acknowledged that a one-time measurement of HbA1c might not necessarily be reflective of the glycemic control over the patients’ diabetes duration. This factor may be particularly relevant in individuals with PDR who may have put more efforts to maintain a good glycemic control after the diagnosis of PDR, owing to a fear of vision loss. Therefore, it is possible that the effect of glycemic control on the genetic susceptibility might be more pronounced if the mean HbA1c over the patient's diabetes duration was used in the analysis. Our study would have been strengthened with the use, in the analysis, of mean HbA1c over the patients’ diabetes duration. However, such information was not available in a large proportion of participants. Future studies examining the effect of glycemic control on the genetic susceptibility should consider the use of mean HbA1c. Fifth, we noticed that the English National Screening guideline was developed to screen for DR, but not for diagnosis or staging. Misclassification was, therefore, possible. However, all STDR cases have been assessed systematically by specialist ophthalmologists to determine the severity of DR. Sixth, the small sample size of the current study, in particular for the stratified analysis, was insufficient to detect SNPs with a modest effect on severe DR. Last, the current study also lacks an independent replication of the findings on association and interaction. Further independent studies with larger sample size would serve to establish more associations and gene–environment interactions with statistical confidence. Nonetheless, one of the strengths of this study was the well-characterized phenotypes of our participants. Furthermore, potential confounding factors that strongly correlate with STDR were taken into account as covariates in the multiple logistic regression model.
In conclusion, our data provided supportive evidence for possible interactions between HbA1c and genetic variants on the risk of severe DR in Chinese patients with T2DM. Our findings may provide novel insight into the pathophysiologic mechanism of DR, which may in turn lead to the design of more effective prevention and treatment strategies.
Supplementary Material
Acknowledgments
The authors thank the research and clinical staffs of the diabetes and ophthalmology clinics of the Queen Mary Hospital, Tseung Kwan O Hospital and United Christian Hospital, and the participants of the current study, for their valuable contributions. This study was supported by the Health and Medical Research Fund (HMRF) of the Food and Health Bureau, HKSAR (Project No. 03144016), to CCYY.
Disclosure: K.K.K. Ng, None; C.Y.Y. Cheung, None; C.-H. Lee, None; C.H.Y. Fong, None; K.H.M. Kwok, None; K.K.W. Li, None; R.A. Gangwani, None; I.Y.H. Wong, None; Y.-C. Woo, None; W.-S. Chow, None; M.M.A. Yuen, None; R.L.C. Wong, None; A. Xu, None; D.S.H. Wong, None; P.-C. Sham, None; K.S.L. Lam, None
References
- 1. Klein BE. Overview of epidemiologic studies of diabetic retinopathy. Ophthalmic Epidemiol. 2007; 14: 179–183. [DOI] [PubMed] [Google Scholar]
- 2. Yau JWY, Rogers SL, Kawasaki R, et al.. Global prevalence and major risk factors of diabetic retinopathy. Diabetes Care. 2012; 35: 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang L, Krzentowski G, Albert A, Lefebvre PJ. Risk of developing retinopathy in Diabetes Control and Complications Trial type 1 diabetic patients with good or poor metabolic control. Diabetes Care. 2001; 24: 1275–1279. [DOI] [PubMed] [Google Scholar]
- 4. Holman RR, Paul SK, Bethel MA, Matthews DR, Neil HA. 10-year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med. 2008; 359: 1577–1589. [DOI] [PubMed] [Google Scholar]
- 5. Manley S. Haemoglobin A1c–a marker for complications of type 2 diabetes: the experience from the UK Prospective Diabetes Study (UKPDS). Clin Chem Lab Med. 2003; 41: 1182–1190. [DOI] [PubMed] [Google Scholar]
- 6. American Diabetes Association. 6. Glycemic targets: standards of medical care in diabetes-2018. Diabetes Care. 2018; 41: S55–S64. [DOI] [PubMed] [Google Scholar]
- 7. Klein R, Klein BE, Moss SE, Cruickshanks KJ. The Wisconsin Epidemiologic Study of Diabetic Retinopathy: XVII. The 14-year incidence and progression of diabetic retinopathy and associated risk factors in type 1 diabetes. Ophthalmology. 1998; 105: 1801–1815. [DOI] [PubMed] [Google Scholar]
- 8. Hallman DM, Huber JC Jr. Gonzalez VH, Klein BE, Klein R, Hanis CL. Familial aggregation of severity of diabetic retinopathy in Mexican Americans from Starr County, Texas. Diabetes Care. 2005; 28: 1163–1168. [DOI] [PubMed] [Google Scholar]
- 9. Fu YP, Hallman DM, Gonzalez VH, et al.. Identification of diabetic retinopathy genes through a genome-wide association study among Mexican-Americans from Starr County, Texas. J Ophthalmol. 2010; 2010: 861291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grassi MA, Tikhomirov A, Ramalingam S, Below JE, Cox NJ, Nicolae DL. Genome-wide meta-analysis for severe diabetic retinopathy. Hum Mol Genet. 2011; 20: 2472–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang YC, Lin JM, Lin HJ, et al.. Genome-wide association study of diabetic retinopathy in a Taiwanese population. Ophthalmology. 2011; 118: 642–648. [DOI] [PubMed] [Google Scholar]
- 12. Sheu WH, Kuo JZ, Lee IT, et al.. Genome-wide association study in a Chinese population with diabetic retinopathy. Hum Mol Genet. 2013; 22: 3165–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin HJ, Huang YC, Lin JM, Wu JY, Chen LA, Tsai FJ. Association of genes on chromosome 6, GRIK2, TMEM217 and TMEM63B (linked to MRPL14) with diabetic retinopathy. Ophthalmologica. 2013; 229: 54–60. [DOI] [PubMed] [Google Scholar]
- 14. Awata T, Yamashita H, Kurihara S, et al.. A genome-wide association study for diabetic retinopathy in a Japanese population: potential association with a long intergenic non-coding RNA. PLoS ONE. 2014; 9: e111715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burdon KP, Fogarty RD, Shen W, et al.. Genome-wide association study for sight-threatening diabetic retinopathy reveals association with genetic variation near the GRB2 gene. Diabetologia. 2015; 58: 2288–2297. [DOI] [PubMed] [Google Scholar]
- 16. Pollack S, Igo RP, Jensen RA, et al.. Multiethnic genome-wide association study of diabetic retinopathy using liability threshold modeling of duration of diabetes and glycemic control. Diabetes. 2018; 68: 441–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Graham PS, Kaidonis G, Abhary S, et al.. Genome-wide association studies for diabetic macular edema and proliferative diabetic retinopathy. BMC Med Genet. 2018; 19: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grassi MA, Tikhomirov A, Ramalingam S, et al.. Replication analysis for severe diabetic retinopathy. Invest Ophthalmol Vis Sci. 2012; 53: 2377–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McAuley AK, Wang JJ, Dirani M, Connell PP, Lamoureux E, Hewitt AW. Replication of genetic loci implicated in diabetic retinopathy. Invest Ophthalmol Vis Sci. 2014; 55: 1666–1671. [DOI] [PubMed] [Google Scholar]
- 20. Hosseini SM, Boright AP, Sun L, et al.. The association of previously reported polymorphisms for microvascular complications in a meta-analysis of diabetic retinopathy. Hum Genet. 2015; 134: 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peng D, Wang J, Zhang R, et al.. Common variants in or near ZNRF1, COLEC12, SCYL1BP1 and API5 are associated with diabetic retinopathy in Chinese patients with type 2 diabetes. Diabetologia. 2015; 58: 1231–1238. [DOI] [PubMed] [Google Scholar]
- 22. Cheung CY, Hui EY, Lee CH, et al.. Impact of genetic loci identified in genome-wide association studies on diabetic retinopathy in Chinese patients with type 2 diabetes. Invest Ophthalmol Vis Sci. 2016; 57: 5518–5524. [DOI] [PubMed] [Google Scholar]
- 23. Woo YC, Cheung BM, Yeung CY, et al.. Cardiometabolic risk profile of participants with prediabetes diagnosed by HbA1c criteria in an urban Hong Kong Chinese population over 40 years of age. Diabet Med. 2015; 32: 1207–1211. [DOI] [PubMed] [Google Scholar]
- 24. Scanlon PH. The English National Screening Programme for diabetic retinopathy 2003-2016. Acta Diabetol. 2017; 54: 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lian JX, Gangwani RA, McGhee SM, Chan CK, Lam CL, Wong DS. Systematic screening for diabetic retinopathy (DR) in Hong Kong: prevalence of DR and visual impairment among diabetic population. Br J Ophthalmol. 2016; 100: 151–155. [DOI] [PubMed] [Google Scholar]
- 26. Purcell S, Neale B, Todd-Brown K, et al.. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001; 125: 279–284. [DOI] [PubMed] [Google Scholar]
- 28. Hampton BM, Schwartz SG, Brantley MA Jr. Flynn HW Jr.. Update on genetics and diabetic retinopathy. Clin Ophthalmol. 2015; 9: 2175–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dunham I, Kundaje A, Aldred SF, et al.. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009; 137: 1194–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burke LJ, Hollemann T, Pieler T, Renkawitz R. Molecular cloning and expression of the chromatin insulator protein CTCF in Xenopus laevis. Mech Dev. 2002; 113: 95–98. [DOI] [PubMed] [Google Scholar]
- 32. Canto-Soler MV, Huang H, Romero MS, Adler R. Transcription factors CTCF and Pax6 are segregated to different cell types during retinal cell differentiation. Dev Dyn. 2008; 237: 758–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tang M, Chen B, Lin T, et al.. Restraint of angiogenesis by zinc finger transcription factor CTCF-dependent chromatin insulation. Proc Natl Acad Sci USA. 2011; 108: 15231–15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mak KM, Png CY, Lee DJ. Type V collagen in health, disease, and Fibrosis. Anat Rec (Hoboken). 2016; 299: 613–629. [DOI] [PubMed] [Google Scholar]
- 35. Spencer M, Unal R, Zhu B, et al.. Adipose tissue extracellular matrix and vascular abnormalities in obesity and insulin resistance. J Clin Endocrinol Metab. 2011; 96: E1990–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vitart V, Bencic G, Hayward C, et al.. New loci associated with central cornea thickness include COL5A1, AKAP13 and AVGR8. Hum Mol Genet. 2010; 19: 4304–4311. [DOI] [PubMed] [Google Scholar]
- 37. Ritelli M, Dordoni C, Venturini M, et al.. Clinical and molecular characterization of 40 patients with classic Ehlers-Danlos syndrome: identification of 18 COL5A1 and 2 COL5A2 novel mutations. Orphanet J Rare Dis. 2013; 8: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gong MT, Li WX, Zhang Q, et al.. Comprehensive analysis of gene expression profiles associated with proliferative diabetic retinopathy. Exp Ther Med. 2018; 16: 3539–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007; 63: 111–119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.