Abstract
Background
A number of researches focused on the study of tumors have concluded that the expression level of lncRNA NKILA was decreased in different tumors. This is an indication that NKILA might influence the start and growth of a cancer. In addition, studies have fatalities and worsening health of cancer patients is associated with a reduced level of NKILA.
Results
The results are the collective screening of nine total studies which included 937 cancer patients. The prognosis of the meta-analysis indicated that cancer patients with a higher expression of NKILA had an overall longer survival (OS) (HR = 0.808, 95% CI: 0.736, 0.887); with regard to the clinical prognosis, the results indicated that reduced NKILA was associated with advanced clinical stage (OR = 0.313, 95% CI: 0.225, 0.434), poor histological grades (OR = 0.833, 95% CI: 0.508, 1.367), positive lymph node metastasis (OR = 0.253, 95% CI: 0.144, 0.444), and additional tumor invasion depth (OR = 0.326, 95% CI: 0.234, 0.454).
Materials and Methods
Related research conducted was accessed by searching in PubMed and Web of Science with the keywords. The accessed material was till the 25th of February, 2020. The present quantitative meta-analysis was done using Stata SE12.0. The aim of the meta-analysis was to investigate the relationship between NKILA expression level and clinical prognosis.
Conclusions
In the result of this meta-analysis, decreased NKILA expression is typical of different kinds of cancer. Moreover, it can perform as a predictive element of prognosis in varied kinds of cancer. Nonetheless, till now, it is deemed essential to carry out larger-size as well as better designed research works for the confirmation of our findings.
1. Introduction
Long noncoding RNAs (lncRNAs) are involved in a number of physiological and pathological processes. They belong to a large class of RNAs with a capacity for non-protein-coding. In other words, they are defined as transcripts with lengths that are greater than two hundred bases [1]. A majority of lncRNAs exhibit similar characteristics when exerting biological functions. They achieve this by regulating various aspects of gene expression [2, 3]. Even though accurate mechanisms by which lncRNAs function has not been explained to date, research has indicated that a majority of the lncRNAs exhibit similar characteristics which are important such as proliferation [4], apoptosis [5], metastasis [6], metabolism [7], senescence [8], and drug resistance [9]. With regard to their functioning mechanism, a model which has been widely proposed is that lncRNAs may be functioning through discrete modules that decoy, guide, or scaffold other regulatory proteins [1].
Some have the potential to result in prooncogene. Contrastingly, other studies have also introduced low expression with regard to the performance of functions that suppress tumors [10–13]. The present study emphasises on the lncRNA, which is known as NKILA, a NF-κB-induced lncRNA; in comparison to others, it is a novel form of lncRNA that maps to chromosome 20q13 [14, 15]. Studies have documented that it has useful functionality in suppressing the growth of cancer genes in a variety of cancers, such as esophageal squamous cell carcinoma [16], laryngeal cancer [17], nasopharyngeal carcinoma [18], rectal cancer [19], tongue squamous cell carcinoma [20], and colorectal cancer [21].
NKILA, whose expression is induced by NF-κB, attaches to NF-κB/IκB and obscures phosphorylation motifs of IκB; as a result, the actions of IKK-mediated IκB phosphorylation and NF-κB are suppressed. Eventually, it assumes the role of suppressing tumors in various cancers, like esophageal squamous cell carcinoma [16], laryngeal cancer [17], nasopharyngeal carcinoma [18], and hepatocellular carcinoma [22]. Moreover, NKILA-mediated NF-κB activation can be regarded as a therapeutic molecule since it inhibits EMT and suppresses the advancement of cancer; this is evident in breast cancer [23], hepatocellular carcinoma [24], osteosarcoma [25], and non-small-cell lung cancer [26]. Meanwhile, it was demonstrated by Liu and Shi that the role of NKILA in the deterioration of NSCLC is dependent on the IL-11/STAT3 signalling [27]. In the meantime, it was also demonstrated that lncRNA was related to the immune evasion of tumors. In addition, an investigation by Huang et al. revealed that NKILA preferentially sensitizes antitumor T cells to cell death following activation by tumor antigens [28]. The study demonstrated that reducing the activity of NKILA intensifies the infiltration of CTL which results in the inhabitation in the expansion and development of the tumor. As a result, it indicates a potential methodology for improving the effectiveness of T cells in adoptive T cell therapy in the treatment of cancers [28].
In order to validate the medical usefulness and effectivity as a biomarker or therapeutic target, it is vital to study the relationship of NKILA expression level to any pathological characteristics. The work done in the present study is aimed at fulfilling an organized review and performing a quantitative meta-analysis to investigate the relationship between the expression level of NKILA and various forms of cancers occurring in humans.
2. Results
2.1. Data Selection and Characteristics
The present study analyses the collective results of nine researches involving 937 cancer patients in total. All participants were accommodated according to the criteria for being suitable participants for the study. All the research work originated from China. Two studies emphasised on hepatocellular carcinoma, one study was related to colorectal cancer, whilst one other emphasised on rectal cancer, whilst another study was associated with osteosarcoma. Apart from these, the other studies, respectively, concentrated on laryngeal cancer, non-small-cell lung cancer, tongue squamous cell carcinoma, esophageal squamous cell carcinoma, and nasopharyngeal carcinoma. Quantitative reverse transcription polymerase chain reaction (RT-qPCR) was applied to detect NKILA. Based on these results, the patients were grouped as having either high NKILA expression or low NKILA expression. Table 1 lists the summarized characteristics of the studies whilst Figure 1 depicts the flow chart of the research study and selection.
Table 1.
Clinical characteristics of the included studies.
| Surname (year) | Country | Cancer type | Sample size | High | Low | Cutoff (high/low) | Detection method | Clinical features | Quality score | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Fei Tao (2018) | China | Rectal cancer | 152 | 76 | 76 | Median | qRT-PCR | C; T; H | 7 | [19] |
| Guodong Zhang (2019) | China | Osteosarcoma | 60 | 30 | 30 | Median | qRT-PCR | C; L | 6 | [25] |
| Tao Yang (2017) | China | Laryngeal cancer | 65 | 33 | 32 | Median | qRT-PCR | C; L; H | 7 | [17] |
| Zhiliang Lu (2017) | China | NSCLC | 110 | 55 | 55 | Median | qRT-PCR | C; H; T; L | 8 | [26] |
| Wei Huang (2016) | China | TSCC | 96 | 44 | 52 | Mean | qRT-PCR | C; H; T | 7 | [20] |
| Peng Jiang (2019) | China | CRC | 173 | 71 | 102 | Mean | qRT-PCR | C; H; T | 6 | [21] |
| Shun Ke (2018) | China | ESCC | 137 | 68 | 69 | Median | qRT-PCR | C; H | 8 | [16] |
| Ronggao Chen (2020) | China | HCC | 90 | 45 | 45 | Median | qRT-PCR | H | 7 | [24] |
| Xiaolan Yu (2018) | China | HCC | 54 | 27 | 27 | Median | qRT-PCR | NA | 7 | [22] |
NSCLC: non-small-cell lung cancer; TSCC: tongue squamous cell carcinoma; CRC: colorectal cancer; ESCC: esophageal squamous cell carcinoma; HCC: hepatocellular carcinoma; C: clinical stage; H: histological grade; L: lymph node metastasis; T: tumor invasion depth; NA: not available.
Figure 1.
Flowchart used for selecting studies for inclusion.
2.2. Association between NKILA Expression and Pathological Features
2.2.1. Clinical Stage
A total of 7 researches offered reports in the relationship between lncRNA NKILA expression and clinical stage (III/IV versus I/II). The heterogeneity of the considered researches was found to be not statistically significant (P > 0.05, I2 = 0.00%). As a result, the fixed-effects model was utilized to perform the calculation of the pooled OR, combined with its 95% CI, which amounted to be significantly different (OR = 0.313, 95% CI (0.225, 0.4234), and P ≤ 0.001) (Figure 2, Table 2). This result implied that a low expression level of NKILA was indeed related with advanced clinical stage.
Figure 2.
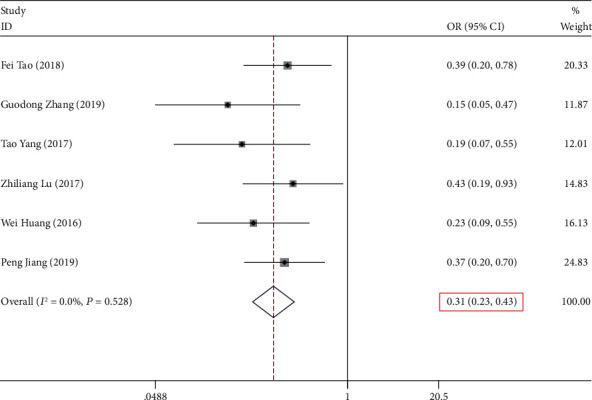
Forest plot for the association between NKILA expression and clinical stage in human cancers.
Table 2.
Meta-analysis results for the association of overexpressed NKILA with clinicopathological parameters.
| Clinicopathological parameters | Studies (n) | Numbers of patients | OR (95% CI) | P value | Heterogeneity | ||
|---|---|---|---|---|---|---|---|
| I 2 | pH | Model | |||||
| Clinical stage (III/IV vs. I/II) | 6 | 652 | 0.313 (0.225, 0.434) | ≤0.001 | 0.00% | >0.05 | Fixed effects |
| Histological grade (poorly/others vs. well/moderately) | 5 | 658 | 0.661 (0.478, 0.914) | ≤0.05 | 17.30% | >0.05 | Fixed effects |
| Lymph node metastasis (+ vs. -) | 3 | 231 | 0.253 (0.144, 0.444) | ≤0.001 | 0.00% | >0.05 | Fixed effects |
| Tumor invasion depth (T3/T4 vs. T1/T2) | 5 | 664 | 0.326 (0.234, 0.454) | ≤0.001 | 0.00% | >0.05 | Fixed effects |
2.2.2. Histological Grade
A collection of five studies reported the relationship between expression and histological grade. It was determined that the heterogeneity in the considered studies was not statistically significant (P > 0.05, I2 = 17.30%). As a result, the random-effects model was utilized to perform the calculation of the pooled OR, combined with its 95% CI, which was significantly different (OR = 0.661, 95% CI (0.478, 0.914), and P ≤ 0.05) (Figure 3, Table 2). This indicates that expression was significantly distinguished with the histological grade.
Figure 3.
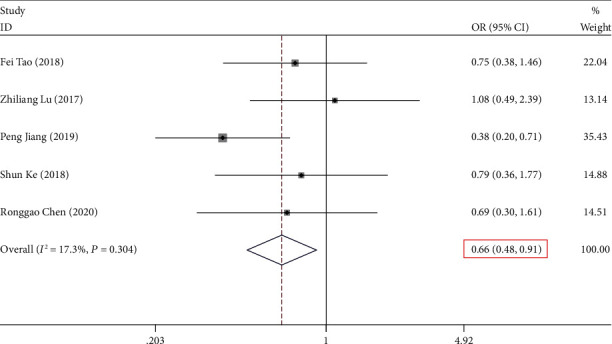
Forest plot for the association between NKILA expression and histological grade in human cancers.
2.2.3. Lymph Node Metastasis
A collection of three studies presented findings on the relationship between NKILA expression and lymph node metastasis. The heterogeneity in the collective results did not appear as statistically significant (P > 0.05, I2 = 0.00%). Consequently, fixed-effects model was utilized to perform the calculation of the pooled OR, combined with its 95% CI, which was significantly different (OR = 0.253, 95% CI (0.144, 0.444), and P ≤ 0.001) (Figure 4, Table 2). The results imply that the cohort of low NKILA expression level was susceptible to a heightened risk of lymph node metastasis in comparison with that of high NKILA expression level.
Figure 4.
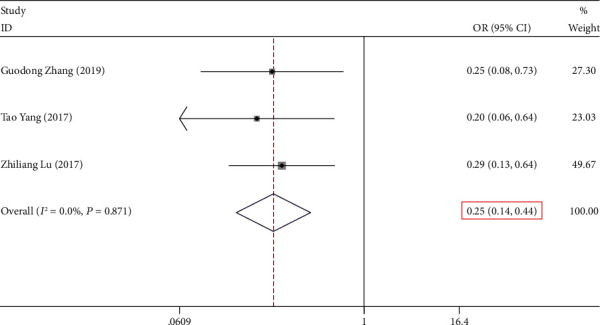
Forest plot for the association between NKILA expression and lymph node metastasis in human cancers.
2.2.4. Tumor Invasion Depth (T)
The collective results of six studies reported the relationship between NKILA expression and tumor invasion depth. The heterogeneity analysis in the collective results did not appear to be statistically significant (P > 0.05, I2 = 0.00%). As a result, the fixed-effects model was utilized to perform the calculation of the pooled OR, combined with its 95% CI; it was determined to be significantly different (OR = 0.326, 95% CI (0.234, 0.454), and P ≤ 0.001) (Figure 5, Table 2). Based on this result, in comparison with that of high NKILA expression level, the cohort of low NKILA expression level possessed a heightened risk of deep tumor attack (T3 stage or above).
Figure 5.
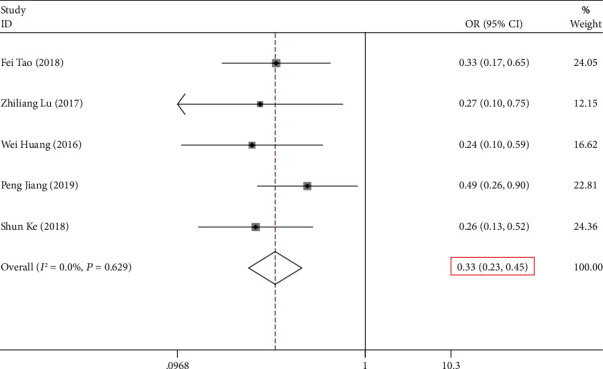
Forest plot for the association between NKILA expression and tumor invasion depth in human cancers.
2.3. Association between NKILA Expression and Survival in Different Types of Cancers
The collective results of seven studies involving 767 patients were used to evaluate the effect of NKILA overexpression on OS in various cancers. Overall survival characteristics of the included studies are shown in Table 3.The results suggested that elevated low NKILA expression projected a deteriorated result for OS with regard to six forms of cancer (pooled HR = 0.808, 95% CI 0.736, 0.887) with heterogeneity (I2 = 61.50%, P ≤ 0.05). Moreover, according to the number of participants in the study, a subgroup analysis was also performed. Evidently, a strong relationship between NKILA low expression and poor OS was observed in the studies associated with a participant number of less than 160 (pooled HR = 0.517, 95% CI 0.404, 0.661) (Figure 6). In comparison with the group of high NKILA expression, the group of low NKILA expression exhibited a substantial statistical reduction in OS whilst exhibiting a correlation with the clinically adverse outcomes.
Table 3.
Overall survival characteristics of the included studies.
| Surname (year) | Country | Cancer type | Survival analysis | HR statistic | Hazard ratios (95% CI) | Follow-up months | Outcome |
|---|---|---|---|---|---|---|---|
| Tao Yang (2018) | China | Laryngeal cancer | Univariate Multivariate |
Data in paper | 0.389 (0.152, 0.993) | 60 | OS |
| Wei Huang (2016) | China | TSCC | Univariate Multivariate |
Data in paper | 0.476 (0.236, 0.975) | 80 | OS, DFS |
| Fei Tao (2018) | China | Rectal cancer | Univariate Multivariate |
Data in paper | 0.540 (0.350, 0.830) | 96 | OS |
| Peng Jiang (2019) | China | CRC | Univariate Multivariate |
Data in paper | 0.870 (0.787, 0.962) | 72 | OS, DFS |
| Xiaolan Yu (2018) | China | HCC | NA | Survival curves | 0.550 (0.180, 1.670) | 60 | OS |
| Shun Ke (2018) | China | ESCC | Univariate Multivariate |
Data in paper | 0.590 (0.360, 0.950) | 100 | OS, DFS |
| Ronggao Chen (2019) | China | HCC | Univariate Multivariate |
Data in paper | 0.454 (0.251, 0.822) | NA | OS |
TSCC: tongue squamous cell carcinoma; CRC: colorectal cancer; HCC: hepatocellular carcinoma; ESCC: esophageal squamous cell carcinoma; NPC: nasopharyngeal carcinoma; Univariate: univariate analysis; Multivariate: multivariate analysis; NA: not available; OS: overall survival; DFS: disease-free survival.
Figure 6.
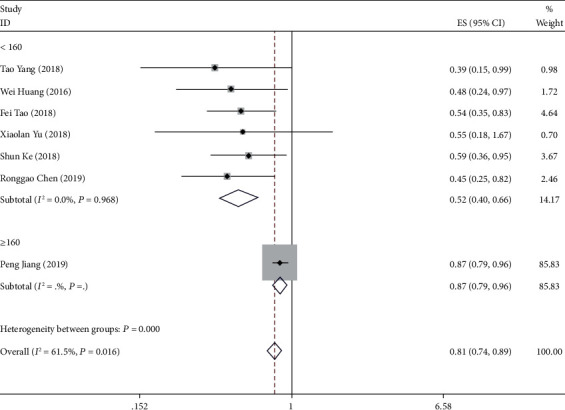
Meta-analysis for the pooled HRs of OS in patients with various cancers.
2.3.1. Assessment of Publication Bias
In order to assess the publication partiality in the current study, a performance of Begg's funnel plot was completed. Following its implementation, there was no notable evidence observed in the publication partiality during the analysis of the clinical stage (P = 0.017), histological grade (P = 0.183), lymph node metastasis (P = 0.234), and also the tumor invasion depth (P = 0.221).
3. Conclusion
In the result of this meta-analysis, decreased NKILA expression is typical of different kinds of cancer. Moreover, it can perform as a predictive element of prognosis in varied kinds of cancer. Nonetheless, till now, it is deemed essential to carry out larger-size as well as better designed research works for the confirmation of our findings.
4. Discussion
Over the recent years, the increased regulation of lncRNAs is performed in various cancers occurring in humans. Apart from its relation with the progression of the disease, lncRNAs such as lncRNA HOTAIR [29] are effective in serving as molecular scaffolds, delivering particular regulatory proteins within proximity of each other. Thus, it is able to function as a distinctly complex structure. It will then have a vital role in the development and progression of a tumor. By regulating the transcription of CDKN1A epigenetically in the nucleus, lncRNA SNHG1 [2] is likely to promote malignancy of cholangiocarcinoma. Thus, it facilitates the survival and metastasis of the cells related to cholangiocarcinoma. Generally, these expressed lncRNAs and are expected to occur as molecular biomarkers. They are useful in developing therapeutic methods which would be effective in the diagnosis and treatment as well as prognosis in various cancers occurring in humans.
The aim of the present meta-analysis was to investigate the relationship between NKILA expression level and pathological attributes in human cancers. A total of 937 patients studied under nine distinct researches were implicated in the end. Based on the algorithmic meta-analysis, the results obtained from the present study indicate that cancer patients with a low level of NKILA expression have shorter OS. In addition, with regard to the clinical prognosis, the results indicated that the group associated with low NKILA expression level had a heightened risk of advanced clinical stage, poor histological grades, positive lymph node metastasis, and extended tumor invasion depth.
However, the present study had a few considerable limitations which are listed below:
There was a geographical limitation since all patients involved in the study were from China. This excludes patients from different nationalities
The number of participating patients was limited since only a small number was considered for each research. In addition, only a few types of cancer were investigated
There was no observed consensus on the cutoff approximated which is needed to distinguish between high and low NKILA expression levels
There was a lack of cohort research which satisfied the inclusion criteria. A combination of an increased sample-size and improved quality in research will further guarantee the results obtained in this study
In summary, a higher expression level of lncRNA NKILA exhibited a close relationship with poorly distinguished grade, deep tumor invasion, and lymph node metastasis. This suggested that it had the capacity to serve as a biomarker of poor prognosis for cancer patients.
5. Materials and Methods
5.1. Literature Search Strategies
Two scholars independently identified the related literature. This was achieved using PubMed and Web of Science. The goal was to source studies related to investigating the relationship between NKILA expression level and pathological characteristics in cancer patients. A mix of keywords (“NKILA”, “cancer or carcinoma or carcinoma or tumor or neoplasm”) was used in the literature search strategy. This was combined with the detection of relevant research work that supplemented the obtained literature.
5.2. Inclusion and Exclusion Criteria
The literature related to the present study was required to be in accordance with the inclusion criteria listed below:
Stated expression levels of NKILA, as figured out with the help of RT-qPCR
Provided the decision
Classified cancer patients according to two groups, high and low expression groups with the use of detailed criteria for NKILA expression levels
Reported findings with regard to the clinicopathological attributes of the patients; at least one of the following pathological attributes should have been reported: histological grade, tumor invasion depth, lymph node metastasis, clinical stage, and distant metastasis
It should be a case-control or cohort study
If the sourced literature for the present study had at least one of the features of the exclusion criteria, it was deemed to be unfit for the purpose of this study:
Researches that were recurring or ones that used patients stated in prior studies
Failure to adequately account for the data
Utilized nonhuman species in the experiments
Reflections, letters, unpublished data, and commentaries
Reports compiled in languages that were not English
The quality of the research was evaluated by two scholars; they studied the title, abstract, and the overall content of each report that referred to the inclusion as well as exclusion criteria.
5.3. Literature Screening and Data Extraction
The data was independently collected by two investigators (Siyuan Tian and Yang Yu). All instances of collected data were in accordance with both the inclusion and exclusion criteria. Any differences were resolved through consensus or discussion with a third investigator (Honghua Huang) before the commencement of analysing the collected data. Data derivation of literature was as follows: first author, publication year, country of origin, kind of cancer, total number of patients, number of patients assigned to both high and low NKILA expression groups, the identification approach, and the cutoff approximations for NKILA expression levels.
5.4. Quality Assessment
Quality assessment of the considered researches was performed using the Newcastle-Ottawa Scale standard; the scores were as follows: including selection (4 points), comparability (2 points), and outcome (3 points). The score was in the range of 0 to 9. Independent evaluation of each article was performed by 2 investigators (Siyuan Tian and Yang Yu); the differences were resolved through consensus or through discussion with a third investigator (Honghua Huang). Each entitled research work was scored in Table 1; a higher score was indicative of an enhanced methodological quality.
5.5. Statistical Analysis
Cochran's and chi-squared-based Q and I2 tests were utilized in order to determine the heterogeneity amongst the considered researches. The performance of the homogeneity test was completed for a significance level of α = 0.1. P values < 0.1 were considered significant, and I2 values > 50% represented the heterogeneity amongst the researches. The fixed-effects model was utilized for analysing the homogenous data. On the other hand, the random-effects model was utilized for analysing the heterogeneous data. Statistical analysis was carried out with the use of Stata SE 12.0 (Stata Corp LP, College Station, Texas, USA); these were also utilized for the assessment of publication partiality.
Acknowledgments
This study was supported by the Key Discipline of Nantong Medical Science (General Surgery).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Siyuan Tian, Yang Yu, and Honghua Huang contributed equally to this work.
References
- 1.Liu B., Sun L., Liu Q., et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27(3):370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Yu Y., Zhang M., Wang N., et al. Epigenetic silencing of tumor suppressor gene CDKN1A by oncogenic long non-coding RNA SNHG1 in cholangiocarcinoma. Cell Death & Disease. 2018;9(7):p. 746. doi: 10.1038/s41419-018-0768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Y., Lian Y., Zhang Y., et al. The long non-coding RNA PVT1 represses ANGPTL4 transcription through binding with EZH2 in trophoblast cell. Journal of Cellular and Molecular Medicine. 2018;22(2):1272–1282. doi: 10.1111/jcmm.13405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L., Lin C., Jin C., et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500(7464):598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huarte M., Guttman M., Feldser D., et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan J. H., Yang F., Wang F., et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Yang F., Zhang H., Mei Y., Wu M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Molecular Cell. 2014;53(1):88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Yap K. L., Li S., Muñoz-Cabello A. M., et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Molecular Cell. 2010;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan Y., Shen B., Tan M., et al. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. The FEBS Journal. 2014;281(7):1750–1758. doi: 10.1111/febs.12737. [DOI] [PubMed] [Google Scholar]
- 10.Li Z., Yu X., Shen J. ANRIL: a pivotal tumor suppressor long non-coding RNA in human cancers. Tumor Biology. 2016;37(5):5657–5661. doi: 10.1007/s13277-016-4808-5. [DOI] [PubMed] [Google Scholar]
- 11.Sun M., Jin F. Y., Xia R., et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14(1):p. 319. doi: 10.1186/1471-2407-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutschner T., Hammerle M., Eissmann M., et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Research. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang E. B., Kong R., Yin D. D., et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5(8):2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hung T., Wang Y., Lin M. F., et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature Genetics. 2011;43(7):621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalvai M., Mondesert O., Bugler B., Manenti S., Ducommun B., Dozier C. Doxorubicin promotes transcriptional upregulation of Cdc25B in cancer cells by releasing Sp1 from the promoter. Oncogene. 2013;32(42):5123–5128. doi: 10.1038/onc.2012.524. [DOI] [PubMed] [Google Scholar]
- 16.Ke S., Li R. C., Meng F. K., Fang M. H. NKILA inhibits NF-κB signaling and suppresses tumor metastasis. Aging. 2018;10(1):56–71. doi: 10.18632/aging.101359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang T., Li S., Liu J., Yin D., Yang X., Tang Q. lncRNA-NKILA/NF-κB feedback loop modulates laryngeal cancer cell proliferation, invasion, and radioresistance. Cancer Medicine. 2018;7(5):2048–2063. doi: 10.1002/cam4.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W., Guo Q., Liu G., et al. NKILA represses nasopharyngeal carcinoma carcinogenesis and metastasis by NF-κB pathway inhibition. PLoS Genetics. 2019;15(8, article e1008325) doi: 10.1371/journal.pgen.1008325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao F., Xu Y., Yang D., et al. LncRNA NKILA correlates with the malignant status and serves as a tumor-suppressive role in rectal cancer. Journal of Cellular Biochemistry. 2018;119(12):9809–9816. doi: 10.1002/jcb.27300. [DOI] [PubMed] [Google Scholar]
- 20.Huang W., Cui X., Chen J., et al. Long non-coding RNA NKILA inhibits migration and invasion of tongue squamous cell carcinoma cells via suppressing epithelial-mesenchymal transition. Oncotarget. 2016;7(38):62520–62532. doi: 10.18632/oncotarget.11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang P., Han X., Zheng Y., Sui J., Bi W. Long non-coding RNA NKILA serves as a biomarker in the early diagnosis and prognosis of patients with colorectal cancer. Oncology Letters. 2019;18(2):2109–2117. doi: 10.3892/ol.2019.10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu X., Tang W., Yang Y., et al. Long noncoding RNA NKILA enhances the anti-cancer effects of baicalein in hepatocellular carcinoma via the regulation of NF-κB signaling. Chemico-Biological Interactions. 2018;285:48–58. doi: 10.1016/j.cbi.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 23.Wu W., Chen F., Cui X., et al. LncRNA NKILA suppresses TGF-β-induced epithelial-mesenchymal transition by blocking NF-κB signaling in breast cancer. International Journal of Cancer. 2018;143(9):2213–2224. doi: 10.1002/ijc.31605. [DOI] [PubMed] [Google Scholar]
- 24.Chen R., Cheng Q., Owusu-Ansah K. G., et al. NKILA, a prognostic indicator, inhibits tumor metastasis by suppressing NF-κB/Slug mediated epithelial-mesenchymal transition in hepatocellular carcinoma. International Journal of Biological Sciences. 2020;16(3):495–503. doi: 10.7150/ijbs.39582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang G. D., Li Y., Liao G. J., Qiu H. W. LncRNA NKILA inhibits invasion and migration of osteosarcoma cells via NF-κB/Snail signaling pathway. European Review for Medical and Pharmacological Sciences. 2019;23(10):4118–4125. doi: 10.26355/eurrev_201905_17913. [DOI] [PubMed] [Google Scholar]
- 26.Lu Z., Li Y., Wang J., et al. Long non-coding RNA NKILA inhibits migration and invasion of non-small cell lung cancer via NF-κB/Snail pathway. Journal of Experimental & Clinical Cancer Research. 2017;36(1):p. 54. doi: 10.1186/s13046-017-0518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu D., Shi X. Long non-coding RNA NKILA inhibits proliferation and migration of lung cancer via IL-11/STAT3 signaling. International Journal of Clinical and Experimental Pathology. 2019;12(7):2595–2603. [PMC free article] [PubMed] [Google Scholar]
- 28.Huang D., Chen J., Yang L., et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nature Immunology. 2018;19(10):1112–1125. doi: 10.1038/s41590-018-0207-y. [DOI] [PubMed] [Google Scholar]
- 29.Tsai M. C., Manor O., Wan Y., et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]


